Abstract
Nucleobases are important compounds that constitute nucleosides and nucleic acids. Although it has long been suggested that specific transporters are involved in their intestinal absorption and uptake in other tissues, none of their molecular entities have been identified in mammals to date. Here we describe identification of rat Slc23a4 as the first sodium-dependent nucleobase transporter (rSNBT1). The mRNA of rSNBT1 was expressed highly and only in the small intestine. When transiently expressed in HEK293 cells, rSNBT1 could transport uracil most efficiently. The transport of uracil mediated by rSNBT1 was sodium-dependent and saturable with a Michaelis constant of 21.2 μm. Thymine, guanine, hypoxanthine, and xanthine were also transported, but adenine was not. It was also suggested by studies of the inhibitory effect on rSNBT1-mediated uracil transport that several nucleobase analogs such as 5-fluorouracil are recognized by rSNBT1, but cytosine and nucleosides are not or only poorly recognized. Furthermore, rSNBT1 fused with green fluorescent protein was mainly localized at the apical membrane, when stably expressed in polarized Madin-Darby canine kidney II cells. These characteristics of rSNBT1 were almost fully in agreement with those of the carrier-mediated transport system involved in intestinal uracil uptake. Therefore, it is likely that rSNBT1 is its molecular entity or at least in part responsible for that. It was also found that the gene orthologous to the rSNBT1 gene is genetically defective in humans. This may have a biological and evolutional meaning in the transport and metabolism of nucleobases. The present study provides novel insights into the specific transport and metabolism of nucleobases and their analogs for therapeutic use.
Keywords: Metabolism/Nucleotide, Nucleoside/Nucleotide/Metabolism, Nucleoside/Nucleotide/Transport, Tissue/Organ Systems/Intestine, Tissue/Organ Systems/Epithelium, Transport, Transport/Drug, Transporter
Introduction
Nucleosides are essential components of DNA and RNA, the genomic memory devices, and also play pivotal roles as energy sources and in signal transductions (1, 2). In mammals, nucleosides can be synthesized via the de novo pathway from small precursor molecules, such as aspartate, glutamine, glycine, and 10-formyltetrahydrofolate, and via the salvage pathway from nucleobases. In the salvage pathway, the supply of nucleobases mainly occurs from extracellular sources, typically those produced by digestion of dietary nucleic acids in the intestinal lumen. There also occurs redistribution of nucleosides/nucleobases via bloodstream from the tissues that are highly active in de novo synthesizing and/or degrading nucleosides. Therefore, carrier-mediated transport systems are needed for the efficient trafficking across cellular membranes of nucleosides and nucleobases, which are hydrophilic and hence can little permeate otherwise (3, 4).
The transporters involved in the cellular uptake of nucleosides have been well characterized in mammals, including humans (5–7). There are two families of transporters, which are sodium-coupled concentrative nucleoside transporters (CNT1/SLC28A1, CNT2/SLC28A2, and CNT3/SLC28A3) and equilibrative nucleoside transporters (ENT1/SLC29A1 and ENT2/SLC29A2). All the members of these transporter families transport nucleosides preferentially, playing roles in maintaining their extracellular and intracellular levels coordinately. In particular, CNT1, which transports pyrimidine nucleosides more preferentially than purine nucleosides, is highly expressed in the epithelial tissues such as small intestine, kidney, and liver (8–11). It is also known that CNT1 is localized at the apical membrane of intestinal epithelial cells, suggesting its physiological and pharmacological roles in the absorption of nucleosides and their analogs in the small intestine.
However, transporters specific for nucleobases have not been identified in mammals, although their presence has long been postulated (3, 4, 12–17). Our earlier study using the everted sacs of the rat small intestine demonstrated that the uptake transport of 5-fluorouracil (5-FU),2 a uracil analog used for cancer chemotherapy, occurs in a saturable and sodium-dependent manner and is inhibited by uracil and thymine, but not by their nucleosides, suggesting the involvement of a sodium-dependent transporter specific for nucleobases (16). Such sodium-dependent transport has also been reported for 5-FU in the rat small intestine by another study using excised tissue rings (12) and for hypoxanthine in the calf small intestine by a study using brush border membrane vesicles (17). Thus, it is likely that sodium-dependent transporters specific for nucleobases are present in the small intestine and, possibly, also in other tissues. In analogy to the situation of nucleosides, equilibrative transporters would also exist for nucleobases (3, 4).
Although no nucleobase transporter has been identified in mammals, several nucleobase transporters have been identified and well characterized in lower organisms, such as bacteria (18), fungi (19–22), and protozoa (23, 24), and in plants (25, 26). They have been categorized into three families of nucleobase-ascorbate transporter (NAT), microbial purine-related transporter, and plant purine-related transporter, based on their molecular and functional characteristics (4). According to bioinformatic analysis, mammals have only one such family of transporters. That is the SLC23 family, of which the members are homologous to NATs (27, 28). SLC23 family has four members and includes SVCT1/SLC23A1 and SVCT2/SLC23A2, which are well characterized as sodium-dependent ascorbate transporters (SVCTs) and interestingly do not have any transport activity for nucleobases (29). SLC23A3 is an orphan transporter with unknown function. This putative transporter, which was cloned before the discoveries of SVCT1/SLC23A1 and SVCT2/SLC23A2, has been reported not to accept nucleobases or ascorbate as substrates (30, 31). Another member of the family has been bioinformatically identified and referred to as SLC23A4. The scarce appearance of its expressed sequence tag in humans in the GenBankTM data base may indicate its only poor or insignificant expression in humans and, in fact, its functional characterization as a solute carrier has not been achieved so far. We, however, found by data base search that SLC23A4 may be expressed in rats. This finding prompted us to clone rat Slc23a4 and examine its possible role as a nucleobase transporter.
Here we describe our successful attempt to clone Slc23a4 from the rat small intestine and characterize it as a sodium-dependent transporter specific for some nucleobases, typically, uracil, thymine, guanine, hypoxanthine, and xanthine. We also assessed its physiological role by examining its activity in the rat small intestine. Because this transporter is the first one of that kind, we herein refer to it as sodium-dependent nucleobase transporter 1 (SNBT1). It is also notable that this transporter, rat SNBT1 (rSNBT1)/Slc23a4, is the first nucleobase transporter identified in mammals. Furthermore, it was found that the gene orthologous to the rSNBT1 gene is genetically defective in humans, suggesting a possibility that its function may have been evolutionarily lost in humans and, possibly, also in other higher primates.
