Abstract
Replication protein A (RPA) is a heterotrimeric protein complex required for a large number of DNA metabolic processes, including DNA replication and repair. An alternative form of RPA (aRPA) has been described in which the RPA2 subunit (the 32-kDa subunit of RPA and product of the RPA2 gene) of canonical RPA is replaced by a homologous subunit, RPA4. The normal function of aRPA is not known; however, previous studies have shown that it does not support DNA replication in vitro or S-phase progression in vivo. In this work, we show that the RPA4 gene is expressed in normal human tissues and that its expression is decreased in cancerous tissues. To determine whether aRPA plays a role in cellular physiology, we investigated its role in DNA repair. aRPA interacted with both Rad52 and Rad51 and stimulated Rad51 strand exchange. We also showed that, by using a reconstituted reaction, aRPA can support the dual incision/excision reaction of nucleotide excision repair. aRPA is less efficient in nucleotide excision repair than canonical RPA, showing reduced interactions with the repair factor XPA and no stimulation of XPF-ERCC1 endonuclease activity. In contrast, aRPA exhibits higher affinity for damaged DNA than canonical RPA, which may explain its ability to substitute for RPA in the excision step of nucleotide excision repair. Our findings provide the first direct evidence for the function of aRPA in human DNA metabolism and support a model for aRPA functioning in chromosome maintenance functions in nonproliferating cells.
Keywords: Cancer, DNA/Recombination, DNA/Repair, DNA/Replication, Gene/Transcription, Replication Protein A
Introduction
Replication protein A (RPA)3 is the major single-stranded DNA-binding protein in human cells (1–3). It is composed of subunits of 70, 32, and 14 kDa (RPA1, the 70-kDa subunit of RPA; RPA2; and RPA3, the 14-kDa subunit of RPA, respectively) and was originally identified as an essential component for simian virus 40 (SV40) replication (1). RPA has since been shown to be essential for DNA replication, DNA repair, recombination, and coordination of the cellular response to DNA damage.
In addition to the three canonical subunits of RPA (RPA1, RPA2, and RPA3), the human genome contains an additional subunit called RPA4 that is 63% similar to RPA2.4 RPA4 was originally identified in a screen for proteins that interact with RPA1 (5). RPA4 is an intronless gene on the X chromosome, and RPA4 homologs with complete coding sequences are only found in primates and horse.4 Initial analysis indicated that at least some human tissues express RPA4 protein, though its role in these tissues was not determined (5). RPA4 protein can substitute for RPA2 in the RPA complex, forming an alternative RPA complex (aRPA) that has biochemical properties similar to canonical RPA (6). Surprisingly, whereas RPA is essential for DNA synthesis in the SV40 replication system, aRPA failed to substitute for RPA and acted in a dominant-negative fashion to inhibit DNA replication in the presence of canonical RPA (6). In addition, studies of RPA2-depleted HeLa cells expressing RPA4 demonstrated that aRPA is not able to support S-phase progression.4 These findings raise the question of whether aRPA has any physiological function commonly associated with RPA. To address this question, we determined the level of expression of the RPA4 gene in normal and transformed human tissues and examined the ability of aRPA to function in nucleotide excision repair and recombination.
Nucleotide excision repair is the main mechanism in humans for the removal from DNA of helix-distorting lesions induced by agents such as ultraviolet (UV) light from the sun (7–9). This multicomponent excision repair reaction requires a core six repair factors that recognize the lesion-containing DNA and make dual incisions bracketing the base adduct to remove (excise) the damaged base(s) in 24–32-nucleotide-long oligonucleotides. The resulting gap is filled and sealed by replicative DNA polymerases and ligases. Importantly, the nucleotide excision repair activity (excision nuclease) has been reconstituted in vitro with purified proteins (10–12), thus providing mechanistic insight into excision repair and allowing the characterization of the specific roles of the six minimal essential factors in the excision reaction.
One of the six core excision repair factors is RPA (10). RPA is thought to participate in multiple steps in excision repair (7, 8, 13). It appears to play an important role in damage recognition because of its higher affinity for damaged DNA than undamaged DNA (13, 14). Both RPA1 and RPA2 subunits also bind to the core repair factor XPA (15–18) though only the RPA1-XPA interaction appears essential for excision repair and survival of UV-irradiated cells (17, 19). RPA and XPA act cooperatively in DNA damage recognition (15, 18, 20), and the presence of RPA in the various “preincision complexes” (20, 21) that can be detected on damaged DNA prior to lesion removal provides additional evidence for a role of RPA in promoting or stabilizing the proper assembly of the excision nuclease. Formation of these complexes may be promoted by the strand separation activity of RPA (22). In addition, RPA participates in the dual incision by stimulating the XPF-ERCC1 endonuclease (23–25). Lastly, RPA has been implicated in the coordination of DNA synthesis after removal of DNA lesions (26). Because RPA appears to have multiple roles in excision repair, we examined whether aRPA could replace the canonical RPA in carrying out the excision reaction by the six-factor ensemble.
Another major repair pathway in human cells, homologous recombination, allows cells to repair double-stranded DNA breaks (27). Recombination depends on proteins in the rad52 epistasis group, including Rad51 and Rad52, and RPA (27). Rad51 is central to this process, forming filaments on single-strand DNA and mediating strand exchange (27, 28). The other rad52 epistasis group proteins (and other mediators) modulate filament formation and regulate recombination (27, 29). RPA interacts with both Rad51 and Rad52 (30–32), and these interactions are thought to mediate the formation of the Rad51 filament needed for efficient recombination (33–35).
We show that RPA4 is expressed in all normal human tissues examined but at different levels in different tissues. RPA4 expression is reduced in cancerous tissues and is very low in human cell lines. In addition, aRPA can support the dual incision/excision reaction, albeit less efficiently and by a different mechanism than RPA. aRPA can also support the initial steps of recombination such as Rad51-dependent DNA strand exchange. These results provide the first evidence for a physiological function of aRPA in human DNA metabolism.
