The structure of the stationary phase survival protein SurE protein from the hyperthermophile Aquifex aeolicus has been solved to 1.5 Å resolution. The divalent-metal-ion-dependent phosphatase active-site pocket is occupied by sulfate ions from the crystallization medium.
Keywords: SurE, Aquifex aeolicus, stationary-phase survival
Abstract
SurE is a stationary-phase survival protein found in bacteria, eukaryotes and archaea that exhibits a divalent-metal-ion-dependent phosphatase activity and acts as a nucleotidase and polyphosphate phosphohydrolase. The structure of the SurE protein from the hyperthermophile Aquifex aeolicus has been solved at 1.5 Å resolution using molecular replacement with one dimer in the asymmetric unit and refined to an R factor of 15.6%. The crystal packing reveals that two dimers assemble to form a tetramer, although gel-filtration chromatography showed the presence of only a dimer in solution. The phosphatase active-site pocket was occupied by sulfate ions from the crystallization medium.
1. Introduction
The SurE gene is part of a stationary-phase survival operon that is found in eubacteria, eukaryotes and archaea and which is induced in cells during high levels of stress or slow growth (Li et al., 1997 ▶; Visick et al., 1998 ▶). Biochemical characterization of the SurE protein in Escherichia coli shows it to be a divalent metal ion-dependent phosphatase with both nucleotidase and exopolyphosphatase activity (Proudfoot et al., 2004 ▶). Initial evidence for phosphatase activity in the SurE family has been provided together with the first crystal structures of the enzyme from the bacterium Thermotoga maritima (Lee et al., 2001 ▶; Zhang et al., 2001 ▶). Subsequently, crystal structures of archaeal SurE from Pyrobaculum aerophilum (Mura et al., 2003 ▶) and SurE from Thermus thermophilus (Iwasaki & Miki, 2007 ▶) have been published. Here, as a target in a high-throughput genomics study, we report the structure of recombinant SurE (AaSurE) from Aquifex aeolicus, a hyperthermophilic bacterium found in hot springs with a growth temperature of 358–368 K (Huber & Stetter, 2001 ▶).
2. Materials and methods
2.1. Cloning, expression and purification
The gene encoding AaSurE protein (aq_832; gi:15606188) was amplified via PCR using A. aeolicus VF5 genomic DNA and was cloned into pET-21a expression vector (Merck Novagen, Darmstadt, Germany). The expression vector was introduced into E. coli BL21-CodonPlus(DE3)-RIL strain (Stratagene, La Jolla, California, USA) and the recombinant strain was cultured in 4.5 l LB medium containing 30 µg ml−1 chloramphenicol and 50 µg ml−1 ampicillin. The harvested cells (19 g) were lysed by sonication in 35 ml 20 mM Tris–HCl buffer pH 8.0 containing 500 mM NaCl, 5 mM β-mercaptoethanol and 1 mM phenylmethylsulfonyl fluoride on ice. The cell lysate was heat-treated at 363 K for 12 min and centrifuged at 15 000g for 30 min at 277 K. The supernatant was desalted by fractionation on a HiPrep 26/10 column (GE Healthcare Biosciences). The sample was applied onto a Toyopearl SuperQ-650M column (Tosoh, Tokyo) equilibrated with 20 mM Tris–HCl buffer pH 8.0 and eluted with a linear (0–0.4 M) gradient of NaCl. The target sample, which eluted in the 0.18 M NaCl fraction, was then applied onto a Resource Q column (GE Healthcare Biosciences) equilibrated with 20 mM Tris–HCl buffer pH 8.0 and eluted with a linear gradient of 0–0.3 M NaCl. The fractions that eluted in 0.14 M NaCl were further purified using a hydroxyapatite CHT20-I column (Bio-Rad Laboratories) with a linear gradient of 0.01–0.5 M potassium phosphate buffer pH 7.0. The target sample, which eluted in the 0.18 M potassium phosphate fraction, was collected and applied onto a HiLoad 16/60 Superdex 200pg column (GE Healthcare Biosciences) equilibrated with 20 mM Tris–HCl buffer pH 8.0 containing 200 mM NaCl. The protein sample was analyzed by SDS–PAGE and was confirmed by N-terminal amino-acid sequencing. After concentration to 27.8 mg ml−1 by ultrafiltration, the protein yield was 42.0 mg from 19 g of cells.
