Abstract
Genetic factors are strongly involved in the development of obesity, likely through the interactions of susceptibility genes with obesigenic environments, such as high-fat, high-sucrose (HFS) diets. Previously, we have established a mouse congenic strain on C57BL/6 J background, carrying an obesity quantitative trait locus (QTL), tabw2, derived from obese diabetic TALLYHO/JngJ mice. The tabw2 congenic mice exhibit increased adiposity and hyperleptinemia, which becomes exacerbated upon feeding HFS diets. In this study, we conducted genome-wide gene expression profiling to evaluate differentially expressed genes between tabw2 and control mice fed HFS diets, which may lead to identification of candidate genes as well as insights into the mechanisms underlying obesity mediated by tabw2. Both tabw2 congenic mice and control mice were fed HFS diets for 10 weeks beginning at 4 weeks of age, and total RNA was isolated from liver and adipose tissue. Whole-genome microarray analysis was performed and verified by real-time quantitative RT–PCR. At False Discovery Rate adjusted P < 0.05, 1026 genes were up-regulated and 308 down-regulated in liver, whereas 393 were up-regulated and 187 down-regulated in adipose tissue in tabw2 congenic mice compared to controls. Within the tabw2 QTL interval, 70 genes exhibited differential expression in either liver or adipose tissue. A comprehensive pathway analysis revealed a number of biological pathways that may be perturbed in the diet-induced obesity mediated by tabw2.
Keywords: Gene expression profiles, QTL, Congenics, Diet-induced obesity, Mice
Introduction
The high prevalence of obesity in our society is currently overwhelming; approximately 1.2 billion people are overweight worldwide and among those at least 300 million people are obese [30]. The related medical complications are life-threatening diseases, including type 2 diabetes, heart disease, hypertension, and many forms of cancer [11]. The etiology of obesity is complex, involving genetic susceptibility, environmental influence, and gene-environmental interactions [23].
Animal models that share both physiologic and genetic similarity with humans have been used to minimize many difficulties encountered in carrying out obesity studies in humans [27]. Polygenic rodent models carrying natural variations have been developed and serve as valuable resources for obesity research, closely mimicking the polygenic inheritance of obesity in humans.
Previously, we have mapped a quantitative trait locus (QTL) linked to body weight on mouse chromosome 6 in a cross between C57BL/6J (B6) and obese diabetic TALLYHO/JngJ (TH) mice [9]. The TH allele was associated with higher body weights, and the QTL is named tabw2 (TALLYHO Associated Body Weight 2). Subsequently, we have constructed a congenic strain that carries a TH-derived genomic segment containing tabw2 on a B6 background. This congenic strain (tabw2 mice) exhibits increased adiposity and hyperleptinemia, and upon feeding high-fat, high-sucrose (HFS) diets, the obesity becomes exacerbated, followed by the development of insulin resistance [9].
The present study sought to investigate the genome-wide gene expression profiles in liver and adipose tissue to elucidate differentially expressed genes between tabw2 and control mice fed HFS diets. This study will identify differentially expressed genes within the congenic region, providing candidate genes for tabw2, as well as other genes involved in common pathways of obesity. The findings will contribute to understanding the gene networks underlying the diet-induced obesity mediated by tabw2.
Materials and methods
Animals and diets
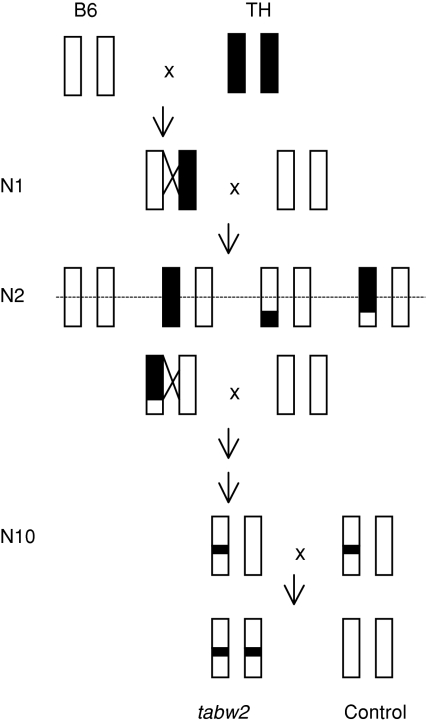
The tabw2 congenic and control mice used in this study were from previously established lines [9]. Briefly, B6 female and TH male mice were crossed to yield F1 (or N1) progeny that were then backcrossed to B6 mice. The resulting N2 progeny were genotyped with flanking markers to select heterozygotes for the tabw2 QTL interval that were then again backcrossed to B6 mice. This procedure was repeated for 10 cycles of backcrossing to achieve more than 99% homogeneity [21] for the B6 genome in the congenic strain at which point two heterozygotes were intercrossed to yield offspring that were either homozygous for the TH alleles (tabw2 mice) or homozygous for the B6 alleles (control mice) (Fig. 1). Homozygous mice were then interbred to maintain the tabw2 and control mice.
Fig. 1.
Construction of a congenic mouse strain carrying the obesity QTL on chromosome 6, named tabw2, derived from TALLYHO/JngJ (TH) mice in the C57BL/6J (B6) background by marker assisted backcrossing. An obese TH male mouse was crossed to a normal B6 female mouse, and the resultant F1 (or N1) mice were backcrossed to B6. Heterozygotes for the QTL were selected using flanking markers (shown as dotted line) and backcrossed again to B6. This procedure was repeated for 10 cycles of backcrossing at which point two heterozygotes were intercrossed to yield offspring that are homozygous for the TH alleles (tabw2 mice) and for the B6 alleles (control mice) across the congenic region
All mice were allowed free access to food and water in a temperature and humidity controlled room with a 12-h light/dark cycle. Mice were weaned onto HFS diets (32% kcal from fat and 25% kcal from sucrose) (12266B, Research Diets, New Brunswick, NJ, USA) at 4 weeks of age. At 14 weeks of age, mice were weighed, then euthanized by CO2 asphyxiation, and liver and adipose tissue (inguinal, epididymal, retroperitoneal, perirenal, and subscapular fat pads) were collected, immediately frozen in liquid nitrogen, and stored at −80°C for RNA isolation. Statistical analysis for body weight data was conducted by ANOVA with StatView 5.0 (Abacus Concepts, Berkeley, CA). All animal studies were carried out with the approval of The University of Tennessee Animal Care and Use Committee.
RNA isolation
Total RNA was isolated from liver and white adipose (combined inguinal, epididymal, retroperitoneal, perirenal, and subscapular fat pads) tissue using RNeasy Lipid Tissue Midi Kit (75842, QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. For adipose tissue, the entire tissue was homogenized and total RNA extracted, whereas approximately 50% of the liver was homogenized. Total RNA was further purified using RNeasy MinElute Cleanup Kit (74204, QIAGEN) for microarray analysis.
Microarray analysis
Hybridizations were performed at Genome Explorations Inc. (Memphis, TN, USA) using GeneChip® Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA, USA) following the standard protocol. The Mouse Genome 430 2.0 Array contains 45,000 probe sets on a single array to analyze the expression level of over 39,000 transcripts and variants from over 34,000 well-characterized mouse genes (Affymetrix). Total RNA isolated from liver and adipose tissue as described previously from 4 male tabw2 mice and 4 male control mice were used for microarray analysis, requiring 16 arrays.
