Abstract
Aprataxin, defective in the neurodegenerative disorder ataxia oculomotor apraxia type 1, resolves abortive DNA ligation intermediates during DNA repair. Here, we demonstrate that aprataxin localizes at sites of DNA damage induced by high LET radiation and binds to mediator of DNA-damage checkpoint protein 1 (MDC1/NFBD1) through a phosphorylation-dependent interaction. This interaction is mediated via the aprataxin FHA domain and multiple casein kinase 2 di-phosphorylated S-D-T-D motifs in MDC1. X-ray structural and mutagenic analysis of aprataxin FHA domain, combined with modelling of the pSDpTD peptide interaction suggest an unusual FHA binding mechanism mediated by a cluster of basic residues at and around the canonical pT-docking site. Mutation of aprataxin FHA Arg29 prevented its interaction with MDC1 and recruitment to sites of DNA damage. These results indicate that aprataxin is involved not only in single strand break repair but also in the processing of a subset of double strand breaks presumably through its interaction with MDC1.
INTRODUCTION
Aprataxin, defective in the human autosomal recessive disorder ataxia oculomotor apraxia type 1 (AOA1), is encoded by the APTX gene (1,2). AOA1 is a neurodegenerative disorder characterized by early onset cerebellar ataxia, oculomotor apraxia, hypoalbuminemia and late peripheral neuropathy (3). It resembles ataxia telangiectasia (A-T) in its neurodegenerative phenotype but lacks the extra-neurological features of A-T that include radiosensitivity, immunodeficiency and cancer predisposition (3,4). ATM, the protein defective in A-T, recognizes and is activated by DNA double strand breaks (DSB) to signal this damage to the DNA repair machinery and activate cell cycle checkpoints (5).
Aprataxin contains three functional domains: an N-terminal FHA domain (6), a central histidine triad (HIT) domain (7) and a C-terminal zinc finger motif. Aprataxin interacts with XRCC1 and PARP-1, two key components of the DNA base excision repair machinery (8–10), suggesting a role for aprataxin in DNA single strand break (SSB) repair. Cells derived from AOA1 patients are sensitive to genotoxic agents that induce SSB in DNA (8–12). Consistent with the involvement of aprataxin in DNA repair, defective SSB repair has been reported in AOA1 cells in response to camptothecin, H2O2 and BSO (11–13). More recently aprataxin was shown to catalyze the nucleophilic release of adenylate groups covalently linked to 5′-termini of DNA molecules (14,15). Ligation of SSB or DSB involves the formation of an AMP-ligase complex, which subsequently transfers the AMP moiety onto the 5′ phosphate of the break site from which it is released after the formation of the phosphodiester bond (16). The AMP hydrolase activity of aprataxin appears to be important in resolving abortive DNA ligation intermediates that can form at ‘dirty’ or ‘complex’ SSB and potentially also at DSB formed by reactive oxygen species attack or clustered damage (15). The involvement of aprataxin in DNA repair is further supported by recent evidence of elevated levels of oxidative DNA damage in AOA1 cells coupled with a reduced expression of PARP-1, apurinic endonuclease 1 (APE1) and OGG1 (17). Furthermore, impaired base excision and gap filling repair efficiencies reveals a synergy between aprataxin, PARP-1, APE-1 and OGG1 in the DNA damage response and highlights both direct and indirect modulating functions for aprataxin on base excision repair (17). Although a role for aprataxin in the repair of abortive ligations at sites of DSB has been suggested by the interaction between aprataxin and XRCC4 (9) and the in vitro binding of aprataxin to DNA double strand ends (15,18), no direct evidence for its involvement in DNA double strand break repair has been reported (19).
In response to DNA damage, many proteins involved in DNA damage signalling/repair, such as Mre11, NBS1 and 53BP1 quickly re-localize to nuclear foci (20,21). These foci co-localize with the phosphorylated form of histone H2AX (γH2AX), a well-established marker for DSB (22). While aprataxin interacts with XRCC1, XRCC4 and PARP-1, we have previously shown that in contrast to the other DNA repair proteins, it does not form detectable nuclear foci after DNA damage exposure induced by either low linear energy transfer (LET) ionizing radiation (IR) or H2O2 (8). The dynamics of recruitment of DNA repair proteins to sites of DNA damage has been investigated by laser irradiation combined with photosensitizers (23–25). Hirano et al. (2007) employed this approach to show that aprataxin was recruited in vivo to SSB. While laser-induced DNA damage produces SSB and DSB, the relative distribution and the density of the damage are greatly influenced by the laser type, energy output and the type of photosensitizer (26). Biological imaging of charged particle tracks represents an alternative approach for investigating the association of repair proteins with chromatin (27,28). This technology offers the advantage of defined intrinsic physical properties of the heavy ion beams and allows the density of lesions along the track to be varied with ion species with different LET values. Using a remote-controlled microscope coupled to the beamline that allows the acquisition of fluorescence images of living cells, in real-time during ion irradiation, we observed that GFP-aprataxin is rapidly localized to DNA damage induced by nickel-ion irradiation (8,28). To gain further insight how aprataxin is recruited to sites of DNA breaks, we investigated the in vivo recruitment of GFP-tagged aprataxin to localized DNA damage induced by high LET heavy ion irradiation. We demonstrate for the first time that aprataxin co-localizes and interacts with mediator of DNA damage checkpoint protein 1 (MDC1), a protein that amplifies ATM-dependent DNA damage signalling (29–35). Interaction between the two proteins is mediated through the FHA domain of aprataxin, which binds to a casein kinase II (CK2)-phosphorylated N-terminal region of MDC1. Mutation of aprataxin Arg29, a key residue conserved within FHA motifs (36), disrupted the interaction between aprataxin and MDC1, and abolished its recruitment to sites of DNA damage. These results together with those of Harris et al. (17) highlight the importance of the aprataxin FHA domain in regulating protein–protein interactions and targeting aprataxin to sites of DNA damage.
MATERIALS AND METHODS
Cell lines, inhibitors
Control lymphoblastoid cell line (LCL) (C3ABR) and cervical adenocarcinoma cells (HeLa) were cultured in RPMI 1640 (GIBCO BRL) and DMEM (GIBCO BRL), respectively containing 10% foetal calf serum (FCS) (Lonza), 2 mM l-glutamine (Life Technologies), 100 U/ml penicillin (GIBCO BRL), 100 U/ml streptomycin (GIBCO BRL) and maintained in a humidified incubator at 37°C/5% CO2. Wild type, Aptx−/− and Mdc1−/− mouse embryonic fibroblasts (MEF), and human AOA1 fibroblasts (FD105) hTERT-transformed (Gift from Prof. Keith Caldecott, University of Sussex, UK) were cultured in DMEM as described earlier. CK2 inhibitors 4,5,6,7-tetrabromotriazole (TBB), and 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT) were purchased from Calbiochem and dissolved in DMSO.
Aprataxin-GST and MDC1-GST pull down assays
Aprataxin-GST proteins have been described earlier (8,37). Pull down assays, were performed as described in Supplementry Data. Proteins were separated on 5% and 10% SDS–PAGE, transferred onto nitrocellulose membranes (Hybond C, Amersham) and detected using the relevant antibody. Aprataxin antibodies have been described earlier (8,37). Sheep polyclonal MDC1 antibody was produced against GST-MDC1 fragments and purified by affinity column against GST only and the antigen using standard protocols. For immunoblotting, antibodies were used at dilutions of: MDC1 (1/2000), aprataxin (1/2000), anti-GST (1/2000) followed by the relevant species-specific HRP-conjugated secondary antibody from Chemicon or Sigma (1/10 000).
In vivo CK2 inhibition and pull downs
HeLa cells were mock treated (DMSO) or treated with a range of concentrations of specific CK2 inhibitor TBB, or DMAT for 8 h in a six well-plate format and then lysed. Pull down was performed as described in Supplementary Data.