EXPERIMENTAL PROCEDURES
Protein Purification
Recombinant RPA and aRPA were expressed in BL21 (DE3) cells and purified as described previously (6, 36, 37). The purification of the core nucleotide excision repair factors XPA, XPC, XPF-ERCC1, XPG, and TFIIH was reported earlier (38). Recombinant DNA Rad51 and Rad52 were purified as described previously (32, 39).
Quantitative PCR
RNA from cell lines was isolated using Qiagen RNeasy mini kit according to the manufacturer's protocol. Normal human RNA was purchased from Ambion as the FirstChoice Human total RNA survey panel, and tumor RNA was purchased from Ambion as FirstChoice Human Tumor RNA. cDNA was generated using TaqMan reverse transcription reagents according to the manufacturer's protocol using oligo(dT)16 and 2 μg total RNA in a 20-μl room temperature reaction.
Quantitative PCR was carried out using a TaqMan Universal PCR Master Mix according to the manufacturer's recommendations using the following primers and probes: glyceraldehyde-3-phosphate dehydrogenase, primers 5′-GCACCACCAACTGCTTAGCA-3′ and 5′-GTCTTCTGGGTGGCACTGATG-3′ and probe 5′-TET-TCGTGGAAGGACTCATGACCACAGTCC-Black Hole Quencher-3′; RPA2, primers TTGTTTGAAGCTCAGAGGGAGAT-3′ and 5′-GGTAGCATCCTTCCAATTCCAT-3′ and probe 5′-6-FAM-CCCACCCTGGATTGCATCCC-Black Hole Quencher-3′; and RPA4, primers 5′-CTCATCAGGAAGGGAAGAGCAT-3′ and 5′- GCCCTCAACGGTCAGATAATCA-3′ and probe 5′-JOE NHS Ester-AGCTCCGGGCTCAGCTCTGC-Black Hole Quencher-3′. Data were analyzed using SDS2.3 software by Applied Biosystems. All data were compared using the comparative CT method (40). All probe pairs amplify their target with equal efficiency (data not shown).
Enzyme-linked Immunosorbent Assay
All incubations were carried out at 25 °C as described previously (37). Wells in microtiter plates were coated with 1 μg of RPA or aRPA or 1 μg of Rad51 or Rad52 in 50 μl of water and incubated for 1 h. Plates were washed with phosphate-buffered saline (PBS) with 0.2% Tween 20 three times to remove unbound protein. Plates were blocked with 300 μl of 5% milk in PBS for 10 min and washed. The indicated amount of XPA, RPA, aRPA, or bovine serum albumin (BSA) was added to each well, incubated for 1 h, and washed. Primary antibodies in PBS with 5% milk for XPA (1:100), RPA/aRPA (1:300) were added to the plates, incubated for 30 min, and washed. Goat α-mouse IgG horseradish peroxidase (1:1,000) in 50 μl of PBS with 5% milk was added to the plates, incubated for 30 min, and washed. Plates were developed using 200 μl of 0.8 mg/ml ο-phenylenediamine in 0.005 m phosphate citrate buffer with 0.03% sodium perborate. A450 was then quantified after 10–60 min using a microtiter plate reader. Background was determined for by using BSA as the secondary protein, and all data shown have these values subtracted. In all assays, the background values were similar and close to zero.
Excision Repair Assay
The assay measures the release of base lesions in the form of 24–32-nucleotide-long oligomers (41). An internally 32P-labeled 140-bp DNA substrate containing a single centrally located (6-4) UV photoproduct was prepared as described (38) by ligating an annealing six oligonucleotides, one of which was radiolabeled and contained a T-T (6-4) photoproduct. The oligomer containing the (6-4) photoproduct was from the Synthetic Organic Chemistry Core at the University of Texas Medical Branch (Galveston, TX). Sequences of the oligomers used are available upon request. Excision assays involved incubation of 5 fmol of substrate in a 10-μl reaction containing the essential excision repair factors (XPA, XPC, XPF-ERCC1, XPG, and TFIIH) and either RPA or aRPA, using conditions described previously (38). Excision products were separated on DNA sequencing gels and detected with a PhosphorImager. The excision repair activity was quantified using ImageQuant 5.2 software (GE Healthcare).
Endonuclease Assay
A “bubble substrate” containing a 20-nt-long bubble in the center of a 70-bp-long duplex was prepared as follows. A 70-mer single-stranded DNA (B20T2) was 3′ end labeled using 3′-[α-32P]dATP and terminal transferase (New England Biolabs). This labeled oligomer was purified from a denaturing polyacrylamide gel and annealed to the 70-mer single-stranded DNA (B20C2), which contains 25 nucleotides (nts) of a complementary sequence flanking a central region of 20 nt of unpaired base pairs. The sequence of the B20T2 oligomer is shown below, where the underlined region indicates the noncomplementary bases: 5′-AGCCAGATCTGCGCCAGCTGGCCACCCTGATTTTTTTTTTTTTTTTTTTTGAGCGCCAAGCTTGGGCTGC-3′. The annealed substrate was purified from a nondenaturing polyacrylamide gel. To examine the structure-specific endonuclease activity of XPF-ERCC1, 5 fmol of substrate was incubated in 10 μl reaction buffer containing 7.5 ng of recombinant XPF-ERCC1 and the indicated amounts of either RPA or aRPA in reaction buffer containing 25 mm HEPES-KOH, pH 7.9, 25 mm KCl, 1.8 mm MgCl2, 0.1 mm MnCl2, 6.5% (v/v) glycerol. Reactions were incubated for 90 min at 37 °C. SDS (0.34%) and proteinase K (20 μg/ml) were then added and incubation continued at 55 °C for 15–30 min. The DNA was then purified by phenol-chloroform extraction and ethanol precipitation. The recovered DNA was resuspended in formamide-dye loading mixture and electrophoresed on a 10% polyacrylamide sequencing gel. Incision products were visualized and quantified by PhosphorImager analysis.
Immunoblotting
Conventional immunoblotting techniques were used to detect the indicated proteins using antibodies that recognize RPA1 (Santa Cruz Biotechnology), RPA2 (Calbiochem), RPA4 (Abnova), and maltose-binding protein (MBP) (Santa Cruz Biotechnology).