The gcRMA (robust multi-array) process in Bioconductor (http://www.bioconductor.org) was used to produce a normalized signal measure for each gene on each array. Data were examined for outliers and consistency of arrays, then statistical analysis was performed using SAS software (SAS Institute Inc., Cary, NC, USA). A mixed ANOVA model [31] for each gene tested factorial treatment effects of genotype and tissue, and used array variation as the experimental error. Genes with significant (P < 0.05) ANOVA interaction and significant pair-wise False Discovery Rate [22] were considered differentially expressed. ANOVA results were used to create volcano plots to help visualize the distribution of differential expression.
Real-time quantitative RT–PCR
Total RNA (2 μg) was reverse-transcribed with SUPERSCRIPT RT (11904-018, Invitrogen, Carlsbad, CA, USA) using oligo d(T)12–18 (18418-012, Invitrogen) as primer to synthesize first-strand cDNA in 20-μl volume according to manufacturer’s instructions. Oligonucleotide primers were synthesized (Sigma–Aldrich, St. Louis, MO, USA) using sequences obtained from Primer Bank (http://pga.mgh.harvard.edu/primerbank) or the published literature (Table 1). The PCR reaction was carried out in a 25-μl volume in 1× SYBR Green PCR core reagents (PA-112, SABiosciences, Frederick, MD, USA) containing 1 μl cDNA template diluate (1:5, v/v) and 6 pmol primers. Real-time PCR was conducted using an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA, USA). For each sample, triplicate amplifications were performed and the average measurements used for data analysis. The difference in average threshold cycle (∆Ct) values between 36B4 gene and a specific gene was calculated for each individual. The data were then presented as relative fold-change using control mice as the reference by equation 2−(∆Ct of tabw2 mice−∆Ct of control mice) [13]. If the difference was negative, the calculation was inverted and made negative, to signify over-expression in tabw2 mice. Mice measured by qRT–PCR were not the same as used in the microarray analysis to increase biological validation (n = 5, male, for each genotype).
Table 1.
Primer sequences for real-time quantitative RT-PCR
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′−3′) | Reference |
|---|---|---|---|
| Acaa1a | TCTCCAGGACGTGAGGCTAAA | CGCTCAGAAATTGGGCGATG | Primer bank |
| Acaca | ATGGGCGGAATGGTCTCTTTC | TGGGGACCTTGTCTTCATCAT | Primer bank |
| Acss2 | AAACACGCTCAGGGAAAATCA | ACCGTAGATGTATCCCCCAGG | Primer bank |
| Arhgdib | ATGACGGAGAAGGATGCACAG | CTCCCAGCAGTGTTTTCTTGTA | Primer bank |
| Ccnd2 | GCGTGCAGAAGGACATCCA | CACTTTTGTTCCTCACAGACCTCTAG | [5] |
| Cyp4a14 | TTTAGCCCTACAAGGTACTTGGA | GCAGCCACTGCCTTCGTAA | Primer bank |
| Daam1 | AGATAGCGGATACCAAATCCAGT | TCTTCGCTTAGGTTGAGGACT | Primer bank |
| Hadhsc | TCAAGCATGTGACCGTCATCG | TGGATTTTGCCAGGATGTCTTC | Primer bank |
| Hsd17b4 | AGGGGACTTCAAGGGAATTGG | GCCTGCTTCAACTGAATCGTAA | Primer bank |
| Klrd1 | TCTAGGATCACTCGGTGGAGA | CACTTGTCCAGGCAAACACAG | Primer bank |
| Lrp6 | TTGTTGCTTTATGCAAACAGACG | GTTCGTTTAATGGCTTCTTCGC | Primer bank |
| Mgll | CGGACTTCCAAGTTTTTGTCAGA | GCAGCCACTAGGATGGAGATG | Primer bank |
| Mup1 | GAAGCTAGTTCTACGGGAAGGA | AGGCCAGGATAATAGTATGCCA | Primer bank |
| Nfatc3 | ACTGCCTCATCACCATCTCC | TCCCAATAATCTCGTTCACATC | [20] |
| Nlk | ACCAAGATGATACCCTGTGACT | AAGAAGTTAGCCAGGAGGATCT | [19] |
| Ret | TTTCTCAAGGGATGCTTACTGGG | CCCGTAGGGCATGGACATAGA | Primer bank |
| Ruvbl1 | AGCTGGGCAGTAAAGTCCCT | CCTCCCCTTCATAAACCTCCT | Primer bank |
| Sfrp5 | CACTGCCACAAGTTCCCCC | TCTGTTCCATGAGGCCATCAG | Primer bank |
| Tcf3 | ACGAGCTGATCCCCTTCCA | CAGGGACGACTTGACCTCAT | Primer bank |
| Tcf7l2 | AACGAACACAGCGAATGTTTCC | CACCTTGTATGTAGCGAACGC | Primer bank |
| Wnt5b | CCAGTGCAGAGACCGGAGATG | GTTGTCCACGGTGCTGCAGTTC | [8] |
| 36B4 | GAGGAATCAGATGAGGATATGGGA | AAGCAGGCTGACTTGGTTGC | [3] |
Primer Bank (http://pga.mgh.harvard.edu/primerbank)
Results
Tabw2 mice fed HFS diets were significantly heavier than control mice [33.4 ± 1.2 (n = 14) vs. 28.0 ± 0.4 (n = 14) g; mean ± SEM; P = 0.0002; male; 14-week old].
Differentially expressed gene profiling overview in liver and adipose tissue from tabw2 and control mice
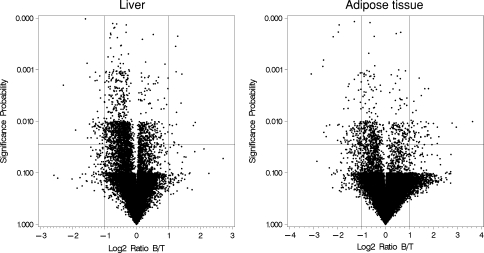
Using a global expression chip, we compared the levels of gene expression in liver and adipose tissue from tabw2 mice and control mice fed HFS diets. Gene expression profiles were visualized by volcano plots (Fig. 2). Overall, large differences in gene expression levels were rare between tabw2 and control mice, which can be deduced from the volcano plots clustered at the center. This may be because the only genomic difference between the tabw2 and control mice is in the congenic region.
Fig. 2.
Volcano plot comparison of gene expression between control (B) and tabw2 (T) mice in liver and adipose tissue. The X-axis indicates the differential expression, plotting the fold-difference ratios on a log-2 scale. The Y-axis indicates log10 statistical significance levels for difference in expression. Vertical reference lines indicate 2-fold expression change, and a horizontal reference line is drawn at P < 0.05
Of over 39,000 transcripts (hereafter referred to as genes), at a significance level of P < 0.05, 1026 genes were up-regulated and 308 down-regulated in liver, whereas 393 were up-regulated and 187 down-regulated in adipose tissue in tabw2 mice compared to control mice. When examined in each tissue for the top 50 (25 up-regulated and 25 down-regulated) genes with the largest effect of genotype (Tables 2 and 3), the most largely changed genes were found in adipose tissue; Sfrp5 (up-regulated in tabw2 mice) and Mup1 (down-regulated in tabw2 mice) (Table 2).
Table 2.