EXPERIMENTAL PROCEDURES
Materials
[3H]Uracil (42.8 Ci/mmol), [3H]adenine (25.0 Ci/mmol), [3H]hypoxanthine (27.0 Ci/mmol), [3H]thymidine (20.0 Ci/mmol), [3H]thymine (65.0 Ci/mmol), [3H]uridine (14.7 Ci/mmol), and [3H]xanthine (12.8 Ci/mmol) were obtained from Moravek Biochemicals (Brea, CA), [14C]inulin (1.9 mCi/g) and [14C]guanine (55 mCi/mmol) were from American Radiolabeled Chemicals (St. Louis, MO), and [14C]ascorbate (8.5 mCi/mmol) and [α-32P]dCTP (3000 Ci/mmol) were from PerkinElmer Life Sciences. Uracil and nitrobenzylthioinosine were obtained from Wako Pure Chemicals (Osaka, Japan), and phlorizin, papaverine, and dipyridamole were from Sigma-Aldrich. Culture medium was obtained from Wako Pure Chemicals, and fetal bovine serum was from Invitrogen. Restriction enzymes were obtained from Toyobo (Osaka, Japan). All other reagents were of analytical grade and commercially obtained.
Cell Culture
HEK293 cells and MDCKII cells were maintained at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Isolation of SNBT1
We conducted a BLAST search of rat nucleotide collection in the GenBank data base, using the amino acid sequence of UpaA, a uracil permease in Aspergillus nidulans, as a query, and identified two cDNA sequences that encode hypothetical proteins with unknown function, XM_231601 and XM_001067886, which had been predicted by computational analysis of the rat genome. However, they seemed to lack the N-terminal and C-terminal regions, respectively, in the deduced amino acid sequences. We therefore deduced the cDNA sequence of a putative open reading frame that has start and stop codons by aligning them.
Thus, deduced cDNA, which was identified to be of rat Slc23a4 and in the present study designated to be rSNBT1 to represent its function as a sodium-dependent nucleobase transporter, was cloned by reverse transcription (RT) and subsequent PCR from total RNA prepared from the small intestine of male Wistar rats by a guanidine isothiocyanate extraction method (32). In brief, an RT reaction was carried out to obtain cDNA mixture, using 1 μg of the total RNA, an oligo(dT) primer, and ReverTra Ace (Toyobo) as a reverse transcriptase. The coding region of rSNBT1 cDNA was amplified by PCR using KOD plus polymerase (Toyobo) and the following primers: forward primer, 5′-TCC AGT TGC CAT GAA CTC TG-3′; reverse primer, 5′-CAC TGT GTG TTC ACC ACG GTA-3′. PCR was performed using the following conditions: 94 °C for 2 min; 33 cycles of 1) 94 °C for 20 s, 2) 56 °C for 20 s, and 3) 72 °C for 2 min. The second PCR was performed using the PCR product as a template and a forward primer containing a XhoI restriction site (underlined), 5′-ATT CTC GAG CTA TGA ACT CTG CAG TCT GCA-3′, and a reverse primer containing an XbaI restriction site (underlined), 5′-GGT CTA GAG TTT TGC ATT TTG CAG GC-3′. Then the amplified product was introduced at the XhoI and XbaI sites into a mammalian expression vector, pCI-neo (Promega, Madison, WI). The sequence of the amplified cDNA product was determined with an automated sequencer (ABI PRISM 3100; Applied Biosystems, Foster City, CA). The plasmid for green fluorescent protein (GFP)-tagged rSNBT1 (GFP-rSNBT1) was generated by transferring the coding region of rSNBT1 into pEGFP-C1 vector (Clontech) at the XhoI and XbaI sites.
Northern Blot Analysis
The tissue distribution of rSNBT1 transcripts in the rat was examined by Northern blots of total RNA samples, as described previously (33), using rSNBT1-specific cDNA probe. The 28 S rRNA was stained with ethidium bromide and used as a reference.
RT-PCR Analysis
Total RNA isolated from various rat tissues was used for cDNA synthesis. PCR reactions for rSNBT1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a reference to confirm the reverse transcription reaction from the cDNAs were performed using TaqDNA polymerase (New England Biolabs Japan, Tokyo, Japan) and the following primers: forward primer for rSNBT1, 5′-CCC CTG ACC CAA AGC TAC C-3′; reverse primer for rSNBT1, 5′-AAC ATG CCT CCA ATC ACG G-3′; forward primer for GAPDH, 5′-CCA TCA CCA TCT TCC AGG AG-3′; reverse primer for GAPDH, 5′-CCT GCT TCA CCA CCT TCT TG-3′. PCR conditions were as follows: 94 °C for 2 min; 35 cycles (rSNBT1) or 30 cycles (GAPDH) of 1) 94 °C for 20 s, 2) 60 °C for 20 s, and 3) 72 °C for 40 s.
The human genomic DNA was isolated from HEK293 cells by proteinase K digestion followed by phenol/chloroform extraction and ethanol precipitation. The purified genomic DNA and cDNAs from human small intestine total RNA (BioChain, Hayward, CA) were subjected to PCR using KOD FX DNA polymerase (Toyobo). The primers were as described above for GAPDH and as follows for hSNBT1: forward primer, 5′-ATG GTC ACT TCC TGT GTC CGG-3′; reverse primer, 5′-GAG ATC CCC ACG GCA GTG ATG-3′. PCR conditions were as follows: 94 °C for 2 min; 35 cycles (hSNBT1) or 30 cycles (GAPDH) of 1) 94 °C for 20 s, 2) 60 °C for 20 s, and 3) 68 °C for 6 min (hSNBT1 from genomic DNA), 60 s (hSNBT1 from cDNAs), or 30 s (GAPDH). PCR products were electrophoresed on a 1.0% agarose gel.
Transport Study in HEK293 Cells Transiently Expressing rSNBT1
HEK293 cells were transiently transfected with the plasmid carrying rSNBT1 cDNA by using HilyMax (Dojindo Laboratories, Kumamoto, Japan) as a transfection reagent, as described in our previous report (34). Transport assays were conducted as described in the same report. In brief, cells in each well of 24-well plates were preincubated for 5 min in 1.5 ml of substrate-free uptake buffer consisting of 140 mm NaCl, 5 mm KCl, 0.4 mm KH2PO4, 0.8 mm MgSO4, 1.0 mm CaCl2, 25 mm glucose, and 10 mm HEPES (pH 7.4), and then transport assays were started by replacing the substrate-free uptake buffer with one containing a radiolabeled substrate (0.25 ml). All the procedures were conducted at 37 °C. Assays were stopped by the addition of ice-cold substrate-free uptake buffer (2 ml), and the cells were washed twice with 2 ml of the same buffer. The cells were solubilized, and the associated radioactivity was determined by liquid scintillation counting, using Clear-sol I (Nakarai Tesque, Kyoto, Japan) as a scintillation fluid for the evaluation of uptake. Cellular protein content was determined by the method of Lowry et al. (35), using bovine serum albumin as the standard. The specific uptake of each substrate by rSNBT1 was estimated by subtracting the uptake in mock cells, which were transfected with empty pCI-neo vector, from that in rSNBT1-transfected cells.
