Abstract
Kinetoplast DNA, the trypanosome mitochondrial genome, is a network of interlocked DNA rings including several thousand minicircles and a few dozen maxicircles. Minicircles replicate after release from the network, and their progeny reattach. Remarkably, trypanosomes have six mitochondrial DNA helicases related to yeast PIF1 helicase. Here we report that one of the six, TbPIF1, functions in minicircle replication. RNA interference (RNAi) of TbPIF1 causes a growth defect and kinetoplast DNA loss. Minicircle replication intermediates decrease during RNAi, and there is an accumulation of multiply interlocked, covalently closed minicircle dimers (fraction U). In studying the significance of fraction U, we found that this species also accumulates during RNAi of mitochondrial topoisomerase II. These data indicate that one function of TbPIF1 is an involvement, together with topoisomerase II, in the segregation of minicircle progeny.
Keywords: DNA Helicase, DNA Replication, DNA Topoisomerase, DNA Topology, Mitochondrial DNA, Trypanosome, PIF1 Helicase, Kinetoplast DNA
Introduction
Trypanosomes and related parasites have a giant DNA network, called kinetoplast DNA (kDNA),3 residing in their single mitochondrion (1, 2). The network is composed of several thousand minicircles (1 kb) and a few dozen maxicircles (23 kb), all of which are interlocked. The network is condensed into a disk-shaped structure that resides in the mitochondrial matrix near the flagellar basal body that is in the cytoplasm. The condensed network is actually linked to the basal body by a transmembrane filament system called the tripartite attachment complex (3). Maxicircles encode rRNAs and mRNAs for a few proteins such as subunits of respiratory complexes. Some maxicircle transcripts are edited by insertion or deletion of uridylate residues at specific sites, thereby creating an open reading frame. Minicircles encode guide RNAs used as templates for editing maxicircle transcripts (4, 5). Thus mitochondrial gene expression is a joint endeavor of both maxicircles and minicircles.
It is not surprising that the unusual kDNA structure has an unconventional mechanism for replication. We will focus here on minicircle synthesis, and the first step is the release by a topoisomerase II of monomeric covalently closed minicircles into an intramitochondrial region between the kDNA network and the membrane near the flagellar basal body. This region, known as the kinetoflagellar zone (KFZ), is the site of replication of the free minicircles. Proteins involved in replication include the universal minicircle sequence-binding protein (UMSBP) and p38 that bind the replication origin (6, 7), primase (8), two DNA polymerases (9), and a type IA topoisomerase (10). These and other proteins not yet identified (such as a DNA helicase) replicate minicircles unidirectionally via θ-type structures. After replication, the progeny are thought to segregate in the KFZ and then migrate to two antipodal sites positioned on opposite sides of the kDNA disk. Within the antipodal sites are enzymes that remove primers (11) and a DNA polymerase β and ligase kβ that repair most but not all of the resulting gaps (12, 13). Finally, the minicircles containing one or more nicks or gaps are reattached to the network periphery by topoisomerase II, which is also positioned in the antipodal sites (14, 15). As replication proceeds and the minicircle copy number increases, the network elongates until all of the minicircles have replicated. Then the double-sized network splits in two, and the remaining minicircle gaps are repaired, apparently by a second DNA polymerase and ligase (polymerase β-PAK and ligase kα) that are localized within the kDNA disk (13, 16).
In a hunt for DNA helicases that may be involved in kDNA replication, we searched the Trypanosoma brucei genome for genes homologous to that encoding Saccharomyces cerevisiae PIF1 mitochondrial helicase. S. cerevisiae in fact has two PIF helicases, ScPif1p and Rrm3p (17). These enzymes have dual localizations, both in the nucleus and the mitochondrion. Nuclear Pif1p and Rrm3p have multiple functions. Pif1p reduces telomerase processivity by dissociating the enzyme from telomeric DNA (18). Rrm3p moves with the replication fork and may remove barriers to fork progression (19). Genetic and biochemical studies also indicate that Pif1p may be involved in Okazaki fragment maturation (20–22). Little is known about the function of mitochondrial Pif1p except that genetic evidence indicates that it may be involved in mitochondrial DNA recombination and repair (23, 24).
We found that the T. brucei genome contains eight genes, named TbPIF1–8, related to the yeast gene encoding ScPif1p and, remarkably, that six of them are mitochondrial (25). Our initial studies indicated that these enzymes play strikingly different roles in mitochondrial DNA metabolism. We demonstrated that TbPIF2 helicase controls maxicircle abundance (25) and that TbPIF5 is involved in the processing of minicircle Okazaki fragments (26).
Here we have investigated TbPIF1 (GenBankTM accession number XP_828762, GeneDB accession number Tb11.02.4730). We found that RNAi of TbPIF1 blocks free minicircle replication and causes kDNA loss. Our most interesting finding was the accumulation of a previously unknown free minicircle species that we named fraction U. We determined that fraction U is a family of multiply interlocked, covalently closed minicircle dimers, and we showed that these molecules also accumulate after RNAi of mitochondrial topoisomerase II. Fraction U is likely derived from unsegregated dimeric replication intermediates, leading us to propose that one function of TbPIF1 is to participate in the segregation of minicircle progeny. We do not yet know whether TbPIF1 also functions at the minicircle replication fork.
EXPERIMENTAL PROCEDURES
Strain, Trypanosome Growth, Transfection, and RNAi
Procyclic strain 29-13 (from G. Cross, Rockefeller University) was used for RNAi. The topoisomerase II RNAi cell line was constructed from a stem-loop vector (15). Procyclic strain 927 was used for the localization experiment. The conditions for cell culture, transfection, and RNAi were described previously (27).
Plasmid Constructs
To make the TbPIF1 RNAi construct, the first 500 bp of the TbPIF1 coding sequence were PCR-amplified using genomic DNA isolated from procyclic strain 427 and inserted into the pJZM vector (27). A BLAST search showed that this sequence has no similarity to other PIFs or any other gene.
Myc Tagging of TbPIF1 and Fluorescence Microscopy
The sequence of the 3′ end of the TbPIF1 coding region (500 bp) and its neighboring 3′-untranslated region were PCR-amplified using primers a–d: primer a, 5′-GACCGGTACCGTTGGTCGTGTCCATCTCGAACTG-3′; primer b, 5′-GCAGCTCGAGTTCCAACGACTCACCAGAGGAGGC-3′; primer c, 5′-GCGGGGATCCAATCGAATTAAAAACTTTGTTTCTTTG-3′; and primer d, 5′-GCATCGCGGCGGCCGCTTCTTCTTTTGTTCTCTCGTCCTTCTG-3′. The PCR products were then inserted into pMOTag33M (28). After digestion with Acc65I and XbaI, the DNA fragments were transfected into procyclic strain 927. Immunostaining for TbPIF1-Myc used 1:100 rabbit anti-Myc polyclonal antibody (Santa Cruz) and 1:500 Alexa Fluor 568-conjugated goat anti-rabbit IgG (Molecular Probes). The conditions for fixing, permeabilizing, and staining cells were described (29).
