Abstract
Hyperhomocysteinemia has long been associated with atherosclerosis and thrombosis and is an independent risk factor for cardiovascular disease. Its causes include both genetic and environmental factors. Although homocysteine is produced in every cell as an intermediate of the methionine cycle, the liver contributes the major portion found in circulation, and fatty liver is a common finding in homocystinuric patients. To understand the spectrum of proteins and associated pathways affected by hyperhomocysteinemia, we analyzed the mouse liver proteome of gene-induced (cystathionine β-synthase (CBS)) and diet-induced (high methionine) hyperhomocysteinemic mice using two-dimensional difference gel electrophoresis and Ingenuity Pathway Analysis. Nine proteins were identified whose expression was significantly changed by 2-fold (p ≤ 0.05) as a result of genotype, 27 proteins were changed as a result of diet, and 14 proteins were changed in response to genotype and diet. Importantly, three enzymes of the methionine cycle were up-regulated. S-Adenosylhomocysteine hydrolase increased in response to genotype and/or diet, whereas glycine N-methyltransferase and betaine-homocysteine methyltransferase only increased in response to diet. The antioxidant proteins peroxiredoxins 1 and 2 increased in wild-type mice fed the high methionine diet but not in the CBS mutants, suggesting a dysregulation in the antioxidant capacity of those animals. Furthermore, thioredoxin 1 decreased in both wild-type and CBS mutants on the diet but not in the mutants fed a control diet. Several urea cycle proteins increased in both diet groups; however, arginase 1 decreased in the CBS+/− mice fed the control diet. Pathway analysis identified the retinoid X receptor signaling pathway as the top ranked network associated with the CBS+/− genotype, whereas xenobiotic metabolism and the NRF2-mediated oxidative stress response were associated with the high methionine diet. Our results show that hyperhomocysteinemia, whether caused by a genetic mutation or diet, alters the abundance of several liver proteins involved in homocysteine/methionine metabolism, the urea cycle, and antioxidant defense.
Homocysteine (Hcy)1 is a thiol-containing amino acid that is produced in every cell of the body as an intermediate of the methionine cycle (Fig. 1, Reactions 1–5) (1). Once formed, the catabolism of homocysteine occurs via three enzymatic pathways. 1) Hcy is remethylated back to methionine using vitamin B12-dependent methionine synthase (Fig. 1, Reaction 4) and/or 2) betaine-homocysteine methyltransferase (BHMT) (Fig. 1, Reaction 5), and 3) Hcy is converted to cysteine via the transsulfuration pathway using CBS and γ-cystathionase (Fig. 1, Reactions 6 and 7). Under normal conditions ∼40–50% of the Hcy that is produced in the liver is remethylated, ∼40–50% is converted to cysteine, and a small amount is exported (1–3). However, when Hcy production is increased (i.e. increased dietary methionine/protein intake) or when Hcy catabolism is decreased (i.e. CBS deficiency or B vitamin deficiencies), excess Hcy is exported into the extracellular space, resulting in hyperhomocysteinemia (1–5).
Fig. 1.
Homocysteine metabolism in liver and kidney. In classical homocystinuria, the initial enzyme of the transsulfuration pathway, CBS (Reaction 6), is deficient. MTHF, methylenetetrahydrofolate; THF, tetrahydrofolate; DHF, dihydrofolate; MeCbl, methylcobalamin; DMG, dimethylglycine; PLP, pyridoxal 5′-phosphate.
Homocystinuria was first described in the 1960s by Carson et al. (6): they observed 10 pediatric patients with severely elevated levels of Hcy in the urine and hypermethioninemia. Normal concentrations of plasma total homocysteine (tHcy) range from 5 to 12 μm (7); however, in homocystinuria, tHcy levels can exceed 100 μm. Homocystinuric patients present with mental retardation, abnormal bone growth, fine hair, malar flush, and dislocation of the lens of the eye, and most die from premature cardiovascular disease (6, 8). Autopsy findings indicate widespread thromboembolism, arteriosclerosis, and fatty livers (6, 8). Mudd et al. (9, 10) identified the cause of homocystinuria as a defect in the enzyme cystathionine β-synthase. A recent study of newborn infants in Denmark estimated the birth prevalence for CBS heterozygosity to be about 1:20,000 (11).
Plasma tHcy concentrations are also directly correlated with dietary methionine/protein intake (12, 13). Guttormsen et al. (13) demonstrated that a protein-rich meal affected tHcy for at least 8–24 h. When normal subjects were fed a low protein-containing breakfast (12–15 g), plasma methionine levels increased slightly after 2 h (22.5–27.5 μm), but tHcy levels did not change significantly. However, when these same subjects were fed a high protein meal (52 g), plasma methionine levels peaked after 4 h (38 μm), and tHcy rose steadily until a maximum level was reached 8 h postmeal (7.6 versus 8.5 μm) (13). Thus, the following questions can be raised. How does the hepatic proteome respond to a hyperhomocysteinemic diet, and are the changes that accompany such a diet the same as or different from those that may be observed in gene-induced hyperhomocysteinemia?
Because hyperhomocysteinemia is a strong independent risk factor for cardiovascular, cerebrovascular, and peripheral vascular disease, most of the current research has focused on the mechanisms involved in Hcy-induced endothelial dysfunction (14–24). The results of those studies have concluded that Hcy induces intracellular oxidative stress by generating ROS, which in turn lead to decreased bioavailable nitric oxide (NO), altered gene expression, increased endoplasmic reticulum stress, and activation of cholesterol biosynthesis. Also, several studies have examined the association between hyperhomocysteinemia and alcoholic liver disease, but few have looked at the effect of Hcy on the non-alcoholic liver even though fatty liver is a constant finding in homocystinuria (6, 8), and the liver is the major source of circulating Hcy (4, 5, 10). We hypothesize that 1) the liver proteome will respond to hyperhomocysteinemia by altering the expression of proteins involved in methionine/homocysteine metabolism and antioxidant defense and that 2) the set of proteins that are expressed when hyperhomocysteinemia is induced by CBS deficiency will differ from those expressed as a result of a high methionine diet. In the present study, we use a well established mouse model of CBS deficiency to study the early changes in the liver proteome that accompany hyperhomocysteinemia (25).
EXPERIMENTAL PROCEDURES
Animals
Colonies of CBS-deficient mice (C57BL/6J background) (25) were bred and housed at the University of Iowa Animal Facilities. All protocols were approved by the University of Iowa's Animal Care Committee. Four experimental groups of animals were studied: 1) wild-type CBS+/+ mice on a control (C) diet, 2) wild-type CBS+/+ mice fed a high methionine (HM) diet, 3) heterozygote CBS+/− mice fed a C diet, and 4) heterozygote CBS+/− mice fed an HM diet. Each experimental group consisted of five to eight animals and included mice of both genders. Upon weaning (3–4 weeks of age), all animals were fed normal mouse chow (LM485, Harlan Teklad Inc., Madison, WI) for 2 weeks. A subgroup of CBS+/+ and CBS+/− mice was administered 0.5% methionine in their drinking water ad libitum. After the 2-week treatment period, EDTA-plasma samples were obtained, the mice were sacrificed, and the livers were removed and snap frozen in liquid N2. One portion of each liver was used for proteomics analysis, and a second portion was used for the determinations of Hcy, Cys, GSH, and Cys-Gly.
Sample Preparation for Thiol Determinations and Thiol Stability
The livers were thawed and dissected into two pieces weighing ∼100 mg each. The portion to be used for total intracellular thiol determinations was homogenized (100 mg of tissue/1 ml of homogenization buffer) in ice-cold 0.1 m KH2PO4 (pH 7.4) containing protease inhibitors (10 μg/ml tosyl lysine chloromethyl ketone, 10 μg/ml soybean trypsin inhibitor, 0.6 mm PMSF, and 3.5 μg/ml aprotinin) for 20 s using a Tissue Tearor homogenizer set at 50% power (Biospec Products, Bartlesville, OK). In these experiments, total thiols were defined as the sum of free reduced thiols, small molecular weight disulfides, and protein-bound thiols. Cysteines that were part of the backbone of the protein were excluded. Because tissue samples were not homogenized under acidic conditions, which preserves thiols from ex vivo oxidizing conditions (26, 27), thiol stability was determined in four normal mouse liver homogenates by spiking the samples with either 50 μm l-Hcy, 250 μm l-Cys, or 500 μm GSH and analyzing the samples 24 and 96 h later. Protein concentrations were determined using the BCA protein assay (Pierce).
Liver Thiol Determinations
Hepatic concentrations of Hcy, Cys, Cys-Gly, and GSH were simultaneously determined in the KH2PO4 homogenates using HPLC with fluorescence detection (28). Briefly, 100 μl of liver homogenate was mixed with 10 μl of 50 mm EDTA containing 1.0 mm dithioerythritol and reduced with 0.3 m sodium borohydride. After neutralization with HCl, the thiols were fluorescently labeled with 0.70 mm monobromobimane and deproteinized with 0.3 m perchloric acid. The perchloric acid supernatant was neutralized with 2 m Tris (pH 8), and a 0.040-ml aliquot was applied to a 4.6 × 150-mm C18 reverse phase HPLC column (Phenomenex Inc., Torrance, CA). The thiol-bimane adducts were resolved using an Agilent Series 1100 HPLC system (Agilent Technologies, Wilmington, DE) with fluorescence detection (λex = 390 nm; λem = 475 nm). A standard curve of each thiol was prepared in normal mouse liver extract. All results were normalized to protein and expressed as nmol of thiol/mg of protein.
NO Determination
The stable end products of NO metabolism, nitrite and nitrate, were determined in a 10-μl aliquot of the KH2PO4 homogenate using a commercial colorimetric assay (Active Motif, Carlsbad, CA). Total nitrate plus nitrite was expressed as nmol/mg of protein.