The 50 genes with largest fold change between tabw2 and control mice in adipose tissue
| Probe set ID | Symbol | Gene name | Chr | Fold |
|---|---|---|---|---|
| Up-regulated in tabw2 | ||||
| 1436075_at | Sfrp5 | Secreted frizzled-related sequence protein 5 | 19 | 8.59 |
| 1436294_at | Ankrd29 | Ankyrin repeat domain 29 | 18 | 6.14 |
| 1418713_at | Pcbdl | Pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nu | 10 | 6.02 |
| 1430596_s_at | 1700110N18Rik | RIKEN cDNA 1700110N18 gene | 16 | 5.97 |
| 1419109_at | Hrc | Histidine rich calcium binding protein | 7 | 5.57 |
| 1441737_s_at | Rassf1 | Ras association (RalGDS/AF-6) domain family 1 | 9 | 5.04 |
| 1438967_x_at | Amhr2 | Anti-Mullerian hormone type 2 receptor | 15 | 4.55 |
| 1426143_at | Trdn | Triadin | 10 | 4.00 |
| 1447851_x_at | Atp10a | ATPase, class V, type 10A | 7 | 3.98 |
| 1455215_at | C530028O21Rik | RIKEN cDNA C530028O21 gene | 6 | 3.92 |
| 1418497_at | Fgf13 | Fibroblast growth factor 13 | X | 3.84 |
| 1422580_at | Myl4 | Myosin, light polypeptide 4 | 11 | 3.82 |
| 1435631_x_at | Exoc6 | Exocyst complex component 6 | 19 | 3.66 |
| 1444089_at | Spnb2 | Spectrin beta 2 | 11 | 3.58 |
| 1436359_at | Ret | Ret proto-oncogene | 6 | 3.46 |
| 1448595_a_at | Rex3 | Reduced expression 3 | X | 3.35 |
| 1429135_at | 1110059M19Rik | RIKEN cDNA 1110059M19 gene | X | 3.31 |
| 1447657_s_at | Synpo2 l | Synaptopodin 2-like | 14 | 3.24 |
| 1429599_a_at | 1110019K23Rik | RIKEN cDNA 1110019K23 gene | 5 | 3.19 |
| 1460010_a_at | Ptdss2 | Phosphatidylserine synthase 2 | 7 | 3.15 |
| 1457021_x_at | Amhr2 | Anti-Mullerian hormone type 2 receptor | 15 | 3.10 |
| 1434797_at | 6720469N11Rik | RIKEN cDNA 6720469N11 gene | 3 | 3.07 |
| 1420143_at | Mnab | Membrane associated DNA binding protein | 2 | 3.06 |
| 1447520_at | Lbp | Lipopolysaccharide binding protein | 2 | 3.05 |
| 1435917_at | Ociad2 | OCIA domain containing 2 | 5 | 3.05 |
| Down-regulated in tabw2 | ||||
| 1434110_x_at | Mup1 | Major urinary protein 1 | 4 | 7.74 |
| 1448229_s_at | Ccnd2 | Cyclin D2 | 6 | 4.67 |
| 1454169_a_at | Epsti1 | Epithelial stromal interaction 1 (breast) | 14 | 4.47 |
| 1422479_at | Acss2 | Acyl-CoA synthetase short-chain family member 2 | 2 | 4.46 |
| 1419480_at | Sell | Selectin, lymphocyte | 1 | 3.70 |
| 1426806_at | 5830411E10Rik | RIKEN cDNA 5830411E10 gene | 1 | 3.49 |
| 1447147_at | Apg7 l | Autophagy-related 7 (yeast) | 6 | 3.46 |
| 1424825_a_at | Glycam1 | Glycosylation dependent cell adhesion molecule 1 | 15 | 3.46 |
| 1460245_at | Klrd1 | Killer cell lectin-like receptor, subfamily D, member 1 | 6 | 3.39 |
| 1426166_at | Mup5 | Major urinary protein 5 | 4 | 3.34 |
| 1435602_at | Sephs2 | Selenophosphate synthetase 2 | 7 | 3.34 |
| 1418126_at | Ccl5 | Chemokine (C–C motif) ligand 5 | 11 | 3.24 |
| 1424931_s_at | Igl-V1 | Immunoglobulin lambda chain, variable 1 | 16 | 3.20 |
| 1436766_at | Luc7l2 | LUC7-like 2 (S. cerevisiae) | 6 | 3.13 |
| 1423371_at | Pole4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | 6 | 3.11 |
| 1451335_at | Plac8 | Placenta-specific 8 | 5 | 3.09 |
| 1460521_a_at | 5830411E10Rik | RIKEN cDNA 5830411E10 gene | 1 | 3.07 |
| 1422411_s_at | Ear1 | Eosinophil-associated, ribonuclease A family, member 1 | 14 | 2.87 |
| 1425137_a_at | H2-Q10 | Histocompatibility 2, Q region locus 10 | 17 | 2.81 |
| 1437636_at | LOC623121 | Similar to Interferon-activatable protein 203 (Ifi-203) (Interferon-inducible protein p203) | 1 | 2.77 |
| 1451644_a_at | H2-Q1 | Histocompatibility 2, Q region locus 1 | 17 | 2.77 |
| 1433827_at | Atp8a1 | ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 | 5 | 2.76 |
| 1451691_at | Ednra | Endothelin receptor type A | 8 | 2.66 |
| 1434152_at | 2210421G13Rik | RIKEN cDNA 2210421G13 gene | 15 | 2.64 |
| 1426159_x_at | Tcrb-V13 | T-cell receptor beta, variable 13 | 6 | 2.61 |
Chr chromosome
Table 3.
The 50 genes with largest fold change between tabw2 and control mice in liver
| Probe set ID | Symbol | Gene name | Chr | Fold |
|---|---|---|---|---|
| Up-regulated in tabw2 | ||||
| 1444438_at | Cib3 | Calcium and integrin binding family member 3 | 8 | 4.89 |
| 1423257_at | Cyp4a14 | Cytochrome P450, family 4, subfamily a, polypeptide 14 | 4 | 3.75 |
| 1455308_at | Tmem16f | Transmembrane protein 16F | 15 | 3.02 |
| 1453462_at | Chst13 | Carbohydrate (chondroitin 4) sulfotransferase 13 | 6 | 2.92 |
| 1452005_at | Dlat | Dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 9 | 2.89 |
| 1437239_x_at | Phc2 | Polyhomeotic-like 2 (Drosophila) | 4 | 2.77 |
| 1449641_at | Adk | Adenosine kinase | 14 | 2.75 |
| 1422076_at | Acot4 | Acyl-CoA thioesterase 4 | 12 | 2.68 |
| 1447227_at | Slc40a1 | Solute carrier family 40 (iron-regulated transporter), member 1 | 1 | 2.61 |
| 1438969_x_at | Dhx30 | DEAH (Asp-Glu-Ala-His) box polypeptide 30 | 9 | 2.56 |
| 1449770_x_at | Tmem191c | Transmembrane protein 191C | 16 | 2.56 |
| 1438617_at | Serpina7 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 | X | 2.53 |
| 1420357_s_at | Xlr3a | X-linked lymphocyte-regulated 3A | X | 2.52 |
| 1431805_a_at | Rhpn2 | Rhophilin, Rho GTPase binding protein 2 | 7 | 2.46 |
| 1417280_at | Slc17a1 | Solute carrier family 17 (sodium phosphate), member 1 | 13 | 2.45 |
| 1438660_at | Gcnt2 | Glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | 13 | 2.45 |
| 1451822_a_at | Scrn2 | Secernin 2 | 11 | 2.45 |
| 1418524_at | Pcm1 | Pericentriolar material 1 | 8 | 2.43 |
| 1438487_s_at | Zzz3 | Zinc finger, ZZ domain containing 3 | 3 | 2.42 |
| 1448385_at | Slc15a4 | Solute carrier family 15, member 4 | 5 | 2.41 |
| 1437983_at | Sall1 | Sal-like 1 (Drosophila) | 8 | 2.37 |
| 1422077_at | Acot4 | Acyl-CoA thioesterase 4 | 12 | 2.35 |