Fluorescent Microscopic Analysis
MDCKII cells were transfected with the plasmid carrying the cDNA of GFP-rSNBT1, using Lipofectamine 2000 (Invitrogen) as a transfection reagent, and Geneticin-resistant clones were selected as described previously (36). The expression of GFP-rSNBT1 was confirmed by fluorescence from GFP-rSNBT1 and transport of [3H]uracil. To examine the localization of GFP-rSNBT1, MDCKII cells stably expressing GFP-rSNBT1 were seeded at a density of 1 × 105 cells on a polycarbonate membrane insert of Transwell and grown to confluence for 5 days. The cells were washed twice with ice-cold phosphate-buffered saline (PBS), fixed in 3% paraformaldehyde, and permeabilized with 0.1% Triton X-100 in PBS for 30 min at room temperature. After washing twice with PBS, the cells were incubated for 10 min at room temperature with 1 μm 4′,6-diamino-2-phenylindole in PBS to stain the nuclei. The cells were washed twice with PBS and then mounted on a glass slide in 9:1 glycerol/PBS. GFP-rSNBT1 and 4′,6-diamino-2-phenylindole-stained nuclei were visualized by using a confocal laser-scanning microscope (LSM510; Carl Zeiss, Jena, Germany).
Transport Study in Everted Intestinal Tissue Sacs
Everted tissue sacs were prepared from the small intestine of male Wistar rats, weighing ∼300 g and not fasted, and uptake experiments were conducted, as reported previously (37). In brief, the everted sacs (2 cm in length) were preincubated for 5 min in substrate-free uptake buffer, that is Krebs-Ringer-bicarbonate buffer (118 mm NaCl, 4.7 mm KCl, 2.5 mm CaCl2, 1.2 mm KH2PO4, 1.2 mm MgSO4, 25 mm NaHCO3) supplemented with 20 mm HEPES and oxygenated with 95% O2, 5% CO2 gas (pH 7.4), and then placed in uptake buffer containing [3H]uracil and [14C]inulin as a non-absorbable marker for the initiation of incubation for uptake. All the incubations were carried out at a temperature of 37 °C and a shaking rate of 100 strokes/min. Uptake was terminated by rinsing the everted sacs briefly with ice-cold saline, and the uptake into the tissue was evaluated by determining the radioactivity by liquid scintillation counting after solubilization of the tissue sample, using 1 ml of Soluene-350 (PerkinElmer Life Sciences) as a tissue solubilizer and 5 ml of Clear-sol I as a scintillation fluid. The experiments were conducted with the approval of the Animal Experiment Ethics Committee of Nagoya City University Graduate School of Pharmaceutical Sciences.
Data Analysis
The saturable transport of each substrate, uracil and xanthine, by rSNBT1 was analyzed by assuming Michaelis-Menten type carrier-mediated transport represented by the following equation: v = Vmax × s/(Km + s). The maximum transport rate (Vmax) and the Michaelis constant (Km) were estimated by fitting this equation to the experimental profile of the uptake rate (v) versus the substrate concentration (s), using a non-linear least-squares regression analysis program, WinNonlin (Pharsight Corp., Mountain View, CA) and the reciprocal of variance as the weight. Stoichiometric coupling between Na+ and uracil was analyzed by using the following Hill equation: v = Vmax,s × [Na+]n/(KNan + [Na+]n). The Hill coefficient (n) was estimated together with the maximum transport rate of uracil at its specified concentration (Vmax,s) and the Na+ concentration that gives half-Vmax,s (KNa) by fitting this equation to the experimental profile of v versus the Na+ concentration in the extracellular medium ([Na+]). The v in the presence of an inhibitor can be expressed as follows: v = v0/(1 + i/IC50). The half-inhibition concentration (IC50) was estimated by fitting this equation to the experimental profile of v versus the inhibitor concentration (i), with v in the absence of inhibitors (v0) fixed at the observed value. In the analysis of uracil transport in the rat small intestine, v was converted to the uptake clearance (CLup) by dividing by s, and the clearance of passive transport by simple diffusion (CLm,d) was added as an additional parameter to be estimated to account for a non-saturable transport component as follows: CLup = Vmax/(Km + s) + CLm,d. A plot of CLup versus logarithmic concentration was used to handle uptake data as it was more suitable for a wider concentration range that was needed in the data analysis. The parameters are presented as the computer-fitted ones with S.E. Experimental data are represented as the means ± S.E. in Figs. 3–5 and 7, and statistical analysis was performed by using Student's t test or, when multiple comparisons were needed, analysis of variance followed by Dunnett's test, with p < 0.05 considered significant (Figs. 4, 5, and 7).
FIGURE 3.
Uptake of several nucleobases and analogs in HEK293 cells transiently expressing rSNBT1. rSNBT1-specific uptake was evaluated by dividing the uptake in rSNBT1-transfected cells by that in mock cells. Uptakes of [3H]adenine (5 nm), [14C]guanine (2 μm), [3H]hypoxanthine (5 nm), [3H]xanthine (10 nm), [3H]thymine (5 nm), [3H]uracil (2 nm), [3H]thymidine (5 nm), [3H]uridine (10 nm), and [14C]ascorbate (5 μm) were evaluated at 37 °C and pH 7.4 for 1 min in both cells. The uptake rates (pmol/min/mg of protein) for unity, which were evaluated in mock cells, were 0.0546 for adenine, 21.5 for guanine, 0.163 for hypoxanthine, 0.0370 for xanthine, 0.0172 for thymine, 0.00740 for uracil, 0.257 for thymidine, 0.170 for uridine, and 19.8 for ascorbate. Data are represented as the means ± S.E. (n = 4).
FIGURE 4.
Functional characteristics of rSNBT1 transiently expressed in HEK293 cells. A, time courses of the uptake of [3H]uracil (2 nm) in rSNBT1-transfected cells (closed circles) and mock cells (open circles). B, effect of extracellular ions on the rSNBT1-specific uptake rate of [3H]uracil (2 nm). NaCl in the control medium was replaced as indicated. C, Na+ dependence of the rSNBT1-specific uptake rate of [3H]uracil. The Hill coefficient was estimated to be 1.07 by analysis using the Hill equation. D, concentration dependence of the rSNBT1-specific uptake rates of [3H]uracil (circles) and [3H]xanthine (squares). The Km and Vmax are 21.2 ± 2.8 μm and 737 ± 23 pmol/min/mg of protein, respectively, for uracil as the computer-fitted parameters with S.E. and 83.0 ± 9.0 μm and 1110 ± 46 pmol/min/mg of protein, respectively, for xanthine. In all experiments, uptake measurements were conducted at 37 °C and pH 7.4 and, in panels B, C, and D, for a 1-min period. Data are represented as the means ± S.E. (n = 4). *, p < 0.05 when compared with the value for control.