In vitro CK2 phosphorylation and pull down
A region of MDC1 containing CK2 sites (amino acids 150–350) was cloned into the bacterial GST expression vector pGEX-6P-1, expressed in E. coli (BL21DE3) and purified as described by the manufacturer. Two micrograms of each GST fusion protein was in vitro phosphorylated by incubation with 200 ng of human recombinant CK2 (New England Biolabs) in buffer P (20 mM Tris–HCl pH 7.5, 50 mM KCl, 10 mM MgCl2, 200 µM ATP) for 1 h at 37°C. Once phosphorylated, GST fusion proteins were used in pull-down assays mixed with 2 mg of HeLa whole cell extracts (WCE) as described earlier.
Immunoprecipitation and immunoblotting
Total cell extracts from control (C3ABR) cells were prepared as described in Supplementary Data and the equivalent of 1 mg of total protein was immunoprecipitated with non-specific serum (1 or 5 µg), aprataxin (1 µg) and MDC1 (5 µg) antibodies overnight at 4°C. 40 µl of protein G-Sepharose beads (Amersham) were added to the immunoprecipitation (IP) and incubated for 1 h on a rotating wheel at 4°C. IP were washed three times with lysis buffer and the beads were resuspended in 20 µl of sample loading buffer. Proteins were separated on 5% and 10% SDS−PAGE, transferred onto nitrocellulose membranes and detected using the relevant primary and secondary antibodies.
Isothermal titration calorimetry
Phosphopeptides were synthesised by Dr W. Mawby (University of Bristol). Aprataxin FHA-phosphopeptide binding was quantified by isothermal titration calorimetry (ITC) using a Microcal Omega VP-ITC calorimeter (MicroCal Inc., Northampton, MA, USA). Protein was dialysed against ITC buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol) and peptides were dissolved in the dialysis buffer. Experiments were carried out at 22°C and involved 30 successive 10 µl injections of 1 mM peptide solution into a sample cell containing 100 µM protein solution. Heats of dilution were subtracted and binding isotherms were plotted and analysed with MicroCal Origin version 7.0, assuming a single-site binding model.
Crystallization and structure determination of the aprataxin FHA domain
Aprataxin FHA domain was crystallized by hanging-drop vapour diffusion. Protein at a concentration of 10 mg/ml in 50 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol was mixed with an equal volume of reservoir solution [0.2 M ammonium fluoride, 20% (w/v) PEG 3350] and incubated at 18°C. Thin plates grew within 1 week and were transferred into cryoprotectant (0.1M ammonium fluoride, 10 % PEG 3350, 25 mM HEPES pH 7.5, 75 mM NaCl, 25% PEG 400) and flash frozen in liquid nitrogen. Data were collected at 100 K on an Raxis-IV detector mounted on a Rigaku MicroMax-007HF rotating anode X-ray source, and integrated and scaled using DENZO and SCALEPACK (38). The structure was solved by molecular replacement using the coordinates of PDB entry 1YJM as a search model with AmoRe (39,40). Subsequent refinement was carried out at 1.65 Å using REFMAC5 (41) and manual model building in Coot (42). Data collection and refinement statistics are summarized in Supplementary Table S2. All structure figures were prepared with PyMol (http://www.pymol.org). Coordinates have been deposited in the Protein Databank: Accession code 3KT9.
High LET heavy ion irradiation procedure
For heavy ion irradiation experiments, cells were seeded on sterilized glass coverslips and irradiated at the UNILAC beamline at GSI (Darmstadt, Germany) with high energy xenon ions (4.5 MeV/nucleon; LET 8815 keV/µm), krypton (5.4 MeV/nucleon; LET 5060 keV/µm), nickel (6.0 MeV/nucleon; LET 3430 keV/µm) or uranium ions (4.2 MeV/nucleon; LET 14925 keV/µm), respectively. Different heavy ion species were used for the recruitment studies since these experiments were carried out over several years and we have had to adhere to the schedule of radiation exposure at the Darmstadt GSI site in Germany. Directly before the irradiation, the coverslips were mounted into sample holders and placed in a medium filled tank. For the ion irradiation, the sample holders were automatically taken from the medium tank, exposed to the ion beam under a small angle of 15° between the axis of the ion beam and the plane of the cell monolayer and placed back again. For the applied fluence of 3 × 106 p/cm2, the whole procedure takes <30 s.
Immunofluorescence and microscopy
For immunostaining, cells grown on coverslips were washed with PBS, fixed in 2% formaladehyde/PBS for 20 min and permeabilized with PT-5 solution [PBS/0.5% Triton X-100] for 5 min. Non-specific binding was blocked by incubating with PTB-5 [PBS/0.5% BSA] for 20 min at room temperature. Coverslips were then incubated with mouse anti-γH2AX antibody (Upstate 1/500 in PTB-5) for 1 h at room temperature. Cells were stained with 5 µg/ml Alexa 568 goat anti-mouse F(ab)2 conjugate (Invitrogen) and the DNA counterstained with 1 µM ToPro3 (Invitrogen). Microscopy was performed on a Leica TCS NT confocal scanner equipped with an ArKr-Laser on the Leica DM IRBE inverted microscope (lens: HCX PlanApo 63x oil/NA1.32). Images were recorded as confocal stacks with optical sections separated by about 0.3 µm. Confocal images were displayed as maximum projections and assembled in Adobe Photoshop 7.0. Heavy ion irradiation procedure is described in Supplementary Data.
In vivo real-time beamline microscopy and heavy ion irradiation
Cells were transfected with Amaxa Nucleofection technology (Amaxa, Koeln, Germany). Transfection of MEFs was performed with Amaxa Nucleofector Kit VDP-1004 and protocol A-023 according to the manufacturer instructions (Amaxa, Germany). Mdc1−/− and Mdc1+/+ cells transiently transfected with GFP-aprataxin were employed for real-time fluorescence intensity measurements. These cells were cultivated on polycarbonate foil (18-mm diameter; 40-µm thickness) and irradiated at the beamline microscope as described earlier (28).
γH2AX foci repair kinetics
Analysis of γH2AX repair foci kinetics was performed as described earlier (43) and is described in Experimental Procedures in Supplementary Data.
Analysis of SSB repair
SSB were induced by exposing confluent cells to low doses of H2O2 (50−100 µM) in RPMI 1640 medium without supplements for 15 min at 37°C. To remove H2O2, cells were washed two times with PBSCMF (140 mM NaCl, 3 mM KCl, 8 mM Na2HPO4 and 1 mM KH2PO4). The numbers of SSB were determined by an alkaline elution assay as described earlier (44). The numbers of SSB in untreated control cells were subtracted in all cases.