XPA Pulldown Assay
MBP-tagged XPA (1 μg) was immobilized on amylose resin (New England Biolabs) in binding buffer (10 mm Tris, pH 7.4, 100 mm NaCl, 10 μg/ml BSA, 10% glycerol, 0.01% Nonidet P-40) and then incubated with either 1 μg of RPA or aRPA in 50 μl of binding buffer at 4 °C overnight. The resin was recovered by centrifugation and washed three times with binding buffer. Proteins were eluted in SDS sample buffer and electrophoresed on a 10% SDS-polyacrylamide gel. The upper portion of the gel was removed and stained with Coomassie Blue to detect MBP-XPA. The lower portion of the gel was transferred to nitrocellulose and analyzed by immunoblotting to detect the RPA and aRPA subunits.
Preparation of Immobilized DNA
pUC19 plasmid DNA was linearized by digestion with EcoRI and then biotinylated with biotin-14-dATP (Invitrogen) and Klenow polymerase (New England Biolabs). The DNA was purified on a Qiagen column and then treated with 300 μm N-acetoxy-2-acetylaminofluorene (AAAF) or ethanol (for unmodified DNA) for 3 h at room temperature in the dark. The AAAF treatment protocol yields ∼1 AAAF-guanine adduct every 200–250 bp of DNA, resulting in 10–14 lesions per plasmid (42). After ether extraction to remove the unincorporated AAAF, the DNA was precipitated in ethanol, dried, and resuspended in TE (10 mm Tris acetate, pH 7.5, 1 mm EDTA). To immobilize the DNA, 300 μl of Dynabeads M-280 streptavidin (Invitrogen) were prewashed in PBS containing 1 mg/ml BSA and 0.05% Nonidet P-40 and then incubated with 15 μg of biotinylated DNA in 10 mm Tris, 1 mm EDTA, 1 m NaCl. Typical yields were ∼40–50 ng of DNA per μl of Dynabeads.
Pulldown Assay with Immobilized DNA
Immobilized DNA was washed twice in reaction buffer (25 mm HEPES-KOH, pH 7.9, 50 mm KCl, 4 mm MgCl2, 0.5 mm dithiothreitol, 10% glycerol, 0.1% Triton X-100, 0.02% Nonidet P-40) before setting up binding reactions. Reactions (25 μl) were prepared in reaction buffer with the indicated combinations of XPA (20 ng), RPA (100 ng), and/or aRPA (100 ng) and either 25, 50, or 100 ng of immobilized DNA. After incubation for 1 h at room temperature, DNA-protein complexes were collected on a magnet and washed three times with reaction buffer. The proteins associating with the DNA were analyzed by SDS-PAGE and immunoblotting. For each experiment, gels were also loaded with standard amounts of XPA, RPA, and aRPA to quantify the relative amount of each protein associating with the DNA. Experiments were repeated six-to-eight times, and the averages and standard deviations were calculated and plotted.
DNA Strand Exchange Assay
DNA strand exchange reaction (20 μl) was performed as described previously (39, 43). Briefly, 15 μm ϕX174 viral (+) strand (nucleotide) DNA was incubated with 3.75 μm Rad51 in buffer containing 25 mm Tris acetate, pH 7.5, 2 mm ATP, 1 mm MgCl2, 2 mm CaCl2, at 37 °C. After 5 min, RPA or aRPA (1 μm) was added and incubation continued for 5 min. The reaction was started by addition of XhoI-linearized 32P-labeled ϕX174 double-stranded DNA (15 μm). After 2 h at 37 °C, the samples were treated with proteinase K (Roche) for 15 min at 37 °C. The reaction products were separated by electrophoresis on a 1% agarose gel (1× TAE (40 mm Tris base, 5 mm sodium acetate, 1 mm EDTA)) at 40 V overnight. The gels were dried and analyzed on a Molecular Dynamics Storm 840 PhosphorImager using ImageQuant Software.
RESULTS
RPA4 mRNA Is Found in Normal Human Tissues
The initial characterization of RPA4 by Keshav et al. (5) examined three human tissues for the presence of RPA4 protein. They showed that RPA4 protein was detectable in placental and colon tissue but not in kidney. If RPA4 is playing a general physiological role in cellular DNA metabolism, it would be expected to be expressed in a variety of tissues. To determine the normal distribution of RPA4, mRNA expression in a panel of human tissues was determined by quantitative PCR.
Because it was not known which tissues normally express RPA4, initial studies were carried out on HeLa cells transiently expressing a plasmid containing RPA4 under control of a cytomegalovirus promoter. These cells express RPA4 protein at high levels.4 PCR amplification of cDNA from untransfected and transfected HeLa cells were compared. mRNA levels for RPA2 and RPA4 were then compared using glyceraldehyde-3-phosphate dehydrogenase as a reference. HeLa cells transfected with the RPA4 plasmid express RPA4 at levels greater than endogenous RPA2 (Fig. 1A, right two columns). In contrast, mock-transfected HeLa cells do not have an appreciable amount of RPA4 mRNA (Fig. 1A). The endogenous level of RPA4 mRNA is close to the level of detection of this assay and may not be statistically significant. We also examined other stable human cell lines (for example HEK-293 and HepG2) and did not find significant expression of RPA4 in any of the lines tested (data not shown).
FIGURE 1.
Quantitative PCR of RPA4 and RPA2 mRNAs from different tissues. Relative mRNA expression of RPA2 (black) and RPA4 (gray) was determined by the comparative Ct method. Errors bars indicate the average of three technical and two experimental replicates. A, cDNA was made from a panel of 20 normal human tissues (Ambion) and HeLa cells were either mock-transformed, and HeLa cells were transformed with GFP-RPA4 fusion protein under the control of the cytomegalovirus promoter.4 B, cDNA made from normal and tumor tissue samples (Ambion).
