Conjugated polyketone reductase C2 from C. parapsilosis IFO 0708 was expressed, purified and crystallized by the sitting-drop vapour-diffusion method. The crystal belonged to space group P212121 and diffracted X-rays to 1.7 Å resolution.
Keywords: conjugated polyketone reductase C, Candida parapsilosis, NAD(P)H-dependent oxidoreductases
Abstract
Conjugated polyketone reductase C2 (CPR-C2) from Candida parapsilosis IFO 0708 is a member of the NADPH-dependent aldo-keto reductase (AKR) superfamily and catalyzes the stereospecific reduction of ketopantoyl lactone to d-pantoyl lactone. A diffraction-quality crystal of recombinant CPR-C2 was obtained by the sitting-drop vapour-diffusion method using PEG 3350 as the precipitant. The crystal diffracted X-rays to 1.7 Å resolution on beamline NW12A of the Photon Factory-Advanced Ring (Tsukuba, Japan). The crystal belonged to space group P212121, with unit-cell parameters a = 55.02, b = 68.30, c = 68.93 Å. The Matthews coefficient (V M = 1.76 Å3 Da−1) indicated that the crystal contained one CPR-C2 molecule per asymmetric unit.
1. Introduction
The aldo-keto reductase (AKR) superfamily consists of approximately 120 members and includes NAD(P)H-dependent oxidoreductases that catalyze the reduction of a wide range of carbonyl compounds such as aliphatic and aromatic aldehydes, ketones, steroids, polycyclic aromatic hydrocarbons and monosaccharides (Jez & Penning, 2001 ▶). AKRs are monomeric enzymes that possess a triose phosphate isomerase (TIM) barrel motif consisting of an eight-stranded parallel β-sheet in the core surrounded by eight α-helices (Jez et al., 1997 ▶). Some of these enzymes play important roles in metabolic pathways such as sugar and steroid catabolism and vitamin C biosynthesis, but the biological functions of many other AKRs have not been identified (Jez & Penning, 2001 ▶).
Conjugated polyketone reductase C2 (CPR-C2) from Candida parapsilosis IFO 0708, first identified as an NADPH-dependent ketopantoyl lactone reductase (Hata et al., 1989 ▶), belongs to the AKR superfamily. It is a monomeric protein of approximately 36 kDa and possesses signature sequences characteristic of AKR-superfamily proteins (Kataoka et al., 2004 ▶). However, it lacks the Ile-Pro-Lys-Ser motif, another signature sequence of the AKR superfamily involved in NAD(P)H recognition (Kostrzynska et al., 1998 ▶). CPR-C2 reduces α-diketones such as ketopantoyl lactone and isatin to d-pantoyl lactone and 3-hydroxy-2-oxoindole, respectively, in a stereospecific manner (Hidalgo et al., 2001 ▶), but it does not reduce typical AKR substrates such as p-nitrobenzaldehyde and pyridine-3-aldehyde. The specific recognition of α-diketones and the stereospecific reduction by CPR-C2 make the enzyme very useful in the production of chiral building blocks for the production of pharmaceuticals. In order to elucidate the molecular mechanism of substrate recognition and stereospecific reduction by CPR-C2 and to improve its substrate specificity, stability and catalytic efficiency by introducing point mutations, we need to determine the high-resolution three-dimensional structure of CPR-C2. Here, we describe the overproduction, purification, crystallization and preliminary X-ray diffraction analysis of CPR-C2 as an initial step towards resolving the first three-dimensional structure of CPR-C2.
2. Materials and methods
2.1. Overproduction and purification of CPR-C2
The gene for CPR-C2 from C. parapsilosis IFO 0708 (gi:38423524) previously cloned into the pET-21a(+) vector (Kataoka et al., 2004 ▶) was amplified by PCR. The PCR primers used were 5′-GGAATTCCATATGACTCAAAGTAACTTACTACC-3′ (including an NdeI site, shown in bold) and 5′-CGGGATCCTTATTACAAATCTTTAAATTGCTCATGG-3′ (including a BamHI site, shown in bold). 30 cycles of PCR were performed using KOD-plus (Toyobo, Osaka, Japan), a high-fidelity thermostable DNA polymerase originally derived from Thermococcus kodakaraensis that is identical to KOD XL DNA (Novagen, Madison, Wisconsin, USA), with a melting phase at 367 K for 30 s, an annealing phase at 325 K for 30 s and a polymerization phase at 341 K for 80 s. The PCR product and the plasmid pET-28a(+) (Novagen) were digested by NdeI and BamHI and ligated with Ligation High (Toyobo). Thus, a 6×His tag followed by a thrombin protease cleavage site, Met-Gly-Ser-Ser-His6-Ser-Ser-Gly-Leu-Val-Pro-Arg-Gly-Ser-His, was added to the N-terminus of CPR-C2. The DNA sequence of the CPR-C2-encoding region