Glycine N-Methyltransferase (GNMT) and S-Adenosylhomocysteine Hydrolase (SAHH) ELISAs
An ELISA for GNMT was developed using rabbit anti-GNMT antibody (Abgent, San Diego, CA) as the capture antibody, and biotinylated rabbit anti-GNMT-streptavidin-horseradish peroxidase was used as the reporter complex. The GNMT protein used to generate the standard curve was a generous gift from Dr. Conrad Wagner (Vanderbilt University). Because no antibodies were available from species other than rabbit, it was necessary to biotinylate the non-labeled rabbit antibody to design a “sandwich” ELISA. A portion of the rabbit anti-GNMT antibody was biotinylated using the EZ-link Micro PEO4 biotinylation kit (Pierce). The concentrations of the samples were quantified using a regression equation derived from the GNMT standard curve (0–100 ng/ml). For SAHH, rabbit anti-SAHH antibody (Novus, Littleton, CO) was used as the capture antibody, and biotinylated rabbit anti-SAHH-streptavidin-horseradish peroxidase was used as the reporter complex. Once again there were no antibodies available from species other than rabbit. Consequently, a portion of the rabbit anti-SAHH was biotinylated using the EZ-link Micro PEO4 biotinylation kit (Pierce). The concentrations of the samples were quantified using a regression equation derived from the SAHH standard curve (0–20 ng/ml).
Sample Preparation and CyDye Labeling for Proteomics Studies
The portion of the liver used for proteomics analysis was homogenized (100 mg of tissue/1 ml of homogenization buffer) in 40 mm Tris, 2% SDS, 80 mm DTT (pH 8.5) containing protease inhibitors (10 μg/ml tosyl lysine chloromethyl ketone, 10 μg/ml soybean trypsin inhibitor, 0.6 mm PMSF, and 3.5 μg/ml aprotinin) for 20 s using a Tissue Tearor homogenizer set at 50% power. Following homogenization, a 100–200-μl aliquot of the SDS homogenate was removed and treated with the 2-D Clean-up kit (GE Healthcare) to remove SDS, salts, lipids, and nucleic acids and to quantitatively precipitate protein. The precipitated proteins were resuspended in 100–200 μl of CyDye labeling buffer (7 m urea, 2 m thiourea, 4% CHAPS, 50 mm Tris (pH 8.6)), and protein concentration was determined using the 2-D Quant kit (GE Healthcare). From this solution, 50 μg of each sample was labeled with 400 pmol of either Cy3 or Cy5 at pH 8.6 for 30 min on ice. A Cy2-labeled internal standard (IS) was prepared by pooling 50 μg of each sample and labeling that pool with 400 pmol of Cy2. The labeling reactions were stopped by addition of excess lysine to quench unreacted N-hydroxysuccinimide groups on the CyDyes. The Cy2 IS was used on each gel for normalization and to correct for gel to gel variability.
Two-dimensional (2-D) DIGE
To detect protein differences between two experimental conditions (i.e. control versus diet or wild type versus mutant), a modified 2-D electrophoresis procedure was used (29). Equal amounts of the three CyDye-labeled samples (50 μg of Cy3-labeled sample, 50 μg of Cy5-labeled sample, and 50 μg of Cy2-labeled IS) were admixed, and DeStreakTM sample buffer (GE Healthcare) containing 1% ampholytes (pH 3–10) was added to the mixture to a final volume of 450 μl. Each analytical gel was loaded with 150 μg of total protein. A preparatory scale gel was loaded with 750 μg of unlabeled IS. IEF was carried out on 24-cm Immobiline DryStrips (pH 3–10 non-linear; GE Healthcare) using the Ettan IPGphor IITM IEF apparatus (GE Healthcare). The sample (450 μl) was added to the IPG DryStrip and actively rehydrated for 11 h at 20 °C using 30 V and 50 μA/strip. Focusing was carried out at 20 °C using the following conditions: step 1: 0.5 h, 0–250 V; step 2: 0.5 h, 250–6000 V; step 3: 5 h, 6000 V; and step 4: 10 h, 6000 V for a total of 78 kV-h. After the focusing step, the strips were equilibrated in 50 mm Tris-HCl, 6 m urea, 2% SDS, 30% glycerol, 2% DTT, 0.002% bromphenol blue (pH 8.8) for 10 min followed by a second 10-min equilibration step in 50 mm Tris-HCl, 6 m urea, 2% SDS, 30% glycerol, 0.002% bromphenol blue, 2% iodoacetamide (pH 8.8). The IPG strips were then electrophoresed on a 24-cm 12.5% precast SDS-polyacrylamide gel (Jule Inc., Milford, CT) for 4 h at constant power (5 watts/strip for 1 h; 20 watts/strip for 3 h) in an Ettan DALTTMtwelve vertical electrophoresis system (GE Healthcare) using the Laemmli buffer system (30). The preparatory scale gel was fixed and stained with GelCode BlueTM Coomassie stain (Pierce).
Image Acquisition and DeCyder Analysis
The gels were analyzed by postrun fluorescence imaging using the Typhoon TrioTM imager (GE Healthcare). The Cy2 images were acquired with a 488 nm excitation laser and a 520 nm emission filter, the Cy3 images used a 532 nm excitation laser with a 580 nm emission filter, and the Cy5 images were acquired with a 633 nm excitation laser and a 670 nm emission filter. The Coomassie-stained preparatory gel was scanned with the Cy5 excitation laser (633 nm) without the use of an emission filter. The resolution was set to 200 μm. After the multiplexed images were acquired, image analysis was performed using DeCyderTM 5.01 software (GE Healthcare) using both the “differential in-gel analysis” module and the “biological variation analysis” module. Protein spots exhibiting a statistically significant (p ≤ 0.05) 2-fold difference in intensity between experimental groups were excised from the preparatory gel and identified by LC-MS-MS as described by Kinter and Sherman (31).
2-D DIGE Validation
Although the manufacturer of the CyDyes reports that they are matched for charge and molecular weight, it was important to experimentally confirm 1) that labeling of any given protein with either of the CyDyes did not change the pI or mobility of that specific protein and 2) that the fluorescence output (pixel intensity) of each dye-labeled protein in co-detected spots was the same. To that end, five mouse liver homogenates were labeled with either Cy2, Cy3, or Cy5, and 2-D DIGE analysis was performed on each admixed sample. Additionally, the intra-assay coefficient of variation was determined by running three 2-D DIGE gels per sample on the same day, and the inter-assay coefficient of variation was determined by running three 2-D DIGE gels per sample on 2 different days. The Cy2 signal data were used for generating the coefficient of variation results.
Identification of Differentially Expressed Proteins by LC-Tandem Mass Spectrometry
The excised spots were minced into 1-mm3 pieces; destained in 50% ethanol, 5% acetic acid; and dehydrated with acetonitrile. The dried gel pieces were digested with 5 μl of trypsin (20 μg/ml) in 50 mm ammonium bicarbonate for 18 h at 23 °C. The samples were microcentrifuged briefly and treated two times with 30 μl of 50% acetonitrile, 5% formic acid to extract the peptides. The extracts were pooled and concentrated to <10 μl using a SpeedVac. The volume was readjusted to 30 μl with 1% acetic acid, and LC-MS-MS analysis was carried out directly on this sample. An linear trap quadrupole (LTQ) ion trap mass spectrometer (Thermo Scientific, San Jose, CA) was used for LC-MS-MS analysis. The HPLC column was a self-packed 8-cm × 75-μm Phenomenex Jupiter C18 reverse phase capillary HPLC column. Samples (10 μl) were injected onto the column and eluted with an acetonitrile, 0.05% acetic acid gradient at a flow rate of 0.2 μl/min. The eluted peptides were directly introduced into the ion source of the mass spectrometer, which was operated at 3.5 kV. The instrument was operated in a data-dependent manner in which full mass spectra (300–2000 Da) were acquired to determine molecular weights, and the four most abundant ions were selected for CID scans. Approximately 3000 CID scans were produced by this mode of analysis. The data from the CID spectra were used to search the 2006 version of the NCBI database and also the Reference Sequence database for matching peptide sequences using the search engine Mascot version 2.0 (Matrix Science, Boston, MA). All identifications were verified by manual inspection of at least three matching CID spectra. All reported identifications had Mascot scores >100.
Database Search of Protein Sequences
Peak lists were extracted using Thermo Electron Excalibur v2.0 subroutine extract.msn.exe. The resultant peak lists were searched with Mascot version 2.0 (Matrix Sciences, London, UK) against the mouse and rat Reference Sequence database (August 25, 2006, 81,049 sequences total). The search parameters were as follows: species, rodent; enzyme, trypsin; number of missed cleavages allowed, 1; fixed modifications, carbamidomethylated cysteine; variable modifications, oxidized methionine; precursor ion mass tolerance, ±2.0 Da; fragment mass tolerance, ± 2.0 Da.
Pathway and Network Analysis
Pathway analysis to identify biological networks associated with hyperhomocysteinemia was performed using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Mountain View, CA). Pathway and network analysis aids the interpretation of the relationships between the 2-D DIGE-identified proteins and facilitates the identification of relatively low abundance proteins that may be missed by 2-D DIGE analysis. The data set consisting of the differentially expressed proteins identified by 2-D DIGE was uploaded into the IPA Knowledge Database, which mapped the proteins (gene/gene products) to global molecular networks to identify focus genes/gene products that are known to directly interact with other gene products in the database. Canonical pathway analysis revealed the biological pathways, networks, and diseases associated with the 2-D DIGE-identified proteins.
Statistics
All results were expressed as mean ± S.D. In all cases, non-paired two-tailed Student's t tests or one-way analysis of variance were used to determine significance of the DIGE results, and false discovery rates were applied to all comparisons. Fischer's exact test was used by IPA to determine the p values for all of the network and pathway analyses. In all cases, a p value ≤0.05 was considered statistically significant.