2.2. Crystallization
Crystallization was performed by the microbatch-under-oil method at 291 K. 0.5 µl crystallization regent, consisting of 0.1 M acetate buffer pH 4.5 containing 2.5 M NaCl and 0.2 M Li2SO4 (Wizard II condition No. 38; Emerald BioSystems), was mixed with 0.5 µl 27.8 mg ml−1 protein solution. The mixture was then covered with 15 µl silicone and paraffin oil. Crystals suitable for X-ray data collection appeared within three weeks and reached final dimensions of 0.2 × 0.2 × 0.03 mm (Fig. 1 ▶). The crystals were flash-cooled in a nitrogen-gas stream at 100 K using 20%(v/v) glycerol as a cryoprotectant for transport to the synchrotron.
Figure 1.
Crystals of A. aeolicus VF5 SurE.
2.3. Data collection and processing
Experiments were performed at the Daresbury Synchrotron Radiation Source (SRS) using the combined crystallography/X-ray absorption beamline 10.1, employing a Si(111) sagittally focused monochromator tuned to a wavelength of 1.04 Å. Diffraction data were recorded at 100 K from a single SurE crystal immersed in reservoir solution with 20% glycerol added as a cryoprotectant. The first image showed evidence in the low-resolution data of the presence of partially ordered ice in the crystal. The initial diffraction resolution was improved by 0.5 Å by annealing the crystal (Ellis et al., 2002 ▶), a procedure that also removed the ice rings and reduced the mosaic spread to 0.28°. Images were recorded using a MAR Mosaic 225 CCD detector and were processed (indexed, integrated and scaled) using HKL-2000 (Otwinowski & Minor, 1997 ▶). The crystal was found to belong to space group P3221, with unit-cell parameters a = b = 65.01, c = 239.19 Å. The estimated solvent content was 52% for two molecules in the asymmetric unit.
2.4. Structure solution and refinement
The structure was solved by molecular replacement with MOLREP (Vagin & Teplyakov, 1997 ▶), using as a search model SurE from Thermotoga maritima with 42% identity and 69% similarity to the target sequence and consisting of one subunit of entry 1j9j (Lee et al., 2001 ▶) from the Protein Data Bank (Abola et al., 1987 ▶). To facilitate structure solution, the search model was truncated by removing the protruding β-hairpin and α-helix belonging to the C-terminus. The initial R factor for two molecules in the asymmetric unit was poor at only 61%, necessitating significant rebuilding with Coot (Emsley & Cowtan, 2004 ▶) during successive cycles of refinement using REFMAC (Murshudov et al., 1997 ▶). As the electron-density maps improved, the sequence for A. aeolicus was gradually introduced into the model to replace the sequence of T. maritima. Water molecules were added to the structure using Coot. The stereochemistry was checked using PROCHECK (Laskowski et al., 1993 ▶). The Ramachandran plot showed 98.0% of residues to be in the core region and 2.0% to be in additionally allowed regions. The R factor and R free for the final model were 15.6% and 19.1%, respectively. Table 1 ▶ summarizes the data-collection and refinement parameters. The stereochemistry of the final model was checked using MolProbity (Davis et al., 2007 ▶). The structure was deposited in the PDB under accession code 2wqk. The protein interfaces, surfaces and assemblies service PISA (Krissinel & Henrick, 2007 ▶) at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) was used to calculate the molecular properties described below.