FIGURE 5.
Effect of various compounds on uracil transport mediated by rSNBT1 transiently expressed in HEK293 cells. A, inhibitory effect of nucleobases on the rSNBT1-specific uptake rate of [3H]uracil (2 nm) evaluated in the presence of varied concentrations of cytosine (open circles), guanine (closed circles), hypoxanthine (open squares), thymine (closed squares), and 5-FU (open triangle). The IC50 values are 80.7 ± 17.1, 17.4 ± 3.1, 12.7 ± 0.6, and 69.0 ± 11.4 μm, respectively, for guanine, hypoxanthine thymine, and 5-FU as the computer-fitted parameters with S.E. B, inhibitory effect of various compounds (0.1 mm) on the rSNBT1-specific uptake rate of [3H]uracil (2 nm). In all experiments, uptake measurements were conducted for a 1-min period at 37 °C and pH 7.4. The control values for normalization were 74.9 and 72.5 fmol/min/mg of protein, respectively, in panels A and B. Data are represented as the means ± S.E. (n = 4). *, p < 0.05 when compared with the value for control. NBMPR, nitrobenzylthioinosine.
FIGURE 7.
rSNBT1 activity in the everted sacs of the rat small intestine. A, time course of the uptake of [3H]uracil (1. 3 nm) in ileal everted sacs. B, Na+ dependence of the uptake clearance of [3H]uracil (1.3 nm) in jejunal and ileal everted sacs. NaCl in the control uptake buffer (closed bars) was replaced with KCl (open bars). C, concentration dependence of the uptake clearance of [3H]uracil in ileal everted sacs. The uptake was evaluated at varied uracil concentrations, which were adjusted by adding unlabeled uracil, and the uptake rate was divided by concentration to estimate the uptake clearance. The values of Vmax, Km, and CLm,d are 1.26 ± 0.09 nmol/min/100 mg of wtw, 40.3 ± 3.1 μm, and 1.34 ± 0.37 μl/min/100 of mg wtw, respectively, as the computer-fitted parameters with S.E. D, effect of various compounds (0.1 mm) on the uptake clearance of [3H]uracil (1.3 nm) in ileal everted sacs. The control value was 31.5 μl/min/100 mg of wtw. In all experiments, uptake measurements were conducted at 37 °C and pH 7.4 and, in panels B, C, and D, for a 2-min period. Data are represented as the means ± S.E. n = 3 (panels A and B) or 6 (panels C and D). *, p < 0.05 when compared with the value for control; †, p < 0.05 when compared with the value for jejunum. NBMPR, nitrobenzylthioinosine.
RESULTS
Identification of rSNBT1
By combining PCR cloning and bioinformatic analysis, we isolated the cDNA of rSNBT1/Slc23a4 expressed in the rat small intestine (GenBank accession number, AB511909), which encodes a putative protein of 614 amino acids (Fig. 1). A multiple alignment analysis of the amino acid sequences among the members of rat Slc23 family demonstrates that rSNBT/Slc23a4 has rather low identity/similarity percentages of 50/67% to SVCT1/Slc23a1, 50/67% to SVCT2/Slc23a2, and 31/49% to Slc23a3. Motif analysis indicated that rSNBT1 has a NAT signature motif ((Q/E/P)NXGXXXXT(R/K/G), where X is a hydrophobic amino acid), which is conserved among the prokaryotic and eukaryotic NAT transporters (38). Hydropathy analyses using the DAS transmembrane prediction server, the SOSUI WWW server, and the TMpred server predicted that rSNBT1 has 12 potential membrane-spanning domains with putative N-glycosylation sites at Asn152 and Asn390, in agreement with the topology models of SVCT1 and SVCT2 and the positions of potential N-glycosylation in them (27).
FIGURE 1.
Sequence analysis of rSNBT1. The deduced amino acid sequence of rSNBT1/Slc23a4 was aligned with those of SVCT1/Slc23a1, SVCT2/Slc23a1, and Slc23a3 of rat, using the program of ClustalW, and processed to visualize using the program BOXSHADE. Identical amino acids and conservative changes are indicated by reversed and shaded characters, respectively. Boxed residues indicate the putative transmembrane domains. Amino acids of the NAT signature motif (Glu397 to Arg406) are indicated by circles, and putative N-glycosylation sites (Asn152 and Asn390) are indicated by asterisks.
Tissue Distribution of rSNBT1 mRNA
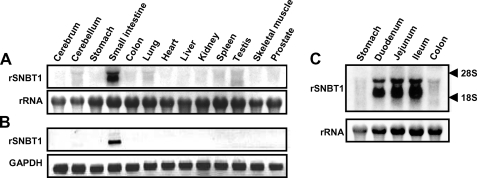
To examine the tissue distribution of rSNBT1, we performed Northern blotting and RT-PCR, using total RNA from various rat tissues (Fig. 2). Northern blot analysis revealed that rSNBT1 mRNA (∼2.5 kb) is highly expressed in the small intestine, being higher in the lower region, whereas it is absent in all the other tissues examined. Consistent with this, RT-PCR analysis also indicated the expression of rSNBT1 only in the small intestine. These results suggest that small intestine is the primary site of expression and operation for rSNBT1.
FIGURE 2.
Analyses of the expression of the mRNA of rSNBT1 in various rat tissues by Northern blot (A) and RT-PCR (B) and in various segments of the rat small intestine by Northern blot (C). Total RNA (10 μg) from each tissue or segment was probed with 32P-labeled cDNA of rSNBT1 in Northern blot analysis. The bands of ethidium bromide-stained 28 S rRNA are also shown as references. Total RNA (1 μg) from each tissue was reverse-transcribed and then amplified by PCR using a set of specific primers for rSNBT1 in RT-PCR analysis. The bands of GAPDH are also shown as references.
Functional Characteristics of rSNBT1
To examine the possibility that rSNBT1 may function as a nucleobase transporter, we first determined the transport activities of several nucleobases and analogs in HEK293 cells transiently expressing rSNBT1 at their trace concentrations and for a 1-min uptake period (Fig. 3). It was found that uracil, a pyrimidine nucleobase, is transported most efficiently, exhibiting about 10-fold greater uptake in the cells expressing rSNBT1 than in mock cells. Thymine, another pyrimidine nucleobase, and guanine, hypoxanthine, and xanthine, which are purine nucleobases, were also found to be transported, although less efficiently. However, adenine, another purine nucleobase, was found not to be transported. Uridine, a uracil-derived nucleoside, was found to be transported, but less efficiently than uracil, suggesting the characteristic of rSNBT1 as a nucleobase transporter. The transport of thymidine, a thymine-derived nucleoside, was barely detected. Furthermore, rSNBT1 did not show any transport activity for ascorbate, although it belongs to SLC23 family, which includes ascorbate transporters (SVCT1 and SVCT2).