RESULTS
Recruitment of aprataxin to DNA damage and co-localization with MDC1 after high LET radiation exposure
A hallmark of DNA repair proteins is their capacity to re-localize in discrete aggregates or foci within the nucleus after DNA damage exposure. We have previously shown that GFP-aprataxin co-localized with XRCC1 along charged particle tracks (8). Irradiation of cells with heavy ions generates multiple damaged sites including DNA lesions like SSB, DSB and base modifications (45). Here, we have focused on sites of DSB that can be identified by the phosphorylation of histone H2AX (γH2AX), the earliest detectable marker for DSB (22) and by recruitment of MDC1 (30). To determine the requirements for aprataxin recruitment to complex DNA damage, cells were transfected with GFP-tagged aprataxin, subjected to heavy ion irradiation at a small angle (<15°) from the horizontal and analysed by confocal fluorescence microscopy (Figure 1A). Expression and localization of GFP-aprataxin and GFP-MDC1 in unirradiated cells is shown in Supplementary Figure S1. High LET heavy ion irradiation at low specific energies produced discrete γH2AX aggregates/foci deposited along the particle trajectory (Figure 1B) as observed earlier (46). GFP-aprataxin was recruited to these sites of DSB as observed by the partial co-localization pattern between GFP-aprataxin and γH2AX (Figure 1B). To further confirm the recruitment of GFP-aprataxin to sites of DSB, we demonstrated co-immunostaining with MDC1 (Figure 1C), another DNA damage response factor that is recruited to DSB through binding to γH2AX (34). As was the case with γH2AX, co-localization was incomplete between aprataxin and MDC1 (Figure 1C). To confirm the recruitment of MDC1 to high LET heavy ion-induced DSB, we used a GFP-MDC1 fusion protein. As expected, GFP-MDC1 completely co-localized with γH2AX staining at sites of heavy ion-induced DSB (Supplementary Figure S2). To address possible artefacts due to either overexpression or the presence of a GFP tag attached to the N-terminus of aprataxin, immunostaining for endogenous aprataxin was also performed (Figure 1D). In this case also, endogenous aprataxin was recruited to sites of DSB as observed by partial co-localization with MDC1 along the particle tracks. To determine whether MDC1 mediates the recruitment of aprataxin to sites of DNA DSB, wild-type and Mdc1−/− MEFs were transiently transfected with GFP-aprataxin and analysed for protein track formation after heavy ion exposure. While aprataxin was recruited to DNA breaks in wild-type MEFs after heavy ion irradiation at 10 min post-irradiation (4.5 MeV/nucleon; LET 8815 keV/µm), no recruitment was observed in Mdc1−/− cells despite the formation of γH2AX tracks (Supplementary Figure S3A). Due to poor transfection efficiency of Mdc1 MEFs, the absence of aprataxin recruitment was only observed in a limited number of cells. Accordingly, to monitor the recruitment of aprataxin in MDC1-deficient cells over shorter time points (up to 4 min), we employed a remote-controlled system coupled to a beamline microscope that allows measurement of GFP fluorescence intensity at sites of DNA breaks in real-time during heavy ion irradiation (28). Both wild-type and Mdc1−/− MEFs were transiently transfected with GFP-aprataxin and subjected to high LET heavy ion irradiation as described earlier (28). Real-time imaging of GFP fluorescence revealed fast recruitment of aprataxin to sites of DNA damage within 1 min after irradiation in both Mdc1−/− and wild-type cells (Supplementary Figure S3B). Interestingly, fluorescence intensity at the sites of DNA damage drops out rapidly in Mdc1−/− compared with wild-type cells suggesting an increased rate of dissociation of GFP-aprataxin from sites of DNA DSB in these cells. This indicates that while MDC1 is not necessary for the initial recruitment of aprataxin to sites of DNA DSB, its absence affects the retention of aprataxin to these sites. On the other hand, loss of XRCC1 prevented any recruitment of aprataxin in cells exposed to high LET radiation (Supplementary Figure S4). Together, these results provide evidence that aprataxin localizes at least in part to sites of DNA DSB, and that MDC1 does not act to recruit aprataxin, but rather as part of a tethering platform for aprataxin and possibly other repair proteins, to ensure effective DNA DSB repair.
Figure 1.
Co-localization of aprataxin with γH2AX and MDC1 at sites of DSB. (A) Methodology employed to study the dynamic recruitment of GFP-tagged fusion proteins (aprataxin and MDC1) to localized DNA damage induced by heavy ion irradiation. Transfection of GFP constructs and irradiation procedures are described in Supplementary Data. (B) Recruitment of GFP-aprataxin to uranium (4.2 MeV/nucleon LET 14925 keV/µm) ions-induced DNA DSB in HeLa cells. DSB are visualized by γH2AX immunostaining and nuclei by ToPro3 staining. (C) Co-localization of GFP-aprataxin with MDC1 along particle tracks after uranium ions irradiation in HeLa cells. (D) Recruitment of endogenous aprataxin to DSB and co-localization with MDC1 along particle tracks after krypton ions irradiation (5.4 MeV/nucleon; LET 5060 keV/µm) revealed by immunostaining with anti-aprataxin and anti-MDC1 antibodies in human fibroblasts.
Interaction between aprataxin and MDC1
The co-localization of MDC1 and aprataxin to sites of DNA breaks prompted us to determine whether these two molecules associate. Using a combination of co-IP and GST pull-down assays we demonstrated an interaction between endogenous aprataxin and MDC1. MDC1 co-immunoprecipitated with aprataxin using a specific anti-aprataxin antibody while non-specific serum (Ig) failed to immunoprecipitate MDC1 (Figure 2A). Three discrete MDC1 bands were detected as has been reported earlier (30,34). This interaction was confirmed in a reverse co-IP with an anti-MDC1 antibody (Figure 2B). To further characterize the interaction between aprataxin and MDC1 and to map the interaction domain on aprataxin, we performed aprataxin GST pull-downs. Using a series of overlapping GST fusion proteins, encompassing aprataxin FHA, HIT and zinc finger domains (Figure 2C), we identified the N-terminal region (amino acid 1−110) of aprataxin as the MDC1 interaction domain (Figure 2D). Binding of MDC1 to the aprataxin FHA domain was retained in the presence of BenzonaseTM suggesting that this was not due to bridging with nucleic acid but rather a direct protein−protein interaction (Figure 2E). To confirm the direct interaction between aprataxin and MDC1, aprataxin GST pull downs were carried out in the presence of ethidium bromide (EtBr) (Supplementary Figure S5). EtBr did not disrupt the interaction between the two proteins. On the contrary, an increase in MDC1 binding was observed in the presence of EtBr which may reflect an increase in available soluble MDC1. Thus the co-localization of aprataxin and MDC1 to sites of DNA damage induced by high LET radiation is supported by evidence of a direct interaction between these two proteins.
Figure 2.
Direct interaction between aprataxin and MDC1. (A) Co-IP of MDC1 and aprataxin using an anti-aprataxin antibody followed by immunoblotting with the respective antibodies. Non-specific serum (Ig) is used as a negative control and the amounts of protein in WCE are shown. (B) Co-IP of aprataxin with an anti-MDC1 antibody. Non-specific serum (Ig) is included and WCE is also shown. (C) Diagram of aprataxin domains and GST-fragments used in the pull-down assays. (D) Protein extracts from control LCL (C3ABR) were either incubated with GST alone or GST-aprataxin fragments. Bound proteins were separated on SDS–PAGE and detected by anti-MDC1 antibodies. Coomassie staining shows equivalent loading of GSTs. (E) Direct binding of aprataxin to MDC1. Aprataxin FHA pull downs in the absence or presence of BenzonaseTM, a potent nuclease that degrades both RNA and DNA.
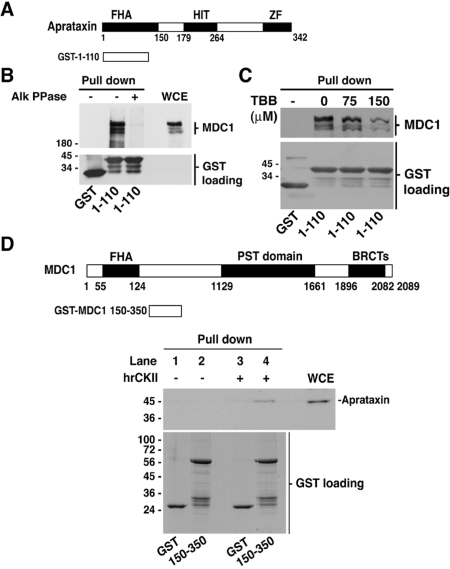
CK2 phosphorylation-dependent interaction of MDC1 with the aprataxin FHA domain
FHA domains are conserved sequences of 65–150 amino acid residues found principally within eukaryotic nuclear proteins that have been shown to mediate phospho-specific protein–protein interactions by binding to phosphopeptides (47,48). Previous reports have demonstrated that the interaction between aprataxin and XRCC1 was dependent on phosphorylation of XRCC1 by CK2 (9,10). Furthermore, XRCC4 which is involved in the repair of DNA DSB via the non-homologous end joining pathway was also shown to bind to the FHA domain of aprataxin (9). To determine whether XRCC4 could also target aprataxin to DNA breaks, we examined the recruitment of aprataxin in Xrcc4−/− cells. Normal recruitment of GFP-aprataxin was observed in Xrcc4−/− cells indicating that XRCC4, albeit binding to the FHA domain of aprataxin, is not involved in its targeting to heavy ion-induced DNA breaks (data not shown). Nevertheless, the interaction between aprataxin and XRCC4 may be important for the assembly of the rejoining complex and/or to ensure effective DNA end ‘clean-up’ during the repair process.