Table 1. Data-collection and refinement parameters.
Values in parentheses are for the highest resolution shell (1.55–1.5 Å).
| Space group | P3221 |
| Unit-cell parameters (Å) | a = b = 65.01, c = 239.19 |
| Resolution (Å) | 36.5–1.5 |
| Unique reflections | 86861 |
| Completeness (%) | 95.9 (68.2) |
| Redundancy | 8.7 (2.1) |
| Rmerge† (%) | 8.1 (19.4) |
| I/σ(I) | 22 (3) |
| R factor‡ (%) | 15.6 |
| Rfree‡ (%) | 19.1 |
| B factors (Å2) | |
| Wilson plot | 19.8 |
| Protein | 15.7 |
| Water | 35.8 |
| Na ion, chain A | 16.7 |
| Na ion, chain B | 16.9 |
| R.m.s. deviations | |
| Bond distances (Å) | 0.015 |
| Bond angles (°) | 1.591 |
| ESU§ (Å) | 0.078 |
R
merge = 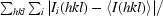
 , where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple symmetry-related observations of that reflection.
, where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple symmetry-related observations of that reflection.
R = 
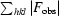 . R
free is the same but calculated for a test set not used in structural refinement.
. R
free is the same but calculated for a test set not used in structural refinement.
Estimated standard uncertainty based on R factor as implemented in REFMAC.
3. Results and discussion
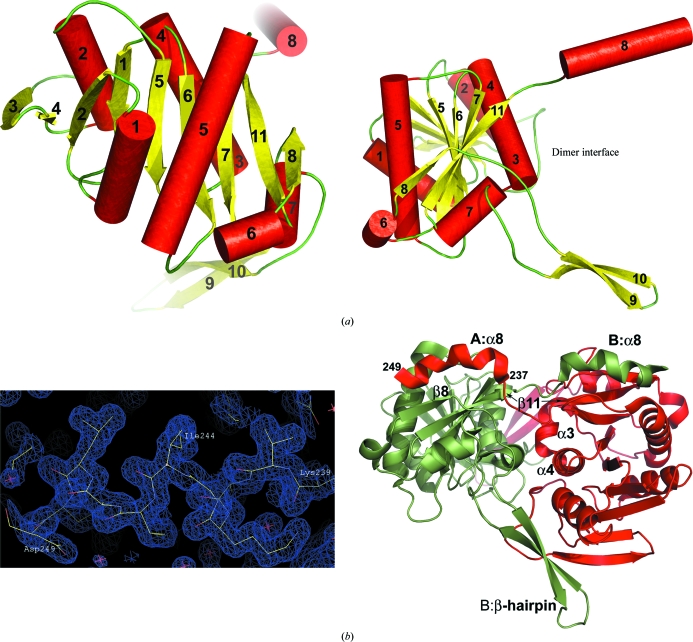
The structure of AaSurE was obtained at 1.5 Å resolution with one dimer (subunits A and B) in the asymmetric unit, comprising 4116 protein atoms, 605 water molecules, two sulfate ions and two sodium ions. For each subunit, 248 residues (of 251) were positioned in the electron-density maps, with only the N-terminal Met1 and C-terminal Ser250 and Pro251 residues missing from the final model. Each subunit comprises a core globular domain containing nine β-strands and six helices in a Rossmann-like fold, with two extended segments at the C-terminal end: a 30-residue β-hairpin linking β-strands 8 and 11 and a 15-residue terminating α-helix, α8 (Fig. 2 ▶ a). The C-terminal extensions are involved in forming the dimer. As in the previously reported structures of bacterial SurE from Thermotoga maritima (Lee et al., 2001 ▶; Zhang et al., 2001 ▶) and Thermus thermophilus (Iwasaki & Miki, 2007 ▶), domain swapping occurs at the C-terminal helix α8. These helices are well ordered in AaSurE and are clearly traceable in the electron-density maps (Fig. 2 ▶ b). Symmetry-related contacts at the dimer interface are provided by residues 40–54 (N-terminal loops), 100–113 (α3 and α4), 174–204 (β8 and β-hairpin) and 227–249 (β11 and α8), yielding a total of 20 salt bridges and more than 30 hydrogen bonds between the two subunits. Dimer formation encompasses up to 25% of the solvent-accessible surface area of each subunit. Experimental evidence from size-exclusion chromatography shows that AaSurE exists as a dimer in solution, while analysis of the protein interfaces (Krissinel & Henrick, 2007 ▶) suggests that both dimeric and tetrameric quaternary structures are stable in solution.