Because rSNBT1 was found to transport uracil most efficiently, we used this nucleobase as a substrate for subsequent functional characterization of this transporter. As shown in Fig. 4A, the uptake of uracil (2 nm) increased rapidly, being in proportion to time up to 2 min in the cells expressing rSNBT1, whereas it remained very low in mock cells. We, therefore, chose 1 min as the uptake period for the evaluation of transport in the initial uptake phase. When NaCl in the uptake buffer was isoosmotically replaced with KCl, choline chloride, or mannitol, the specific uptake of uracil mediated by rSNBT1 was almost completely inhibited (Fig. 4B), indicating that Na+ is essential for the transport function. The Hill coefficient, which indicates the number of Na+ ions required to transport uracil, was estimated to be 1.07 by analysis using the Hill equation (Fig. 4C), suggesting that one Na+ ion is co-transported with each uracil molecule. Kinetic analysis indicated that the rSNBT1-mediated transport of uracil was saturable with a Km of 21.2 μm and a Vmax of 737 pmol/min/mg of protein (Fig. 4D). For xanthine, about 4-fold greater Km (83.0 μm), indicating a lower affinity, and a comparable Vmax (1110 pmol/min/mg of protein) were indicated.
To investigate the substrate specificity of rSNBT1, we first compared the IC50 values of several nucleobases for inhibition of rSNBT1-mediated uracil transport (Fig. 5A). It was found that guanine, hypoxanthine, thymine, and 5-FU, a uracil derivative, are potent inhibitors in the following order of decreasing IC50 (increasing affinity): guanine (80.7 μm) > 5-FU (69.0 μm) > hypoxanthine (17.4 μm) > thymine (12.7 μm). Assuming competitive inhibition by these potential substrates, these IC50 values should be nearly equivalent to the inhibition constants because the experiments were conducted at a uracil concentration (2 nm) much lower than its Km (21.2 μm) for rSNBT1. These IC50 values were in a range comparable with the Km values for uracil and xanthine, indicating comparable affinities for rSNBT1. Although uracil is a pyrimidine nucleobase, cytosine, another pyrimidine nucleobase, did not exhibit any inhibitory activity. This may suggest a role of a keto group at the fourth position (C-4) of pyrimidine ring in substrate recognition because the keto group in uracil is replaced with a primary amino group in cytosine. Supporting this suggestion, adenine, a purine nucleobase that was found not to be transported by rSNBT1 (Fig. 3), also has a primary amino group at the corresponding position of pyrimidine ring in its purine structure. On the other hand, the other purine nucleobases of guanine, hypoxanthine, and xanthine and another pyrimidine nucleobase of thymine, which were found to be recognized by rSNBT1, all have a keto group at that position. Interestingly, 5-FU, which is a uracil analog substituted with fluorine at the C-5 position, was found to be a weaker inhibitor than uracil and thymine. It has generally been suggested that fluorine substitution alters the electronic property of a molecule extensively due to the high electronegativity of fluorine, although changes in its size and shape are minimal. Therefore, the electron-withdrawing effect of fluorine at the C-5 position may be a factor involved in lowering the affinity for rSNBT1, and the electronic status of the pyrimidine ring may be important in the substrate binding to rSNBT1.
We then examined the effect of various compounds (0.1 mm) on the specific uptake of uracil (2 nm) mediated by rSNBT1 (Fig. 5B). The most extensive inhibition was observed for thymine, and the second most extensive one was found for 5-FU. Mercaptopurine and oxypurinol, which are purine derivatives, also inhibited rSNBT1-specific uracil uptake, although less extensively. However, the other purine derivatives, azathioprine and allopurinol, and purine did not. Inhibitions by the nucleosides of adenosine, guanosine, cytidine, thymidine, and uridine were only modest or insignificant, consistent with the weak transport activities of rSNBT1 for thymidine and uridine (Fig. 3). Furthermore, dipyridamole exhibited an inhibitory effect. Nitrobenzylthioinosine is a specific inhibitor of ENTs, which are reportedly able to transport hypoxanthine. However, this compound did not inhibit rSNBT1-specific uracil uptake. Papaverine and phlorizin have been reported to inhibit a nucleobase transport system in LLC-PK1 cells (13). The finding that these compounds do not inhibit rSNBT1-specific uracil uptake suggests that rSNBT1 is not the molecular entity of the transport system.
Localization of rSNBT1 in MDCKII Cells
To examine the membrane localization of rSNBT1, GFP-rSNBT1, which has GFP tagged at the C terminus, was stably expressed in MDCKII cells, and its localization was determined by confocal laser scanning microscopy. As shown in Fig. 6, GFP-rSNBT1 was mainly localized at the apical membrane of MDCKII cells, which were polarized on the permeable membrane filter. Because rSNBT1 is highly expressed in the small intestine, it is most likely that this transporter is expressed at the brush border membrane of intestinal epithelial cells, where the presence of a sodium-dependent nucleobase transport system for uracil and analogs has long been suggested (12, 16).
FIGURE 6.
Cellular localization of GFP-rSNBT1 stably expressed in polarized MDCKII cells. The confocal laser-scanning microscopic image shows predominant apical membrane localization of GFP-rSNBT1 (green) and nuclei stained with 4′,6-diamino-2-phenylindole (white).
rSNBT1 Activity in Rat Small Intestine
Although a sodium-dependent nucleobase transport system for uracil and analogs has been suggested to be present in the small intestine, information about the characteristics of the transport system is scarce. We therefore examined the activity of the nucleobase transport system using the everted sacs of the rat small intestine to assess the role of rSNBT1 as its molecular entity. The uptake of uracil (1.3 nm) was examined for a 2-min period for the evaluation of transport in the initial phase where uptake was in proportion to time, as shown in Fig. 7A for ileal uptake. The uptake of uracil was extensively reduced when Na+ was removed by replacement with K+ (Fig. 7B), indicating the involvement of sodium-dependent transport. It was also observed that uracil uptake was greater in the ileum than in the jejunum (Fig. 7B), in agreement with the higher expression of rSNBT1 mRNA in the former (Fig. 2C). Therefore, we examined uracil uptake in the ileum in subsequent experiments. The uptake of uracil in ileal everted sacs was highly saturable, as indicated by a decrease in the uptake clearance with an increase in the concentration (Fig. 7C). Kinetic analysis assuming Michaelis-Menten type carrier-mediated transport indicated a Km of 40.3 μm, which was comparable with that of rSNBT1 expressed in HEK293 cells (21.2 μm), and Vmax of 1.26 nmol/min/100 mg of wet tissue weight (wtw). A non-saturable transport component was not negligible, but the CLm,d for that was as small as 1.34 μl/min/100 mg of wtw, suggesting that uracil can permeate only minimally by simple diffusion and that the prevailing sodium-dependent transport observed at a low concentration of 1.3 nm (Fig. 7B) is equivalent to the saturable transport.
