The MIF4G domain of DAP5 was crystallized in two distinct crystal forms. Diffraction patterns have been analyzed and preliminary analysis, including molecular replacement, is presented here.
Keywords: DAP5, eIF4G, internal ribosomal entry site, cap-independent translation initiation, translational regulation, eIF4A, eIF3
Abstract
Death-associated protein 5 (DAP5) is a member of the eIF4G family of scaffolding proteins that mediate cap-independent translation initiation by recruiting the translational machinery to internal ribosomal entry sites (IRESs) on mRNA. The MIF4G domain of DAP5 directly interacts with the eukaryotic initiation factors eIF4A and eIF3 and enhances the translation of several viral and cellular IRESs. Here, the crystallization and preliminary X-ray diffraction analysis of the MIF4G domain of DAP5 is presented.
1. Introduction
Initiation of eukaryotic protein translation can occur by two distinct mechanisms: (i) cap-dependent translation and (ii) cap-independent or IRES-mediated translation (Holcik & Sonenberg, 2005 ▶). Cap-dependent translation involves the recruitment of the pre-initiation complex to the 5′-m7GTP-cap structure of mRNAs. Here, the ribosome is brought into close proximity with the mRNA via the initiation factor eIF4F, a complex that consists of the cap-binding protein eIF4E, the ATP-dependent RNA-helicase eIF4A and the scaffolding protein eIF4G. The scaffolding protein eIF4G not only co-localizes eIF4E and eIF4A, but also interacts with the ribosome-associated initiation-factor complex eIF3, thereby bringing the ribosome into the picture. eIF4G also confers circularization of the mRNA, a process that dramatically increases translation efficiency, by simultaneously interacting with the polyA tail-binding protein (PABP) found at the 3′-end of mRNAs and with eIF4E at the 5′-end (Derry et al., 2006 ▶; Wells et al., 1998 ▶).
In contrast, cap-independent translation entails the recruitment of the translational machinery directly to the mRNA by structural elements within the 5′-UTR, the so-called internal ribosomal entry sites (IRESs). In this case, eIF4G directly recognizes the IRES element of an mRNA, thus eliminating the need for interactions with the 5′-cap structure. As a result, IRES-mediated translation generally requires the involvement of fewer initiation factors in the process, although IRES binding is enhanced in the presence of eIF4A (Lomakin et al., 2000 ▶).
The interactions made by eIF4G with eIF4A and eIF3, as well as with RNA, are mediated by a segment of approximately 30 kDa in the middle of the protein termed the MIF4G domain. This domain is central to the function of eIF4G and has in fact been shown to be sufficient to drive cap-independent translation independently in vivo (De Gregorio et al., 1998 ▶, 1999 ▶). The crystal structure of MI4FG from human eIF4GII revealed a domain consisting of five consecutive HEAT repeats stacked together to form a superhelical crescent-shaped structure with two distinct surfaces (Marcotrigiano et al., 1999 ▶). Subsequently, the structure of the MIF4G domain from yeast eIF4G in complex with eIF4A was also determined, revealing that two regions on the convex side of the crescent make contact with the N- and C-terminal domains of eIF4A (Schütz et al., 2008 ▶). The site of RNA interaction on MIF4G has not yet been unambiguously mapped.
Another scaffolding protein called death-associated protein 5 (DAP5; also known as p97 or NAT1) shows a high degree of homology to the central part of eIF4G containing the MIF4G domain and the regions C-terminal of this domain. DAP5 was independently identified by four groups (Imataka et al., 1997 ▶; Levy-Strumpf et al., 1997 ▶; Shaughnessy et al., 1997 ▶; Yamanaka et al., 1997 ▶). It received the name DAP5 owing to its identification in a screen based on transfections of expression cDNA libraries and selection of cells resistant to apoptosis (Levy-Strumpf et al., 1997 ▶). DAP5 lacks the N-terminal region of eIF4G that enables it to bind eIF4E and consequently can only support cap-independent translation initiation. It can drive IRES-mediated translation of a number of cellular mRNAs (Henis-Korenblit et al., 2002 ▶; Hundsdoerfer et al., 2005 ▶; Lewis et al., 2008 ▶; Warnakulasuriyarachchi et al., 2004 ▶).
Human DAP5 is a 907-residue protein that also contains an MIF4G domain (DAP5M) by virtue of its 29% sequence identity to eIF4G in the corresponding middle region. Despite the rather low similarity, the MIF4G domain of DAP5 (referred to in this manuscript as DAP5M) retains the ability to engage eIF4A and eIF3. Like MIF4G, DAP5M can also bind to IRESs, but in a distinct manner. eIF4G has been shown to interact strongly with the IRES of the encephalomyocarditis virus (EMCV) in vitro, whereas DAP5 cannot (Lomakin et al., 2000 ▶). In contrast, DAP5 enhances the translation of a number of cellular mRNAs in vivo, including its own mRNA, which eIF4G cannot (Marash et al., 2008 ▶).