| 1441141_at | Amn1 | Antagonist of mitotic exit network 1 homolog (S. cerevisiae) | 6 | 2.33 |
| 1426350_at | Mgat2 | Mannoside acetylglucosaminyltransferase 2 | 12 | 2.32 |
| 1444810_at | Acaca | Acetyl-Coenzyme A carboxylase alpha | 11 | 2.29 |
| Down-regulated in tabw2 | ||||
| 1421447_at | Onecut1 | One cut domain, family member 1 | 9 | 3.57 |
| 1452431_s_at | H2-Aa | Histocompatibility 2, class II antigen A, alpha | 17 | 3.43 |
| 1453007_at | 3110082I17Rik | RIKEN cDNA 3110082I17 gene | 5 | 2.67 |
| 1421571_a_at | Ly6c | Lymphocyte antigen 6 complex, locus C | 15 | 2.62 |
| 1457619_at | BC015286 | cDNA sequence BC015286 | 8 | 2.44 |
| 1417025_at | H2-Eb1 | Histocompatibility 2, class II antigen E beta | 17 | 2.38 |
| 1456524_at | Nrg1 | Neuregulin 1 | 8 | 2.37 |
| 1439256_x_at | Tm 7sf1 | Transmembrane 7 superfamily member 1 | 13 | 2.36 |
| 1422754_at | Tmod1 | Tropomodulin 1 | 4 | 2.33 |
| 1424186_at | 2610001E17Rik | RIKEN cDNA 2610001E17 gene | 16 | 2.28 |
| 1447870_x_at | 1110002E22Rik | RIKEN cDNA 1110002E22 gene | 3 | 2.25 |
| 1450839_at | D0H4S114 | DNA segment, human D4S114 | 18 | 2.17 |
| 1432620_at | Ttn | Titin | 2 | 2.05 |
| 1428549_at | Ccdc3 | Coiled-coil domain containing 3 | 2 | 1.98 |
| 1425167_a_at | Gngt1 | Guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 | 6 | 1.97 |
| 1455307_at | BC037112 | cDNA sequence BC037112 | 5 | 1.97 |
| 1423028_at | Ifna2 | Interferon alpha family, gene 2 | 4 | 1.92 |
| 1421226_at | Trem2 | Triggering receptor expressed on myeloid cells 2 | 17 | 1.85 |
| 1425358_at | Riok1 | RIO kinase 1 (yeast) | 13 | 1.85 |
| 1457023_at | 5830411G16Rik | RIKEN cDNA 5830411G16 gene | 6 | 1.83 |
| 1427048_at | Smo | Smoothened homolog (Drosophila) | 6 | 1.81 |
| 1450068_at | Baz1b | Bromodomain adjacent to zinc finger domain, 1B | 5 | 1.80 |
| 1429144_at | Prei4 | Preimplantation protein 4 | 2 | 1.80 |
| 1456093_at | Zfp536 | Zinc finger protein 536 | 7 | 1.77 |
| 1450843_a_at | Serpinh1 | Serine (or cysteine) peptidase inhibitor, clade H, member 1 | 7 | 1.73 |
Chr chromosome
Differentially expressed genes located within the tabw2 QTL interval
Using congenic mice, the microarray analysis strategy has been useful in identification of QTLs [1, 28]. In an attempt to select attractive positional candidate genes for tabw2, we examined the gene expression levels located within the tabw2 congenic interval on chromosome 6, based on the hypothesis that the genetic alteration of tabw2 may cause dysregulation of the gene expression. Forty-five genes in liver and 32 genes in adipose tissue located within the congenic interval (47.0–137.3 Mb) were differentially expressed between tabw2 and control mice (Table 4); 7 genes, including Znrf2, Pole4, Isy1, Frmd4b, Tmcc1, Ccnd2, and Lrp6, appeared in both tissues. Of these 70 genes, seven (5830411G16Rik and Chast13 in liver and Pole4, Ret, C530028O21Rik, Ccnd2, and Klrd1 in adipose tissue) were present in the top 50 genes with the largest fold change between tabw2 and control mice (boldface entries Table 4).
Table 4.
Differentially expressed genes between tabw2 and control mice in liver and adipose tissue (fat) that are located within the congenic interval on chromosome 6
| Probe set ID | Symbol | Gene name | Mb | Tissue | Fold |
|---|---|---|---|---|---|
| 1441727_s_at | Zfp467 | Zinc finger protein 467 | 48.4 | Fat | −1.85 |
| 1434043_a_at | Repinl | Replication initiator 1 | 48.5 | Fat | −1.73 |
| 1424375_s_at | Gimap4 | GTPase, IMAP family member 4 | 48.6 | Fat | 1.64 |
| 1420365_a_at | Hnrpa2b1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 51.4 | Liver | −1.16 |
| 1428922_at | 1200009O22Rik | RIKEN cDNA 1200009O22 gene | 53.8 | Fat | −2.20 |
| 1434016_at | Znrf2 | Zinc and ring finger 2 | 54.8 | Fat | 1.77 |
| 1444735_at | Liver | −1.28 | |||
| 1423784_at | Gars | Glycyl-tRNA synthetase | 55.0 | Liver | −1.26 |
| 1418697_at | Inmt | Indolethylamine N-methyltransferase | 55.1 | Fat | 2.26 |
| 1418656_at | Lsm5 | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) | 56.7 | Liver | −1.23 |
| 1457023_at | 5830411G16Rik | Riken cDNA 5830411G16 gene | 56.7 | Liver | 1.83 |
| 1432026_a_at | Herc5 | Hect domain and RLD 5 | 57.4 | Liver | 1.32 |
| 1429194_at | Tigd2 | Tigger transposable element derived 2 | 59.2 | Liver | −1.41 |
| 1449519_at | Gadd45a | Growth arrest and DNA-damage-inducible 45 alpha | 67.0 | Liver | −1.74 |
| 1427860_at | LOC100047162 | Similar to Ig kappa chain V–V region MPC11 precursor | 70.4 | Fat | 1.54 |
| 1425335_at | Cd8a | CD8 antigen, alpha chain | 71.3 | Fat | 1.96 |
| 1443830_x_at | Rnf103 | Ring finger protein 103 | 71.5 | Liver | −1.39 |
| 1420289_at | T25656 | Expressed sequence T25656 | 71.6 | Liver | 1.11 |
| 1424716_at | Restsat | Retinol saturase (all trans retinol 13, 14 reductase) | 72.5 | Liver | −1.58 |
| 1450117_at | Tcf3 | Transcription factor 3 | 72.6 | Fat | −1.64 |
| 1448895_a_at | Ctnna2 | Catenin (cadherin associated protein), alpha 2 | 76.8 | Liver | 1.38 |
| 1432286_at | Pole4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | 82.6 | Liver | 1.26 |
| 1423371_at | Fat | 3.11 | |||
| 1436618_at | Sfxn5 | Sideroflexin 5 | 85.2 | Liver | −1.67 |
| 1432969_at | 4933423K11Rik | RIKEN cDNA 4933423K11 gene | 85.3 | Liver | 1.09 |
| 1418013_at | Cml1 | Camello-like 1 | 85.9 | Fat | −1.51 |
| 1447277_s_at | Pcyox1 | Prenylcysteine oxidase 1 | 86.3 | Liver | −1.27 |
| 1418229_s_at | Nfu1 | NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) | 87.0 | Liver | −1.32 |
| 1453132_a_at | Gkn2 | Gastrokine2 | 87.3 | Liver | 1.38 |
| 1459728_at | Isy1 | ISY1 splicing factor homolog (S. cerevisiae) | 87.8 | Fat | −2.46 |
| Liver | −1.35 | ||||
| 1416244_a_at | Cnbp | Cellular nucleic acid binding protein | 87.8 | Liver | −1.17 |