Fig. 7D shows the effect of various compounds (0.1 mm) on the uptake of uracil at its trace concentration of 1.3 nm. It was found that guanine, hypoxanthine, and thymine inhibit uracil uptake extensively. 5-FU and oxypurinol also inhibited it, although less extensively. All the other compounds, including nucleosides such as thymidine and uridine, did not affect uracil uptake significantly, but dipyridamole tended to reduce it. This profile of inhibition of uracil uptake was mostly in agreement with that observed for rSNBT1 (Fig. 5).
Thus, all the characteristics of the nucleobase transport system involved in intestinal uracil uptake are almost fully in agreement with those of rSNBT1. Therefore, it is likely that rSNBT1 is its molecular entity or at least in part responsible for that.
Human Ortholog of rSNBT1
To identify the human ortholog of rSNBT1, we performed a BLAST search using the amino acid sequence of rSNBT1 as a query. The search revealed that a genomic region on chromosome 7q33 appears to encode the putative human ortholog of SNBT1/SLC23A4 (hSNBT1). However, as shown in Fig. 8A, the hSNBT1 gene was found to contain only seven exons, whereas the rSNBT1 gene contains 12 exons. Notably, the hSNBT1 gene lacks the counterparts of exons 6, 7, and 8 of the rSNBT1 gene, which encode the transmembrane domains that are highly conserved among rat Slc23 family members. It was also found that the SNBT1 gene of chimpanzee is similarly defective, but functional orthologs of SNBT1 seem to be present in many non-primate mammals and non-mammalian vertebrates, such as zebrafish (Danio rerio; NP_001013353), chicken (Gallus gallus; XP_416178), mouse (Mus musculus; EDL13672), dog (Canis familiaris; XP_539823), and horse (Equus caballus; XP_001497573). Thus, it is most likely that the SNBT1 gene is a pseudogene in humans and, possibly, also in other higher primates, although a functional version of the gene is conserved in other mammals and non-mammalian vertebrates.
FIGURE 8.
Comparison of the SNBT1 gene locus between rat and human. A, relative position and spanning of exons that encode SNBT1 and its mRNA. The scales are set for exons, introns, and mRNA as indicated. B, RT-PCR analysis for the expression of the mRNA of hSNBT1 in the human intestine. cDNAs reverse-transcribed from human small intestine total RNA (1 μg) and human genomic DNA isolated from HEK293 cells were amplified by PCR using a set of specific primers for hSNBT1.
To confirm that the hSNBT1 gene is a pseudogene, which does not produce transcripts, we carried out RT-PCR analysis with a set of specific primers designed from the potential exons, using human small intestine total RNA (Fig. 8B). As expected, no PCR product was detected at the position of 0.85 kb, which was for the hSNBT1 cDNA predicted from the rSNBT1 cDNA by assuming the identical length of coding region. The specificity of the primer set was confirmed by the amplification of the expected fragment of a size of 6.8 kb from the human genomic DNA. This result supports the suggestion that hSNBT1 is a pseudogene.
DISCUSSION
Although the involvement of carrier-mediated transport systems in the cellular uptake of nucleobases has long been suggested in the small intestine and other tissues (3, 4), none of their molecular entities have been identified in mammals to date. Here we have described our successful identification of rSNBT1 as a sodium-dependent nucleobase transporter, which is highly and specifically expressed in the small intestine. rSNBT1 was found to specifically transport or recognize several nucleobases and analogs, such as uracil, thymine, guanine, hypoxanthine, and xanthine, but have little affinity for nucleosides (Fig. 3). Although it did not transport ascorbate, it is notable that its substrate specificity is overall similar to those of NAT transporters in lower organisms (4, 28), suggesting that it is a paralog of them. Such characteristics of its function and small intestine-specific expression suggest its role in the active absorption of nucleobases derived from dietary nucleic acids by hydrolytic digestion. Indeed, the characteristics of the carrier-mediated transport system involved in uracil uptake in the rat small intestine were found to be almost fully in agreement with those of rSNBT1, strongly suggesting that rSNBT1 is responsible for the transport system. It is also of interest that rSNBT1 recognizes a group of purine/pyrimidine nucleobases, which consist of guanine and hypoxanthine/uracil and thymine, whereas it does not recognize another group, which consist of adenine/cytosine. Because the other members of SLC23 family reportedly do not transport nucleobases (29), there would be at least one more nucleobase transporter, which is for the group of adenine and cytosine, in some other transporter family. Because rSNBT1 is little expressed in tissues other than small intestine, there would be some more concentrative (sodium-dependent) and/or equilibrative types of nucleobase transporters in those extraintestinal tissues.
Sodium-dependent nucleobase transport systems have been described in several tissues and cell lines other than the rat small intestine. Previous studies using renal brush border membrane vesicles (BBMVs) of guinea pigs (14), intestinal BBMVs of calves (17), and LLC-PK1 cells, a renal epithelial cell line derived from the porcine kidney (13), demonstrated the presence of sodium-dependent hypoxanthine transport systems, which can be inhibited by nucleobases such as uracil, thymine, and guanine, but not by adenine and nucleosides. Such characteristics of hypoxanthine (nucleobase) transport systems are similar to those of rSNBT1 observed in this study. With regard to stoichiometry, the Na+/hypoxanthine stoichiometry has been reported to be 1:1 for the Na+-dependent hypoxanthine transport system in LLC-PK1 cells (13) and 2:1 for the one in renal BBMVs of guinea pigs (14). Therefore, rSNBT1, for which the Na+/uracil stoichiometry was suggested to be 1:1 from the estimated Hill coefficient of 1.07 (Fig. 4C), has the same stoichiometric characteristic as the hypoxanthine transport system in LLC-PK1 cells. However, the Km values of those transport systems for hypoxanthine, 2.1 μm in guinea pig renal BBMVs, 0.33 μm in calf intestinal BBMVs, and 0.79 μm in LLC-PK1 cells, were more than an order of magnitude smaller than the IC50 value (17.4 μm) of hypoxanthine for rSNBT1, indicating their much higher affinities. In addition, hypoxanthine uptake in calf intestinal BBMVs is reported to be much greater in the proximal jejunum than in the distal part (17), in contrast to the higher expression and transport activity of rSNBT1 in the distal than in the proximal. Possibilities are that the orthologs of SNBT1 may have different affinities for hypoxanthine and tissue distribution in guinea pigs, calves, and porcines than in rats and/or that transporters distinct from SNBT1 orthologs may be involved.