RNA from 20 different tissues was analyzed for RPA2 and RPA4 expression. In agreement with the protein studies by Keshav et al. (5), RPA4 mRNA was detected at levels above RPA2 mRNA in placental tissue (Fig. 1A). RPA4 mRNA was also detected at levels similar to or above RPA2 mRNA in a number of tissues including bladder, colon, esophagus, lung, and prostate. In other tissues (brain, kidney, ovary, and spleen), RPA4 was expressed at levels <20% of the total middle subunit mRNA (RPA2 mRNA and RPA4 mRNA; Fig. 1A). The remaining tissues expressed RPA4 at intermediate levels. These results are consistent with an initial analysis of RPA4 protein levels in placental and colon tissues (data not shown) and the analysis performed by Keshav et al. (5). Similar variations were observed for RPA2 mRNA (Fig. 1A). For example, heart, liver, and skeletal muscle all have low amounts of RPA2 mRNA compared with ovary, spleen, testes, and thyroid, which have the most RPA2 mRNA in the tissues sampled. We conclude that all normal tissues examined transcribe RPA4 at significant levels, and, although there is tissue specific variation, in many tissues, RPA4 mRNA levels are comparable with RPA2 mRNA.
To determine whether RPA4 was also expressed in cancerous tissues, RNA from several types of tumors was examined. RPA4 mRNA was expressed at reduced levels in tumors from cervix, colon, kidney, and liver when compared with nonmatched normal tissue (Fig. 1B). In three of the four tissues compared, the levels of RPA2 mRNA increased. This is in agreement with the literature that has found increased expression of RPA in metastatic cancers (44, 45). These data, together with the finding that RPA4 is not expressed at significant levels in stable cultured cell lines, suggests that RPA4 is down-regulated in transformed cells. This supports the hypothesis that RPA4 plays a role in normal tissues but not in tissues with a large fraction of proliferating cells.
aRPA Supports Rad51-dependent Strand Exchange
We next examined the ability of aRPA to interact with Rad51 and Rad52, which are required for homologous recombination. aRPA interacted with Rad52 at a level similar to RPA, even though Rad52-RPA interactions are mediated through both RPA1 and RPA2 (Fig. 2A) (32). In contrast to Rad52, aRPA exhibited a decreased interaction with Rad51 when compared with RPA (Fig. 2A). Rad51 also interacts with both RPA1 and RPA2 (30, 46). To explore domains involved in the altered interactions, we also examined interactions with a mutant form of RPA1 composed solely of two copies of a fragment of the core DNA binding domain, AA-His (containing residues 177–303 of RPA1) (47). This fragment interacts with Rad51 to the same level as aRPA (Fig. 2B). We conclude that in aRPA, the interaction between RPA2 and Rad51 was lost but that Rad51-RPA1 interaction was retained.
FIGURE 2.
aRPA interactions with Rad51 and Rad52 and stimulates strand exchange. Enzyme linked immunosorbent assay in which interactions were measured between different forms of RPA and either Rad52 (A) or Rad51 (B). Forms of RPA used: RPA (open diamonds), aRPA (black squares), and AA-His (gray triangles). Errors bars indicate the average of two or more independent replicates. BSA was used to determine nonspecific background in each assay; BSA values, generally <0.1 OD were subtracted. C, schematic of DNA strand exchange between circular single-stranded DNA (ss) and homologous linear double-stranded DNA (ds) to produce joint molecules (JM) and nicked circular DNA (NC). The asterisk shows the 32P-label on each strand. D, DNA strand exchange assay where ϕX174 (+) strand was incubated with Rad51 followed by RPA/aRPA and 32P-labeled XhoI linearized ϕX174 double-stranded DNA. Samples were deproteinized and reaction products were separated by electrophoresis through a 1.0% agarose gel. The positions of joint molecules, nicked circular DNA (NC DNA), double-stranded DNA (dsDNA), and displaced single-stranded DNA (ssDNA) are indicated.
To investigate whether the altered interactions with Rad51 affected function, Rad51-dependent DNA strand exchange assays were performed. It has been shown that RPA can stimulate DNA strand exchange by Rad51 in vitro (also shown schematically in Fig. 2C) (48). When compared with RPA, aRPA can stimulate the Rad51 DNA strand exchange as well as RPA. Both RPA and aRPA extensively stimulate formation of nicked circular double-stranded DNA and the slower migrating joint molecules of which none are detected in the absence of RPA or aRPA (Fig. 2D). These data suggest that aRPA can support the central steps of recombination.
aRPA Substitutes for RPA in Nucleotide Excision Repair
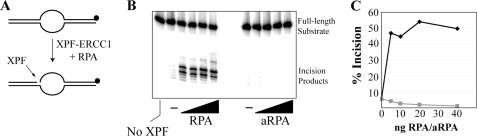
To determine whether the RPA4-containing aRPA protein supports nucleotide excision repair, we incubated aRPA or RPA (Fig. 3A) in reactions containing the other five excision repair factors (XPA, XPC-HR23B, TFIIH, XPF-ERCC1, and XPG) and an internally 32P-labeled 140-bp DNA substrate containing a site-specific (6-4) UV photoproduct (Fig. 3B). The excision assay involves damage recognition and dual incisions of the damaged strand at 20 ± 5 nt 5′ and 6 ± 3 nt 3′ to the damage, resulting in the release of damage-containing oligomers 24–32 nt in size that can be visualized on a denaturing polyacrylamide gel (38, 41). As seen in Fig. 3C (lanes 1–4) and in agreement with previous reports (10–12), the excision exhibits absolute requirement for RPA. Significantly, we find that aRPA can be substituted for RPA in the excision reaction (Fig. 3C, lane 7). However, at equimolar concentrations, aRPA is less effective than canonical RPA (Fig. 3C, lanes 3–4 and 5–6) and ∼3–4-fold higher concentration of aRPA is required to achieve similar levels of excision as the reaction reconstituted with RPA. Further increase in aRPA concentration did not increase the excision efficiency and actually had a modest inhibitory effect (Fig. 3C, lanes 8 and 9). These results indicated that even though aRPA can substitute for RPA in the excision reaction, it does so with lower efficiency. Next, we carried out a kinetic experiment to determine whether aRPA affected the rate or the extent of the excision reaction. We used the concentrations that were determined to be optimal for RPA and aRPA in the excision assays in Fig. 3C for the kinetic assays. The results shown in Fig. 3, D and E, show that under these conditions, the rate of excision by aRPA-reconstituted excision nuclease is approximately two times slower than the rate with canonical RPA. Taking into account that the optimal aRPA concentration for the excision assay is ∼3-fold higher than that of RPA, it can be stated that aRPA exhibits 5–8-fold lower activity in reconstituting excision nuclease. It should be noted that the lower activity of aRPA compared with RPA was seen with two independent preparations of aRPA and RPA and thus must reflect the intrinsic properties of these proteins. However, even though there is a difference in activity, it is clear that aRPA can substitute for RPA in this important cellular process. This finding provides additional direct evidence for aRPA functioning in cellular DNA metabolism.