RESULTS
Total Plasma Homocysteine in CBS-deficient Mice
As expected both the HM diet and CBS deficiency resulted in significantly elevated tHcy (Fig. 2A). A small, but significant increase was seen in the CBS-deficient mice on the control diet (CBS+/−, C) versus wild-type animals (CBS+/+, C) (4.27 ± 0.27 μm (n = 4) versus 3.15 ± 0.35 μm (n = 6), respectively, p ≤ 0.05). However, exposure to the HM diet resulted in a 3-fold increase in tHcy in wild-type animals (CBS+/+, HM = 10.7 ± 2.24 μm (n = 6), p ≤ 0.05), and ≈20-fold increase in tHcy in CBS+/− mice (CBS+/−, HM = 61.45 ± 17.46 μm (n = 4), p ≤ 0.05) (Fig. 2A). Thus, there is a dramatic gene-nutrient effect on plasma tHcy levels in this model.
Fig. 2.
Effect of CBS genotype and HM diet on plasma homocysteine, liver thiols, and liver nitrite/nitrate concentrations. A, tHcy was measured in CBS+/+ and CBS+/− mice after 14 days on a C diet or a diet supplemented with 0.5% methionine in the drinking water (HM diet). Total liver concentrations of Hcy (B), Cys (C), GSH (D), Cys-Gly (E), and nitrite/nitrate (NOx; F) were determined on tissue homogenates from the same animals. Results were normalized to protein concentration and are expressed as mean ± S.D. *, p ≤ 0.05 versus (+/+) C; n = 4–6 animals per group. Error bars indicate ± 1 S.D.
It is important to point out that all four animal groups in this study were composed of both male and female animals. Because the number of each gender within each group was low (between n = 2 and n = 4) any specific sex-related influence on plasma or liver Hcy could have been masked. Recently, it was reported that female mice have higher levels of plasma tHcy compared with males, and this difference was attributed to lower CBS activities in the female kidney (32). In the current study, there was a non-significant trend (p = 0.18) toward higher plasma tHcy levels in the female mice; however, larger animal numbers are needed to confirm this observation. When plasma tHcy levels were measured in a larger colony of CBS+/+ and CBS+/− mice (n = 96) fed either the control or HM diet, the effect of sex on tHcy was somewhat stronger but still not statistically significant (p = 0.07; data not shown).
Total Liver Homocysteine, Cysteine, Glutathione, and Cysteinyl-glycine in CBS-deficient Mice
Liver concentrations of Hcy were significantly elevated (Fig. 2B) in the CBS-deficient mice regardless of diet (CBS+/+, C = 1.39 ± 0.15 nmol/mg of protein (n = 6) versus CBS+/−, C = 2.68 ± 0.44 nmol/mg of protein (n = 4) and CBS+/−, HM = 2.7 ± 0.22 nmol/mg of protein (n = 4); p ≤ 0.05). The hepatic levels of Hcy did not change in the wild-type CBS+/+ mice on the HM diet. Neither Cys (Fig. 2C) nor Cys-Gly (Fig. 2E) levels changed in any group. However, GSH levels were significantly decreased in the CBS-deficient mice on the control diet (CBS+/+, C = 332 ± 68 nmol/mg of protein (n = 6) versus CBS+/−, C = 266 ± 5 nmol/mg of protein (n = 4); p ≤ 0.05) (Fig. 2D). Surprisingly, when the CBS+/− mice were administered the HM diet, GSH levels normalized, suggesting that even when only one CBS allele is mutated there is still an increased flux of Hcy through the transsulfuration pathway, allowing for increased GSH production (Fig. 2D).
Thiol Stability
The percentage of l-Cys, l-Hcy, and GSH that was recovered 24 h posthomogenization (and two freeze-thaw cycles) was 88, 72, and 99%, respectively, (supplemental Fig. S1). After 96 h of storage and three freeze-thaw cycles, there was no additional loss in l-Cys or l-Hcy. GSH showed a small, but non-significant additional loss in the 96-h sample compared with the 24-h sample (92 versus 99%). Perhaps the losses were less than anticipated (26, 27) because immediately following homogenization the samples were centrifuged and the aliquot to be used for total thiols was removed, treated with 50 mm disodium EDTA containing 1 mm dithioerythritol, and stored at −80 °C.
2-D DIGE Validation
Approximately 2500 spots were detected on each of five gels. The fluorescent images from a representative gel are shown in supplemental Fig. S2. Twenty-four random spots (1% of the total spots detected) were selected for statistical analysis of their Cy2, Cy3, and Cy5 pixel intensities (supplemental Fig. S3). Graphs of the raw pixel volumes and normalized spot volumes for the 24 spots are shown in supplemental Figs. S4 and S5. Importantly, there is no significant difference between the fluorescence emission, expressed either as pixel volume or normalized volume, for any particular protein (supplemental Table S1). However, in several instances, the Cy5 output was slightly (although not significantly) greater than the Cy3 output for the same spot (supplemental Figs. S4 and S5). The Cy5 versus Cy3 variation is due to a small difference in the extinction coefficient for Cy5. For this reason, the manufacturer recommends that within any particular experimental group some samples should be labeled with Cy3 and some should be labeled with Cy5. The intra-assay coefficient of variation was found to be 18–19%, and the interassay coefficient of variation varied from 9 to 19% (supplemental Table S2). These results confirm that the Cy2-, Cy3-, or Cy5-labeled spots from the same sample are exactly overlaid and co-detected, and there is no significant difference in the fluorescence output between any given spot pair.
Identification of Differentially Expressed Proteins in Livers of CBS-deficient Mice by 2-D DIGE
2-D DIGE coupled with LC-MS-MS identified nine proteins whose expression levels were significantly changed (p ≤ 0.05 and ± 2-fold change) as a result of genotype, 27 proteins whose expression levels changed solely as a function of diet, and 14 proteins whose levels changed when both the CBS mutation and HM diet were present (Tables I and II). The positions and identifications of the differentially expressed proteins are shown in Fig. 3, and the IPA-identified metabolic pathways associated with those proteins are shown in Fig. 4. The CBS+/− genotype altered the expression of proteins involved in 1) homocysteine metabolism (SAHH), 2) arginine and nitrogen metabolism (Nit protein 2 (NIT2), arginase 1 (ARG1), and carbamoyl-diphosphate synthetase 1 (CPS1)), 3) selenoamino acid metabolism (SELENBP1), 4) glutamate metabolism (CPS1), 5) proteolysis (cytosolic nonspecific dipeptidase 2 (CNDP2)), 6) lipid transport (phosphatidylethanolamine-binding protein 1 (PEBP1)), and 7) transcription (enolase (ENO1)) (Figs. 3 and 4 and Table III). The HM diet further increased the number of proteins involved in the metabolism of almost every amino acid as well as carbohydrate and lipid metabolism (Fig. 4 and Table III). Two additional enzymes involved in homocysteine metabolism were up-regulated in response to diet, namely BHMT and GNMT. The HM diet also up- and down-regulated the expression of proteins involved in the response of the cell to oxidative stress (peroxiredoxins 1 (PRDX1) and 2 (PRDX2), thioredoxin (TXN), and epoxide hydrolase 2 (EPHX2)), protein folding and proteolysis (heat shock protein 90, α (HSP90AA) and proteosome 28 subunit α (PSME1)), and molecular transport (SELENBP1, PEBP1, and albumin (ALB)) (Table III). Although the expression of ARG1 was down-regulated in the CBS+/− mice on the C diet, its expression was unchanged when those animals were placed on the HM diet (Table I). A similar normalization occurred with proteins involved in transport and the response of the cell to oxidative stress (Table I). Importantly, TXN was down-regulated in both HM diet groups compared with wild-type controls (Table I), and fatty acid-binding protein 1 (FABP1) showed a 4-fold decrease in expression in the CBS+/− animals on the HM diet. Although FABP1 expression was normal in the CBS+/− mice on the control diet, the lipid transport protein PEBP1 was significantly reduced in that group. Western blot analysis and ELISAs for SAHH, BHMT, and GNMT were performed to validate the 2-D DIGE results and are described below.
Table I. Differentially expressed proteins in mouse livers as a result of CBS genotype and/or HM diet.
Only proteins with a significant (p ≤ 0.05) 2-fold difference are included.
| Protein name | Gene | gi (accession no.) | -Fold change |
||
|---|---|---|---|---|---|
| CBS+/+vs. CBS+/− | CBS+/+vs. CBS+/+ HM | CBS+/+vs. CBS+/− HM | |||
| SAHH | AHCY | 15929766 | 2.07 | 2.24 | 2.22 |
| Nit protein 2 | NIT2 | 12963555 | 2.26 | 2.35 | 3.13 |
| Selenium-binding protein 1 | SELENBP1 | 285016 | 2.53 | 2.09 | |
| Enolase 1 | ENO1 | 12963491 | 2.2 | ||
| Cytosolic nonspecific dipeptidase 2 | CNDP2 | 31981273 | 2.15 | ||
| Carbamoyl-diphosphate synthetase 1 | CPS1 | 62533156 | 2.83 | 2 | |
| Arginase 1 | ARG1 | 7106255 | −2.7 | ||
| Phosphatidylethanolamine-binding protein 1 | PEBP1 | 53236978 | −2.38 | 2.87 | |
| Major urinary protein | LOC100129193 | 13654245 | −2.4 | ||
| Argininosuccinate lyase | ASL | 19526986 | 2.45 | 2.7 | |
| Aldehyde dehydrogenase 9 | ALDH9A1 | ALDH9A1 | 2.64 | 2.5 | |
| Aldehyde dehydrogenase 2 | ALDH2 | 13529509 | 2.7 | 2.52 | |
| Epoxide hydrolase 2 | EPHX2 | 31982393 | 2.14 | 3.53 | |
| Fructose-1,6-bisphosphatase 1 | FBP1 | 6688689 | 3.05 | 2.42 | |
| Glycine N-methyltransferase | GNMT | 15679953 | 2.18 | 2.02 | |
| Betaine-homocysteine methyltransferase | BHMT | 62533211 | 2.1 | 2 | |
| 10-Formyltetrahydrofolate dehydrogenase | ALDH1L1 | 20380027 | 2.7 | 3.36 | |
| Malate dehydrogenase 1 | MDH1 | 319837 | 2.31 | 2.02 | |
| Proteosome 28 subunit α | PSME1 | 6755212 | 2.18 | 2.17 | |
| Albumin | ALB | 3647327 | 2 | ||
| Regucalcin | RGN | 6677739 | 2.43 | ||
| Annexin A5 | ANXA1 | 6753060 | 2.3 | ||
| Ferritin light chain 1 | FTL | 6753914 | 3.09 | ||
| Peroxiredoxin 1 | PRDX1 | 56103807 | 2.87 | ||
| Peroxiredoxin 2 | PRDX2 | 31560539 | 2 | ||
| Farnesyl-diphosphate synthetase | FDPS | 182398 | 2.17 | ||
| Thioredoxin 1 | TXN | 6755911 | −2.2 | −2.17 | |
| β-Actin | ACTB | 6671509 | 2.35 | ||
| Glyoxalase 1 | GLO1 | 31981282 | 2.04 | ||
| Heat shock protein 90, α | HSP90AA | 6754254 | 2.12 | ||
| 6-Phosphogluconolactonase | PGLS | 13384778 | 2.45 | ||
| Fatty acid-binding protein 1 | FABP1 | 2894517 | −4.87 | ||
Table II. Tandem mass spectrometry Mascot results.