of the resulting plasmid was verified. The N-terminally 6×His-tagged CPR-C2 was overexpressed in Escherichia coli Rosetta (DE3) (Novagen) grown in 1 l LB medium at 310 K. Protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM when the OD600 reached ∼0.6 and the culture was grown for another 15 h at 298 K. The harvested cells were resuspended in 50 mM Tris–HCl pH 8.0, 500 mM NaCl and 20 mM imidazole and then disrupted by sonication on ice. The lysate was centrifuged at 40 000g at 277 K for 30 min. The following purification procedures were performed at room temperature (∼295 K) unless stated otherwise. The supernatant was loaded onto an Ni Sepharose 6 Fast Flow (GE Healthcare Biosciences, Uppsala, Sweden) column equilibrated with the same buffer as above. The column was washed with the same buffer to remove nonspecifically bound proteins; 6×His-tagged CPR-C2 was then eluted in a stepwise manner with 50 mM Tris–HCl pH 8.0, 500 mM NaCl and 500 mM imidazole. The eluate was dialyzed against 10 mM Tris–HCl pH 8.0 at 277 K. The desalted crude 6×His-tagged CPR-C2 was concentrated to 5 mg ml−1 using a 20 ml Vivaspin concentrator (10 kDa cutoff; Sartorius, Göttingen, Germany) and then applied onto a Superdex 75 HR 10/30 (GE Healthcare Biosciences) column equilibrated with 10 mM Tris–HCl pH 8.0. The purified 6×His-tagged CPR-C2 was then concentrated to approximately 15 mg ml−1 using a 20 ml Vivaspin concentrator (10 kDa cutoff). Protein concentrations were measured with a Bio-Rad protein-assay kit (Bio-Rad, Foster City, California, USA) with bovine serum albumin as the standard (Bradford, 1976 ▶). The enzyme activity was measured as described previously (Hata et al., 1989 ▶).
2.2. Crystallization, data collection and preliminary X-ray analysis
Initial crystallization screening was performed using commercially available kits, namely Crystal Screen HT, Index HT (Hampton Research, Laguna Niguel, California, USA) and Wizard Screens I and II (Emerald BioSystems, Bainbridge Island, Washington, USA), at 293 K by high-throughput crystallization screening using 96-well Intelli-Plates (Art Robbins, Sunnyvale, California, USA). A crystallization drop was prepared by mixing 1 µl protein solution and 1 µl reservoir solution and was equilibrated against 70 µl reservoir solution. Index HT condition D9 [0.1 M Tris–HCl pH 8.5 and 25%(w/v) PEG 3350] gave tiny needle-like crystals. This condition was optimized to obtain crystals of better quality by changing the precipitant concentration and the buffer pH by the sitting-drop vapour-diffusion method using 24-well Cryschem Plates (Hampton Research). A crystallization drop was prepared by mixing 1 µl protein solution and 1 µl reservoir solution and was equilibrated against 500 µl reservoir solution. A diffraction-quality crystal was obtained with a reservoir composition of 0.1 M Tris–HCl pH 8.1 and 23%(w/v) PEG 3350. A crystal was picked up in a mounting loop and frozen in a cold nitrogen-gas stream using a cryoprotectant solution consisting of 10%(v/v) PEG 400 in 0.1 M Tris–HCl pH 8.1 and 23%(w/v) PEG 3350. X-ray diffraction data were obtained on beamline NW12A of the Photon Factory-Advanced Ring (Tsukuba, Japan). A data set comprised of 360 images was collected with a wavelength of 1.0000 Å, a crystal-to-detector distance of 154.3 mm, a rotation angle of 0.5° and an exposure time of 3 s per image using an ADSC Quantum 210 detector. The data were indexed and scaled using the XDS program package (Kabsch, 1993 ▶).
3. Results and discussion
The recombinant 6×His-tagged CPR-C2 was expressed in Escherichia coli Rosetta (DE3) (Novagen) and purified by two steps of column chromatography: Ni Sepharose 6 Fast Flow (GE Healthcare Biosciences) and Superdex 75 HR 10/30 (GE Healthcare Biosciences). Approximately 15 mg purified 6×His-tagged CPR-C2 with an N-terminal 6×His tag was obtained from 1 g wet E. coli cell pellet. The purified 6×His-tagged CPR-C2 gave a single band on SDS–PAGE (Fig. 1 ▶). Table 1 ▶ summarizes the purification. The specific activity of the recombinant 6×His-tagged CPR-C2 was comparable to that of native CPR-C2 purified from C. parapsilosis (Hidalgo et al., 2001 ▶). This result indicates that the additional sequence in the N-terminus does not affect the activity of CPR-C2.
Figure 1.

SDS–PAGE of purified 6×His-tagged CPR-C2. The molecular masses of the markers (kDa) are indicated on the left. The purified protein is indicated by the arrow on the right.