Electron Microscopy (EM)
For fraction U from TbPIF1 RNAi cells, free minicircle replication intermediates (from 8 × 109 cells, after 3 days of RNAi) were fractionated by sucrose gradient centrifugation. Fractions enriched in fraction U were pooled. DNA was phenol-extracted, ethanol-precipitated, and subjected to EM (6, 30). For fraction U from topoisomerase II RNAi cells, DNA samples (from the sucrose gradients) were further treated with 20 μg/ml RNase A (37 °C for 20 min) followed by 0.5% SDS and 100 μg/ml proteinase K treatment (55 °C for 1 h). The samples were then cleaned by size exclusion chromatography with 6% agarose beads (Agarose Bead Technologies, Tampa, FL). The deproteinized, and purified DNA was prepared for EM using the aqueous drop spreading method as described (31). In brief, the DNA was mixed with a dilute solution of cytochrome c in a buffer of 0.25 m ammonium acetate. A 50-μl drop was placed on Parafilm, and the DNA was picked up on a parlodion-covered EM grid, dehydrated, air-dried, and rotary shadowcast with platinum:palladium (80%:20%) at an angle of 8° in a high vacuum. The samples were examined in an FEI (Hillsboro, OR) Tecnai 12 TEM, and the images were recorded using a Gatan Inc. (Pleasanton, CA) Ultrascan400 CCD camera. EM of isolated kDNA networks was performed as described (32).
Expression and Purification of Recombinant Protein
Recombinant TbPIF1 was expressed in Escherichia coli with a tag of six histidines at its N terminus. Briefly, the coding sequence (minus the first 34 amino acids that constitute a predicted mitochondrial targeting signal) was PCR-amplified (forward primer, 5′-GCCCTCTAGAGCACTGATGCCTGTTGAC-3′; reverse primer, 5′-CCCGCTCGAGCTATTCCAACGACTCACCAG-3′), cloned into pET28a (Novagen), and transformed into the E. coli RosettaTM (DE3) pLysS strain (Novagen). The cells were cultured in LB medium (500 ml, containing 34 μg/ml chloramphenicol and 30 μg/ml kanamycin) and grown at 37 °C to an A600 nm of 0.6. After the addition of 1 mm isopropyl β-d-thiogalactopyranoside, the culture was incubated for another 3 h at 25 °C. The cells were harvested by centrifugation (8000 × g for 10 min), and the cell pellet was resuspended in 20 ml of buffer A (50 mm sodium phosphate, 300 mm NaCl, 10 mm imidazole, pH 8.0). After lysis by sonication, the suspension was centrifuged (10,000 × g for 30 min), and the supernatant was mixed gently with 2 ml of nickel-nitrilotriacetic acid slurry (Qiagen) (1 h at 4 °C). The nickel-nitrilotriacetic acid beads were then washed four times with 2 ml of buffer B (50 mm sodium phosphate, pH 8.0, 300 mm NaCl, 20 mm imidazole). The proteins were eluted three times with 0.5 ml of buffer C (50 mm sodium phosphate, 300 mm NaCl, 250 mm imidazole, pH 8.0). The eluates were dialyzed overnight at 4 °C against buffer D (25 mm Tris-HCl, 300 mm NaCl, 1 mm dithiothreitol, pH 7.5). The samples were loaded onto a 0.5-ml heparin-Sepharose FF (Bioscience Healthcare) column equilibrated with the same buffer. Recombinant protein was eluted at 0.8 m NaCl and dialyzed against buffer E (25 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm dithiothreitol). Recombinant TbPIF1 (∼125 μg) was purified to ∼90% homogeneity as judged by SDS-PAGE from a 500-ml E. coli culture.
ATPase and Helicase Assay
ATPase and helicase assays were carried out as described (33). See the legend of Fig. 1 for more details of these experiments.
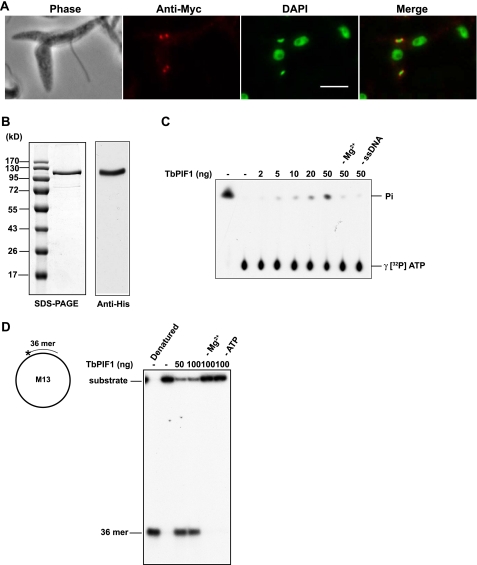
FIGURE 1.
Localization and enzymatic activity of TbPIF1. A, localization. Anti-Myc is in red, and DAPI is in green. Bar, 5 μm. B, left panel, SDS-PAGE of 1 μg of recombinant TbPIF1; the gel was stained with Coomassie Blue. Right panel, Western blot of TbPIF1 using anti-His antibody. C, ATPase assay of TbPIF1. Recombinant TbPIF1 was incubated (20-μl reaction, 10 min, 37 °C) with 8.25 nm [γ-32P]ATP (6000 Ci/mmol), 150 μm nonradioactive ATP, 50 mm Tris-HCl, pH 8.5, 50 mm NaCl, 2 mm dithiothreitol, 2 mm MgCl2, 0.25 mg/ml bovine serum albumin, and 50 ng of M13mp18 single-stranded DNA. The samples (1 μl) were spotted onto a polyethyleneimine-cellulose plate (J. T. Baker) and developed in 1.0 m formic acid, 0.5 m LiCl followed by autoradiography. The same amount of [γ-32P]ATP was converted to [32P]Pi by boiling 5 min in 1 m HCl; the [32P]Pi was used as a standard (left-most lane). D, helicase assay of TbPIF1. To make the substrate, a 36-mer oligonucleotide (5′-CGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATT-3′) was 5′ end-labeled with [γ-32P]ATP and annealed to M13mp18 single-strand DNA circles. Recombinant TbPIF1 was incubated with 10 fmol of DNA in 20 μl of reaction buffer (50 mm Tris-HCl, pH 8.5, 50 mm NaCl, 2 mm dithiothreitol, 2 mm MgCl2, 2 mm ATP, 0.25 mg/ml bovine serum albumin) for 15 min at 37 °C. The reactions products were subjected to 12% polyacrylamide gel electrophoresis. The gel was dried and autoradiographed.