| Protein name | Gene | gi (accession no.) | No. of unique peptides identified | Sequence coverage | Mascot score |
|---|---|---|---|---|---|
| % | |||||
| SAHH | AHCY | 15929766 | 19 | 49 | 1157 |
| Nit protein 2 | NIT2 | 12963555 | 5 | 24 | 286 |
| Selenium-binding protein 1 | SELENBP1 | 285016 | 23 | 59 | 1567 |
| Enolase 1 | ENO1 | 12963491 | 12 | 38 | 744 |
| Cytosolic nonspecific dipeptidase 2 | CNDP2 | 31981273 | 3 | 10 | 132 |
| Carbamoyl-diphosphate synthetase 1 | CPS1 | 62533156 | 5 | 14 | 205 |
| Arginase 1 | ARG1 | 7106255 | 11 | 39 | 833 |
| Phosphatidylethanolamine-binding protein 1 | PEBP1 | 53236978 | 10 | 79 | 601 |
| Major urinary protein | LOC100129193 | 13654245 | 3 | 16 | 133 |
| Argininosuccinate lyase | ASL | 19526986 | 18 | 46 | 1089 |
| Aldehyde dehydrogenase 9 | ALDH9A1 | ALDH9A1 | 4 | 7 | 192 |
| Aldehyde dehydrogenase 2 | ALDH2 | 13529509 | 14 | 42 | 979 |
| Epoxide hydrolase 2 | EPHX2 | 31982393 | 9 | 18 | 508 |
| Fructose-1,6-bisphosphatase 1 | FBP1 | 6688689 | 17 | 65 | 1463 |
| Glycine N-methyltransferase | GNMT | 15679953 | 8 | 39 | 431 |
| Betaine-homocysteine methyltransferase | BHMT | 62533211 | 8 | 22 | 301 |
| 10-Formyltetrahydrofolate dehydrogenase | ALDH1L1 | 20380027 | 16 | 18 | 936 |
| Malate dehydrogenase 1 | MDH1 | 319837 | 14 | 44 | 817 |
| Proteosome 28 subunit α | PSME1 | 6755212 | 3 | 13 | 169 |
| Albumin | ALB | 3647327 | 37 | 67 | 2310 |
| Regucalcin | RGN | 6677739 | 20 | 65 | 1316 |
| Annexin A5 | ANXA1 | 6753060 | 20 | 68 | 1417 |
| Ferritin light chain 1 | FTL | 6753914 | 3 | 20 | 183 |
| Peroxiredoxin 1 | PRDX1 | 56103807 | 4 | 23 | 175 |
| Peroxiredoxin 2 | PRDX2 | 31560539 | 5 | 26 | 328 |
| Farnesyl-diphosphate synthetase | FDPS | 182398 | 9 | 42 | 564 |
| Thioredoxin 1 | TXN | 6755911 | 4 | 40 | 224 |
| β-Actin | ACTB | 6671509 | 5 | 23 | 341 |
| Glyoxalase 1 | GLO1 | 31981282 | 9 | 48 | 539 |
| Heat shock protein 90, α | HSP90AA | 6754254 | 3 | 6 | 133 |
| 6-Phosphogluconolactonase | PGLS | 13384778 | 7 | 37 | 372 |
| Fatty acid-binding protein 1 | FABP1 | 2894517 | 2 | 25 | 104 |
Fig. 3.
Coomassie-stained 2D gel of differentially expressed proteins. One hundred micrograms of each sample was pooled, and 750 μg of total pooled protein was run on a single 2-D gel. Following electrophoresis, the gel was fixed, stained with GelCode Blue Coomassie stain (Pierce), and imaged on the Typhoon Trio imager (GE Healthcare). The gel image was matched to the analytical gel images, and differentially regulated protein bands were identified by DeCyder software (GE Healthcare). The identified bands were cut from the gel and identified by LC-MS-MS. The positions and identifications of the differentially expressed proteins are shown in the figure, and the -fold change of each protein is given in Table I. RGN, regucalcin; FTL, ferritin light chain 1; PGLS, 6-phosphogluconolactonase; ACTB, β-actin.
Fig. 4.
Regulated canonical pathways associated with hyperhomocysteinemia. The data set of identified proteins was loaded into IPA for pathway analysis. The top canonical pathways that were significantly (p ≤ 0.05) associated with the CBS+/− genotype are indicated by the filled black bars, those associated with the wild-type mice on the HM diet are indicated by the white bars, and the those associated with the CBS+/− mice on the HM diet are shown by the filled gray bars. The horizontal line represents the threshold value for a significance of p ≤ 0.05.
Table III. Biological and molecular functions of differentially expressed proteins.
| Protein name | Biological function | Molecular function |
|---|---|---|
| SAH hydrolase | Hcy metabolism | Hydrolase, copper binder |
| Nit protein 2 | Nitrogen metabolism | Nitrilase |
| Selenium-binding protein 1 | Protein transport | Selenium binder |
| Enolase 1 | Negative regulator of transcription | Transcription factor |
| Cytosolic nonspecific dipeptidase 2 | Proteolysis | Carboxypeptidase, zinc binder |
| Carbamoyl-diphosphate synthetase 1 | Urea cycle | Nucleotide binder |
| Arginase 1 | Urea cycle | Hydrolase, manganese binder |
| Phosphatidylethanolamine-binding protein 1 | Transporter | Lipid binder, nucleotide binder |
| Major urinary protein | Transporter | Pheromone binding, odorant |
| Argininosuccinate lyase | Urea cycle | Catalytic lyase |
| Aldehyde dehydrogenase 9 | Aldehyde metabolism | Oxidoreductase |
| Aldehyde dehydrogenase 2 | Carbohydrate metabolism | Oxidoreductase |
| Epoxide hydrolase 2 | Prostaglandin synthesis, oxidative stress response | Hydrolase |
| Fructose-1,6-bisphosphatase 1 | Carbohydrate metabolism | Phosphatase |
| Glycine N-methyltransferase | Protein modification | Transferase, folate binder |
| Betaine-homocysteine methyltransferase | Hcy metabolism | Methyltransferase |
| 10-Formyltetrahydrofolate dehydrogenase | One-carbon metabolism | Methyltransferase |
| Malate dehydrogenase 1 | Carbohydrate metabolism | Oxidoreductase, NAD binder |
| Proteosome 28 subunit α | Proteosome activator | Protein binder |
| Albumin | Transport | Binding protein |
| Regucalcin | Calcium homeostasis | Calcium binder, enzyme activity regulator |
| Annexin A5 | Antiapoptosis, blood coagulation | Phospholipase inhibitor |
| Ferritin light chain 1 | Iron transport and hemostasis | Oxidoreductase, iron binder |
| Peroxiredoxin 1 | Oxidative stress response | Oxidoreductase, antioxidant |
| Peroxiredoxin 2 | Oxidative stress response | Oxidoreductase, antioxidant |
| Farnesyl-diphosphate synthetase | Cholesterol biosynthesis | Transferase |
| Thioredoxin 1 | Redox homeostasis | Thiol disulfide exchange, ribonucleotide reduction |
| β-Actin | Motility | Cytoskeletal structure |
| Glyoxalase 1 | Carbohydrate metabolism | Lyase |
| Heat shock protein 90, α | Protein folding, mitochondrial transport | Nucleotide binder |
| 6-Phosphogluconolactonase | Carbohydrate metabolism | Hydrolase |
| Fatty acid-binding protein 1 | Fatty acid metabolism | Lipid transport |
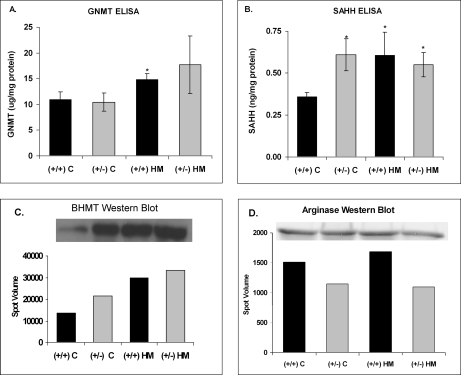
Glycine N-Methyltransferase and Betaine-homocysteine Methyltransferase
GNMT and BHMT were found to co-migrate in one of the differentially up-regulated protein spots (Figs. 3 and 5). To determine which of the two proteins was responsible for the increased expression, the level of GNMT was determined by ELISA, and BHMT was analyzed by Western blot (Fig. 6). The ELISA results demonstrated that GNMT was significantly elevated in the wild-type mice on the HM diet (Fig. 6A). GNMT was also elevated (but not significantly) in the CBS-deficient mice on the HM diet; however, genotype alone did not result in a significant increase in GNMT (Fig. 6A). Western blot analysis of the pooled samples from each group showed an increase in BHMT protein in response to diet and genotype when compared with wild-type control mice (Fig. 6C). Thus, GNMT expression increased only in response to diet, whereas BHMT expression increased in response to diet and CBS deficiency.