Table 1. Purification of CPR-C2 from 1 g of wet E. coli cell pellet.
| Step | Total protein (mg) | Total activity (U) | Specific activity (U mg−1) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell-free extract | 149 | 1457 | 9.81 | 100 | 1 |
| Ni Sepharose 6 FF | 39.9 | 908 | 22.8 | 62.3 | 2.32 |
| Superdex 75 HR 10/30 | 15.4 | 562 | 36.5 | 38.6 | 3.72 |
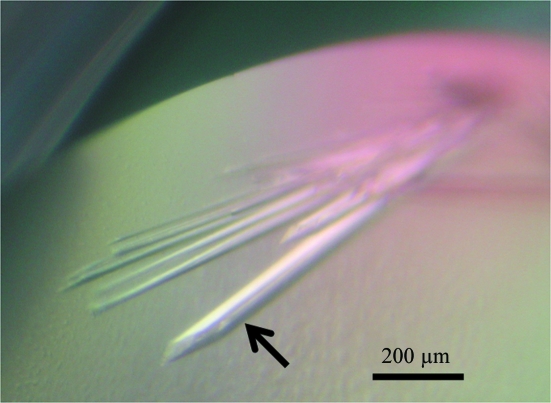
A diffraction-quality crystal of 6×His-tagged CPR-C2 was obtained by the sitting-drop vapour-diffusion method using the condition 0.1 M Tris–HCl pH 8.1 and 23%(w/v) PEG 3350. The crystal grew to final dimensions of 0.06 × 0.06 × 0.8 mm within 10 d at 293 K (Fig. 2 ▶). The crystal diffracted X-rays to 1.7 Å resolution on beamline NW12A of the Photon Factory-Advanced Ring (Tsukuba, Japan; Fig. 3 ▶). The data-collection statistics are summarized in Table 2 ▶. The Matthews coefficient (V M = 1.76 Å3 Da−1; Matthews, 1968 ▶) indicated that the crystal contained one 6×His-tagged CPR-C2 molecule per asymmetric unit. Structure determination of 6×His-tagged CPR-C2 is currently under way by the molecular-replacement method using the atomic coordinates of the 2,5-diketo-d-gluconic acid reductases (2,5-DKGRs) from Thermotoga maritima (37% sequence identity; PDB code 1vp5; Joint Center for Structural Genomics, unpublished work), Corynebacterium sp. (36% sequence identity; PDB code 1hw6; Sanli & Blaber, 2001 ▶) and E. coli (34% sequence identity; PDB code 1mzr; Jeudy et al., 2006 ▶) as search models. In these structures, the Ile-Pro-Lys-Ser motif (Ile-Pro-Lys-Thr in 1vp5 and Phe-Pro-Lys-Ser in 1hw6) is located at the NADPH-binding site and the Lys residue in the motif (in bold) binds to the 2′-phosphate of NADPH. Since the corresponding sequence in CPR-C2 is Val-Thr-Thr-Ser (residues 259–262), CPR-C2 would possess a different mechanism of NADPH binding. The crystal structure of 6×His-tagged CPR-C2 will reveal the structural basis of the unique substrate specificity and NADPH recognition of CPR-C2.
Figure 2.
A diffraction-quality crystal of 6×His-tagged CPR-C2 (indicated by an arrow) together with thin crystals.
Figure 3.
A diffraction image of the crystal of 6×His-tagged CPR-C2.
Table 2. Summary of data-collection statistics.
Values in parentheses are for the highest resolution shell.
| X-ray source | Photon Factory-Advanced Ring beamline NW12A |
| Wavelength (Å) | 1.0000 |
| Resolution range (Å) | 20.0–1.70 (1.74–1.70) |
| No. of observed reflections | 209050 |
| No. of unique reflections | 29200 |
| Data completeness (%) | 99.8 (99.9) |
| Rmerge† | 0.044 (0.525) |
| 〈I〉/〈σ(I)〉 | 28.91 (3.84) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 55.02, b = 68.30, c = 68.93 |
R
merge = 
 .
.
Acknowledgments
The synchrotron-radiation experiments were performed at Photon Factory beamline AR-NW12A (Tsukuba, Japan) with the approval of the Japan Synchrotron Radiation Research Institute (Proposal No. 2008S2-001). This work was supported by the Targeted Proteins Research Program (TPRP) of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Hata, H., Shimizu, S., Hattori, S. & Yamada, H. (1989). Biochim. Biophys. Acta, 990, 175–181. [DOI] [PubMed]
- Hidalgo, A.-R. G. D., Akond, M. A., Kita, K., Kataoka, M. & Shimizu, S. (2001). Biosci. Biotechnol. Biochem.65, 2785–2788. [DOI] [PubMed]
- Jeudy, S., Monchois, V., Maza, C., Claverie, J. M. & Abergel, C. (2006). Proteins, 62, 302–307. [DOI] [PubMed]
- Jez, J. M., Bennett, M. J., Schlegel, B. P., Lewis, M. & Penning, T. M. (1997). Biochem. J.326, 625–636. [DOI] [PMC free article] [PubMed]
- Jez, J. M. & Penning, T. M. (2001). Chem. Biol. Interact.130–132, 499–525. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Kataoka, M., Delacruz-Hidalgo, A. R., Akond, M. A., Sakuradani, E., Kita, K. & Shimizu, S. (2004). Appl. Microbiol. Biotechnol.64, 359–366. [DOI] [PubMed]
- Kostrzynska, M., Sopher, C. R. & Lee, H. (1998). FEMS Microbiol. Lett.159, 107–112. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Sanli, G. & Blaber, M. (2001). J. Mol. Biol.309, 1209–1218. [DOI] [PubMed]