Miscellaneous Methods
DNA and RNA purification, gel electrophoresis, Southern blotting, Northern blotting, topoisomerase reactions, and sucrose gradient sedimentation were performed as described previously (6).
RESULTS
Localization of TbPIF1
In a previous survey of TbPIF1–8 gene products, we found that a TbPIF1-green fluorescent protein fusion protein, expressed ectopically from a strong procyclin promoter, localized throughout the mitochondrion (25). Overexpression may saturate its normal localization site, causing it to spill over into the mitochondrial matrix. To determine more reliably the localization of endogenous TbPIF1, we tagged one allele of TbPIF1 with three copies of the Myc epitope at its C terminus; this tagged protein would more likely be expressed at its normal level and be localized in its normal site. In fact, immunofluorescence revealed that TbPIF1-Myc localizes to the antipodal sites flanking the kinetoplast (Fig. 1A). More than 80% of the cells in an asynchronous log phase culture had this localization, and virtually no cells had a different localization, Because only ∼30% of cells in a log phase culture are in the S phase, it is likely that TbPIF1 undergoes little or no change in its position in the mitochondrion during the cell cycle.
TbPIF1 Is an ATP-dependent DNA Helicase
We expressed a His-tagged TbPIF1 in E. coli and purified it to near homogeneity (Fig. 1B). The mobility of this protein on SDS-PAGE in comparison with standards is consistent with its molecular mass (101 kDa) predicted from its sequence. This recombinant protein had ATPase activity, removing the γ-phosphate, in a reaction dependent on single-stranded DNA and Mg2+ (Fig. 1C). The specific activity is 122 mol of hydrolyzed ATP/mol enzyme/min, which is comparable with that of TbPIF5 (26). We then found that TbPIF1 has helicase activity in that it could unwind a 36-mer oligonucleotide annealed to a region of an M13 single-strand circle (Fig. 1D). The unwinding activity was dependent on ATP and Mg2+. We concluded that TbPIF1 is an ATP-dependent DNA helicase.
TbPIF1 RNAi Stops Cell Growth and Causes kDNA Loss
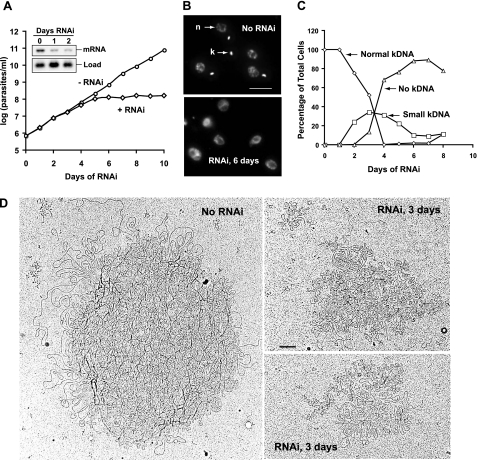
To study the function of TbPIF1, we inserted a fragment of its coding sequence into the pZJM RNAi vector. Within pZJM, the TbPIF1 sequence is flanked by opposing tetracycline-inducible T7 promoters (27). This construct was then transfected into 29-13 cells that constitutively express both the T7 RNA polymerase and the tetracycline repressor (34). After cloning a stable cell line, addition of tetracycline to the culture induced production of double-stranded RNA, thereby knocking down TbPIF1 expression. Although only ∼70% TbPIF1 mRNA was degraded after 2 days of RNAi (Fig. 2A, inset), the cells stopped growing after 4 days (Fig. 2A). TbPIF1 RNAi also caused the shrinkage and disappearance of kDNA as judged by DAPI staining of uninduced cells and those that had undergone RNAi for 6 days (Fig. 2B). Fig. 2C shows the kinetics of kDNA loss, with kinetoplast shrinkage starting at day 2 and with almost 80% of the cells having no detectable kDNA at day 4. Visualization of DAPI-stained isolated networks clearly demonstrated the shrinkage of the kDNA networks (Fig. 3E, upper panels), and EM of isolated networks after 3 days of RNAi confirmed this conclusion (Fig. 2D).
FIGURE 2.
Effect of TbPIF1 RNAi on growth and kDNA size. A, effect of TbPIF1 RNAi on cell growth. The value of parasites/ml on the y axis is the measured value times the dilution factor. Inset, Northern blot showing level of TbPIF1 mRNA (∼3.5 kb) without or with RNAi. B, DAPI staining of cells with or without TbPIF1 RNAi (6 days). n, nucleus; k, kinetoplast. Bar, 5 μm. C, kinetics of kDNA loss as determined by visual analysis of DAPI-stained cells (>200 randomly selected cells were analyzed each day). D, electron micrographs of kDNA isolated from cells with (right panels) or without (left panel) TbPIF1 RNAi.
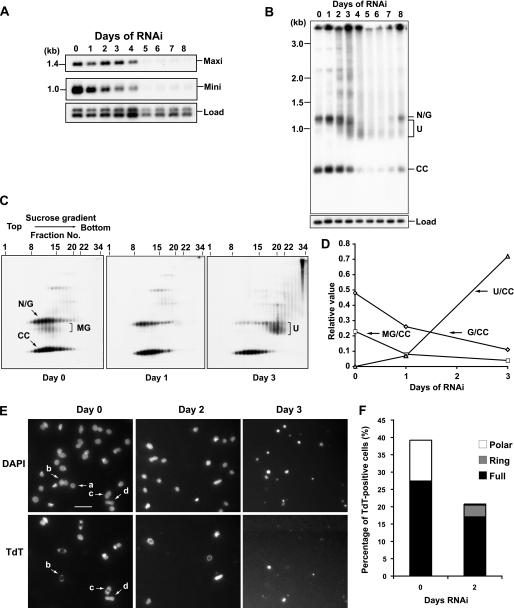
FIGURE 3.