Fig. 5.
The effect of hyperhomocysteinemia on methionine cycle proteome. A, 2-D DIGE images of the spots identified by LC-MS-MS as SAHH, BHMT, and GNMT. BHMT and GNMT co-migrated as a single spot. Quantitation of the normalized spot volumes for SAHH (B) and BHMT/GNMT (C) is shown. SAHH was significantly increased in response to CBS genotype and HM diet, whereas the BHMT/GNMT spot was increased only in the HM diet groups. The Cy3 and/or Cy5 spot volumes of each sample were normalized to the spot volume for the Cy2-labeled internal standard and are expressed as mean ± S.D. *, p ≤ 0.05 versus (+/+) C; n = 4–6 animals per group. Error bars indicate ± 1 S.D.
Fig. 6.
Validation of DIGE results using ELISA and Western blot analysis. GNMT (A) and SAHH (B) concentrations were measured by ELISA in liver homogenates from wild-type CBS+/+ and heterozygote CBS+/− mice after 14 days on a normal diet (C diet) or a diet supplemented with 0.5% methionine in the drinking water (HM diet). Results were normalized to protein concentration and are expressed as mean ± S.D. *, p ≤ 0.05 versus (+/+) C; n = 4–6 animals per group. In C and D, the pooled homogenates of each experimental group were electrophoresed by 12% SDS-PAGE, and BHMT and arginase were detected by Western blot, respectively. Error bars indicate ± 1 S.D.
S-Adenosylhomocysteine Hydrolase
Proteomics analysis demonstrated that SAHH was up-regulated in all three experimental groups, suggesting that SAHH is regulated by both genotype and diet (Table I and Fig. 5). To confirm the DIGE results, SAHH levels were measured using an ELISA as described under “Experimental Procedures” (Fig. 6). ELISA confirmed the results found by 2-D DIGE whereby hepatic SAHH levels were significantly increased in response to diet and genotype (Fig. 6B).
Arginase and Nitric Oxide
As mentioned above, the proteomics analysis showed that ARG1 was significantly decreased by 2.7-fold in the CBS+/− mice on the normal diet. Western blot analysis confirmed the 2-D DIGE results; however, the blots also indicated that ARG1 was decreased in the CBS+/− mice on the HM diet (Fig. 6D), and this result was not seen in the proteomics analysis. Total nitrate and nitrite were significantly increased in the CBS+/− mice (Fig. 2F); this may be the result of the decrease in ARG1 expression.
Network Analysis
To identify signaling pathways involved in hyperhomocysteinemia, we carried out network analysis on the differentially expressed proteins using IPA software. The predominant network associated with the CBS+/− genotype is shown in Fig. 7A. Only the thyroid hormone receptor/retinoid X receptor (RXR) pathway was significantly associated with the CBS genotype, whereas the HM diet was associated with four different signaling cascades, namely the NRF2-mediated oxidative stress response, aryl hydrocarbon receptor signaling, lipopolysaccharide/IL-1 mediated inhibition of RXR function, and xenobiotic metabolism signaling (Fig. 7B). RXR is involved in xenobiotic metabolism by regulating the transcription of genes involved in GSH biosynthesis and utilization (33). Interestingly, when the CBS+/− mice were placed on the HM diet, the lipopolysaccharide/IL-1 signaling cascade, which is an inhibitor of thyroid hormone receptor/RXR activation, was activated (33) (Fig. 7B).
Fig. 7.
Network analysis of CBS-regulated proteins. A, direct (solid lines) and indirect (dashed lines) protein-protein interactions between the up-regulated (gray) and down-regulated (black) hepatic proteins of the CBS+/− mice and members of the extracellular signal-regulated protein/mitogen-activated protein kinase (ERK/MAPK) signaling cascade. Solid lines indicate simple binding associations between proteins, whereas arrowed lines indicate that the protein nearest to the arrowhead is being acted upon by the protein furthest from the arrowhead. B, the top function(s) associated with the CBS genotype (black bar), the HM diet (white bar), and CBS+/− mice on the HM diet (gray bar). TR, thyroid hormone receptor.
DISCUSSION
In classical homocystinuria produced by mutations in the CBS gene, fatty liver disease is a common finding (6, 8, 34, 35). However, fatty liver disease is also associated with non-gene-induced hyperhomocysteinemia (36–40). Although liver disease has been associated with alterations in methionine metabolism since the 1940s (41, 42) and with homocysteine metabolism since the 1960s (6, 8), the exact mechanism(s) involved in homocysteine-induced liver injury is still unclear despite many years of study in experimental animal models and in subjects with chronic liver disease (36–38, 43–49). There is considerable evidence suggesting that homocysteine-induced endoplasmic reticulum and oxidative stress mediates the liver injury by promoting apoptosis, inflammation, insulin resistance, and dysregulated lipid metabolism (46, 47, 50). In this study, we used expression proteomics and pathway analysis to dissect and differentiate the metabolic and signaling pathways involved in gene-induced and/or diet-induced hyperhomocysteinemia. We found that hyperhomocysteinemia, whether caused by CBS deficiency or HM diet, alters the abundance of several liver proteins and metabolites involved in homocysteine/methionine metabolism, nitrogen and lipid metabolism, and oxidative stress.
In CBS-deficient animals on a control diet, six proteins were identified with increased expression (SAHH, NIT2, SELENBP1, ENO1, CNDP2, and CPS1), and three were identified with decreased expression (ARG1, PEBP1, and LOC100129193). Surprisingly, SAHH was the only methionine cycle enzyme whose expression was changed in the CBS+/− mice. SAHH is the only reversible enzyme in the cycle, and based on the equilibrium constant (Keq ≈ 1 μm) for this reaction, S-adenosylhomocysteine (SAH) formation should be favored over SAH hydrolysis (51). However, SAH hydrolysis occurs more frequently in vivo because the breakdown products, Hcy and adenosine, are rapidly removed downstream by methionine synthase, BHMT, CBS, adenosine deaminase, and adenosine kinase. In the CBS+/− mice, however, the transsulfuration pathway is compromised, resulting in increased levels of intracellular Hcy and plasma tHcy (Fig. 2A). Thus, it is postulated that the liver responds to increased intracellular Hcy by inducing the expression of SAHH. In fact, SAHH levels were increased in all three groups of hyperhomocysteinemic animals regardless of the underlying cause of hyperhomocysteinemia. S-Adenosylmethionine (SAM), SAH, and adenosine were not measured in the present study but need to be determined to correlate the SAHH expression with the actual SAH levels and SAM/SAH ratio because SAH is a feedback inhibitor of its own synthesis by SAHH (52, 53). Furthermore, SAH is an end product inhibitor of a number of methyltransferases that are involved in its production from SAM, thereby regulating the SAM/SAH ratio (54). Finally, adenosine can dramatically affect SAHH activity as demonstrated by the adenosine analogs, which are potent inhibitors of SAHH activity (55) and are widely used as antiviral, antiparasitic, antiarthritic, and chemotherapeutic drugs because of their effect on gene expression, cell cycle, differentiation, and apoptosis (51, 55). Interestingly, IPA confirmed that proteins involved in cell cycle and differentiation were most affected by hyperhomocysteinemia.
The liver proteome of the wild-type and CBS-deficient animals on the HM diet was dramatically different from those on a control diet. When CBS+/+ mice were fed the HM diet for 2 weeks, 26 proteins were up-regulated, and one protein was down-regulated. In addition to SAHH, two additional enzymes of the methionine cycle were up-regulated, namely GNMT and BHMT. GNMT is an abundant hepatic protein whose physiological roles include regulation of the SAM/SAH ratio (56) and control of methyl group synthesis via the folate-dependent one-carbon pool (56–58). Its activity is directly proportional to the level of methionine in the diet (59–61) and inversely proportional to the level of dietary folate (62, 63). It is allosterically inhibited by 5-methyltetrahydrofolate pentaglutamate (56–58). In the present study, both the C and HM diets contained adequate supplies of folate. Thus, the increase in GNMT was a direct consequence of the HM diet. Interestingly, a second major folate-binding protein was up-regulated in the mice on the HM diet, namely 10-formyltetrahydrofolate dehydrogenase, suggesting the possibility that GNMT levels were up-regulated because their inhibitor, 5-methyltetrahydrofolate pentaglutamate, was bound to 10-formyltetrahydrofolate dehydrogenase. BHMT comprises ∼1% of total liver protein (64), and paradoxically its activity has been shown to increase in response to both a methionine-deficient and a methionine-rich diet (65). However, Park and Garrow (66) noted that BHMT activity is influenced more by dietary choline and/or betaine than methionine. In the present study, the dietary contents of choline and betaine were sufficient; therefore, the BHMT up-regulation was solely the result of the HM diet.
The methionine cycle proteome responded to gene-induced hyperhomocysteinemia (CBS mutation) by up-regulating a single enzyme of the cycle, SAHH, whereas administration of an HM diet resulted in the up-regulation of SAHH, BHMT, GNMT, and 10-formyltetrahydrofolate dehydrogenase. Although the same four enzymes were up-regulated in both wild-type and CBS+/− mice on the HM diet, only the wild-type animals were able to maintain a normal level of intracellular Hcy. Furthermore, the levels of tHcy in the CBS+/− mice on the HM diet were 6-fold greater than wild-type animals on the HM diet. Taken together, these results suggest that even when one CBS allele is absent Hcy catabolism is dramatically impaired, producing the need for compensatory increases in SAHH, GNMT, and BHMT. However, when Hcy catabolism by these enzymes is maxed out, export is the only recourse that the cell has to protect itself from excess Hcy.