| 1416585_at | Ruvbl1 | RuvB-like protein 1 | 88.4 | Liver | −1.30 |
| 1442560_at | Mgll | Monoglyceride lipase | 88.7 | Liver | −2.01 |
| 1453462_at | Chst13 | Carbohydrate (chondroitin 4) sulfotransferase 13 | 90.3 | Liver | −2.92 |
| 1451229_at | Hdac11 | Histone deacetylase 11 | 91.1 | Liver | −1.27 |
| 1456879_at | C130022K22Rik | RIKEN cDNA C130022K22 gene | 91.8 | Liver | −1.50 |
| 1416911_a_at | 6330407G11Rik | RIKEN cDNA 6330407G11 gene | 92.0 | Liver | −2.10 |
| 1449194_at | Mrps25 | Mitochondrial ribosomal protein S25 | 92.1 | Liver | −1.24 |
| 1439933_at | B430316J06Rik | RIKEN cDNA B430316J06 gene | 93.9 | Liver | −1.74 |
| 1443231_at | AW544 786 | Expressed sequence tag | 94.2 | Fat | −2.22 |
| 1433671_at | A130022J15Rik | RIKEN cDNA A130022J15 gene | 97.1 | Liver | 1.49 |
| 1452123_s_at | Frmd4b | FERM domain containing 4B | 97.2 | Liver | −1.83 |
| 1426594_at | Fat | 1.34 | |||
| 1421111_at | Rybp | RING1 and YY1 binding protein | 100.1 | Liver | 1.18 |
| 1428137_at | Arl8b | ADP-ribosylation factor-like 8b | 108.7 | Liver | −1.50 |
| 1443954_at | Rad18 | RAD18 homolog (S. cerevisiae) | 112.6 | Fat | 1.27 |
| 1423189_at | 6720456B07Rik | RIKEN cDNA 6720456B07 gene | 113.5 | Liver | −1.37 |
| 1444806_at | AK054191 | Expressed sequence tag | 113.5 | Fat | 1.98 |
| 1447147_at | AI747732 | Expressed sequence tag | 114.8 | Fat | 3.46 |
| 1440028_at | 4631423B10Rik | RIKEN cDNA 4631423B10 gene | 114.8 | Fat | −1.70 |
| 1437677_at | AI449595 | Expressed sequence tag | 114.9 | Liver | 1.11 |
| 1416078_s_at | Rafl | v-Raf-1 leukemia viral oncogene 1 | 115.5 | Liver | −1.37 |
| 1440384_at | Tmccl | Transmembrane and coiled-coil domains 1 | 115.9 | Liver | −1.94 |
| Fat | −1.88 | ||||
| 1417574_at | Cxcl12 | Chemokine (C-X-C motif) ligand 12 | 117.1 | Fat | 2.25 |
| 1436359_at | Ret | Ret proto-oncogene | 118.1 | Fat | −3.46 |
| 1422602_a_at | Wnt5b | Wingless-related MMTV integration site 5B | 119.3 | Liver | 1.19 |
| 1417407_at | Fbxl14 | F-box and leucine-rich repeat protein 14 | 119.4 | Liver | −1.50 |
| 1424247_at | Ercl | ELKS/RAB6-interacting/CAST family member 1 | 119.5 | Fat | −1.71 |
| 1434221_at | BC030863 | cDNA sequence BC030863 | 120.8 | Liver | −1.20 |
| 1425951_a_at | Clec4n | C-type lectin domain family 4, member n | 123.1 | Fat | 1.68 |
| 1426770_at | Pex5 | Peroxisome biogenesis factor 5 | 124.3 | Liver | −1.33 |
| 1422106_a_at | Spsb2 | SplA/ryanodine receptor domain and SOCS box containing 2 | 124.7 | Fat | −1.12 |
| 1455215_at | C530028O21Rik | RIKEN cDNA C530028O21 gene | 124.9 | Fat | −3.92 |
| 1455785_at | Kcnal | Potassium voltage-gated channel, shaker-related subfamily, member 1 | 126.5 | Fat | 1.26 |
| 1448229_s_at | Ccnd2 | Cyclin D2 | 127.0 | Fat | 4.67 |
| 1434745_at | Liver | −1.69 | |||
| 1460245_at | Klrdl | Killer cell lectin-like receptor, subfamily D, member 1 | 129.5 | Fat | 3.39 |
| 1446155_at | AK078025 | Expressed sequence tag | 133.0 | Fat | 2.32 |
| 1415968_a_at | Kap | Kidney androgen-regulated protein | 133.7 | Fat | −2.87 |
| 1440982_at | BB209400 | Expressed sequence tag | 134.0 | Fat | −1.23 |
| 1451022_at | Lrp6 | Low density lipoprotein receptor-related protein 6 | 134.4 | Fat | −1.92 |
| Liver | −1.29 | ||||
| 1435085_at | Crebl2 | CAMP responsive element binding protein-like 2 | 134.8 | Liver | −1.39 |
| 1434045_at | Cdkn1b | Cyclin-dependent kinase inhibitor 1B | 134.8 | Liver | −1.89 |
| 1426454_at | Arhgdib | Rho, GDP dissociation inhibitor (GDI) beta | 136.8 | Liver | 1.58 |
Genes in bold are present in the top 50 genes with the largest fold change between tabw2 and control mice
Mb, mega-base; ‘−’ indicates up-regulation and ‘no sign’ indicates down-regulation in tabw2 mice compared to control mice
Except for a few genes, such as Mgll, the differentially expressed genes within the congenic interval had mostly unknown connections with obesity. Monoglyceride lipase (Mgll) hydrolyzes the monoglycerides formed during the hydrolysis of triglycerides [24]. The gene expression of Mgll was increased in liver of tabw2 mice. In agreement with this, hepatic increases in protein and activity of Mgll have previously been reported in obese mice fed high-fat diets, whereas little changes in adipose tissue occurred [2].
Another interesting finding was the down-regulation of Arhgdib gene in liver of tabw2 mice. ARHGDIB (also known as Rho GDIβ or D4/Ly GDI) negatively regulates Rho small GTP-binding protein by inhibiting dissociation of GDP from Rho protein. The Arhgdib gene is usually largely expressed in hematopoietic cells and known to be involved in immune response regulation [12, 32]. In the context of immune functions, a significant decrease in the expression of the Klrd1 gene was also exhibited in adipose tissue of tabw2 mice. KLRD1 (also known as CD94) associates with a member of the NKG2 family and regulates natural killer cell functions [6].
Biochemical pathways differentially regulated in tabw2 and control mice
In order to elucidate a biochemical differentiation between tabw2 and control mice, we conducted a pathway analysis. All the differentially expressed genes were examined for known pathway networks using the Database for Annotation, Visualization, and Integration Discovery Bioinformatics Resources 2008 (DAVID) Functional Annotation Tool (http://david.abcc.ncifcrf.gov/). Through the biochemical pathways of the Kyoto Encyclopedia of Genes and Genomes (KEGG), 70 genes were assigned to 13 known pathways in liver, whereas 32 genes were assigned to 9 known pathways in adipose tissue with Expression Analysis Systematic Explorer (EASE) threshold of 0.1 and a minimum of 2 genes present for the corresponding pathway (Table 5).
Table 5.