Effect of TbPIF1 RNAi on kDNA replication. A, effect of TbPIF1 RNAi on minicircle and maxicircle abundance. Total maxicircles (Maxi) and minicircles (Mini) were detected by probing a Southern blot after total DNA was digested with HindIII/XbaI followed by fractionation on an agarose gel. The maxicircle probe detects a 1.4-kb fragment, and the minicircle probe detects the 1.0-kb linearized minicircle (other minicircle fragments, arising from the heterogeneous minicircle population, declined during RNAi with the same kinetics observed with the 1-kb fragment). Hexose transporter genes, encoded by nuclear DNA, was probed as a loading control (Load). B, free minicircle analysis. Total DNA (106 cell equivalents/lane) was fractionated on a 1.5% agarose gel in TBE buffer (both the gel and running buffer contained 1 μg/ml ethidium bromide). Following transfer to the membrane, the blot was probed for minicircles. U, fraction U. C, sedimentation of free minicircle intermediates in a 5–20% sucrose gradient (34 ml) (6). Fractions (1 ml) were collected from the top, and 10 μl of each fraction (fractions 1–22, 24, 26, 28, 30, 32, and 34) were subjected to electrophoresis, and a Southern blot was probed for minicircles. D, quantitation of changes in level of free minicircle species as determined by scanning blots from Fig. 3C (fraction 8–22) with a PhosphorImager. The ratio of each free minicircle species to covalently closed minicircles (nicked/gapped (G/CC), multiply gapped (MG/CC), and fraction U (U/CC)) was determined for each time of RNAi. E, TdT labeling of isolated kDNA networks. kDNA was isolated during the course of an RNAi experiment and labeled with fluorescein-12-dUTP using terminal deoxyuridylate transferase followed by DAPI staining (35). Network a is TdT-negative and could be pre- or post-replication. Networks b and c are TdT-positive with polar labeling and are at early and later stages of replication, respectively. Network d is a uniformly TdT-labeled network, indicating that it has completed replication and is ready to segregate. Bar, 8 μm. F, more than 500 kDNA networks were quantified for TdT labeling. Polar, polar labeling; Full, full labeling; Ring, ring-shaped labeling.
We further assessed kDNA loss by Southern blotting of total DNA digested with restriction enzymes. Both minicircles and maxicircles were barely detectable after 5 days of RNAi (Fig. 3A). However, minicircle loss started at day 1 and then progressed gradually to near completion at day 5. In contrast, maxicircles declined more abruptly, mostly starting at day 4 when minicircle loss was nearly complete. This difference suggested strongly that TbPIF1 functions in minicircle replication.
Effect of TbPIF1 RNAi on Minicircle Replication
As mentioned in the Introduction, minicircle synthesis in wild type cells involves their individual release from the network. Then the free minicircles, all of which are covalently closed, undergo unidirectional replication as θ-structures. The products are a singly gapped molecule containing the newly synthesized leading strand and a multiply gapped circle containing unligated Okazaki fragments. Most of these gaps are repaired in the antipodal sites just prior to attachment to the network; thus the multiply gapped minicircles are converted to nicked or gapped molecules (2). As shown in Fig. 3B, day 0, the free minicircles in cells uninduced for RNAi, like wild type, contain roughly equal quantities of covalently closed (CC, the replication precursors) and nicked and gapped species (N/G, the replication products that migrate very similarly on this gel). Just below N/G, the multiply gapped (MG) circles run in a faint smear that usually shows up only in longer exposures (Fig. 3C, left panel, for better view of MG minicircles).
We then analyzed the effect of TbPIF1 RNAi on free minicircle species (Fig. 3B). Using a one-dimensional agarose-ethidium gel, we found that RNAi caused both nicked/gapped and covalently closed minicircle abundance to decline in parallel with the shrinking of the kDNA network. More interestingly, we detected at day 2 a smear that probably includes oligomeric minicircle species running from the top of the gel to the position of nicked/gapped minicircles. There was also a more abundant smear, just below the nicked/gapped minicircles, that ran in the position expected for multiply gapped minicircles. In the following experiment we will demonstrate that this smear, named fraction U, is not multiply gapped minicircles but is a new free minicircle species characterized for the first time in these RNAi cells.
To better resolve and quantify the RNAi-mediated change in the different free minicircle species, we first fractionated total DNA (containing free minicircles) by sedimentation velocity on a sucrose gradient; then we resolved each gradient fraction on an agarose-ethidium gel (Fig. 3C). This procedure clearly demonstrated that fraction U (Fig. 3C, right panel) differs from MG minicircles; although they co-migrated on the agarose-ethidium gel, fraction U sedimented almost twice as fast on the sucrose gradient. In addition, the sucrose gradient experiment showed that during the first 3 days of RNAi there was only a relatively small change in the level of covalently closed minicircles (the precursor of replication), but there was a much more rapid decline, starting at day 1, of the segregated products including MG and N/G minicircles (Fig. 3, B–D). Thus we conclude that during the first 3 days of RNAi, the covalently closed minicircles continued to be released from the network at a fairly normal rate, thus explaining the network shrinkage mentioned above. In contrast, the replication products (nicked/gapped and multiply gapped minicircles) declined and were replaced by fraction U and the oligomeric species that run slower than nicked/gapped minicircles on the agarose-ethidium gel. These data raised the possibility that fraction U was a replication intermediate or derived from an intermediate that was upstream of the nicked/gapped and multiply gapped minicircle progeny that are found in wild type cells.
If the production of nicked/gapped free minicircles is reduced, then the number of these molecules on the network should also be reduced. To test this possibility, we isolated kDNA networks from cells that had undergone RNAi for 1–3 days. We then labeled nicked/gapped minicircles with fluorescein-12-dUTP using terminal deoxynucleotidyltransferase (TdT labeling; Fig. 3E). TdT adds the fluorescent nucleotides to 3′ hydroxyl groups on the newly synthesized nicked/gapped circles. During kDNA replication of wild type T. brucei, all of the newly synthesized minicircles (containing at least one gap or nick) reattach to the polar regions of the network adjacent to the antipodal sites (35). As replication proceeds, polar regions containing nicked/gapped minicircles increase in size, and the central region (from which the covalently closed minicircles are removed) undergoes shrinkage. Eventually the network becomes double-sized, and all of its minicircles are nicked/gapped (2). See examples of the various stages in replication under day 0 in Fig. 3E (these stages are described in the Fig. 3 legend). As shown in Fig. 3F, after 2 days of RNAi, the percentage of TdT-positive networks dropped from 39 to 20%, as expected from a drop in attachment of newly synthesized minicircles. Moreover, the pattern of TdT labeling changed (Fig. 3, E and F), with a disappearance of networks with the typical polar-labeling pattern. One new pattern of TdT labeling was a peripheral ring, present in 3% of the total networks. We previously observed TdT-labeled rings following RNAi of two other kDNA replication proteins, structure-specific endonuclease I and p38 (see Ref. 35 for speculation on the origin and significance of these rings). Although the percentage of the TdT-positive network further decreased after 3 days RNAi, it was impossible to quantify accurately because some networks were too small for detection of TdT fluorescence (Fig. 3E). Thus the TdT experiments are consistent with an RNAi-mediated block in replication prior to the step in which nicked/gapped minicircle progeny reattach to the network.