FIGURE 3.
aRPA supports nucleotide excision repair. A, RPA and aRPA were separated by SDS-PAGE and visualized by Coomassie Blue staining. The position of each subunit is indicated along with approximate molecular mass (kDa). B, schematic of the nucleotide excision repair assay. An internally 32P-labeled (circle) 140-bp duplex DNA substrate containing a single (6-4) photoproduct (triangle) is incubated with purified excision repair factors, which results in dual incisions and release of 24–32-nt-long damage-containing oligomers. C, damage-containing substrate was incubated with 60 nm XPA, 9 nm XPC, 4 nm XPF-ERCC1, 3 nm XPG, 12.5 nm THIIH supplemented with indicated amounts of RPA or aRPA where indicated. The location of the excision products is indicated to the right. The percent of substrate in each reaction undergoing excision is indicated. D, time course of reconstituted excision repair reactions containing optimal amounts of either aRPA (530 ng) or RPA (150 ng). E, quantification of excision repair assays. Results indicate the average and S.D. from three independent experiments.
aRPA Does Not Stimulate the XPF-ERCC1 Endonuclease
RPA plays multiple roles in nucleotide excision repair, including stimulation of XPF-ERCC1 nuclease activity, binding to XPA to aid in cooperative recognition of DNA damage, and finally directly recognizing damaged DNA. To gain an insight into the lower activity of aRPA in the overall excision reaction we tested aRPA for each of these partial excision reactions.
The role of XPF-ERCC1 in excision repair is to make the 5′ incision of the dual incision reaction. We previously found that RPA highly stimulates the structure-specific endonuclease activity of XPF-ERCC1 using a 32P-labeled substrate containing a stretch of 30 unpaired nucleotides (23, 24). This bubble structure mimics the unwound state of DNA generated around the lesion by the helicase activity of the excision repair factor TFIIH (12, 49). To determine whether aRPA functions like RPA in the stimulation of XPF-ERCC1, we incubated a 3′ end-labeled 70-mer DNA substrate containing a 20-nt bubble in the center with recombinant XPF-ERCC1 and either RPA or aRPA (Fig. 4A). As expected, RPA stimulated the junction cutting activity of XPF-ERCC1 (Fig. 4B, lanes 2–6). No stimulation was observed with aRPA (Fig. 4B, lanes 7–11), and at high concentrations of the protein, the intrinsic junction cutting activity of XPF-ERCC1 was inhibited by aRPA (Fig. 4, B and C). We conclude that aRPA does not confer or stimulate the structure-specific activity of XPF-ERCC1. This difference between RPA and aRPA may contribute to the lower efficiency of aRPA in promoting the dual incision reaction.
FIGURE 4.
aRPA does not confer structure-specific endonuclease activity to XPF-ERCC1. A, schematic of XPF junction cutting assay. A 3′-labeled (circle) 70-mer DNA containing a central 20-nt unpaired region was incubated in reactions containing RPA or aRPA and XPF-ERCC1. Junction cutting activity by the XPF endonuclease was detected by electrophoresis of substrate on a denaturing polyacrylamide gel. B, sample incision assay containing XPF-ERCC1 (7.5 ng) and increasing amounts of either RPA or aRPA (0, 5, 10, 20, and 40 ng). C, quantification of data from panel B.
aRPA Does Not Stably Bind XPA
Next we determined whether aRPA bound to the excision repair factor XPA. It is known that XPA, along with RPA and XPC, is involved in the initial steps of damage recognition of DNA lesions inducing distortion to the DNA duplex (9, 49) and that both RPA1 and RPA2 interact with XPA (16, 17), enabling cooperative binding of RPA and XPA to damaged DNA (14, 15, 20). Enzyme-linked immunosorbent assays were done to examine the direct interaction between purified aRPA and XPA. As shown in Fig. 5A, RPA shows a strong interaction with XPA as reported previously (15, 16, 18). However, aRPA has a reduced interaction with XPA when compared with RPA.
FIGURE 5.
aRPA has altered interactions with XPA. A, enzyme-linked immunosorbent assay with RPA (open diamonds) or aRPA (black squares) and XPA. Errors bars indicate the average of two or more independent replicates. BSA was used to determine nonspecific background in each assay; BSA values, generally <0.1 OD were subtracted. B, immunoblot analysis of RPA and aRPA showing that anti-RPA2 and anti-RPA4 antibodies specifically recognize the appropriate subunits in RPA and aRPA, respectively. C, MBP-tagged XPA immobilized on amylose resin was incubated with RPA or aRPA overnight at 4 °C and then analyzed by SDS-PAGE and Western blotting with a mixture of the indicated antibodies. The recovered MBP-XPA was stained with Coomassie Blue after SDS-PAGE. Input represents 10% (100 ng) of RPA and aRPA and 1 μg of MBP-XPA.