Biological pathways associated with differentially expressed genes between tabw2 and control mice through KEGG pathway using DAVID
| Term | KEGG ID | Count | EASE score | Gene |
|---|---|---|---|---|
| Adipose tissue | ||||
| ErbB signaling pathway | mmu04012 | 10 | 0.0038 | Gabl, Akt3, Mapk9, Camk2g, Pak4, Bad, Cdknla, Map2k7, Gsk3b, Camk2d |
| Pyruvate metabolism | mmu00620 | 6 | 0.016 | Acaca, Akr1b3, Acat2, Acacb, Dlat, Acss2 |
| Propanoate metabolism | mmu00640 | 5 | 0.022 | Acaca, Acat2, Acacb, Mut, Acss2 |
| Prostate cancer | mmu05215 | 8 | 0.038 | Tcf3, Igf1, Akt3, Igflr, Bad, Cdkn1a, Pdgfc, Gsk3b |
| Melanoma | mmu05218 | 7 | 0.041 | Igf1, Akt3, Igflr, Bad, Fgf13, Cdknla, Pdgfc |
| Glioma | mmu05214 | 6 | 0.074 | Igfl, Akt3, Igf1r, Camk2g, Cdkn1a, Camk2d |
| Olfactory transduction | mmu04740 | 4 | 0.080 | Clca1, Clca2, Camk2g, Camk2d |
| Wnt signaling pathway | mmu04310 | 10 | 0.082 | Tcf3,Ccnd2, Mapk9, Camk2g, Nfatc3, Daaml,Lrp6, Sfrp5, Gsk3b, Camk2d |
| Focal adhesion | mmu04510 | 12 | 0.090 | Itgal, Thbsl, Igfl, Akt3, Ccnd2, Igflr, Mapk9, Pak4, Bad, Pdgfc, Itgb5, Gsk3b |
| Liver | ||||
| Long-term potentiation | mmu04720 | 10 | 0.019 | Camk2b, Ppp3c, Rps6ka2, Gnaq,Rafl, Prkacb, Ppplrl2a, Braf Itpr2, Crebbp |
| Fatty acid metabolism | mmu00071 | 8 | 0.019 | Gcdh, Cyp4al0, Hsdl7b4, Ehhadh, Acaala, Cyp4al4, Acaalb, Hadh |
| Wnt signaling pathway | mmu04310 | 17 | 0.025 | Nlk, Camk2b, Ppp3ca, Mapk8, Prkacb, MapklO,Ccnd2, Tcf7l2,Lrp6, Fzd7,Wnt5b, Csnk2a2, Mmp7, Ruvbll, Nfatc3, Nfat5, Crebbp |
| Caprolactam degradation | mmu00930 | 4 | 0.038 | Sirt5, Hsdl 7b4, Ehhadh, Hadh |
| Fatty acid biosynthesis | mmu00061 | 3 | 0.053 | Oxsm, Acaca, Mcat |
| Geraniol degradation | mmu00281 | 3 | 0.053 | Hsdl7b4, Acaalb, Hadh |
| Prostate cancer | mmu05215 | 11 | 0.054 | Creb3l2, Igfl,Raf1, Chuk, Cdknlb, Sos2, Braf, Pten, Nfkbl, Tcf7l2, Crebbp |
| Lysine degradation | mmu00310 | 7 | 0.057 | Gcdh, Ogdh, Hsdl 7b4, Ehhadh, Aadat, Hadh, Aass |
| Gap junction | mmu04540 | 11 | 0.065 | Gnaq,Raf1, Prkacb, Sos2, Tubb2a, Itpr2, Gjal, Prkgl, Htr2b, Tubb2b, Gnai3 |
| Acute myeloid leukemia | mmu05221 | 8 | 0.074 | Sfpil,Raf1, Chuk, Sos2, Braf, Nfkbl, Tcf7l2, Rps6kbl |
| Melanogenesis | mmu04916 | 11 | 0.087 | Camk2b, Ednrb, Creb3l2, Gnaq,Raf1, Prkacb, Tcf7l2, Fzd7,Wnt5b, Crebbp, Gnai3 |
| MAPK signaling pathway | mmu04010 | 23 | 0.092 | Map3kl, Nlk, Gadd45a, Ppp3ca, Mapk8,Raf1, Sos2, MapklO, Prkacb, Chuk, Braf, Cacnb3, Stk4, Elk4, Rps6ka2, Dusp3, Cdl4, Rasal, Mapkl2, Nfkbl, Rapgef2, Map3k7ip2, Fgfr3 |
| Citrate cycle (TCA cycle) | mmu00020 | 5 | 0.094 | Pcx, Ogdh, Sdhd, Idh3a, Aco1 |
Genes in bold are in the congenic interval
DAVID, Database for Annotation, Visualization and Integration Discovery Bioinformatics Resources 2008 (http://david.abcc.ncifcrf.gov/); KEGG, Kyoto Encyclopedia of Genes and Genomes; Term, enriched terms (pathways) associated with the gene list; Count, the number of genes involved in the term; EASE (Expression Analysis Systematic Explorer) score, Modified Fisher Exact P-value (smaller means more enriched)
While only 6 genes included in the pathways (boldface entries) were located within the tabw2 congenic region, five (Tcf3, Ccnd2, Lrp6, Wnt5b, and Ruvbl1) out of the 6 genes were involved in the Wnt signaling pathway in either liver or adipose tissue.
Multiple genes were present in pathways associated with intermediary metabolism. These include genes required for fatty acid oxidation, such as Hadhsc (mitochondrial β-oxidation), Acaa1a and Hsd17b4 (peroxisomal β-oxidation), and Cyp4a14 (microsomal ω-oxidation) and lipogenic enzymes, such as Acss2 and Acaca. Acyl-CoA synthetase short-chain family member 2 (Acss2) catalyzes the production of acetyl-CoA from CoA and acetate, producing a key molecule in multiple metabolic pathways [14, 26]. Acetyl-CoA carboxylase alpha (Acaca) catalyzes the carboxylation of acetyl-CoA to produce malonyl-CoA that is used as a building block in the de novo long-chain fatty acid synthesis [29].
Microarray validation by real-time qRT–PCR
Changes of gene expression elucidated by microarray analysis were further verified with selected genes by real-time qRT-PCR. We chose to validate 21 genes of interest from the list of genes found in the top 50 genes with the largest effect of genotype, located on the tabw2 interval, or involved in Wnt signaling or intermediary metabolism (Table 6). The qRT-PCR results from the 21 selected genes showed close agreement with microarray fold-changes (r = 0.81, P < 0.001). Few genes including Ccnd2, Lrp6, and Nfatc3 in adipose tissue and Ruvbl1 and Nlk in liver were outside the qRT-PCR confidence interval.
Table 6.