Characterization of Fraction U
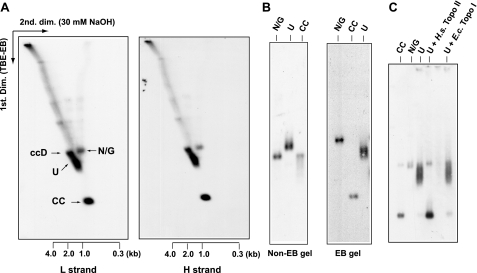
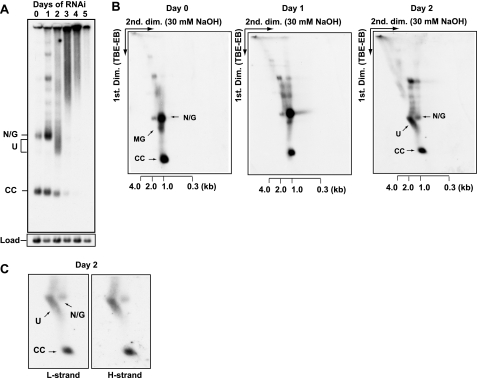
To understand the function of TbPIF1, it was essential to characterize this novel free minicircle species. Therefore, we isolated total DNA from cells undergoing TbPIF1 RNAi (for 3 days) and then fractionated it on a neutral/alkaline two-dimensional gel. The first dimension was a standard agarose-ethidium bromide gel in TBE buffer, and the second dimension was in 30 mm NaOH to denature the minicircle species. We probed the blots separately for the L-strand (the leading strand) and the H-strand (the lagging strand) (Fig. 4A). Fraction U minicircles contain both L- and H-strands in the 1–2-kb range (estimated from denatured double-stranded linear markers). Because both probes had similar specific radioactivities, the two strands of fraction U must be in an approximately equimolar ratio. We detected no small fragments derived from fraction U (such as Okazaki fragments), even with a 5-fold longer autoradiographic exposure (data not shown).
FIGURE 4.
Characterization of fraction U. A, total DNA from 3 × 107 induced cells (3 days of RNAi) were fractionated on a neutral/alkaline two-dimensional agarose gel. The first dimension was in TBE buffer with 1 μg/ml ethidium bromide (TBE-EB) (conditions were same as those for the gel in Fig. 3B). The second dimension was in alkali (30 mm NaOH). Strand-specific oligonucleotide probes detected light strand (L strand) and heavy strand (H strand). U, fraction U; ccD, covalently closed dimer with single interlock. The scales below the panels indicated the sizes of alkali-denatured linear markers in the second dimension. B, gel-purified fraction U (from 2 × 107 cells) was run on a 1.5% agarose gel with or without ethidium bromide (1 μg/ml) and detected by probing a Southern blot. Gel-purified N/G and CC minicircles were used as markers. C, gel-purified fraction U was treated with or without topoisomerase. The first three lanes contain markers: CC minicircles, N/G minicircles, and fraction U (U). The two right-hand lanes contain fraction U treated with human (H.s.) topoisomerase II (USB) and fraction U treated with E. coli (E.c.) topoisomerase I (NEB).
We next gel-purified fraction U and ran it on an agarose gel with or without ethidium bromide (Fig. 4B). Without ethidium bromide, fraction U ran slower than the nicked/gapped or covalently closed monomers (Fig. 4B, left panel), suggesting that it is larger in size than a minicircle monomer. In the presence of ethidium bromide, fraction U ran faster than nicked/gapped minicircles, suggesting that it may be covalently closed and supertwisted by ethidium (Fig. 4B, right panel). We next treated gel-purified fraction U with several topoisomerases (Fig. 4C). Topoisomerase II (Fig. 4C, fourth lane) converted fraction U to a species co-migrating with covalently closed minicircle monomers, suggesting that fraction U is a covalently closed minicircle dimer. The fact that treatment with either E. coli topoisomerase I (Fig. 4C, fifth lane) or wheat germ topoisomerase I (data not shown) had no effect on fraction U mobility suggests that this novel molecule does not contain a significant number of supertwists.
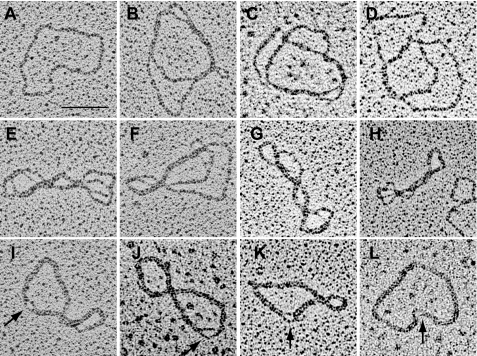
Our most powerful tool for identifying the structure of fraction U minicircles was EM using protein-free spreading (30). Panel A of Fig. 5 shows as a control a relaxed minicircle monomer, and panels B–L show fraction U molecules. Although some molecules could not be interpreted unambiguously, most appear to be catenated minicircle dimers with varying numbers of interlocks. Amazingly, we found some examples that contained a very high number of interlocks, possibly near the maximum that are sterically allowed (Fig. 5, I–L); in these molecules the individual double helices could be distinguished in only a small region (marked by arrow).
FIGURE 5.
Electron micrographs of fraction U from TbPIF1 RNAi cells. A, relaxed minicircle monomer. B–L, fraction U dimers, which differ in number of interlocks. The arrows in I–L mark the small regions where individual double helices are visible. Bar, 500 nm.
Does Fraction U Derive from Networks?
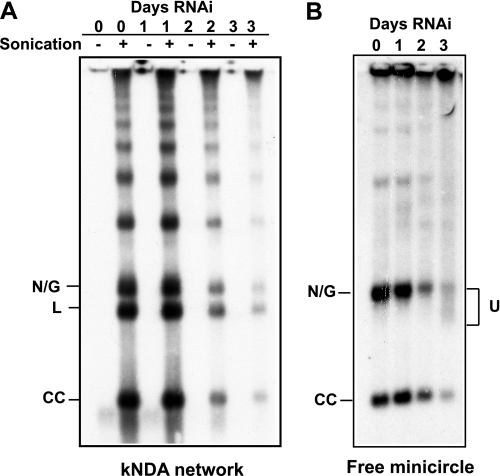
Fraction U could derive from a replication intermediate, and we will address this and other possibilities under “Discussion.” Here we shall examine whether multiply interlocked minicircles are present on the network in RNAi cells. If they were present, fraction U could be released from the network as it shrunk in size. Studies on kDNA from Crithidia fasciculata revealed that a nonreplicating network contains covalently closed relaxed minicircles that are connected by single interlocks (36) to an average of three other minicircles (37). The same is likely true in T. brucei. To test whether RNAi cells contain multiply interlocked minicircles linked to networks that could be released as fraction U, we briefly sonicated kDNA networks isolated from cells following 3 days of RNAi. We then analyzed the products by gel electrophoresis and Southern blotting (Fig. 6). Many minicircles are hit once by sonication and thus linearized. These minicircles are released from the network together with any neighboring minicircle monomer or oligomers. This treatment should release some fraction U if present. However, we did not detect fraction U even from a large amount of sonicated kDNA (from 2 × 107 cells; Fig. 6A). In contrast, we easily observed the accumulation of fraction U as a component of free minicircles isolated from the same cells used for sonication of the network (but using only 2 × 106 cells; Fig. 6B). We also examined the sonicated kDNA networks by EM and never observed any molecule resembling fraction U (data not shown). We concluded that fraction U does not derive from networks.