To investigate the stability of the aRPA-XPA interaction, MBP-tagged XPA was immobilized on amylose resin and then incubated with either RPA or aRPA. The resin was separated from the solvent by centrifugation and the resin-bound proteins were detected by SDS-PAGE and Western blotting using antibodies recognizing RPA1, RPA2, and RPA4. Although RPA2 and RPA4 show significant sequence homology the antibodies against these proteins do not cross-react (Fig. 5B). Consistent with the well described interaction of RPA with XPA (15, 16, 18), RPA binds to the MBP-XPA resin, as indicated by the presence of both RPA1 and RPA2 subunits in the XPA pulldown (Fig. 5C). In contrast, neither RPA1 nor RPA4 were pulled down with the MBP-XPA resin when aRPA was used in the binding experiment (Fig. 5C, lanes 3 and 6). These results indicate that the RPA2 subunit of RPA plays a critical role in stabilizing the interaction of the heterotrimeric complex with XPA, consistent with earlier observations (14, 19). These data indicate that aRPA can interact with XPA but that the complex is less stable in vitro than the RPA-XPA complex. This difference may contribute to the reduced efficiency of aRPA in excision nuclease reconstitution (Fig. 3). However, the finding that aRPA can support excision in the reconstituted reaction indicates that the RPA-XPA interaction, although important for efficient excision nuclease activity, is not essential for the assembly of the holoenzyme dual incision complex on DNA.
aRPA Binds Damaged DNA with Higher Affinity than RPA
Even though the direct binding assay failed to detect a stable interaction between XPA and aRPA, we considered the possibility that damaged DNA may facilitate such an interaction as both XPA and RPA are known to have specific affinity for damaged DNA (7, 8). Moreover, it has been shown that XPA and RPA bind to damaged DNA cooperatively (14, 15, 17, 20). To determine whether aRPA cooperates with XPA in the recognition of DNA damage, we incubated XPA and RPA or aRPA with plasmid DNA containing AAAF-guanine adducts and immobilized on magnetic beads. Consistent with previous reports, RPA stimulated the association of XPA with damaged DNA (Fig. 6A, lanes 1–3 and 13–15, and Fig. 6B). Similarly, the binding of RPA to damaged DNA was markedly enhanced in the presence of XPA (Fig. 6A, lanes 7–9 and 13–15, and Fig. 4B). Then, we examined the association of aRPA with the AAAF-damaged DNA in the absence or presence of XPA. Interestingly, we observed significantly more aRPA than canonical RPA associating with the damaged DNA (Fig. 4A, lanes 4–6 and 7–9). However, XPA did not stimulate aRPA binding to damaged DNA (Fig. 6A, lanes 4–6 and 10–12, and Fig. 6B), and aRPA failed to stabilize the XPA-DNA complex (Fig. 6A, lanes 1–3 and 10–12). These results support the data that aRPA and XPA do not stably associate with one another in the absence of DNA (Fig. 4) and furthermore suggest that DNA does not promote their association with one another.
FIGURE 6.
Binding of XPA, RPA, and aRPA to damaged DNA. A, linearized, biotinylated pUC19 plasmid DNA treated with AAAF was immobilized on streptavidin-coated magnetic beads and then incubated for 1 h at room temperature with the indicated proteins. Beads were washed, and associated proteins were analyzed by SDS-PAGE and Western blotting (WB) with anti-MBP and anti-RPA1 antibodies. Reactions contained 25, 50, or 100 ng of immobilized DNA, 20 ng of XPA and 100 ng of RPA or aRPA. B, quantification of experiments performed as in A. Western blot signals were normalized to a standard amount of XPA, RPA, or aRPA in each experiment to determine the percentage of each protein associating with the DNA. Data represent averages and S.D. from six-to-eight independent experiments. C, unmodified or AAAF-treated pUC19 plasmid DNA (25, 50, or 100 ng) immobilized on magnetic beads was incubated with RPA or aRPA for 1 h at room temperature. The associated RPA and aRPA was detected by SDS-PAGE and Western blotting with an anti-RPA1 antibody. The graph represents the average amount of RPA1 observed on the 100 ng DNA sample in two independent experiments. Data were normalized to the RPA1 signal from aRPA in both experiments.
Because of the apparently higher affinity of aRPA than RPA to damaged DNA (Fig. 6A, lanes 4–6 and 7–9) we wished to find out whether aRPA has overall higher affinity to DNA compared with RPA. To this end, we determined the binding of the two forms of RPA to undamaged and damaged DNA in parallel reactions using immobilized DNA. As shown in Fig. 6C, in fact, aRPA has a slightly lower affinity for undamaged DNA than RPA in agreement with a recent analysis of affinities of RPA and aRPA to short single-stranded DNA oligomers (6). In contrast, whereas RPA shows only a modest preference (∼2-fold) for damaged over undamaged DNA under these experimental conditions, consistent with earlier measurements (13, 20), ∼20-fold more aRPA associated with the AAAF-damaged DNA compared with undamaged DNA (Fig. 6C). This increased affinity of aRPA to damaged DNA may account for its ability to support NER, even though it appears to lack some of the other nucleotide excision repair-related interactions.
DISCUSSION
Our results provide the first direct evidence for a physiological function for human aRPA. We show that aRPA is able to support the dual incision/excision steps of nucleotide excision repair and support Rad51-dependent DNA strand exchange. These results indicate that aRPA plays a role in cellular DNA maintenance.
We also show that RPA4 is expressed in normal human tissues and that, while expression varies between tissues, RPA4 mRNA levels are in the same range as RPA2 in many tissues. (The relative mRNA levels in tissues range from RPA2 being several times higher than RPA4 to RPA4 being several times higher than RPA2.) Biochemical analysis of recombinant protein indicates that aRPA forms with similar efficiency to canonical RPA.5 We also have shown previously that the stability of the RPA2 and RPA4 proteins are similar when expressed in tissue culture cells.4 Thus, it is likely that the ratio of RPA and aRPA complexes in cells will be proportional to their respective messenger RNA levels.
RPA4 mRNA expression is decreased in tumors relative to relative to normal tissue. RPA4 expression is also very low in proliferating cell lines. These findings are consistent with the original analysis of RPA4 that suggested that it was primarily expressed in nonproliferating tissues (5). Together, these results are consistent with aRPA functioning in DNA maintenance in predominantly nonproliferating cells.