To determine whether the interaction between MDC1 and aprataxin is also dependent on phosphorylation of MDC1, GST pull downs with the aprataxin FHA domain were performed in the presence of protein phosphatase (Figure 3A and B). Alkaline phosphatase treatment disrupted the binding of MDC1 to the aprataxin FHA domain demonstrating a phosphorylation-dependent interaction between the two proteins (Figure 3B). Since CK2 was found to phosphorylate XRCC1 to mediate the aprataxin–XRCC1 interaction (9,10), we therefore investigated whether CK2 could also phosphorylate MDC1 and thus mediate the interaction between the two proteins. As shown in Figure 3C, inhibition of CK2 activity in vivo with increasing concentration of the specific CK2 inhibitor, 4,5,6,7-Tetrabromotriazole (TBB), resulted in decreased binding of MDC1 to the aprataxin FHA domain. Use of another CK2-specific inhibitor, 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT), also showed a reduction in MDC1 binding to aprataxin FHA (Supplementary Figure S6). Analysis of MDC1 protein sequence using GPS2.0 TM, NetPhos2.0TM, ScansiteTM prediction programs and cross-reference with the Phospho-ELMTM database identified several putative CK2 phosphorylation sites within the N-terminal region of MDC1 (Supplementary Figure S7). To confirm the CK2 phosphorylation-dependent interaction between aprataxin and MDC1, we made a GST fusion protein corresponding to the N-terminal region of MDC1 residues (150–350) that contains nine potential sites for CK2, three of which are predicted to be pT and required for FHA binding. The GST-fusion protein (150–350) was in vitro phosphorylated with recombinant human CK2 and pull downs were performed. As shown in Figure 3D, MDC1 fragment 150–350 pulled down endogenous aprataxin from total cell extracts (Figure 3D, Lane 4) after in vitro CK2 phosphorylation, indicating that the CK2-phosphorylated 150–350 region is the aprataxin binding domain on MDC1.
Figure 3.
CK2-mediated phosphorylation-dependent binding of MDC1 to the aprataxin FHA domain. (A) Diagram of aprataxin GST fusion used in the pull-down assays. (B) Effect of protein phosphatase activity on MDC1 binding to the aprataxin FHA domain. Whole cell extracts from HeLa cells were mock treated or treated with alkaline phosphatase (Alk PPase: 2500 U) for 1 h at room temperature and subsequently used for pull-down assays using GST only, and GST-aprataxin FHA domain. (C) Reduced binding of MDC1 to the aprataxin FHA domain following CK2 inhibitor treatment. HeLa cells were treated for 8 h with increasing concentrations of CK2 inhibitor TBB, lysed and WCE were used for GST pull-down assays. (D) Schematic of MDC1 protein sequence showing the GST fusion protein (150–350) used in the pull-down assays. In vitro binding of CK2-phosphorylated (150–350) region of MDC1 with HeLa WCE. Binding endogenous aprataxin to CK2-phosphorylated MDC1 (150–350) fragment was revealed by immunoblotting.
Binding of aprataxin to CK2-phosphorylated MDC1 pSDpTD sites
To confirm and quantify the interaction between the aprataxin FHA domain and CK2-phosphorylated MDC1, binding experiments were performed by ITC using recombinant aprataxin FHA domain and phosphorylated MDC1 peptides. FHA domains typically target pT residues (47,48), and within the first 500 residues of MDC1 there are seven threonine residues found in putative CK2 phosphorylation sites. Six of these are present in highly conserved sequences with the consensus D/N-S-D-T-D-x-E/D-E/D-E/D with additional preferences for small hydrophobic residues (Ala/Val) in the pT +2 position, and medium hydrophobic residues (Leu/Ile) in the pT−4 site (Figure 4A and Supplementary Figure S7). Whilst the threonine sites are predicted to be the best CK2 substrates, the preceding serines (S) are also putative phosphorylation sites for CK2. Therefore, both mono- and di-phosphorylated peptides were tested in ITC experiments (Figure 4B and C and Supplementary Table S1). A 14-residue peptide encompassing S329 and T331 was chosen, as these residues have been detected in the phosphorylated form of MDC1 in vivo (49–52). As shown in Figure 4B, the peptide phosphorylated only on T331 bound the aprataxin FHA domain stoichiometrically, albeit with an affinity of 58 µM. However, introduction of a second phosphate on residue S329 increased binding affinity ∼10-fold to 5.6 µM (Figure 4C) which is well within the range of binding affinities previously observed for FHA domains in other signalling contexts and similar to the 4.1 µM affinity observed by ITC for binding of polynucleotide kinase (PNK) FHA to CK2-phosphorylated XRCC4 (53). Importantly, no detectable binding to peptides phosphorylated only on S329 was observed even at high concentration in the ITC experiment (Figure 4B), demonstrating an absolute dependence of binding on threonine phosphorylation. A shorter di-phosphorylated peptide, terminating after the pT+3 position, bound aprataxin with comparable affinity to the longer version, possibly echoing the unusual lack of sequence selectivity in residues C-terminal to the pT observed by in vitro library screening of the PNK FHA domain (53; Supplementary Table S1). Thus, these data clearly demonstrate that aprataxin binds to a confirmed CK2 phosphorylation site in MDC1 in a pT-dependent fashion, its affinity is significantly elevated in the di-phospho form and supports the MDC1/aprataxin interaction data described in Figure 3. To substantiate ITC binding data, aprataxin GST pull downs were performed using GFP-MDC1 mutant constructs (Figure 4E). Wild-type (SDTD) MDC1 sequence, a mutant where the SDTD region was deleted (ΔSDTD), and a mutant where the key Thr residue was changed to Ala (SDAD)2 were expressed in HeLa cells and GST pull downs using aprataxin FHA domain were carried out. As expected, wild-type MDC1 was efficiently pulled down, while both the (ΔSDTD) and (SDAD)2 were not (Figure 4F). Together, these data suggest that the aprataxin FHA is able to detect varying phosphorylation levels that directly modulate its affinity for MDC1. Given the similarity in sequences, it seems highly likely that the aprataxin FHA domain can bind to any of the six sequences in MDC1 that contain the core SDTD motif. Since both aprataxin and Nbs1 bind to the same CK2-phosphorylated region of MDC1 (51,52,54), we determined whether Nbs1 could be in a complex with aprataxin and MDC1, and whether it was required for the recruitment of aprataxin to sites of DNA damage. Using Nbs1 defective cells, we demonstrate that both aprataxin binding to MDC1 and its recruitment to DNA breaks was independent of Nbs1 (Supplementary Figure S8). Thus, this suggests that MDC1 interacts with multiple proteins through a common CK2-phosphorylated region, and that it may reside in several distinct protein complexes within the cell. The presence of multiple phosphorylated SDTD motifs would provide a high local concentration of binding sites for aprataxin, Nbs1 and, potentially, other pS/T-dependent binding proteins.
Figure 4.