The methionine cycle enzymes were not the only proteins whose expression levels were differentially regulated by hyperhomocysteinemia. Several enzymes involved in nitrogen metabolism and the urea cycle also changed in response to the CBS mutation and/or HM diet. High protein diets generally result in significant increases in the concentrations of the urea cycle enzymes (67) to catabolize the excess nitrogen provided by the diet. Homocystinurics may also be in a state of excess nitrogen as a consequence of their hypermethioninemia. Thus, it seems only natural that the urea cycle enzymes would be up-regulated in all of the groups in the present study. CPS1, an important regulator of the urea cycle, and argininosuccinate lyase (ASL) were both up-regulated in CBS+/− and CBS+/+ mice on the HM diet. However, ARG1 was decreased by −2.7-fold in the CBS+/− animals, and this down-regulation was abolished when those animals were administered the HM diet. Hepatic levels of nitrate and nitrite, the stable end products of NO metabolism, increased in the CBS-deficient animals, and those levels normalized when the CBS+/− mice were placed on the HM diet (Fig. 2F). Because ARG1 competes with nitric-oxide synthase (NOS) for available arginine, the decrease in ARG1 in the CBS+/− animals should result in increased NOS activity and NO synthesis. But is increased hepatic NO beneficial or detrimental? The general view is that NO protects the vasculature by promoting vascular relaxation and preventing endothelial dysfunction, but in the liver it has been shown to potentiate oxidative injury via the formation of peroxynitrite in ischemia-reperfusion injury (68–70), alcoholic liver disease (71), and acetaminophen-induced hepatotoxicity (72). Thus, the decrease in ARG1 and increase in NO in the CBS+/− mice may be indicative of early liver damage.
The increase in hepatic NO in the current study is in direct contrast to what has been described in the vasculature. In studies using cultured endothelial cells, bioavailable NO was significantly decreased in Hcy-treated cells (17, 73, 74). Also, in aortic rings from hyperhomocysteinemic mice, Lentz and co-workers (18, 75, 76) demonstrated that maximal relaxation was significantly impaired in CBS+/− mice fed a low folate or high methionine diet. One explanation is that homocysteine alters the cellular redox status, resulting in the generation of ROS such as hydrogen peroxide and superoxide, which in turn oxidatively inactivate endothelium-derived NO (15, 17, 18, 22). Tyagi et al. (74) proposed a mechanism for homocysteine-induced oxidative stress in microvascular endothelial cells whereby Hcy activates protease-activated receptor 4, which then increases the expression of NADPH oxidase, reduces the expression of TXN, and results in the production of reactive oxygen species. Studies by Loscalzo and co-workers (22, 77) have confirmed that Hcy is associated with increased superoxide generation and a decrease in the antioxidant enzyme glutathione peroxidase 1 (GPx-1). In the present study, GPx-1 expression was not changed; however, total GSH was significantly decreased in the CBS+/− mice.
Because hyperhomocysteinemia is an important source of oxidative stress and because hepatic GSH was significantly decreased in the CBS+/− mice, one might predict the need for up-regulation of the antioxidant enzymes. However, none of the usual candidate enzymes (i.e. GPx-1, superoxide dismutase, or catalase) were changed in the CBS+/− mice. On the other hand, the expression levels of three antioxidant enzymes, namely PRDX1, PRDX2, and TXN, were altered in response to the HM diet. PRDX1 and -2 were significantly increased by 2.87- and 2-fold, respectively, in the wild-type mice, whereas TXN was significantly down-regulated by −2.2- and −2.17-fold in the CBS+/+ and CBS+/− on the HM diet, respectively. TXN is a 12-kDa oxidoreductase that can reduce, form, or isomerize protein disulfide bonds as well as scavenge singlet oxygen and hydroxyl radical (78, 79). PRDX1 and -2 belong to the antioxidant family of peroxidases, and they protect the cell from ROS by reducing a wide range of hydroperoxides. TXN maintains PRDX1 and -2 in their active reduced states by serving as an electron donor to reduce their corresponding oxidized forms. As mentioned above, Tyagi et al. (74) showed that TXN expression was reduced in Hcy-treated microvascular endothelial cells, and this reduction was associated with a concomitant increase in protease-activated receptor 4, NADPH oxidase, ROS, and inducible NOS, and a reduction in endothelial NOS. Also, wild-type CBS mice on the HM diet were able to up-regulate the expression of PRDX1 and -2, possibly as a consequence to the decreased TXN expression, but the CBS+/− mice were unable to increase either PRDX, although they too exhibited low TXN levels. Thus, the CBS+/− mice may be at an even greater risk for oxidative liver injury. It is possible that the plasma tHcy levels were too low in the CBS+/− mice on the control diet (tHcy = 4.27 μm) to trigger this response, whereas plasma tHcy in the wild-type and CBS+/− mice on the HM diet (tHcy: C (+/+) = 10.7 μm; C (+/−) = 61.45 μm) were sufficiently elevated to initiate the events leading to the oxidative stress response. Interestingly, IPA analysis confirmed that the wild-type mice on the HM diet did activate an NFR2-mediated oxidative stress response, whereas the CBS+/− mice did not (Fig. 7B).
In summary, we used a quantitative proteomics approach to identify hepatic proteins whose expression levels were changed as a consequence of hyperhomocysteinemia, whether the hyperhomocysteinemia was induced by a genetic mutation in CBS or by a high methionine diet. Although there were a few similarities between the two proteomes, there were many more differences. Nine proteins were changed in response to genotype, whereas 27 proteins changed in response to diet. SAHH was up-regulated in both groups, but the HM diet caused a further increase in GNMT and BHMT. Many proteins involved in nitrogen and urea metabolism were up-regulated in the diet group; however and importantly, ARG1 was down-regulated and NO was increased in the CBS mutants. Finally, several proteins involved in the response of the cell to oxidative stress were up-regulated in wild-type mice in response to diet (PRDX1, PRDX2, and HSP90AA) but not in the CBS mutants, and total GSH levels were significantly decreased in the CBS+/− mice. Taken together, these results suggest that in heterozygous CBS-deficient mice the liver may be at an increased risk for homocysteine-induced oxidative damage, and this is further compounded when an HM diet is introduced.
Supplementary Material
* This work was supported, in whole or in part, by National Institutes of Health Grants HL 52234 (to D. W. J.) and HL 063943 and NS024621 (to S. R. L.) from the NHLBI. This work was also supported by American Heart Association Grant AHA 0860052Z (to S. D.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
1 The abbreviations used are:
- Hcy
- homocysteine
- tHcy
- plasma total homocysteine
- SAM
- S-adenosylmethionine
- SAH
- S-adenosylhomocysteine
- CBS
- cystathionine β-synthase
- BHMT
- betaine-homocysteine methyltransferase
- GNMT
- glycine N-methyltransferase
- SAHH
- S-adenosylhomocysteine hydrolase
- NIT2
- Nit protein 2
- ARG1
- arginase 1
- ASL
- argininosuccinate lyase
- CPS1
- carbamoyl-diphosphate synthetase 1
- SELENBP1
- selenoamino acid-binding protein 1
- CNDP2
- cytosolic nonspecific dipeptidase 2
- PEBP1
- phosphatidylethanolamine-binding protein 1
- ENO1
- enolase 1
- PRDX
- peroxiredoxin
- TXN
- thioredoxin
- EPHX2
- epoxide hydrolase 2
- HSP90AA
- heat shock protein 90, α
- PSME1
- proteosome 28 subunit α
- ALB
- albumin
- FABP1
- fatty acid-binding protein 1
- RXR
- retinoid X receptor
- NRF2
- nuclear factor erythroid 2-related factor 2
- GPx-1
- glutathione peroxidase 1
- NOS
- nitric-oxide synthase
- HM
- high methionine
- C
- control
- ROS
- reactive oxygen species
- NO
- nitric oxide
- 2-D
- two-dimensional
- IPA
- Ingenuity Pathway Analysis
- IS
- internal standard.