The ability of aRPA to substitute for RPA in a reconstituted nucleotide excision repair system is unambiguous, as this repair system has an absolute requirement for RPA for the excision reaction (7, 8). However, aRPA exhibits lower activity, compared with RPA in the excision assay. This may be explained by the reduced stability of the aRPA-XPA complex or its failure to stimulate the activity of the repair endonuclease XPF-ERCC1. Interestingly, aRPA appears to have higher affinity for damaged DNA than canonical RPA, and this property of aRPA may partially compensate for its apparent lack of interactions with XPA and XPF-ERCC1. These findings suggest that both RPA and aRPA can support nucleotide excision repair in cells expressing RPA4.
Our data indicate that although an interaction of the RPA2 subunit of RPA with XPA may aid in dual incision, the interaction is not essential for repair. This conclusion is consistent with a previous report that showed that XPA-deficient cells expressing an N-terminal truncated form of XPA that is unable to interact with RPA2 shows little or no defect in nucleotide excision repair, as evidenced by essentially the same UV survival as cells expressing full-length XPA (17, 19). Thus, an RPA1-XPA interaction, which would be expected to be shared in both the canonical and alternative forms of RPA, is likely sufficient for excision repair.
aRPA is also able to interact with two proteins essential for DNA recombination, Rad52 and Rad51. A decrease in the interaction with Rad51 similar to that with XPA was observed. However, we show that aRPA is still able to support the Rad51-dependent DNA strand exchange, despite the reduced interaction, indicating that the interaction between RPA1 and Rad51 is sufficient for this reaction. These results suggest aRPA may also be able to support recombination. The model that aRPA functions in nonproliferating cells predicts that the primary role of aRPA in recombination would be recombination-mediated double-strand brake repair.
Protein interactions are essential for the function of canonical RPA. Several domains of RPA including the C terminus of RPA2 have been found to interact with multiple protein partners (3). Structural studies have indicated that the C terminus of RPA2 makes direct contacts with similar motifs in the repair and recombination factors XPA, UNG2, and Rad52 (50). SV40 large T antigen also interacts with this domain but through a different motif (51). Strikingly, interactions between aRPA and these different protein partners vary considerably. An early study found that RPA2 but not RPA4 interacted with UNG2 in a yeast two-hybrid analysis (52). We show here that aRPA has a reduced interaction with XPA but that the Rad52-aRPA interaction is the same as the Rad52-RPA interaction. In addition, aRPA has reduced interactions with Rad51 (Fig. 3) but unchanged interactions with SV40 T antigen (6). Rad51, Rad52, T antigen, and XPA all interact with both RPA1 and RPA2 (3). Thus, multiple domains contribute to interactions between aRPA and protein partners, and it appears that there is redundancy in the interactions essential for function.
It is also important to note that RPA is an abundant protein in human cells. It has been estimated that the concentration of canonical RPA in a normal cell is high enough to make single-stranded DNA-binding stoichiometric under physiological conditions (1). Thus, RPA and aRPA are probably not limiting under most conditions in vivo. So, even if aRPA has reduced protein interactions or supports repair less efficiently, this may not significantly limit these processes in the cell.
aRPA does not support SV40 DNA replication in vitro (6). It is currently not known what step(s) in replication are not supported. It is possible that aRPA may similarly inhibit DNA synthesis associated with DNA repair. Similarly, it remains to be determined whether the excision gaps covered with aRPA are as effective as those containing RPA in activating the ATR-mediated DNA damage response signaling (4, 53). Clearly, additional work will be necessary to better understand the differences in mechanisms by which RPA and aRPA contribute to many DNA maintenance reactions that govern genomic stability.
Acknowledgments
We thank Aishwarya Prakash for the gift of Rad52 and Kerry Brader for the gift of Rad51 protein. We also thank the members of the Wold lab for evaluation of data and critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Research Grants GM32833 (to A. S.), GM62653 (to S. C. K.), and GM44721 (to M. S. W.).
Haring, S. J., Humphreys, T. D., and Wold, M. S. (2010) Nucleic Acids Res. 37, in press.
A. C. Mason, unpublished data.
- RPA
- human replication protein A
- aRPA
- alternative RPA
- MBP
- maltose-binding protein
- TF
- transcription factor
- AAAF
- N-acetoxy-2-acetylaminofluorene
- nt
- nucleotide
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin.
REFERENCES
- 1.Wold M. S. (1997) Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 2.Iftode C., Daniely Y., Borowiec J. A. (1999) CRC Crit. Rev. Biochem. 34, 141–180 [DOI] [PubMed] [Google Scholar]
- 3.Fanning E., Klimovich V., Nager A. R. (2006) Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 5.Keshav K. F., Chen C., Dutta A. (1995) Mol. Cell. Biol. 15, 3119–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason A. C., Haring S. J., Pryor J. M., Staloch C. A., Gan T. F., Wold M. S. (2009) J. Biol. Chem. 284, 5324–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sancar A. (1996) Annu. Rev. Biochem. 65, 43–81 [DOI] [PubMed] [Google Scholar]
- 8.Wood R. D. (1997) J. Biol. Chem. 272, 23465–23468 [DOI] [PubMed] [Google Scholar]