Aprataxin binds to a conserved di-phosphorylated CK2 motif in MDC1. (A) MDC1 contains six conserved motifs with a core SDTD motif that is di-phosphorylated in vivo (48–50). (B) ITC binding isotherms for interaction of wild-type aprataxin FHA with pSDTD and SDpTD phosphopeptides (C, D) ITC binding isotherms for the titration of pSDpTD phosphopeptide into wild-type and K38A aprataxin FHA, respectively. (E) Diagram of GFP-MDC1 mutants used in the pull-down assays. (F) Aprataxin FHA GST pull downs from HeLa cells expressing wild-type GFP-MDC1, a mutant containing a deletion of the SDTD region (ΔSDTD), and a mutant where the Thr residues contained in the SDTD sequence were replaced by Ala (SDAD)2.
Structure of the aprataxin FHA domain and a model for pSDpTD interactions
To further investigate the molecular basis of the phosphopeptide aprataxin FHA interaction, we attempted to crystallize the aprataxin FHA domain both with and without bound phosphopeptide. The apo protein crystallized readily and the structure was determined by molecular replacement using PNK FHA domain as the search model (1YJM). Residues 1–102 of the aprataxin FHA domain were modelled into the density and the structure was refined to an Rfree value of 23.7% at 1.65Å, with excellent stereochemistry (Supplementary Table S2). As expected the aprataxin FHA domain is structurally similar to that of PNK, with which it shares 39% sequence identity, and both adopt the classical FHA β-sandwich topology. The two structures superimpose with an overall root mean-square deviation in alpha carbon atoms of 0.6 Å for a core of 82 matched Cα’s, with largest deviations occurring in the loops connecting β1–β2 and β6–β7 (Figure 5A), consistent with the pattern of sequence conservation (Figure 5B). Despite extensive screening, the aprataxin FHA domain has, thus far, failed to crystallize in complex with either the mono- or di-phosphorylated MDC1 peptide. However, although the PNK and aprataxin FHA domains have only 39% sequence identity, they do show a remarkably high structural conservation at and around the phosphopeptide binding site. We were, therefore, able to model the pSDpTD peptide into the apo structure, using the PNK FHA domain structure with an XRCC4 phosphopeptide bound as a guide (55; Figure 5C). As has been observed in all previously characterized FHA–phosphopeptide interactions, pT recognition is provided by Arg29 and Ser41 (equivalent to PNK Arg35 and Ser47).
Figure 5.
Structural comparison of the aprataxin and PNK FHA domains and modelling of the aprataxin–MDC1 interaction. (A) Superposition of Cα backbone structures of the aprataxin FHA domain (green) with that of the PNK FHA/XRCC4 phosphopeptide complex (grey). (B) Sequence alignment of the core FHA domains residues from aprataxin and PNK. Identical positions are shown in red with the highly conserved arginine and serine residues that make canonical pT contacts in all available FHA-phosphopeptide structures are boxed. Black dots highlight the five Arg/Lys residues conserved between the two proteins. (C) Modelling of the aprataxin FHA–pSDpTD MDC1 phospho-motif complex (left) based on the crystal structure of the aprataxin FHA domain and the PNK–XRCC4 complex structure (right).
The weakly conserved isoleucine at the pT−4 position of the MDC1 peptide is in contact with Pro31, which is conserved in PNK FHA and which forms a platform for interaction with a pT−4 tyrosine in the PNK/XRCC4 complex (55). Arg42 (PNK Arg48) occupies a position that is weakly conserved in the FHA domain family, and is suitably positioned for pT interaction. Most importantly, the model suggests that selectivity for pS in the pT–2 position is provided by a basic surface created by Arg29 and Lys38. In support of this, mutation of Arg44 in PNK, which is structurally equivalent to aprataxin Lys38, abolishes PNK binding to XRCC4 phosphopeptides containing a glutamate rather than pS at the –2 position (55). To test this hypothesis ITC experiments were repeated with Arg29Ala (R29A) and Lys38Ala (K38A) mutant FHA domains. Consonant with its highly conserved role in pT interactions in all FHA domains, the R29A mutant reduced binding to the di-phosphorylated MDC1 peptide to undetectable levels. For K38A, both the monothreonine phosphorylated and di-phosphorylated peptides bind to the mutant with affinities comparable to that observed for wild-type FHA interaction with the monothreonine phosphorylated ligand (Figure 4D and Supplementary Table S1). Thus, substitution of Lys38 effectively eliminates the additional binding affinity provided by accessory serine phosphorylation, consistent with our structural model. Overall, the functional significance of the extended binding surface evident from these experiments is exemplified by the fact that Lys38, Lys39, Arg29 and Arg42 constitute four of only five basic residues conserved in PNK from a total of 15 present in the aprataxin FHA (Figure 5B).
Aprataxin FHA Arg29Ala mutant ablates the phospho-dependent aprataxin–MDC1 interaction
ITC data using purified aprataxin FHA and synthetic peptides, together with the crystallographic structure of aprataxin FHA and in silico modelling of MDC1 peptide binding demonstrated the importance of basic residues which include Arg29 and Lys38 for the binding of di-phosphorylated MDC1 peptide. To confirm these data, aprataxin GST pull downs using both Arg29Ala (R29A) and Lys38Ala (K38A) mutants were performed with total cell extracts. While MDC1 bound to the wild-type FHA sequence, R29A FHA mutant ablated MDC1 binding (Figure 6A), in agreement with the ITC binding and modelling data. Similarly, substitution of Lys38 effectively eliminates the additional binding affinity provided by accessory serine phosphorylation (Figure 4D) and, consistent with the extremely low affinity (∼80 µM), GST pull downs with the K38A FHA mutant also did not show any significant interaction with MDC1 (Figure 6A).
Figure 6.
Key role of aprataxin FHA domain in targeting aprataxin to sites of DNA damage in vivo. (A) Substitution of Arg29 and Lys38 to Ala abrogated the interaction between aprataxin and MDC1 as determined by pull-down assays as described earlier. (B) Effect of Arg29Ala and Lys38Ala mutations on the recruitment of aprataxin to high LET heavy ion-induced DSB in HeLa cells. HeLa cells were transiently transfected with wild-type GFP-aprataxin, GFP-aprataxin R29A or GFP-aprataxin K38A mutants, and cells were subjected to nickel ions irradiation (6.0 MeV/nucleon; LET 3430 keV/µm). DSB are visualized by γH2AX immunostaining and nuclei by ToPro3 staining.
Recruitment of aprataxin to sites of DNA damage is abolished by Arg29Ala mutation
To investigate the functional importance of the aprataxin FHA domain as a protein–protein interaction platform and its potential role in targeting aprataxin to DNA damage, we examined the effect of Arg29Ala and Lys38Ala FHA mutations on the in vivo recruitment of aprataxin to sites of DNA breaks. Recruitment of aprataxin to heavy ion-induced DNA damage was visualized using GFP-aprataxin containing either the wild-type sequence, R29A or K38A substitutions. GFP-aprataxin carrying the wild-type sequence was effectively recruited to DNA breaks as demonstrated by the co-localization with γH2AX along the particle tracks (Figure 6B) and as previously shown in Figure 1. Consistent with the ITC experiments, no recruitment to sites of DNA damage was observed with the R29A FHA mutant, while recruitment was reduced in the K38A aprataxin mutant (Figure 6B). Similar results were obtained with laser irradiation (data not shown). These data demonstrate the importance of the FHA domain in mediating phosphorylation-dependent protein–protein interactions and in targeting aprataxin to sites of DNA breaks.