Interestingly, we found that the gene encoding the SNBT1 ortholog is defective in humans (Fig. 8), although it does not seem to be the case in vertebrates other than higher primates. Therefore, it is likely that the SNBT1 gene was silenced during evolution in higher primates. An analysis of the human genomic sequence has revealed that more than 33 genes are pseudogenes (39). It is also known that there are some more pseudogenes in humans. They include the uricase gene, which is well known to be mutationally defective in humans and some other higher primates (40, 41). Due to the lack of this enzyme, which catalyzes the conversion of urate to allantoin, the elimination pathway of urate is limited to renal excretion in humans, and hence, the urate concentration in serum is maintained higher in humans than in, for example, rodents, which have dual pathways of renal excretion and uricase-mediated metabolism for urate elimination (42). In humans, the homeostatically maintained high serum urate concentration may be easily disturbed, due to the low capacity of the elimination pathway, by a massive and transient input of urate from exogenous sources such as diet. Because metabolic enzymes responsible for the conversion of purine nucleosides/nucleobases to urate are known to be abundantly present in the epithelial cells of the small intestine (43–45), that would occur if a highly efficient transporter such as SNBT1 were present and nucleobases from dietary sources were taken up rapidly and extensively. In fact, it has been reported that the ingestion of a large amount of nucleobases, such as hypoxanthine and guanine, leads to some increase in the appearance of urate in the blood in humans (46–48). Therefore, we speculate that an evolutionary pressure to suppress the absorption of nucleobases may have contributed to the defect of the SNBT1 gene in humans and, possibly, also in other higher primates. It would be of interest to investigate whether the genetic defect of SNBT1 might have any association with the defect of uricase. Future studies would also be needed on the relationship between the transport and metabolism of nucleobases.
Although it was thus found that humans do not have SNBT1, 5-FU has been known to be absorbed fairly well after oral administration (49, 50). Therefore, we cannot exclude the possibility that an alternative transporter, which would not be as efficient as SNBT1, might be present in the human small intestine to absorb nucleobases to some extent. It is even more likely that humans have as yet unidentified nucleobase transporters in extraintestinal tissues, where they would be needed for trafficking of nucleobases across cellular membranes. To date, only two other transporters, ENT2 and organic anion transporter 2 (OAT2), have been reported to be able to transport nucleobases (51, 52). However, they are primarily for nucleosides and organic anions, respectively, and have only modest transport activities for nucleobases. Therefore, it is unlikely that they would be involved physiologically in the transport of nucleobases.
In conclusion, we identified rSNBT1, which is the first nucleobase transporter in mammals, in the present study. The functional characteristics of rSNBT1 were found to be almost fully in agreement with those of the carrier-mediated transport system involved in intestinal uracil uptake. Therefore, it is likely that rSNBT1 is its molecular entity or at least in part responsible for that. Another finding that humans and, possibly, also other higher primates lack SNBT1 may have a biological and evolutional meaning in the transport and metabolism of nucleobases. Knowing the fact and further elucidating the underlying mechanisms may help us to optimize the therapeutic use of nucleobase analogs.
This work was supported in part by a grant-in-aid for young scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and a grant-in-aid for research in Nagoya City University.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) AB511909.
- 5-FU
- 5-fluorouracil
- rSNBT1
- rat SNBT1
- hSNBT1
- human SNBT1
- SVCT
- sodium-dependent ascorbate transporter
- CNT
- concentrative nucleoside transporter
- ENT
- equilibrative nucleoside transporter
- NAT
- nucleobase-ascorbate transporter
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFP
- green fluorescent protein
- BBMV
- brush border membrane vesicle
- HEK
- human embryonic kidney
- MDCK
- Madin-Darby canine kidney
- PBS
- phosphate-buffered saline
- RT
- reverse transcription
- wtw
- wet tissue weight.
REFERENCES
- 1.Zöllner N. (1982) Proc. Nutr. Soc. 41, 329–342 [DOI] [PubMed] [Google Scholar]
- 2.Burnstock G. (2007) Physiol. Rev. 87, 659–797 [DOI] [PubMed] [Google Scholar]
- 3.Plagemann P. G., Wohlhueter R. M., Woffendin C. (1988) Biochim. Biophys. Acta 947, 405–443 [DOI] [PubMed] [Google Scholar]
- 4.de Koning H., Diallinas G. (2000) Mol. Membr. Biol. 17, 75–94 [DOI] [PubMed] [Google Scholar]
- 5.Gray J. H., Owen R. P., Giacomini K. M. (2004) Pflugers Arch. 447, 728–734 [DOI] [PubMed] [Google Scholar]
- 6.Baldwin S. A., Beal P. R., Yao S. Y., King A. E., Cass C. E., Young J. D. (2004) Pflugers Arch. 447, 735–743 [DOI] [PubMed] [Google Scholar]
- 7.Molina-Arcas M., Trigueros-Motos L., Casado F. J., Pastor-Anglada M. (2008) Nucleosides Nucleotides Nucleic Acids 27, 769–778 [DOI] [PubMed] [Google Scholar]
- 8.Huang Q. Q., Yao S. Y., Ritzel M. W., Paterson A. R., Cass C. E., Young J. D. (1994) J. Biol. Chem. 269, 17757–17760 [PubMed] [Google Scholar]
- 9.Ritzel M. W., Yao S. Y., Huang M. Y., Elliott J. F., Cass C. E., Young J. D. (1997) Am. J. Physiol. 272, C707–C714 [DOI] [PubMed] [Google Scholar]