REFERENCES
- 1.Finkelstein J. D., Martin J. J. (1984) Methionine metabolism in mammals. Distribution of homocysteine between competing pathways. J. Biol. Chem 259, 9508–9513 [PubMed] [Google Scholar]
- 2.Reed M. C., Nijhout H. F., Sparks R., Ulrich C. M. (2004) A mathematical model of the methionine cycle. J. Theor. Biol 226, 33–43 [DOI] [PubMed] [Google Scholar]
- 3.Storch K. J., Wagner D. A., Burke J. F., Young V. R. (1988) Quantitative study in vivo of methionine cycle in humans using [methyl-2H3]- and [1–13C]methionine. Am. J. Physiol. Endocrinol. Metab 255, E322–E331 [DOI] [PubMed] [Google Scholar]
- 4.Stead L. M., Brosnan M. E., Brosnan J. T. (2000) Characterization of homocysteine metabolism in the rat liver. Biochem. J 350, 685–692 [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen B., Refsum H., Vintermyr O., Ueland P. M. (1991) Homocysteine export from cells cultured in the presence of physiological or superfluous levels of methionine: methionine loading of non-transformed, transformed, proliferating, and quiescent cells in culture. J. Cell. Physiol 146, 52–62 [DOI] [PubMed] [Google Scholar]
- 6.Carson N. A., Dent C. E., Field C. M., Gaull G. E. (1965) Homocystinuria: clinical and pathological review of ten cases. J. Pediatr 66, 565–583 [DOI] [PubMed] [Google Scholar]
- 7.Jacobsen D. W., Gatautis V. J., Green R., Robinson K., Savon S. R., Secic M., Ji J., Otto J. M., Taylor L. M., Jr. (1994) Rapid HPLC determination of total homocysteine and other thiols in serum and plasma: sex differences and correlation with cobalamin and folate concentrations in healthy subjects. Clin. Chem 40, 873–881 [PubMed] [Google Scholar]
- 8.McCully K. S. (1969) Vascular pathology of homocysteinemia: implications for the pathogenesis of arteriosclerosis. Am. J. Pathol 56, 111–128 [PMC free article] [PubMed] [Google Scholar]
- 9.Mudd S. H., Finkelstein J. D., Irreverre F., Laster L. (1964) Homocystinuria: an enzymatic defect. Science 143, 1443–1445 [DOI] [PubMed] [Google Scholar]
- 10.Mudd S. H., Levy H. L., Kraus J. B. (2001) Disorders of transsulfuration, in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Sly W. S., Valle D.) McGraw-Hill Inc., New York [Google Scholar]
- 11.Refsum H., Fredriksen A., Meyer K., Ueland P. M., Kase B. F. (2004) Birth prevalence of homocystinuria. J. Pediatr 144, 830–832 [DOI] [PubMed] [Google Scholar]
- 12.Mansoor M. A., Svardal A. M., Schneede J., Ueland P. M. (1992) Dynamic relation between reduced, oxidized, and protein-bound homocysteine and other thiol components in plasma during methionine loading in healthy men. Clin. Chem 38, 1316–1321 [PubMed] [Google Scholar]
- 13.Guttormsen A. B., Schneede J., Fiskerstrand T., Ueland P. M., Refsum H. M. (1994) Plasma concentrations of homocysteine and other aminothiol compounds are related to food intake in healthy human subjects. J. Nutr 124, 1934–1941 [DOI] [PubMed] [Google Scholar]
- 14.Stühlinger M. C., Tsao P. S., Her J. H., Kimoto M., Balint R. F., Cooke J. P. (2001) Homocysteine impairs the nitric oxide synthase pathway: role of asymmetric dimethylarginine. Circulation 104, 2569–2575 [DOI] [PubMed] [Google Scholar]
- 15.Lentz S. R., Rodionov R. N., Dayal S. (2003) Hyperhomocysteinemia, endothelial dysfunction, and cardiovascular risk: the potential role of ADMA. Atheroscler. Suppl 4, 61–65 [DOI] [PubMed] [Google Scholar]
- 16.Wang S., Wright G., Harrah J., Touchon R., McCumbee W., Geng W., Fultz M. E., Abdul-Jalil M. N., Wright G. L. (2000) Short-term exposure to homocysteine depresses rat aortic contractility by an endothelium-dependent mechanism. Can. J. Physiol. Pharmacol 78, 500–506 [PubMed] [Google Scholar]
- 17.Upchurch G. R., Jr., Welch G. N., Fabian A. J., Freedman J. E., Johnson J. L., Keaney J. F., Jr., Loscalzo J. (1997) Homocyst(e)ine decreases bioavailable nitric oxide by a mechanism involving glutathione peroxidase. J. Biol. Chem 272, 17012–17017 [DOI] [PubMed] [Google Scholar]
- 18.Dayal S., Arning E., Bottiglieri T., Böger R. H., Sigmund C. D., Faraci F. M., Lentz S. R. (2004) Cerebral vascular dysfunction mediated by superoxide in hyperhomocysteinemic mice. Stroke 35, 1957–1962 [DOI] [PubMed] [Google Scholar]
- 19.Dayal S., Wilson K. M., Leo L., Arning E., Bottiglieri T., Lentz S. R. (2006) Enhanced susceptibility to arterial thrombosis in a murine model of hyperhomocysteinemia. Blood 108, 2237–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kokame K., Kato H., Miyata T. (1996) Homocysteine-respondent genes in vascular endothelial cells identified by differential display analysis. GRP78/BiP and novel genes. J. Biol. Chem 271, 29659–29665 [DOI] [PubMed] [Google Scholar]
- 21.Handy D. E., Zhang Y., Loscalzo J. (2005) Homocysteine down-regulates cellular glutathione peroxidase (GPx1) by decreasing translation. J. Biol. Chem 280, 15518–15525 [DOI] [PubMed] [Google Scholar]
- 22.Lubos E., Loscalzo J., Handy D. E. (2007) Homocysteine and glutathione peroxidase-1. Antioxid. Redox Signal 9, 1923–1940 [DOI] [PubMed] [Google Scholar]
- 23.Outinen P. A., Sood S. K., Pfeifer S. I., Pamidi S., Podor T. J., Li J., Weitz J. I., Austin R. C. (1999) Homocysteine-induced endoplasmic reticulum stress and growth arrest leads to specific changes in gene expression in human vascular endothelial cells. Blood 94, 959–967 [PubMed] [Google Scholar]
- 24.Werstuck G. H., Lentz S. R., Dayal S., Hossain G. S., Sood S. K., Shi Y. Y., Zhou J., Maeda N., Krisans S. K., Malinow M. R., Austin R. C. (2001) Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J. Clin. Investig 107, 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe M., Osada J., Aratani Y., Kluckman K., Reddick R., Malinow M. R., Maeda N. (1995) Mice deficient in cystathionine beta-synthase: animal models for mild and severe homocyst(e)inemia. Proc. Natl. Acad. Sci. U.S.A 92, 1585–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen R. E., Roth D., Winther J. R. (2009) Quantifying the global cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U.S.A 106, 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon B. M., Heath S. H., Kim R., Suh J. H., Hagen T. M. (2008) Assessment of endoplasmic reticulum glutathione redox status is confounded by extensive ex vivo oxidation. Antioxid. Redox Signal 10, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobsen D. W., Gatautis V. J., Green R. (1989) Determination of plasma homocysteine by high-performance liquid chromatography with fluorescence detection. Anal. Biochem 178, 208–214 [DOI] [PubMed] [Google Scholar]
- 29.Unlü M., Morgan M. E., Minden J. S. (1997) Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18, 2071–2077 [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 31.Kinter M., Sherman N. E. (2000) Protein Sequencing and Identification Using Tandem Mass Spectrometry, Wiley-Interscience, New York [Google Scholar]
- 32.Vitvitsky V., Prudova A., Stabler S., Dayal S., Lentz S. R., Banerjee R. (2007) Testosterone regulation of renal cystathionine beta-synthase: implications for sex-dependent differences in plasma homocysteine levels. Am. J. Physiol. Renal Physiol 293, F594–F600 [DOI] [PubMed] [Google Scholar]
- 33.Wu Y., Zhang X., Bardag-Gorce F., Robel R. C., Aguilo J., Chen L., Zeng Y., Hwang K., French S. W., Lu S. C., Wan Y. J. (2004) Retinoid X receptor alpha regulates glutathione homeostasis and xenobiotic detoxification processes in mouse liver. Mol. Pharmacol 65, 550–557 [DOI] [PubMed] [Google Scholar]
- 34.Robert K., Nehmé J., Bourdon E., Pivert G., Friguet B., Delcayre C., Delabar J. M., Janel N. (2005) Cystathionine beta synthase deficiency promotes oxidative stress, fibrosis, and steatosis in mice liver. Gastroenterology 128, 1405–1415 [DOI] [PubMed] [Google Scholar]
- 35.Hamelet J., Demuth K., Paul J. L., Delabar J. M., Janel N. (2007) Hyperhomocysteinemia due to cystathionine beta synthase deficiency induces dysregulation of genes involved in hepatic lipid homeostasis in mice. J. Hepatol 46, 151–159 [DOI] [PubMed] [Google Scholar]
- 36.García-Tevijano E. R., Berasain C., Rodríguez J. A., Corrales F. J., Arias R., Martín-Duce A., Caballería J., Mato J. M., Avila M. A. (2001) Hyperhomocysteinemia in liver cirrhosis: mechanisms and role in vascular and hepatic fibrosis. Hypertension 38, 1217–1221 [DOI] [PubMed] [Google Scholar]
- 37.Ventura P., Rosa M. C., Abbati G., Marchini S., Grandone E., Vergura P., Tremosini S., Zeneroli M. L. (2005) Hyperhomocysteinaemia in chronic liver diseases: role of disease stage, vitamin status and methylenetetrahydrofolate reductase genetics. Liver Int 25, 49–56 [DOI] [PubMed] [Google Scholar]
- 38.Gulsen M., Yesilova Z., Bagci S., Uygun A., Ozcan A., Ercin C. N., Erdil A., Sanisoglu S. Y., Cakir E., Ates Y., Erbil M. K., Karaeren N., Dagalp K. (2005) Elevated plasma homocysteine concentrations as a predictor of steatohepatitis in patients with non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol 20, 1448–1455 [DOI] [PubMed] [Google Scholar]