Figure 2.
(a) Two orthogonal orientations of the AaSurE subunit showing the secondary structure and overall fold. The core globular domain consists of nine β-strands (yellow) and six helices (red cylinders), with helix α7 linking the C-terminal β-hairpin to the main body of the molecule. The dimer interface is located on the subunit edge where the β-hairpin (strands β9 and β10) and helix α8 project away from the main body of the molecule. (b) The AaSurE dimer (right) as found in the asymmetric unit, indicating the secondary-structure elements involved in forming salt bridges and/or hydrogen bonds between the two subunits A (red) and B (green). The domain-swapped α8 helices are well ordered. The quality of the electron-density map (left) is shown for A α8 at the 1σ contour level. The overall fold of AaSurE compared with the SurE dimers from other organisms give root-mean-square deviations for Cα atoms of 2.9 Å (PDB code 1j9j, the molecular-replacement search model) and 2.6 Å for 1ilv (both from Thermotoga maritima), 2.1 Å for 1l5x from P. aerophilum and 6.7 Å for 2e6e from Thermus thermophilus. This figure was drawn using Coot (Emsley & Cowtan, 2004 ▶) and PyMOL (DeLano, 2008 ▶).
The biologically relevant molecular assembly in solution was initially identified as a dimer in Thermotoga maritima SurE (Lee et al., 2001 ▶) and P. aerophilum SurE (Mura et al., 2003 ▶), while a functional tetramer was proposed for T. maritima SurE on the basis of crystal packing and size-exclusion chromatography (Zhang et al., 2001 ▶). More recently, Thermus thermophilus SurE has been crystallized with one or more tetramers in the asymmetric unit in space groups P3121 and P212121, with additional evidence for a dimer–tetramer equilibrium being obtained from sedimentation-equilibrium experiments (Iwasaki & Miki, 2007 ▶). A tetramer of AaSurE, obtained by examining the crystal packing, is shown in Fig. 3 ▶(a). The two dimers interact primarily through contacts between the β-hairpins of adjacent subunits, interfaces A–A′ and B–B′, burying 4.8% of the solvent-accessible surface area of each subunit at each interface. Only eight additional potential hydrogen bonds are created at each interface by tetramerization. These observations suggest that the association of dimers into tetramers is relatively weak and may explain why only dimers were found in solutions of AaSurE.
Figure 3.
(a) A tetramer of AaSurE, generated by applying symmetry operations to the dimer, is shown in two orthogonal views. The main contacts forming the tetramer interfaces occur between the β-hairpin strands of adjacent A (red) subunits and adjacent B (green) subunits. The metal-binding site that gives the enzyme its phosphatase activity is shown for each subunit, with a water molecule or Na ion (magenta sphere) occupying the position usually taken by a divalent metal ion and with a sulfate ion (red and yellow spheres) filling the active-site pocket. (b) Close-up of the active site of subunit A (red sticks) compared with the equivalent sites in SurE from Thermotoga maritima with Mg2+ (PDB entry 1j9j, green) or water (1ilv, magenta) at the metal-binding site, from Thermus thermophilus with water and a sulfate ion in the active site (2e69, cyan) or an empty active site (2e6e, yellow) and from P. aerophilum (1l5x, blue) with water at the active site. The numbering of the active-site ligands follows the sequence of AaSurE.