Normal repair of IR-induced DSB in aprataxin-deficient cells
The interaction between aprataxin, XRCC4 and MDC1 and its recruitment to heavy ion irradiation-induced DNA damage suggest a role for aprataxin in the repair of DSB. To get further insight on the functional relevance of aprataxin recruitment to DNA breaks and its interaction with MDC1, we transfected both wild-type and FHA R29A mutant full length APTX cDNAs in AOA1 (FD105) fibroblasts and measured the repair kinetics of DSB in these cells. Repair of DSB was monitored by imaging the disappearance of γH2AX foci, a surrogate marker for DSB formation and rejoining, after low LET IR. Similar DSB repair kinetics were observed in AOA1 and AOA1 complemented with wild-type GFP-aprataxin, indicating that aprataxin is not necessary for the repair of DSB induced by low LET irradiation (Figure 7A). AOA1 cells transfected with GFP-aprataxin R29A exhibited a similar DSB repair kinetic (Figure 7A). While aprataxin has been implicated in DSB processing through its interaction with XRCC4, and the interaction with MDC1 described here, we failed to detect any significant difference in the repair kinetics of low LET-induced DSB in the presence or absence of aprataxin. Similar results were obtained for H2O2-induced DSB (Supplementary Figure S9). While surprising to some extent, these data are in agreement with previous reports which failed to detect a significant DSB repair defect in AOA1 after IR and H2O2 treatments (9,19). Not surprisingly, Mdc1−/− cells display some defect in the repair of DNA DSB induced by low LET IR (Figure 7B). These data are in agreement with the reported role of MDC1 in the signalling and repair of IR-induced DNA DSB (30–34,56). While high LET heavy ion radiation, IR and H2O2 are all capable of inducing DSB, they also induce a variety of DNA lesions that include base damage and SSB (45). Although MDC1 has been solely implicated in DSB recognition and repair, the induction of SSB by high LET heavy ion radiation prompted us to investigate whether MDC1 may also participate in SSB repair. In contrast to Aptx−/− cells where there is evidence for a defect in the repair of DNA SSB, normal SSB repair was observed in Mdc1−/− cells (Supplementary Figure S10), indicating that MDC1 is not required for the repair of SSB, and that the interaction described here between aprataxin and MDC1 may only be relevant to a subset of DSB, those resulting from abortive DNA ligation attempts.
Figure 7.
Normal repair of IR-induced DSB in AOA1 cells. (A) γH2AX foci repair kinetic after low LET ionizing radiation (IR; 2 Gy). AOA1 fibroblasts (FD105) were transfected with either GFP-aprataxin (wild-type) or GFP-aprataxin (R29A) mutant. Untransfected cells were used as control. Graph represents average numbers of foci per cell ± SD. Foci were counted in 50–60 individual cells per time point. DSB repair kinetics were similar regardless of the presence of absence of aprataxin indicating that aprataxin is not required for low LET IR-induced DSB repair. (B) Repair kinetics of DSB (γH2AX foci) after low LET irradiation (IR) in wild-type and Mdc1−/− cells. Graph represents average number of foci per cell ± SD. Foci were counted in 50–60 individual cells per time points a significant difference was observed at 6 and 8 h post-irradiation (*P < 0.05, Student’s t-test).
DISCUSSION
Previous reports have shown that aprataxin interacts with XRCC1, Ligase III and PARP-1, implicating it in the repair of SSB (8–10). Further evidence for its role in repair of single strand interruptions in DNA was provided by Ahel et al. (2006) who showed that it resolves abortive DNA ligation intermediates. We have provided additional evidence here for a defect in SSB repair in AOA1 cells after exposure to H2O2 which supports the specific role of aprataxin in repairing only a subgroup of relatively ‘slowly processed’ SSB carrying abortive ligation intermediates, since the reduced rate of repair in Aptx−/− cells is only significant at later time points (>15 min). Importantly, a delay in the repair of DNA SSB was only detected in non-proliferating cells, consistent with the results of Reynolds et al. (2009) where a defect in SSB repair was observed in AOA1 cells after inhibiting proliferation with aphidicolin (57). These data are in agreement with the SSB repair delay observed in AOA1 fibroblasts caused by intracellular accumulation of reactive oxygen species, where this repair defect was also evident after 1 h post-treatment (12). Notwithstanding these observations there exists some disagreement in the literature on the capacity of AOA1 cells to repair SSB (9,19). This may be explained by the specific mutation in AOA1, the cell type used (proliferating versus non-proliferating) or the repair assay, alkaline comet versus the alkaline elution assay.
More recently, Reynolds et al. (2009) demonstrated that short-patch SSB repair in AOA1 cell extracts bypasses the point of aprataxin action at oxidative breaks, resulting in the accumulation of adenylated DNA nicks. This defect results from insufficient levels of non-adenylated DNA ligase, and short-patch SSB repair could be restored in AOA1 extracts by the addition of recombinant DNA ligase (57). In addition, aphidicolin inhibition of long-patch repair uncovered a significant defect in SSB repair in Aptx−/− neural astrocytes while it did not affect SSB repair rates in wild-type mouse neural astrocytes (57). Most recently, Harris et al. (2009) provided evidence for reduced expression of key base excision repair proteins (PARP-1, APE1 and OGG1) in AOA1 cells, and revealed the presence of elevated levels of oxidative DNA damage in these cells coupled with reduced base excision and gap filling repair efficiencies (17). Thus, a synergy between aprataxin, PARP-1, APE-1 and OGG1 in the DNA damage response uncovered both direct and indirect modulating functions for aprataxin in base excision repair (17). In addition, we also have shown here that aprataxin was not recruited to DNA breaks in the absence of XRCC1. Together, these data demonstrate that aprataxin participates in chromosomal SSB repair in vivo, that it modulates base excision repair efficiency and suggest that short-patch SSB repair is arrested in AOA1 because of insufficient non-adenylated DNA ligase.
Clements et al. (2004) identified XRCC4 as a binding partner for aprataxin using a yeast 2-hybrid screen and confirmed this by demonstrating co-IP with DNA ligase IV, a constitutive binding partner. Since XRCC4/Ligase IV is an essential component of the non-homologous end-joining (NHEJ) pathway these data suggested that aprataxin might also be involved in proof-reading or processing of DSB. We provide here compelling evidence that aprataxin is recruited to sites of DNA damage and interacts with MDC1, an important component of the DNA damage response. MDC1 is required for efficient phosphorylation of H2AX, for recruitment and retention of ATM and MRN adjacent to break sites (29,30,34,35) and for the recruitment of the ubiquitin ligase RNF8 to ubiquitinate H2AX at damaged sites (58–61). This protein appears to act as a scaffold to bring together several DNA damage response proteins to mediate and enhance the recognition and signalling of DSB (58–66). Our data add an additional DNA repair protein, aprataxin, to the list of proteins interacting with MDC1. MDC1 interacts with DNA damage response proteins in a number of different ways. Interaction with γH2AX and the anaphase-promoting complex occurs through its C-terminal, BRCT domain (33,34,65), while it binds DNA-PK through an internal set of repeat sequences (32). It also possesses an FHA domain (amino acids 55–124) that mediates interaction with Chk2 and Rad51 (33,66). It employs an S/TQ rich region for ATM-dependent interaction with the ubiquitin ligase RNF8 (58).
We have described here a novel interaction between MDC1 and aprataxin, involving an extended cluster of CK2 phosphorylation sites (S-D-T-D) located downstream from the MDC1 FHA domain (amino acids 150–350). This was confirmed by the use of CK2 inhibitors, binding studies after in vitro CK2 phosphorylation, GST pull downs and modelling studies based on binding of mono- and di-phosphorylated peptides to the aprataxin FHA domain and the X-ray crystal structure of this domain. We provide here compelling evidence for a role for CK2 activity in the interaction of this region of MDC1 with aprataxin and show that a phosphopeptide containing a core pSDpTD motif corresponding to one of these sites binds to the FHA domain of aprataxin with high affinity. While a single threonine phosphorylation is both necessary and sufficient for binding, affinity is substantially increased by secondary serine phosphorylation. This, in turn, appears to provide for accessory interactions with an extended FHA phosphopeptide binding surface not previously documented. The data presented here uncover another important role for CK2 in mediating the interaction between aprataxin and the DSB repair protein MDC1.