- 39.Herrero J. I., Quiroga J., Sangro B., Beloqui O., Pardo F., Cienfuegos J. A., Prieto J. (2000) Hyperhomocysteinemia in liver transplant recipients: prevalence and multivariate analysis of predisposing factors. Liver Transpl 6, 614–618 [DOI] [PubMed] [Google Scholar]
- 40.Woo C. W., Prathapasinghe G. A., Siow Y. L., O K. (2006) Hyperhomocysteinemia induces liver injury in rat: protective effect of folic acid supplementation. Biochim. Biophys. Acta 1762, 656–665 [DOI] [PubMed] [Google Scholar]
- 41.Kinsell L. W., Harper H. A., Barton H. C., Michaels G. D., Weiss H. A. (1947) Rate of disappearance from plasma of intravenously administered methionine in patients with liver damage. Science 106, 589–590 [DOI] [PubMed] [Google Scholar]
- 42.Kinsell L. W., Harper H. A., Barton H. C., Hutchin M. E., Hess J. R. (1948) Studies in methionine and sulfur metabolism. I. The fate of intravenously administered methionine in normal individuals and in patients with liver damage. J. Clin. Investig 27, 677–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplowitz N., Ji C. (2006) Unfolding new mechanisms of alcoholic liver disease in the endoplasmic reticulum. J. Gastroenterol. Hepatol 21, Suppl. 3, S7–S9 [DOI] [PubMed] [Google Scholar]
- 44.Lu S. C., Tsukamoto H., Mato J. M. (2002) Role of abnormal methionine metabolism in alcoholic liver injury. Alcohol 27, 155–162 [DOI] [PubMed] [Google Scholar]
- 45.Ji C., Shinohara M., Vance D., Than T. A., Ookhtens M., Chan C., Kaplowitz N. (2008) Effect of transgenic extrahepatic expression of betaine-homocysteine methyltransferase on alcohol or homocysteine-induced fatty liver. Alcohol. Clin. Exp. Res 32, 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji C. (2008) Dissection of endoplasmic reticulum stress signaling in alcoholic and non-alcoholic liver injury. J. Gastroenterol. Hepatol 23, Suppl 1, S16–S24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaplowitz N., Than T. A., Shinohara M., Ji C. (2007) Endoplasmic reticulum stress and liver injury. Semin. Liver Dis 27, 367–377 [DOI] [PubMed] [Google Scholar]
- 48.Sazci A., Ergul E., Aygun C., Akpinar G., Senturk O., Hulagu S. (2008) Methylenetetrahydrofolate reductase gene polymorphisms in patients with nonalcoholic steatohepatitis (NASH). Cell Biochem. Funct 26, 291–296 [DOI] [PubMed] [Google Scholar]
- 49.Hirsch S., Poniachick J., Avendaño M., Csendes A., Burdiles P., Smok G., Diaz J. C., de la Maza M. P. (2005) Serum folate and homocysteine levels in obese females with non-alcoholic fatty liver. Nutrition 21, 137–141 [DOI] [PubMed] [Google Scholar]
- 50.Ji C., Kaplowitz N. (2003) Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology 124, 1488–1499 [DOI] [PubMed] [Google Scholar]
- 51.Prigge S. T., Chiang P. K. (2001) S-Adenosyl hydrolase, in Homocysteine in Health and Disease (Carmel R., Jacobsen D. W.) pp. 79–91, Cambridge University Press, Cambridge, UK [Google Scholar]
- 52.Finkelstein J. D. (1990) Methionine metabolism in mammals. J. Nutr. Biochem 1, 228–237 [DOI] [PubMed] [Google Scholar]
- 53.Finkelstein J. D. (2001) Regulation of homocysteine metabolism, in Homocysteine in Health and Disease (Carmel R., Jacobsen D. W.) pp. 92–99, Cambridge University Press, Cambridge, UK [Google Scholar]
- 54.Kerr S. J. (1972) Competing methyltransferase systems. J. Biol. Chem 247, 4248–4252 [PubMed] [Google Scholar]
- 55.Chiang P. K. (1998) Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacol. Ther 77, 115–134 [DOI] [PubMed] [Google Scholar]
- 56.Cook R. J., Wagner C. (1984) Glycine N-methyltransferase is a folate binding protein of rat liver cytosol. Proc. Natl. Acad. Sci. U.S.A 81, 3631–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner C., Briggs W. T., Cook R. J. (1985) Inhibition of glycine N-methyltransferase activity by folate derivatives: implications for regulation of methyl group metabolism. Biochem. Biophys. Res. Commun 127, 746–752 [DOI] [PubMed] [Google Scholar]
- 58.Luka Z., Pakhomova S., Loukachevitch L. V., Egli M., Newcomer M. E., Wagner C. (2007) 5-Methyltetrahydrofolate is bound in intersubunit areas of rat liver folate-binding protein glycine N-methyltransferase. J. Biol. Chem 282, 4069–4075 [DOI] [PubMed] [Google Scholar]
- 59.Ogawa H., Fujioka M. (1982) Induction of rat liver glycine methyltransferase by high methionine diet. Biochem. Biophys. Res. Commun 108, 227–232 [DOI] [PubMed] [Google Scholar]
- 60.Rowling M. J., McMullen M. H., Chipman D. C., Schalinske K. L. (2002) Hepatic glycine N-methyltransferase is up-regulated by excess dietary methionine in rats. J. Nutr 132, 2545–2550 [DOI] [PubMed] [Google Scholar]
- 61.Cook R. J., Horne D. W., Wagner C. (1989) Effect of dietary methyl group deficiency on one-carbon metabolism in rats. J. Nutr 119, 612–617 [DOI] [PubMed] [Google Scholar]
- 62.Balaghi M., Horne D. W., Woodward S. C., Wagner C. (1993) Pancreatic one-carbon metabolism in early folate deficiency in rats. Am. J. Clin. Nutr 58, 198–203 [DOI] [PubMed] [Google Scholar]
- 63.Nieman K. M., Hartz C. S., Szegedi S. S., Garrow T. A., Sparks J. D., Schalinske K. L. (2006) Folate status modulates the induction of hepatic glycine N-methyltransferase and homocysteine metabolism in diabetic rats. Am. J. Physiol. Endocrinol. Metab 291, E1235–E1242 [DOI] [PubMed] [Google Scholar]
- 64.Garrow T. A. (2001) Betaine-dependent remethylation, in Homocysteine in Health and Disease (Carmel R., Jacobsen D. W.) pp. 145–152, Cambridge University Press, Cambridge, UK [Google Scholar]
- 65.Luka Z., Cerone R., Phillips J. A., 3rd, Mudd H. S., Wagner C. (2002) Mutations in human glycine N-methyltransferase give insights into its role in methionine metabolism. Hum. Genet 110, 68–74 [DOI] [PubMed] [Google Scholar]
- 66.Park E. I., Garrow T. A. (1999) Interaction between dietary methionine and methyl donor intake on rat liver betaine-homocysteine methyltransferase gene expression and organization of the human gene. J. Biol. Chem 274, 7816–7824 [DOI] [PubMed] [Google Scholar]
- 67.Zaragozá R., Renau-Piqueras J., Portolés M., Hernández-Yago J., Jordá A., Grisolía S. (1987) Rats fed prolonged high protein diets show an increase in nitrogen metabolism and liver megamitochondria. Arch. Biochem. Biophys 258, 426–435 [DOI] [PubMed] [Google Scholar]
- 68.Chen T., Zamora R., Zuckerbraun B., Billiar T. R. (2003) Role of nitric oxide in liver injury. Curr. Mol. Med 3, 519–526 [DOI] [PubMed] [Google Scholar]
- 69.Isobe M., Katsuramaki T., Hirata K., Kimura H., Nagayama M., Matsuno T. (1999) Beneficial effects of inducible nitric oxide synthase inhibitor on reperfusion injury in the pig liver. Transplantation 68, 803–813 [DOI] [PubMed] [Google Scholar]
- 70.Lee V. G., Johnson M. L., Baust J., Laubach V. E., Watkins S. C., Billiar T. R. (2001) The roles of iNOS in liver ischemia-reperfusion injury. Shock 16, 355–360 [DOI] [PubMed] [Google Scholar]
- 71.Zima T., Fialová L., Mestek O., Janebová M., Crkovská J., Malbohan I., Stípek S., Mikulíková L., Popov P. (2001) Oxidative stress, metabolism of ethanol and alcohol-related diseases. J. Biomed. Sci 8, 59–70 [DOI] [PubMed] [Google Scholar]
- 72.Hinson J. A., Michael S. L., Ault S. G., Pumford N. R. (2000) Western blot analysis for nitrotyrosine protein adducts in livers of saline-treated and acetaminophen-treated mice. Toxicol. Sci 53, 467–473 [DOI] [PubMed] [Google Scholar]
- 73.Zhang X., Li H., Jin H., Ebin Z., Brodsky S., Goligorsky M. S. (2000) Effects of homocysteine on endothelial nitric oxide production. Am. J. Physiol. Renal Physiol 279, F671–F678 [DOI] [PubMed] [Google Scholar]
- 74.Tyagi N., Sedoris K. C., Steed M., Ovechkin A. V., Moshal K. S., Tyagi S. C. (2005) Mechanisms of homocysteine-induced oxidative stress. Am. J. Physiol. Heart Circ. Physiol 289, H2649–H2656 [DOI] [PubMed] [Google Scholar]
- 75.Dayal S., Bottiglieri T., Arning E., Maeda N., Malinow M. R., Sigmund C. D., Heistad D. D., Faraci F. M., Lentz S. R. (2001) Endothelial dysfunction and elevation of S-adenosylhomocysteine in cystathionine beta-synthase-deficient mice. Circ. Res 88, 1203–1209 [DOI] [PubMed] [Google Scholar]
- 76.Lentz S. R., Erger R. A., Dayal S., Maeda N., Malinow M. R., Heistad D. D., Faraci F. M. (2000) Folate dependence of hyperhomocysteinemia and vascular dysfunction in cystathionine beta-synthase-deficient mice. Am. J. Physiol. Heart Circ. Physiol 279, H970–H975 [DOI] [PubMed] [Google Scholar]
- 77.Eberhardt R. T., Forgione M. A., Cap A., Leopold J. A., Rudd M. A., Trolliet M., Heydrick S., Stark R., Klings E. S., Moldovan N. I., Yaghoubi M., Goldschmidt-Clermont P. J., Farber H. W., Cohen R., Loscalzo J. (2000) Endothelial dysfunction in a murine model of mild hyperhomocyst(e)inemia. J. Clin. Investig 106, 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ottaviano F. G., Handy D. E., Loscalzo J. (2008) Redox regulation in the extracellular environment. Circ. J 72, 1–16 [DOI] [PubMed] [Google Scholar]
- 79.Das K. C., Das C. K. (2000) Thioredoxin, a singlet oxygen quencher and hydroxyl radical scavenger: redox independent functions. Biochem. Biophys. Res. Commun 277, 443–447 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.