Microarray vs. real-time quantitative RT-PCR (qRT-PCR) for selected genes in liver and adipose tissue (fat) from tabw2 and control mice
| Microarray | qRT-PCR | ||||
|---|---|---|---|---|---|
| Probe set ID | Symbol | Gene name | Tissue | Fold | Fold (CI) |
| 1416946_a_at | Acaala | Acetyl-Coenzyme A acyltransferase 1A | Liver | −1.32 | −1.08 (−3.08, 2.60) |
| 1434185_at | Acaca | Acetyl-Coenzyme A carboxylase alpha | Fat | 2.03 | 3.21 (−1.19, 12.34) |
| 1422479_at | Acss2 | Acyl-CoA synthetase short-chain family member 2 | Fat | 4.46 | 2.35 (1.19, 4.66) |
| 1426454_at | Arhgdib | Rho, GDP dissociation inhibitor (GDI) beta | Liver | 1.58 | 1.42 (−2.60, 4.21) |
| Fat | 1.43 (1.05, 2.17) | ||||
| 1448229_s_at | Ccnd2 | Cyclin D2 | Liver | −1.69 | −1.71 (−9.33, 3.18) |
| 1434745_at | Fat | 4.67 | 1.04 (−1.40, 1.52) | ||
| 1423257_at | Cyp4a14 | Cytochrome P450, family 4, subfamily a, polypeptide 14 | Liver | −3.75 | −1.15 (−61.22, 46.19) |
| 1431035_at | Daaml | Dishevelled associated activator of morphogenesis 1 | Fat | −1.38 | −1.27 (−1.80, 1.10) |
| 1436756_x_at | Hadhsc | L-3-hydroxyacyl-Coenzyme A dehydrogenase, short chain | Liver | −1.54 | −1.36 (−2.06, 1.11) |
| 1455777_x_at | Hsd17b4 | Hydroxysteroid (17-beta) dehydrogenase 4 | Liver | −1.29 | −1.06 (−2.04, 1.80) |
| 1460245_at | Klrdl | Killer cell lectin-like receptor, subfamily D, member 1 | Fat | 3.39 | 6.22 (2.97, 13.03) |
| 1451022_at | Lrp6 | Low density lipoprotein receptor-related protein 6 | Liver | −1.29 | −1.06 (−2.60, 2.31) |
| Fat | −1.92 | 1.70 (−1.09, 3.19) | |||
| 1442560_at | Mgll | Monoglyceride lipase | Liver | −2.01 | −3.63 (−14.2, 1.07) |
| 1434110_x_at | Mup1 | Major urinary protein 1 | Fat | 7.74 | 5.44 (2.40, 12.29) |
| 1419976_s_at | Nfatc3 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 | Liver | −1.50 | −1.05 (−2.89, 2.62) |
| Fat | −1.52 | −1.03 (−1.30, 1.21) | |||
| 1419112_at | Nlk | Nemo like kinase | Liver | −1.53 | −1.13 (−1.50, 1.15) |
| 1436359_at | Ret | Ret proto-oncogene | Fat | −3.46 | −4.16 (−13.91, −1.24) |
| 1416585_at | Ruvbl1 | RuvB-like protein 1 | Liver | −1.30 | 1.59 (−1.16, 2.97) |
| 1436075_at | Sfrp5 | Secreted frizzled-related sequence protein 5 | Fat | −8.59 | −3.98 (−13.07, −1.21) |
| 1450117_at | Tcf3 | Transcription factor 3 | Fat | −1.64 | 1.04 (−2.05, 2.23) |
| 1429428_at | Tcf7l2 | Transcription factor 7-like 2, T-cell specific, HMG-box | Liver | −1.33 | −1.59 (−3.39, 1.33) |
| 1422602_a_at | Wnt5b | Wingless-related MMTV integration site 5B | Liver | 1.19 | 1.01 (−3.63, 3.75) |
| Fat | 2.56 (1.04, 6.85) | ||||
‘−’ indicates up-regulation and ‘no sign’ indicates down-regulation in tabw2 mice compared to control mice; CI=95%, confidence interval (lower limit, upper limit)
Discussion
We applied oligonucleotide microarray analysis accompanied by real-time qRT-PCR to evaluate changes in gene expression in diet-induced obesity mediated by tabw2 QTL. By using the tabw2 congenic mice and control mice fed a HFS diet, we were able to elucidate gene networks that may be perturbed by tabw2.
Emerging evidence indicates that Wnt signaling is involved in adipogenesis, as well as in glucose and lipid metabolism [18]. In our study, we detected changes in gene expression of a Wnt member, Wnt5b, and several regulators and effectors of Wnt signaling, including Sfrp5 that prevents Wnts binding to frizzed receptors, in tabw2 mice. A large increase in gene expression levels of Sfrp5 was also previously reported in diet-induced obesity in mice [10]. Recently, the WNT5B gene has been reported to be associated with risk of type 2 diabetes in the Japanese populations [8] and Caucasian subjects [25].
Obesity is often concomitant with alterations in the rhythmic regulations of biological systems. For example, blunted diurnal variations and dampened ultradian pulsatility of circulating hormones, such as leptin and ghrelin, were observed in obese humans [7]. Gene expression of Mup, the lipocalin family, is regulated in liver by a pulsatile stimulus of growth hormone [16]. Interestingly, decreased MUP levels in urine were exhibited in obese mice [15]. Although the role of MUP in adipose tissue is unknown, we speculate that the significant decrease of the Mup1 gene expression in adipose tissue of tabw2 mice (Table 2) might reflect alterations in endocrine rhythmicity in these mice.
Given that fat mass is significantly increased in tabw2 mice, it was surprising to observe that expression of genes involved in fatty acid oxidation systems (Acaa1a, Cyp4a14, Hadhsc, and Hsd17b4) was up-regulated in liver, and expression of lipogenic genes (Acss2 and Acaca) was down-regulated in adipose tissue of tabw2 mice (Table 6). A decreased expression of lipogenic genes in adipose tissue was previously reported in obese human subjects [4, 17]. A possible reason for the paradoxical findings is that the decreased expression of lipogenic genes reflects a late and adaptive process; i. e., when the adipose tissue was sampled, the subjects were at a late stage of obesity and no longer expanding fat mass [4]. Observations in the present study do not rule out the possibility of an increase in lipogenic gene expression in adipose tissue at younger ages when the process of fat storing might be more rapid and dynamic than at 14 weeks of age.
Seventy of the differentially expressed genes were located within the congenic interval, which provides the possibility that a polymorphism/mutation in one of these genes could be responsible for the obesity phenotype attributed to tabw2. Our microarray data will assist candidate gene selections when the tabw2 interval is fine mapped.
In summary, we have provided a genome-wide overview of changes in gene expression that may contribute to diet-induced obesity mediated by tabw2. Our genomic profiling increased our understanding of dysregulated biological systems in tabw2 mice that will lead to targeted metabolic and molecular studies. These data may contribute to understanding the mechanisms of gene-by-diet interactions in the development of obesity, which potentially provides insights into mechanisms for human obesity.
Acknowledgments
This work was supported in part by American Heart Association Grants 0235345 N and 0855300E, NIH/National Institute of Diabetes and Digestive and Kidney Disease Grant 1R01DK077202-01A2, funding from the Center of Genomics and Bioinformatics, and a pilot and feasibility grant from the University of Tennessee Obesity Research Center to J.H.Kim.
Conflict of interest statement Authors declare not to have any conflict of interest.