- 10.Valdés R., Ortega M. A., Casado F. J., Felipe A., Gil A., Sánchez-Pozo A., Pastor-Anglada M. (2000) Gastroenterology 119, 1623–1630 [DOI] [PubMed] [Google Scholar]
- 11.Govindarajan R., Bakken A. H., Hudkins K. L., Lai Y., Casado F. J., Pastor-Anglada M., Tse C. M., Hayashi J., Unadkat J. D. (2007) Am. J. Physiol. Regul. Integr. Comp. Physiol. 293, R1809–R1822 [DOI] [PubMed] [Google Scholar]
- 12.Bronk J. R., Hastewell J. G. (1987) J. Physiol. 382, 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffith D. A., Jarvis S. M. (1993) J. Biol. Chem. 268, 20085–20090 [PubMed] [Google Scholar]
- 14.Griffith D. A., Jarvis S. M. (1994) Biochem. J. 303, 901–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Washington C. B., Giacomini K. M. (1995) J. Biol. Chem. 270, 22816–22819 [DOI] [PubMed] [Google Scholar]
- 16.Yuasa H., Matsuhisa E., Watanabe J. (1996) Biol. Pharm. Bull. 19, 94–99 [DOI] [PubMed] [Google Scholar]
- 17.Theisinger A., Grenacher B., Scharrer E. (2003) J. Comp. Physiol. B 173, 165–170 [DOI] [PubMed] [Google Scholar]
- 18.Andersen P. S., Frees D., Fast R., Mygind B. (1995) J. Bacteriol. 177, 2008–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diallinas G., Scazzocchio C. (1989) Genetics 122, 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber E., Rodriguez C., Chevallier M. R., Jund R. (1990) Mol. Microbiol. 4, 585–596 [DOI] [PubMed] [Google Scholar]
- 21.Gorfinkiel L., Diallinas G., Scazzocchio C. (1993) J. Biol. Chem. 268, 23376–23381 [PubMed] [Google Scholar]
- 22.Cecchetto G., Amillis S., Diallinas G., Scazzocchio C., Drevet C. (2004) J. Biol. Chem. 279, 3132–3141 [DOI] [PubMed] [Google Scholar]
- 23.Burchmore R. J., Wallace L. J., Candlish D., Al-Salabi M. I., Beal P. R., Barrett M. P., Baldwin S. A., de Koning H. P. (2003) J. Biol. Chem. 278, 23502–23507 [DOI] [PubMed] [Google Scholar]
- 24.Ortiz D., Sanchez M. A., Pierce S., Herrmann T., Kimblin N., Archie Bouwer H. G., Landfear S. M. (2007) Mol. Microbiol. 64, 1228–1243 [DOI] [PubMed] [Google Scholar]
- 25.Gillissen B., Bürkle L., André B., Kühn C., Rentsch D., Brandl B., Frommer W. B. (2000) Plant Cell 12, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argyrou E., Sophianopoulou V., Schultes N., Diallinas G. (2001) Plant Cell 13, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takanaga H., Mackenzie B., Hediger M. A. (2004) Pflugers Arch. 447, 677–682 [DOI] [PubMed] [Google Scholar]
- 28.Gournas C., Papageorgiou I., Diallinas G. (2008) Mol. Biosyst. 4, 404–416 [DOI] [PubMed] [Google Scholar]
- 29.Tsukaguchi H., Tokui T., Mackenzie B., Berger U. V., Chen X. Z., Wang Y., Brubaker R. F., Hediger M. A. (1999) Nature 399, 70–75 [DOI] [PubMed] [Google Scholar]
- 30.Guimarães M. J., Bazan J. F., Zlotnik A., Wiles M. V., Grimaldi J. C., Lee F, McClanahan T. (1995) Development 121, 3335–3346 [DOI] [PubMed] [Google Scholar]
- 31.Faaland C. A., Race J. E., Ricken G., Warner F. J., Williams W. J., Holtzman E. J. (1998) Biochim. Biophys. Acta 1442, 353–360 [DOI] [PubMed] [Google Scholar]
- 32.Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 33.Ohta K. Y., Inoue K., Hayashi Y., Yuasa H. (2006) Drug Metab. Dispos. 34, 1868–1874 [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto S., Inoue K., Ohta K. Y., Fukatsu R., Maeda J. Y., Yoshida Y., Yuasa H. (2009) J. Biochem. 145, 437–443 [DOI] [PubMed] [Google Scholar]
- 35.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 36.Nakai Y., Inoue K., Abe N., Hatakeyama M., Ohta K. Y., Otagiri M., Hayashi Y., Yuasa H. (2007) J. Pharmacol. Exp. Ther. 322, 469–476 [DOI] [PubMed] [Google Scholar]
- 37.Inoue K., Nakai Y., Ueda S., Kamigaso S., Ohta K. Y., Hatakeyama M., Hayashi Y., Otagiri M., Yuasa H. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 294, G660–G668 [DOI] [PubMed] [Google Scholar]
- 38.Diallinas G., Valdez J., Sophianopoulou V., Rosa A., Scazzocchio C. (1998) EMBO J. 17, 3827–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Human Genome Sequencing Consortium (2004) Nature 431, 931–94515496913 [Google Scholar]
- 40.Wu X. W., Lee C. C., Muzny D. M., Caskey C. T. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 9412–9416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oda M., Satta Y., Takenaka O., Takahata N. (2002) Mol. Biol. Evol. 19, 640–653 [DOI] [PubMed] [Google Scholar]
- 42.Anzai N., Kanai Y., Endou H. (2007) Curr. Opin. Rheumatol. 19, 151–157 [DOI] [PubMed] [Google Scholar]
- 43.Witte D. P., Wiginton D. A., Hutton J. J., Aronow B. J. (1991) J. Cell Biol. 115, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohamedali K. A., Guicherit O. M., Kellems R. E., Rudolph F. B. (1993) J. Biol. Chem. 268, 23728–23733 [PubMed] [Google Scholar]
- 45.Moriwaki Y., Yamamoto T., Higashino K. (1999) Histol. Histopathol. 14, 1321–1340 [DOI] [PubMed] [Google Scholar]
- 46.Clifford A. J., Riumallo J. A., Young V. R., Scrimshaw N. S. (1976) J. Nutr. 106, 428–434 [Google Scholar]
- 47.Choi H. K., Atkinson K., Karlson E. W., Willett W., Curhan G. (2004) N. Engl. J. Med. 350, 1093–1103 [DOI] [PubMed] [Google Scholar]
- 48.Choi H. K., Liu S., Curhan G. (2005) Arthritis Rheum. 52, 283–289 [DOI] [PubMed] [Google Scholar]
- 49.Diasio R. B., Harris B. E. (1989) Clin. Pharmacokinet. 16, 215–237 [DOI] [PubMed] [Google Scholar]
- 50.Baker S. D., Khor S. P., Adjei A. A., Doucette M., Spector T., Donehower R. C., Grochow L. B., Sartorius S. E., Noe D. A., Hohneker J. A., Rowinsky E. K. (1996) J. Clin. Oncol. 14, 3085–3096 [DOI] [PubMed] [Google Scholar]
- 51.Yao S. Y., Ng A. M., Vickers M. F., Sundaram M., Cass C. E., Baldwin S. A., Young J. D. (2002) J. Biol. Chem. 277, 24938–24948 [DOI] [PubMed] [Google Scholar]
- 52.Cropp C. D., Komori T., Shima J. E., Urban T. J., Yee S. W., More S. S., Giacomini K. M. (2008) Mol. Pharmacol. 73, 1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]