The divalent metal ion-binding site consists of residues that are highly conserved among SurE genes (Lee et al., 2001 ▶). The preferred metal cofactor is Mg2+ in Thermotoga maritima (Lee et al., 2001 ▶; Zhang et al., 2001 ▶) and Thermus thermophilus (Iwasaki & Miki, 2007 ▶) and Co2+ in the hyperthermophilic P. aerophilum (Mura et al., 2003 ▶). The locations of the four identical active sites in the AaSurE tetramer are shown in Fig. 3 ▶(a). A superposition of the active sites of subunit A with those of Thermotoga maritima, Thermus thermophilus and P. aerophilum are shown in Fig. 3 ▶(b). In AaSurE, the active-site pocket is occupied by a sulfate ion, presumably from the crystallization medium, with electron density at the metal-binding site adequate for modelling with either solvent or a metal ion.
Supplementary Material
PDB reference: SurE, 2wqk, r2wqksf
Acknowledgments
We thank Mr Yoshiaki Kitamura and Dr Akio Ebihara for help in sample preparation. This work was supported in part by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was supported by the Synchrotron Radiation Department at the Science and Technology Facilities Council, Daresbury Laboratory UK and X-ray data were collected on beamline 10.1 at the Synchrotron Radiation Source, which was supported by Biotechnology and Biological Sciences Research Council Grant BB/E001971 (to SSH and RWS).
References
- Abola, A., Bernstein, F. C., Bryant, S. H., Koetzle, T. F. & Weng, J. (1987). Crystallographic Databases – Information Content, Software Systems, Scientific Applications, edited by F. H. Allen, G. Bergerhoff & R. Sievers, pp. 107–132. Bonn/Cambridge/Chester: Data Commission of the International Union of Crystallography.
- DeLano, W. L. (2008). PyMOL Molecular Viewer. DeLano Scientific, Palo Alto, California, USA. http://www.pymol.org.
- Ellis, M. J., Antonyuk, S. & Hasnain, S. S. (2002). Acta Cryst. D58, 456–458. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Huber, R. & Stetter, K. O. (2001). Methods Enzymol.330, 11–24. [DOI] [PubMed]
- Iwasaki, W. & Miki, K. (2007). J. Mol. Biol.371, 123–136. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol.372, 774–797. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Lee, J. Y., Kwak, J. E., Moon, J., Eom, S. H., Liong, E. C., Pedelacq, J. D., Berendzen, J. & Suh, S. W. (2001). Nature Struct. Biol.8, 789–794. [DOI] [PubMed]
- Li, C., Wu, P. Y. & Hsieh, M. (1997). Microbiology, 143, 3513–3520. [DOI] [PubMed]
- Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B. III, Snoeyink, J., Richardson, J. S. & Richardson, D. C. (2007). Nucleic Acids Res.35, W375–W383. [DOI] [PMC free article] [PubMed]
- Mura, C., Katz, J. E., Clarke, S. G. & Eisenberg, D. (2003). J. Mol. Biol.326, 1559–1575. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Proudfoot, M., Kuznetsova, E., Brown, G., Rao, N. N., Kitagawa, M., Mori, H., Savchenko, A. & Yakunin, A. F. (2004). J. Biol. Chem.279, 54687–54694. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Visick, J. E., Ichikawa, J. K. & Clarke, S. (1998). FEMS Microbiol. Lett.167, 19–25. [DOI] [PubMed]
- Zhang, R.-G., Skarina, T., Katz, J. E., Beasley, S., Khachatryan, A., Vyas, S., Arrowsmith, C. H., Clarke, S., Edwards, A., Joachimiak, A. & Savchenko, A. (2001). Structure, 9, 1095–1106. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: SurE, 2wqk, r2wqksf
PDB reference: SurE, 2wqk, r2wqksf