FIGURE 6.
Was fraction U derived from a kDNA network? kDNA and total DNA were isolated from TbPIF1 RNAi cells (3 days after induction). A, isolated kDNA (from 2 × 107 cells) was subjected to a brief sonication, fractionated on an agarose gel with ethidium bromide, and detected by probing a Southern blot (36). B, total DNA (from 2 × 106 cells) was fractionated for free minicircle analysis. U, fraction U.
Fraction U Is Also Formed Following RNAi of the Mitochondrial Topoisomerase II
We wondered whether fraction U may have formed during RNAi of any other kDNA replication enzymes. We therefore searched the literature for data that might suggest a previous appearance of fraction U. In a published study on the mitochondrial topoisomerase II (15), we had conducted a free minicircle analysis similar to that in Fig. 3B. After 4 days of RNAi, we detected a smear below the nicked/gapped minicircles that we had attributed to partial minicircle degradation. Because this smear runs in the position of fraction U, we now have revisited the topoisomerase II RNAi experiment to see whether fraction U was indeed formed. A one-dimensional gel, showing free minicircles during a course of RNAi (Fig. 7A), closely resembles the previously published experiment, except fraction U appeared much sooner after RNAi induction. We do not know the reason for the differing kinetics. We used strain 29-13 trypanosomes carrying the same stem-loop RNAi construct for topoisomerase II RNAi that had been developed in our previous study (15). However, our cell line had been passaged independently for several years; its doubling time was 6 h, whereas the previously published value was 12 h. Here we observed a striking increase at day 1 of nicked/gapped minicircles, whereas previously we had detected the increase in this species during days 1–3. Again we saw the fraction U-like smear that appeared at day 2, whereas previously it emerged at day 4 (Fig. 7A).
FIGURE 7.
Fraction U was formed after topoisomerase II RNAi. A, free minicircle analysis as in Fig. 3B except topoisomerase II was knocked down by RNAi. B, neutral/alkaline two-dimensional gel electrophoresis of free minicircles isolated from topoisomerase II RNAi cells (days 0–2). C, two-dimensional gels of free minicircles (also after RNAi for 2 days) as in B. Strand-specific hybridizations with synthetic oligonucleotide probes detected light strand (L strand) and heavy strand (H strand) of free minicircle species. U, fraction U. The scales below the panels indicate the sizes of linear markers in the second dimension.
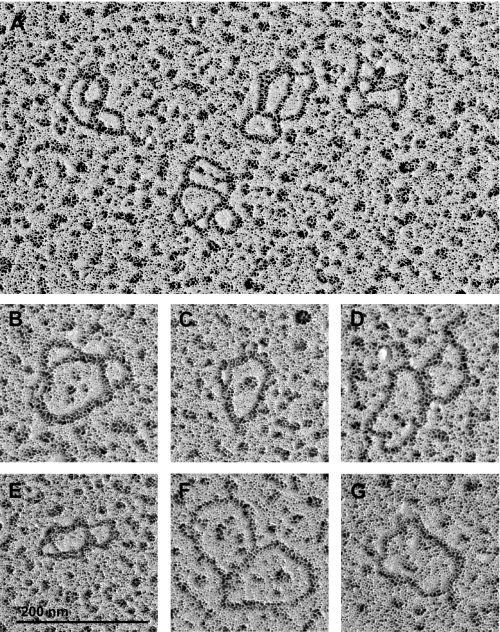
We next used two-dimensional gels, like those in Fig. 4, to characterize the nicked/gapped species and determine whether the smear had the characteristic pattern of fraction U (Fig. 7B). Prior to RNAi (day 0), there are comparable levels of CC and N/G minicircles (day 0; Fig. 7B). However, the ratio of nicked/gapped circles to that of covalently closed circles changed to ∼2.5:1 after 1 day of RNAi induction, consistent with our previous study showing that nicked/gapped minicircles accumulated after topoisomerase II RNAi (15). After 2 days of RNAi, the nicked/gapped minicircles decreased abruptly, and there appeared a smear migrating as an arc that is very similar to the pattern of fraction U from TbPIF1 RNAi cells (Fig. 4A). Strand-specific hybridization further confirmed that the two strands of the smear are approximately equimolar (Fig. 7C). Finally, we conducted EM on fraction U, in this case using the Kleinschmidt method in which the DNA is coated by cytochrome c. These experiments provided strong evidence that fraction U molecules are also multiply interlocked dimers that differ in the number of interlocks (Fig. 8). Thus we conclude that fraction U accumulates not only following RNAi of TbPIF1 helicase but also following RNAi of topoisomerase II.
FIGURE 8.
Electron micrographs of fraction U from topoisomerase II RNAi cells. A, a field containing four fraction U dimers. B–F, enlarged images of fraction U dimers. G, relaxed minicircle monomer. Bar, 200 nm.
DISCUSSION
Here we localized TbPIF1 in the antipodal sites and demonstrated that a recombinant form of this protein has ATP-dependent DNA helicase activity. RNAi knockdown of TbPIF1 resulted in kDNA loss, indicating that this helicase is essential for kDNA replication or maintenance. RNAi of TbPIF1 reduced the minicircle level starting at day 1, and the abundance decreased progressively until they were virtually undetectable by day 5. It is striking that a 3-day course of RNAi had only small effects on the abundance of covalently closed free minicircles, indicating that release of these minicircles from the network continues unabated (Fig. 3, B and C). However, there was a much more prominent decline in nicked/gapped minicircles, the monomeric progeny of free minicircle replication (seen more clearly in Fig. 3D). These latter species were replaced by fraction U. An attractive interpretation of these findings is that fraction U is an intermediate in minicircle replication or is derived from an intermediate. If so, then the RNAi experiments would suggest that the TbPIF1 helicase (and topoisomerase II) could be required for conversion of fraction U or a fraction U-related molecule to monomeric nicked/gapped minicircles.