- 9.Reardon J. T., Sancar A. (2005) Prog. Nucleic Acid Res. Mol. Biol. 79, 183–235 [DOI] [PubMed] [Google Scholar]
- 10.Mu D., Park C. H., Matsunaga T., Hsu D. S., Reardon J. T., Sancar A. (1995) J. Biol. Chem. 270, 2415–2418 [DOI] [PubMed] [Google Scholar]
- 11.Mu D., Hsu D. S., Sancar A. (1996) J. Biol. Chem. 271, 8285–8294 [DOI] [PubMed] [Google Scholar]
- 12.Evans E., Moggs J. G., Hwang J. R., Egly J. M., Wood R. D. (1997) EMBO J. 16, 6559–6573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reardon J. T., Sancar A. (2003) Genes Dev. 17, 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M., Mahrenholz A., Lee S. H. (2000) Biochemistry 39, 6433–6439 [DOI] [PubMed] [Google Scholar]
- 15.He Z., Henricksen L. A., Wold M. S., Ingles C. J. (1995) Nature 374, 566–569 [DOI] [PubMed] [Google Scholar]
- 16.Matsuda T., Saijo M., Kuraoka I., Kobayashi T., Nakatsu Y., Nagai A., Enjoji T., Masutani C., Sugasawa K., Hanaoka F., Yasui A., Tanaka K. (1995) J. Biol. Chem. 270, 4152–4157 [DOI] [PubMed] [Google Scholar]
- 17.Li L., Lu X., Peterson C. A., Legerski R. J. (1995) Mol. Cell. Biol. 15, 5396–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stigger E., Drissi R., Lee S. H. (1998) J. Biol. Chem. 273, 9337–9343 [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto I., Miura N., Niwa H., Miyazaki J., Tanaka K. (1992) J. Biol. Chem. 267, 12182–12187 [PubMed] [Google Scholar]
- 20.Wakasugi M., Sancar A. (1999) J. Biol. Chem. 274, 18759–18768 [DOI] [PubMed] [Google Scholar]
- 21.Wakasugi M., Sancar A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6669–6674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patrick S. M., Turchi J. J. (1999) J. Biol. Chem. 274, 14972–14978 [DOI] [PubMed] [Google Scholar]
- 23.Matsunaga T., Park C. H., Bessho T., Mu D., Sancar A. (1996) J. Biol. Chem. 271, 11047–11050 [DOI] [PubMed] [Google Scholar]
- 24.Bessho T., Sancar A., Thompson L. H., Thelen M. P. (1997) J. Biol. Chem. 272, 3833–3837 [DOI] [PubMed] [Google Scholar]
- 25.De Laat W. L., Appeldoorn E., Sugasawa K., Weterings E., Jaspers N. G., Hoeijmakers J. H. (1998) Genes Dev. 12, 2598–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shivji M. K., Podust V. N., Hübscher U., Wood R. D. (1995) Biochemistry 34, 5011–5017 [DOI] [PubMed] [Google Scholar]
- 27.San Filippo J., Sung P., Klein H. (2008) Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 28.West S. C. (2003) Nat. Rev. Mol. Cell. Biol. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 29.Sung P., Klein H. (2006) Nature reviews 7, 739–750 [DOI] [PubMed] [Google Scholar]
- 30.Golub E. I., Gupta R. C., Haaf T., Wold M. S., Radding C. M. (1998) Nucleic Acids Res. 26, 5388–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park M. S., Ludwig D. L., Stigger E., Lee S. H. (1996) J. Biol. Chem. 271, 18996–19000 [DOI] [PubMed] [Google Scholar]
- 32.Jackson D., Dhar K., Wahl J. K., Wold M. S., Borgstahl G. E. (2002) J. Mol. Biol. 321, 133–148 [DOI] [PubMed] [Google Scholar]
- 33.Sugiyama T., Kowalczykowski S. C. (2002) J. Biol. Chem. 277, 31663–31672 [DOI] [PubMed] [Google Scholar]
- 34.Eggler A. L., Inman R. B., Cox M. M. (2002) J. Biol. Chem. 277, 39280–39288 [DOI] [PubMed] [Google Scholar]
- 35.Sugiyama T., Kantake N. (2009) J. Mol. Biol. 390, 45–55 [DOI] [PubMed] [Google Scholar]
- 36.Henricksen L. A., Umbricht C. B., Wold M. S. (1994) J. Biol. Chem. 269, 11121–11132 [PubMed] [Google Scholar]
- 37.Binz S. K., Dickson A. M., Haring S. J., Wold M. S. (2006) Methods Enzymol. 409, 11–38 [DOI] [PubMed] [Google Scholar]
- 38.Reardon J. T., Sancar A. (2006) Methods Enzymol. 408, 189–213 [DOI] [PubMed] [Google Scholar]
- 39.Carreira A., Hilario J., Amitani I., Baskin R. J., Shivji M. K., Venkitaraman A. R., Kowalczykowski S. C. (2009) Cell 136, 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J. C., Svoboda D. L., Reardon J. T., Sancar A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3664–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi J. H., Lindsey-Boltz L. A., Sancar A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13301–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bugreev D. V., Mazin A. V. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9988–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Givalos N., Gakiopoulou H., Skliri M., Bousboukea K., Konstantinidou A. E., Korkolopoulou P., Lelouda M., Kouraklis G., Patsouris E., Karatzas G. (2007) Mod. Pathol. 20, 159–166 [DOI] [PubMed] [Google Scholar]
- 45.Tomkiel J. E., Alansari H., Tang N., Virgin J. B., Yang X., VandeVord P., Karvonen R. L., Granda J. L., Kraut M. J., Ensley J. F., Fernández-Madrid F. (2002) Clin. Cancer Res. 8, 752–758 [PMC free article] [PubMed] [Google Scholar]
- 46.Stauffer M. E., Chazin W. J. (2004) J. Biol. Chem. 279, 25638–25645 [DOI] [PubMed] [Google Scholar]
- 47.Wyka I. M., Dhar K., Binz S. K., Wold M. S. (2003) Biochemistry 42, 12909–12918 [DOI] [PubMed] [Google Scholar]
- 48.Sugiyama T., Zaitseva E. M., Kowalczykowski S. C. (1997) J. Biol. Chem. 272, 7940–7945 [DOI] [PubMed] [Google Scholar]
- 49.Shuck S. C., Short E. A., Turchi J. J. (2008) Cell research 18, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mer G., Bochkarev A., Gupta R., Bochkareva E., Frappier L., Ingles C. J., Edwards A. M., Chazin W. J. (2000) Cell 103, 449–456 [DOI] [PubMed] [Google Scholar]
- 51.Arunkumar A. I., Klimovich V., Jiang X., Ott R. D., Mizoue L., Fanning E., Chazin W. J. (2005) Nat. Struct. Mol. Biol. 12, 332–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagelhus T. A., Haug T., Singh K. K., Keshav K. F., Skorpen F., Otterlei M., Bharati S., Lindmo T., Benichou S., Benarous R., Krokan H. E. (1997) J. Biol. Chem. 272, 6561–6566 [DOI] [PubMed] [Google Scholar]
- 53.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]