References
- 1.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, St Lezin E, Kurtz TW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 2.Chon SH, Zhou YX, Dixon JL, Storch J. Intestinal monoacylglycerol metabolism: developmental and nutritional regulation of monoacylglycerol lipase and monoacylglycerol acyltransferase. J Biol Chem. 2007;282:33346–33357. doi: 10.1074/jbc.M706994200. [DOI] [PubMed] [Google Scholar]
- 3.Dalen KT, Ulven SM, Bamberg K, Gustafsson JA, Nebb HI. Expression of the insulin-responsive glucose transporter GLUT4 in adipocytes is dependent on liver X receptor alpha. J Biol Chem. 2003;278:48283–48291. doi: 10.1074/jbc.M302287200. [DOI] [PubMed] [Google Scholar]
- 4.Diraison F, Dusserre E, Vidal H, Sothier M, Beylot M. Increased hepatic lipogenesis but decreased expression of lipogenic gene in adipose tissue in human obesity. Am J Physiol Endocrinol Metab. 2002;282:E46–E51. doi: 10.1152/ajpendo.2002.282.1.E46. [DOI] [PubMed] [Google Scholar]
- 5.Glassford J, Vigorito E, Soeiro I, Madureira PA, Zoumpoulidou G, Brosens JJ, Turner M, Lam EW. Phosphatidylinositol 3-kinase is required for the transcriptional activation of cyclin D2 in BCR activated primary mouse B lymphocytes. Eur J Immunol. 2005;35:2748–2761. doi: 10.1002/eji.200425812. [DOI] [PubMed] [Google Scholar]
- 6.Hoare HL, Sullivan LC, Clements CS, Ely LK, Beddoe T, Henderson KN, Lin J, Reid HH, Brooks AG, Rossjohn J. Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors. J Mol Biol. 2008;377:1297–1303. doi: 10.1016/j.jmb.2008.01.098. [DOI] [PubMed] [Google Scholar]
- 7.Kalra SP, Bagnasco M, Otukonyong EE, Dube MG, Kalra PS. Rhythmic, reciprocal ghrelin and leptin signaling: new insight in the development of obesity. Regul Pept. 2003;111:1–11. doi: 10.1016/S0167-0115(02)00305-1. [DOI] [PubMed] [Google Scholar]
- 8.Kanazawa A, Tsukada S, Sekine A, Tsunoda T, Takahashi A, Kashiwagi A, Tanaka Y, Babazono T, Matsuda M, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakamura Y, Maeda S. Association of the gene encoding wingless-type mammary tumor virus integration-site family member 5B (WNT5B) with type 2 diabetes. Am J Hum Genet. 2004;75:832–843. doi: 10.1086/425340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JH, Stewart TP, Zhang W, Kim HY, Nishina PM, Naggert JK. Type 2 diabetes mouse model TallyHo carries an obesity gene on chromosome 6 that exaggerates dietary obesity. Physiol Genomics. 2005;22:171–181. doi: 10.1152/physiolgenomics.00197.2004. [DOI] [PubMed] [Google Scholar]
- 10.Koza RA, Nikonova L, Hogan J, Rim JS, Mendoza T, Faulk C, Skaf J, Kozak LP. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2:e81. doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence VJ, Kopelman PG. Medical consequences of obesity. Clin Dermatol. 2004;22:296–302. doi: 10.1016/j.clindermatol.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Lelias JM, Adra CN, Wulf GM, Guillemot JC, Khagad M, Caput D, Lim B. cDNA cloning of a human mRNA preferentially expressed in hematopoietic cells and with homology to a GDP-dissociation inhibitor for the rho GTP-binding proteins. Proc Natl Acad Sci USA. 1993;90:1479–1483. doi: 10.1073/pnas.90.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Luong A, Hannah VC, Brown MS, Goldstein JL. Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. J Biol Chem. 2000;275:26458–26466. doi: 10.1074/jbc.M004160200. [DOI] [PubMed] [Google Scholar]
- 15.Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA, Hilton DJ, Alexander WS. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature. 2000;405:1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- 16.Norstedt G, Palmiter R. Secretory rhythm of growth hormone regulates sexual differentiation of mouse liver. Cell. 1984;36:805–812. doi: 10.1016/0092-8674(84)90030-8. [DOI] [PubMed] [Google Scholar]
- 17.Ortega FJ, Mayas D, Moreno-Navarrete JM, Catalán V, Gómez-Ambrosi J, Esteve E, Rodriguez-Hermosa JI, Ruiz B, Ricart W, Peral B, Fruhbeck G, Tinahones FJ, Fernández-Real JM (2009) The gene expression of the main lipogenic enzymes is downregulated in visceral adipose tissue of obese subjects. Obesity (Silver Spring). doi:10.1088/oby.2009.202 [DOI] [PubMed]
- 18.Prestwich TC, Macdougald OA. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol. 2007;19:612–617. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prosniak M, Zborek A, Scott GS, Roy A, Phares TW, Koprowski H, Hooper DC. Differential expression of growth factors at the cellular level in virus-infected brain. Proc Natl Acad Sci USA. 2003;100:6765–6770. doi: 10.1073/pnas.0430999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramakrishna S, Kim IM, Petrovic V, Malin D, Wang IC, Kalin TV, Meliton L, Zhao YY, Ackerson T, Qin Y, Malik AB, Costa RH, Kalinichenko VV. Myocardium defects and ventricular hypoplasia in mice homozygous null for the Forkhead Box M1 transcription factor. Dev Dyn. 2007;236:1000–1013. doi: 10.1002/dvdy.21113. [DOI] [PubMed] [Google Scholar]
- 21.Rapp JP, Deng AY. Detection and positional cloning of blood pressure quantitative trait loci: is it possible? Hypertension. 1995;25:1121–1128. doi: 10.1161/01.hyp.25.6.1121. [DOI] [PubMed] [Google Scholar]
- 22.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 23.Romao I, Roth J. Genetic and environmental interactions in obesity and type 2 diabetes. J Am Diet Assoc. 2008;108:S24–S28. doi: 10.1016/j.jada.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 24.Saario SM, Laitinen JT. Monoglyceride lipase as an enzyme hydrolyzing 2-arachidonoylglycerol. Chem Biodivers. 2007;4:1903–1913. doi: 10.1002/cbdv.200790158. [DOI] [PubMed] [Google Scholar]
- 25.Salpea KD, Gable DR, Cooper JA, Stephens JW, Hurel SJ, Ireland HA, Feher MD, Godsland IF, Humphries SE. The effect of WNT5B IVS3C>G on the susceptibility to type 2 diabetes in UK Caucasian subjects. Nutr Metab Cardiovasc Dis. 2009;19(2):140–145. doi: 10.1016/j.numecd.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Sone H, Shimano H, Sakakura Y, Inoue N, Amemiya-Kudo M, Yahagi N, Osawa M, Suzuki H, Yokoo T, Takahashi A, Iida K, Toyoshima H, Iwama A, Yamada N. Acetyl-coenzyme A synthetase is a lipogenic enzyme controlled by SREBP-1 and energy status. Am J Physiol Endocrinol Metab. 2002;282:E222–E230. doi: 10.1152/ajpendo.00189.2001. [DOI] [PubMed] [Google Scholar]
- 27.Speakman J, Hambly C, Mitchell S, Krol E. Animal models of obesity. Obes Rev. 2007;8(1):55–61. doi: 10.1111/j.1467-789X.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- 28.Stylianou IM, Clinton M, Keightley PD, Pritchard C, Tymowska-Lalanne Z, Bunger L, Horvat S. Microarray gene expression analysis of the Fob3b obesity QTL identifies positional candidate gene Sqle and perturbed cholesterol and glycolysis pathways. Physiol Genomics. 2005;20:224–232. doi: 10.1152/physiolgenomics.00183.2004. [DOI] [PubMed] [Google Scholar]
- 29.Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilborn C, Beckham J, Campbell B, Harvey T, Galbreath M, La Bounty P, Nassar E, Wismann J, Kreider R. Obesity: prevalence, theories, medical consequences, management, and research directions. J Int Soc Sports Nutr. 2005;2:4–31. doi: 10.1186/1550-2783-2-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolfinger RD, Gibson G, Wolfinger E, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS. Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- 32.Yin L, Schwartzberg P, Scharton-Kersten TM, Staudt L, Lenardo M. Immune responses in mice deficient in Ly-GDI, a lymphoid-specific regulator of Rho GTPases. Mol Immunol. 1997;34:481–491. doi: 10.1016/S0161-5890(97)00053-9. [DOI] [PubMed] [Google Scholar]