In three recent reports constitutive phosphorylation of the N-terminal SDT repeats of MDC1 by CK2 has been demonstrated in the recruitment of the MRN complex to sites of DSB through direct interactions with the FHA domain of Nbs1 (51,52,54). Spycher et al. (54) reported that doubly phosphorylated pSDpT motifs regulate the accumulation and retention of the MRN complex in the DSB-flanking chromatin, and depletion of CK2 disrupted the MDC1–MRN complex and DNA damage-induced Nbs1 foci formation. Moreover, disruption of the SDT phosphoacceptor sites on MDC1 prevented the recruitment of Nbs1 to damaged chromatin (51,52,54). We have demonstrated that Nbs1 is not required for the binding of aprataxin to MDC1 and its recruitment to sites of DNA damage. Therefore, MDC1 interacts with multiple proteins through a common CK2-phosphorylated region and may reside in several distinct protein complexes within the cell. It is likely that the interaction between MDC1/Nbs1 and MDC1/aprataxin is influenced or regulated by the relative abundance of these proteins in the cells. These data together with previous findings that identified a direct role for CK2 in the repair of chromosomal DNA strand breaks (67), the CK2-dependent aprataxin/XRCC1 (9,10) and aprataxin/XRCC4 (9) interactions provide growing evidence for a role for CK2 in maintaining genome integrity by contributing to protein–protein complexes formation and/or recruitment of DNA damage regulators/repair factors at sites of DNA lesions.
As shown here, aprataxin re-localizes rapidly to discrete foci along charged particle tracks that overlap with γH2AX foci, a marker of DSB after exposure of cells to high LET radiation which gives rise to complex damage in DNA (22). The recruitment and co-localization of aprataxin to sites of γH2AX-labelled DNA breaks has previously been observed using different heavy ion species (8,17,28, this work). However, while MDC1 completely co-localized with γH2AX in the DSB-flanking microenvironment as reported earlier (68), aprataxin only partially co-localized with MDC1 and γH2AX in smaller foci (microfoci). This suggests that aprataxin may be recruited only to the DSB or a sub-group of DNA breaks directly within sites of complex or clustered damage or that its association with DNA damaged sites may be more transient. This transient behaviour has been described for Nbs1 in the absence of MDC1 (26,52). In addition, we provide evidence here that the lack of MDC1 doesn’t affect the rapid recruitment of aprataxin to sites of high LET-induced DNA DSB, but in contrast affects the retention of aprataxin at sites of DNA breaks, supporting a role for MDC1 as a scaffold during the DNA damage response. Proteins such as SMC1 and DNA-PKcs that are clearly involved in the DNA damage response cannot be localized to sites of DNA damage except after exposure to extremely high doses of radiation (68). Spatial-co-localization studies revealed that unlike γH2AX and MDC1, phosphorylated DNA-PKcs was localized only at very specific regions, presumably representing sites of DSB within the tracks (69). The presence of microfoci surrounded by regions of γH2AX-modified chromatin after laser-induced DNA damage has been reported (68). Several factors involved in DNA repair including Nbs1 and Mre11 are present in microfoci located in the centre of microirradiated tracks and surrounded by vast regions of γH2AX-modified chromatin (68,70). In the present context these microirradiated tracks coincide with MDC1-modified chromatin. The heterogeneity of staining may be explained by the presence of significant amounts of ssDNA adjacent to DSB and/or the presence of clustered or complex DNA damage (45,46). Exposure of cells to low LET radiation (IR) or H2O2 failed to localize aprataxin to sites of damage (8). This may be similar to that observed for SMC1 or DNA-PK where extremely high levels of damage are required to see such localization (68). Low LET radiation is more sparsely ionizing and thus less damaging to DNA than the complex forms of damage arising from high LET exposure which could explain the failure to see aprataxin foci after low LET radiation (45). The difference in capacity of aprataxin to localize to sites of DNA damage after the two types of IR correlates well with the extent of repair of DNA DSB. The rate of repair of DNA DSB in response to low LET radiation was unaffected by aprataxin status of the cells. Similar to previous reports (9,19), the failure to detect any significant DSB repair defect in the presence or absence of aprataxin after low LET irradiation or high doses of H2O2 suggests that aprataxin recruitment to DNA breaks may only be relevant for a sub-group of breaks, possibly more frequently/effectively induced by high energy and high LET heavy ions irradiation. This sub-group of breaks could represent those where abortive ligation was occurring, a property of AOA1 cells (14).
The interaction of aprataxin with DNA damage recognition and repair proteins suggest that aprataxin participates in the processing of some SSB and DSB to resolve abortive DNA ligations. The model described in Figure 8 provides an integrated approach as to how aprataxin may function at sites of damage (DSB and SSB) in DNA. This involves recruitment to damaged sites by XRCC1, MDC1 and other unidentified factors, followed by interaction with either XRCC1/LigaseIII or XRCC4/LigaseIV to assist in the DNA end-processing and resealing of SSB and DSB, respectively. In summary, it is likely that aprataxin contributes to the repair of both DNA SSB and DSB in its capacity to resolve abortive ligation intermediates. This is most evident when cells are exposed to agents that form complex damage in DNA.
Figure 8.
A model for the dual role of aprataxin in SSB and DSB repair. The FHA domain of aprataxin, through which it interacts with MDC1, XRCC1 and XRCC4, is shown three times to represent a series of dynamic interactions with these three proteins or to represent individual aprataxin molecules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by grants from the Australian National Health & Medical Research Council (NHMRC) (M.F.L.); and by the Medical Research Council, UK (S.J.S.). Funding for open access charge: Australian National Health & Medical Research Council (NHMRC).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Farrell for excellent cell culture assistance, Drs M. Goldberg, R. Chapman and S. Jackson (The Wellcome Trust/Cancer Research, UK) for the kind gift of the GFP-MDC1 vectors, and Prof. Keith Caldecott (Genome Damage and Stability Centre, University of Sussex, UK) for the AOA1 fibroblast line (FD105).