Support for this model was provided by identification of fraction U as a family of covalently closed minicircle dimers with varying numbers of interlocks. Multiply interlocked dimers are conventional intermediates in the late stages of θ-type replication. Unwinding of parental strands causes accumulation of positive supertwists in front of the replication fork, and these supertwists can be removed by a topoisomerase. As replication nears completion, the unreplicated, positive supertwisted region becomes too short for the topoisomerase to bind and remove the supertwists. Instead, the positive supertwists diffuse across the replication fork into the replicated region. Therefore, the two newly synthesized double strands interwine with each other, producing multiply interlocked dimers. Topoisomerase II can resolve the interlocks, thereby completing the segregation of the two daughter DNA circles (38). Multiply interlocked dimers (with up to 30 interlocks) were first observed in SV40 DNA from cells cultured in a hypertonic medium (the cellular topoisomerase II is less active under this condition) (39, 40). Similar multiply interlocked intermediates have also been detected for trypanosome kDNA minicircles when the mitochondrial topoisomerase II was inhibited by etoposide (41). Finally, our finding that RNAi knockdown of topoisomerase II results in accumulation of fraction U fits perfectly with this model. It is clear why RNAi knockdown of a topoisomerase II would prevent resolution of multiply interlocked dimers, but it is not readily apparent why a helicase deficiency would have this effect. We suggest two possible explanations for the accumulation of multiply interlocked dimers in TbPIF1 RNAi cells. One is that a normal function of TbPIF1 is to dissociate a protein from the minicircle that inhibits topoisomerase II-mediated segregation (see Ref. 18 for a discussion of PIF-mediated protein dissociation). Alternatively, TbPIF1 may be a co-factor (possibly independent of its helicase activity) that allows topoisomerase II to decatenate the dimer. It is even possible that dimer decatenation requires an assembly of proteins of which TbPIF1 is a member and topoisomerase II provides the catalytic activity.
This explanation for the production of fraction U following TbPIF1 RNAi is far more attractive than the alternatives. We presented evidence that fraction U does not derive from networks (Fig. 6), and we think it is even more unlikely that topoisomerase II, acting on covalently closed free minicircles, would be able to dimerize these molecules with the introduction of large numbers of interlocks (also, the fact that fraction U forms even when mitochondrial topoisomerase II is knocked down would rule out that possibility).
We therefore think that fraction U is produced during minicircle replication. If it is an intermediate in replication or derived from an intermediate, then it should be possible to label it metabolically with DNA precursors. Unfortunately we cannot use [3H]thymidine because tracer concentrations of this precursor are not taken up by procyclic trypanosomes, possibly because there is no high affinity nucleoside transporter that recognizes thymidine. However, it is possible to label procyclic DNA with high concentrations (50 μm) of bromodeoxyuridine (BrdUrd), a thymidine analog, but the assay is not sensitive or quantitative and is cumbersome to carry out (BrdU is detected by an antibody on blots only after denaturing the DNA) (6, 42). We were initially surprised that a 40-min metabolic labeling with BrdU (after 3 days of RNAi) failed to reveal incorporation into fraction U under conditions where there was incorporation into nicked/gapped minicircles, which are well characterized replication intermediates (6, 43) (data not shown). We think that this result does not rule out the possibility that fraction U derives from a replication product. If the efficiency of production of fraction U was low, and it accumulated gradually over a course of more than a day (Fig. 3B), then little BrdU would be incorporated into fraction U during the 40-min pulse. Thus we think it most likely that fraction U derives from a product of minicircle replication.
There are other interesting questions about fraction U. One concerns the mechanism by which fraction U minicircles become covalently closed. In wild type cells, minicircle closure does not occur until after minicircles are attached to the network and all have replicated. It could be that free minicircle closure indeed occurs, but very slowly. Wild type minicircles do not close because the monomeric products of replication are quickly reattached to the network, where closure does not occur until after replication is complete. If dimeric minicircles cannot be attached to the network, they would have a much longer time for closure to occur. We also wondered why so many interlocks formed. Possibly many interlocks also exist transiently in wild type cells, but they have not been observed because their number is rapidly reduced by topoisomerase II action. Another question concerns the nature of the oligomeric species that accumulate starting on day 2. Because these species also migrate as an arc on neutral/alkaline two-dimensional gels (Fig. 4A) and are sensitive to topoisomerase II treatment (data not shown), we believe that they are multiply interlocked oligomers. These species could be replication intermediates of minicircle oligomers that were accumulated when the conversion to monomers is blocked by TbPIF1 RNAi.
The topoisomerase II data is of great interest. We previously assessed the population of free minicircles in topoisomerase RNAi cells only by one-dimensional gel electrophoresis under conditions like that shown in Fig. 7A (15). We noted a striking increase in nicked/gapped minicircles that we concluded was due to their failure to attach to the network periphery in the absence of topoisomerase II. Following the rise in nicked/gapped minicircles, we also noted the appearance of a smear that we then attributed to minicircle degradation. However, based on the two-dimensional gel in Fig. 7B (day 2), we now know that this smear is fraction U. Thus we can now draw important new conclusions about the topoisomerase II RNAi phenotype. In the early stages of RNAi, nicked/gapped monomeric minicircles accumulate, presumably in the antipodal sites, because the knockdown of topoisomerase II prevents their reattachment to the network. Subsequently, fraction U appears. As discussed above, we believe that these multiply interlocked circles derive from an intermediate just upstream of nicked/gapped circles. Thus the topoisomerase II in wild type cells must form nicked/gapped minicircles by decatenating multiply interlocked dimers and then immediately attaching them to the network. In topoisomerase II RNAi cells, the switch from nicked/gapped accumulation (on day 1) to fraction U accumulation (on day 2) can be explained by declining levels of topoisomerase II during RNAi. In the initial stages of RNAi (day 1 in Fig. 7B), there is likely a higher level of topoisomerase II that permits the conversion of multiply interlocked minicircles to nicked/gapped species but not the attachment of the nicked/gapped minicircles to the network (the latter reaction would require a higher level of topoisomerase II). After 2 days of RNAi, there could be a further reduction of the topoisomerase II level, and both reactions would be stopped.
It is not clear where in the mitochondrion the conversion of multiply interlocked minicircles to nicked/gapped monomers takes place. According to the current replication model presented in the Introduction, segregation of minicircle replication products takes place in the KFZ. However, evidence for some features of this model, including the site of segregation of minicircle dimers, are not yet compelling. It is significant that both TbPIF1 and topoisomerase II, the two enzymes that are apparently involved in segregation, are predominantly localized in the antipodal sites. Either a small percentage of these enzymes is situated permanently in the KFZ or small amounts migrate from the antipodal sites to the KFZ; alternatively, segregation may occur in the antipodal sites. The same problem arises with p38, which binds to the minicircle replication origin and functions in replication initiation (6). p38 also localizes predominantly in the antipodal sites but may function in the KFZ.