REFERENCES
- 1.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, et al. The gene mutated in ataxia-ocular apraxia type 1 encodes the new HIT/Zn-finger protein aprataxin. Nat. Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 2.Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat. Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 3.Aicardi J, Barbosa C, Andermann E, Andermann F, Morcos R, Ghanem Q, Fukuyama Y, Awaya Y, Moe P. Ataxia-ocular motor apraxia: asyndrome mimicking ataxia-telangiectasia. Ann. Neurol. 1988;4:497–502. doi: 10.1002/ana.410240404. [DOI] [PubMed] [Google Scholar]
- 4.Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 5.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6:931–942. doi: 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 6.Caldecott KW. DNA single-strand break repair and spinocerebellar ataxia. Cell. 2003;112:7–10. doi: 10.1016/s0092-8674(02)01247-3. [DOI] [PubMed] [Google Scholar]
- 7.Brenner C, Bieganowski P, Pace HC, Huebner K. The histidine triad superfamily of nucleotide-binding proteins. J. Cell Physiol. 1999;181:179–187. doi: 10.1002/(SICI)1097-4652(199911)181:2<179::AID-JCP1>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gueven N, Becherel OJ, Kijas AW, Chen P, Howe O, Rudolph JH, Gatti R, Date H, Onodera O, Taucher-Scholz G, et al. Aprataxin, a novel protein that protects against genotoxic stress. Hum. Mol. Genet. 2004;13:1081–1093. doi: 10.1093/hmg/ddh122. [DOI] [PubMed] [Google Scholar]
- 9.Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW. The ataxia-oculomotor apraxia type 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair. 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 10.Luo H, Chan DW, Yang T, Rodriguez M, Chen BP, Leng M, Mu JJ, Chen D, Songyang Z, Wang Y, et al. A new XRCC1-containing complex and its role in cellular survival of methyl methane sulfonate treatment. Mol. Cell. Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosesso P, Piane M, Palitti F, Pepe G, Penna S, Chessa L. The novel human gene aprataxin is directly involved in DNA single-strand break repair. Cell. Mol. Life Sci. 2005;62:485–491. doi: 10.1007/s00018-004-4441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano M, Yamamoto A, Mori T, Lan L, Iwamoto TA, Aoki M, Shimada K, Furiya Y, Kariya S, Asai H, et al. DNA single-strand break repair is impaired in aprataxin-related ataxia. Ann. Neurol. 2007;61:162–174. doi: 10.1002/ana.21078. [DOI] [PubMed] [Google Scholar]
- 13.Gueven N, Chen P, Nakamura J, Becherel OJ, Kijas AW, Grattan-Smith P, Lavin MF. A subgroup of spinocerebellar ataxias defective in DNA damage responses. Neuroscience. 2007;145:1418–1425. doi: 10.1016/j.neuroscience.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 15.Rass U, Ahel I, West SC. Actions of aprataxin in multiple DNA repair pathways. J. Biol. Chem. 2007;282:9469–9474. doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- 16.Lehman IR. DNA ligase: structure, mechanism, and function. Science. 1974;186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 17.Harris JL, Jakob B, Taucher-Scholz G, Becherel OJ, Lavin MF. Aprataxin combines with Poly-ADP Ribose Polymerase 1 (PARP-1) and Apurinic Endonuclease 1 (APE1) to protect the genome against oxidative damage. Hum. Mol. Genet. 2009;18:4102–4117. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- 18.Kijas AW, Harris JL, Harris JM, Lavin MF. Aprataxin forms a discrete branch in the HIT (histidine triad) superfamily of proteins with both DNA/RNA binding and nucleotide hydrolase activities. J. Biol. Chem. 2006;281:13939–13948. doi: 10.1074/jbc.M507946200. [DOI] [PubMed] [Google Scholar]
- 19.El-Khamisy SF, Katyal S, Patel P, Ju L, McKinnon PJ, Caldecott KW. Synergistic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair. 2009;8:760–766. doi: 10.1016/j.dnarep.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 21.Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 23.Okano S, Kanno S, Nakajima S, Yasui A. Cellular responses and repair of single-strand breaks introduced by UV damage endonuclease in mammalian cells. J. Biol. Chem. 2000;275:32635–32641. doi: 10.1074/jbc.M004085200. [DOI] [PubMed] [Google Scholar]
- 24.Limoli CL, Ward JF. A new method for introducing double-strand breaks into cellular DNA. Radiat. Res. 1993;134:160–169. [PubMed] [Google Scholar]
- 25.Katsumi S, Kobayashi N, Imoto K, et al. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J. Invest. Dermatol. 2001;117:1156–1161. doi: 10.1046/j.0022-202x.2001.01540.x. [DOI] [PubMed] [Google Scholar]
- 26.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakob B, Scholz M, Taucher-Scholz G. Biological imaging of heavy charged-particle tracks. Radiat. Res. 2003;159:676–684. doi: 10.1667/0033-7587(2003)159[0676:biohct]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Jakob B, Rudolph JH, Gueven N, Lavin MF, Taucher-Scholz G. Live cell imaging of heavy-ion-induced radiation responses by beamline microscopy. Radiat. Res. 2005;163:681–690. doi: 10.1667/rr3374. [DOI] [PubMed] [Google Scholar]
- 29.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg M, Stucki M, Falck J, D'A;mours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 31.Lou Z, Chini CC, Minter-Dykhouse K, Chen J. Mediator of DNA damage checkpoint protein 1 regulates BRCA1 localization and phosphorylation in DNA damage checkpoint control. J. Biol. Chem. 2003;278:13599–13602. doi: 10.1074/jbc.C300060200. [DOI] [PubMed] [Google Scholar]
- 32.Lou Z, Chen BP, Asaithamby A, Minter-Dykhouse K, Chen DJ, Chen J. MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J. Biol. Chem. 2004;279:46359–46362. doi: 10.1074/jbc.C400375200. [DOI] [PubMed] [Google Scholar]
- 33.Lou Z, Minter-Dykhouse K, Wu X, Chen J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature. 2003;421:957–961. doi: 10.1038/nature01447. [DOI] [PubMed] [Google Scholar]
- 34.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 35.Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl Acad. Sci. USA. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahajan A, Yuan C, Lee H, Chen ES, Wu PY, Tsai MD. Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal. 2008;1:re12. doi: 10.1126/scisignal.151re12. [DOI] [PubMed] [Google Scholar]
- 37.Becherel OJ, Gueven N, Birrell GW, Schreiber V, Suraweera A, Jakob B, Taucher-Scholz G, Lavin MF. Nucleolar localization of aprataxin is dependent on interaction with nucleolin and on active ribosomal DNA transcription. Hum. Mol. Genet. 2006;15:2239–2249. doi: 10.1093/hmg/ddl149. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr., 50, 760–763. [Google Scholar]
- 40.Navaza J, Saludjian P, Charles WC., Jr AMoRe: an automated molecular replacement program package. Methods Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Suraweera A, Becherel OJ, Chen P, Rundle N, Woods R, Nakamura J, Gatei M, Criscuolo C, Filla A, Chessa L, et al. Senataxin defective in ataxia oculomotor apraxia type 2 is involved in the defence against oxidative DNA damage. J. Cell Biol. 2007;177:969–979. doi: 10.1083/jcb.200701042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Epe B, Hegler J. Oxidative DNA damage: endonuclease fingerprinting. Methods Enzymol. 1994;234:122–131. doi: 10.1016/0076-6879(94)34083-8. [DOI] [PubMed] [Google Scholar]
- 45.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 46.Jakob B, Splinter J, Taucher-Scholz G. Positional stability of damaged chromatin domains along radiation tracks in mammalian cells. Radiat. Res. 2009;171:405–418. doi: 10.1667/RR1520.1. [DOI] [PubMed] [Google Scholar]
- 47.Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 48.Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- 49.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 51.Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11–RAD50–NBS1 complex to damaged chromatin. J. Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galcia S, Koch CA, Cass CE, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 56.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 57.Reynolds JJ, El-Khamisy SF, Katyal S, Clements P, McKinnon PJ, Caldecott KW. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia 1. Mol. Cell. Biol. 2009;29:1354–1362. doi: 10.1128/MCB.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 60.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakasai R, Tibbetts R. RNF8-dependent and RNF8-independent regulation of 53BP1 in response to DNA damage. J. Biol. Chem. 2008;283:13549–13555. doi: 10.1074/jbc.M710197200. [DOI] [PubMed] [Google Scholar]
- 62.Eliezer Y, Argaman L, Rhie A, Doherty AJ, Goldberg M. The direct interaction between 53BP1 and MDC1 is required for the recruitment of 53BP1 to sites of damage. J. Biol. Chem. 2008;284:426–435. doi: 10.1074/jbc.M807375200. [DOI] [PubMed] [Google Scholar]
- 63.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 64.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 65.Coster G, Hayouka Z, Argaman L, Strauss C, Friedler A, Brandeis M, Goldberg M. The DNA damage response mediator MDC1 directly interacts with the anaphase-promoting complex/cyclosome. J. Biol. Chem. 2007;282:32053–32064. doi: 10.1074/jbc.M705890200. [DOI] [PubMed] [Google Scholar]
- 66.Zhang J, Ma Z, Treszezamsky A, Powell SN. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat. Struct. Mol. Biol. 2005;12:902–909. doi: 10.1038/nsmb991. [DOI] [PubMed] [Google Scholar]
- 67.Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, Sarno S, Meggio F, Pinna LA, Caldecott KW. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell. 2004;117:17–28. doi: 10.1016/s0092-8674(04)00206-5. [DOI] [PubMed] [Google Scholar]
- 68.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asaithamby A, Uematsu N, Chatterjee A, Story MD, Burma S, Chen DJ. Repair of HZE-particle-induced DNA double-strand breaks in normal human fibroblasts. Radiat. Res. 2008;169:437–446. doi: 10.1667/RR1165.1. [DOI] [PubMed] [Google Scholar]
- 70.Lisby M, Antunez de Mayolo A, Mortensen UH, Rothstein R. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle. 2003;2:479–483. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.