A critical question is whether TbPIF1 is the helicase functioning at the minicircle replication fork. The fact that some replication (e.g. the formation of fraction U) occurs during RNAi might suggest that TbPIF1 functions only at the end of replication to help segregate progeny minicircles. However, it may be that its helicase function at the fork requires only low levels of TbPIF1 that might be present after RNAi. Its function in segregating progeny may require much higher levels, and therefore this step might be selectively sensitive to RNAi. Thus future experiments will be needed to establish whether PIF1 functions at the replication fork.
Acknowledgments
We thank Mike Delannoy for help with EM of isolated kDNA and Dr. Wade Gibson for the use of the PhosphorImager. We thank Dr. Tao-Shih Hsieh and all the members of our lab for helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants AI058613 (to P. T. E.) and GM31819 (to J. D. G.).
- kDNA
- kinetoplast DNA
- RNAi
- RNA interference
- KFZ
- kinetoflagellar zone
- EM
- electron microscopy
- DAPI
- 4′,6-diamidino-2-phenylindole
- CC
- covalently closed
- N/G
- nicked and gapped
- MG
- multiply gapped
- TdT
- terminal deoxynucleotidyltransferase
- BrdUrd
- bromodeoxyuridine.
REFERENCES
- 1.Shlomai J. (2004) Curr. Mol. Med. 4, 623–647 [DOI] [PubMed] [Google Scholar]
- 2.Liu B., Liu Y., Motyka S. A., Agbo E. E., Englund P. T. (2005) Trends Parasitol. 21, 363–369 [DOI] [PubMed] [Google Scholar]
- 3.Ogbadoyi E. O., Robinson D. R., Gull K. (2003) Mol. Biol. Cell 14, 1769–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson L., Aphasizhev R., Gao G., Kang X. (2004) RNA 10, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stuart K. D., Schnaufer A., Ernst N. L., Panigrahi A. K. (2005) Trends Biochem. Sci 30, 97–105 [DOI] [PubMed] [Google Scholar]
- 6.Liu B., Molina H., Kalume D., Pandey A., Griffith J. D., Englund P. T. (2006) Mol. Cell. Biol. 26, 5382–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abu-Elneel K., Robinson D. R., Drew M. E., Englund P. T., Shlomai J. (2001) J. Cell Biol. 153, 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C., Englund P. T. (1997) J. Biol. Chem. 272, 20787–20792 [DOI] [PubMed] [Google Scholar]
- 9.Klingbeil M. M., Motyka S. A., Englund P. T. (2002) Mol. Cell 10, 175–186 [DOI] [PubMed] [Google Scholar]
- 10.Scocca J. R., Shapiro T. A. (2008) Mol. Microbiol. 67, 820–829 [DOI] [PubMed] [Google Scholar]
- 11.Engel M. L., Ray D. S. (1998) Nucleic Acids Res. 26, 4733–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torri A. F., Englund P. T. (1995) J. Biol. Chem. 270, 3495–3497 [DOI] [PubMed] [Google Scholar]
- 13.Downey N., Hines J. C., Sinha K. M., Ray D. S. (2005) Eukaryot. Cell 4, 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melendy T., Sheline C., Ray D. S. (1988) Cell 55, 1083–1088 [DOI] [PubMed] [Google Scholar]
- 15.Wang Z., Englund P. T. (2001) EMBO J. 20, 4674–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxowsky T. T., Choudhary G., Klingbeil M. M., Englund P. T. (2003) J. Biol. Chem. 278, 49095–49101 [DOI] [PubMed] [Google Scholar]
- 17.Boulé J. B., Zakian V. A. (2006) Nucleic Acids Res. 34, 4147–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulé J. B., Vega L. R., Zakian V. A. (2005) Nature 438, 57–61 [DOI] [PubMed] [Google Scholar]
- 19.Azvolinsky A., Dunaway S., Torres J. Z., Bessler J. B., Zakian V. A. (2006) Genes Dev. 20, 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budd M. E., Reis C. C., Smith S., Myung K., Campbell J. L. (2006) Mol. Cell. Biol. 26, 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu G. H., Tanaka H., Kim D. H., Kim J. H., Bae S. H., Kwon Y. N., Rhee J. S., MacNeill S. A., Seo Y. S. (2004) Nucleic Acids Res. 32, 4205–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi M. L., Pike J. E., Wang W., Burgers P. M., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foury F., Kolodynski J. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahaye A., Stahl H., Thines-Sempoux D., Foury F. (1991) EMBO J. 10, 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B., Wang J., Yaffe N., Lindsay M. E., Zhao Z., Zick A., Shlomai J., Englund P. T. (2009) Mol. Cell 35, 490–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B., Wang J., Yildirir G., Englund P. T. (2009) PLoS Pathog. 5, e1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z., Morris J. C., Drew M. E., Englund P. T. (2000) J. Biol. Chem. 275, 40174–40179 [DOI] [PubMed] [Google Scholar]
- 28.Oberholzer M., Morand S., Kunz S., Seebeck T. (2006) Mol. Biochem. Parasitol 145, 117–120 [DOI] [PubMed] [Google Scholar]
- 29.Kulikowicz T., Shapiro T. A. (2006) J. Biol. Chem. 281, 3048–3056 [DOI] [PubMed] [Google Scholar]
- 30.Griffith J. D., Christiansen G. (1978) Annu. Rev. Biophys. Bioeng. 7, 19–35 [DOI] [PubMed] [Google Scholar]
- 31.Thresher R., Griffith J. (1992) Methods Enzymol. 211, 481–490 [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Morga D. L., Englund P. T. (1993) Nucleic Acids Res. 21, 1327–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka H., Ryu G. H., Seo Y. S., Tanaka K., Okayama H., MacNeill S. A., Yuasa Y. (2002) Nucleic Acids Res. 30, 4728–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Englund P. T. (2007) Mol. Microbiol. 64, 676–690 [DOI] [PubMed] [Google Scholar]
- 36.Rauch C. A., Perez-Morga D., Cozzarelli N. R., Englund P. T. (1993) EMBO J. 12, 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J., Rauch C. A., White J. H., Englund P. T., Cozzarelli N. R. (1995) Cell 80, 61–69 [DOI] [PubMed] [Google Scholar]
- 38.Wang J. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 39.Sundin O., Varshavsky A. (1981) Cell 25, 659–669 [DOI] [PubMed] [Google Scholar]
- 40.Sundin O., Varshavsky A. (1980) Cell 21, 103–114 [DOI] [PubMed] [Google Scholar]
- 41.Shapiro T. A. (1994) Mol. Cell. Biol. 14, 3660–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson D. R., Gull K. (1994) J. Cell Biol. 126, 641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan K. A., Englund P. T. (1989) Mol. Cell. Biol. 9, 3212–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]