The only structural information available for DAP5 is a crystal structure of its very C-terminal domain (residues 730–898), which is involved in recruiting the Ser/Thr kinase Mnk1 (Liberman et al., 2008 ▶). Our interest lies in the structural basis of the interactions made by the DAP5M domain; here, we present the crystallization and preliminary diffraction analysis of this domain.
2. Materials and methods
2.1. Construction of expression vectors for the production of recombinant DAP5
Based on secondary-structure analysis by the PredictProtein server (Rost et al., 2004 ▶) and sequence alignments with eIF4G, a construct encompassing DAP5M (residues 61–323) was amplified by PCR using the following primers: forward, 5′-AGACGAGGATCCAACTCCGCAGCAAACAAC-3′; reverse, 5′-TTGACGGAATTCCTAGATCGTCTTTAATCC-3′. The PCR product was cloned into the BamHI and EcoRI restriction sites of the pProEX HTb vector (Invitrogen, Carlsbad, USA) for bacterial expression of a fusion protein with an N-terminal hexahistidine tag which is cleavable with the tobacco etch virus (TEV) protease. Owing to cloning artifacts, the cleaved protein contains an additional five amino acids (sequence GAMGS) at the N-terminus.
2.2. Expression and purification of recombinant protein
pProEX HTb containing DAP5M was transformed into Escherichia coli (Rosetta 2 strain) and grown on LB agar plates supplemented with 50 µg ml−1 ampicillin. A single colony was used to inoculate 50 ml LB medium supplemented with 50 µg ml−1 ampicillin for overnight growth at 310 K to produce a starter culture. 10 ml starter culture was then used to inoculate 1 l LB medium supplemented with 50 µg ml−1 ampicillin. Cultures were grown to an optical density at 600 nm (OD600) of 0.6–0.8, at which point the temperature was reduced to 303 K. After 30 min, protein production was induced by the addition of a 1 mM final concentration of isopropyl β-d-1-thiogalactopyranoside (IPTG) and cultures were grown for a further 4 h. Bacteria were harvested at 2500g for 10 min at 277 K and bacterial pellets were resuspended in buffer NiA (25 mM Tris pH 8.0, 500 mM NaCl, 10 mM imidazole, 0.5 mM phenylmethylsulfonyl fluoride and 5% glycerol). Cells were homogenized by two passes through a High-Pressure EmulsiFlex-C3 homogenizer (Avestin, Mannheim, Germany) at a homogenizing pressure of 103 MPa. The lysate was cleared by centrifugation at 48 000g for 45 min at 277 K. Cleared lysate was applied onto a HisTrap FF column (GE Healthcare, Little Chalfont, England) equilibrated in buffer NiA using an ÄKTA FPLC system (GE Healthcare, Little Chalfont, England). Bound protein was eluted using a 50 ml linear gradient to buffer NiB (NiA supplemented with 500 mM imidazole). Following overnight TEV cleavage (using approximately 1 mg TEV protease per 20 mg crude protein) and dialysis against buffer NiA at 277 K with a 3.5 kDa molecular-weight cutoff cellulose membrane, TEV protease (hexahistidine-tagged) was removed by applying samples onto a HisTrap FF column and collecting the flowthrough. Proteins were further purified using size-exclusion chromatography (Superdex 75 10/300; GE Healthcare, Little Chalfont, England) in buffer containing 25 mM Tris pH 8.0, 150 mM NaCl, 5% glycerol and 1 mM DTT. DAP5M fractions were pooled and concentrated. Tris(2-carboxyethyl)phosphine (TCEP; 1 mM final concentration) was added to protein samples before use in crystallization trials. Purified proteins were sent to the Centre for Biological Applications of Mass Spectrometry (CBAMS) at Concordia University to assess their mass and homogeneity.
2.3. Crystallization and diffraction data collection
Initial screening of crystallization conditions was conducted at 293 K with protein concentrations of 10 and 20 mg ml−1 in 0.2 µl drops using the sitting-drop vapour-diffusion method (mixing 0.1 µl protein solution with 0.1 µl reservoir solution and equilibrating against 100 µl reservoir volume) with a Phoenix crystallization robot (Art Robbins, Sunnyvale, USA) on an Intelli-Plate 96 (Art Robbins, Sunnyvale, USA). The Classics I, Classics II and PEG-Ion crystallization suites (Qiagen, Germantown, USA) were screened. After the identification of initial hit conditions, manual two-dimensional grid screens around the hit conditions were carried out for optimization of crystal growth using the hanging-drop vapour-diffusion technique with 2–4 µl drops equilibrated against 1 ml reservoir solution. Crystals suitable for structure determination were grown at 291 K using a protein concentration of 15–20 mg ml−1 in drops of 2–4 µl volume (1–2 µl protein solution mixed with 1–2 µl reservoir solution). Crystal form A was grown using a reservoir solution consisting of 0.1 M HEPES pH 7.5, 0.2 M ammonium sulfate and 18–20%(w/v) polyethylene glycol 5000 monomethyl ether (PEG 5000 MME); the reservoir solution for crystal form B was 0.1 M HEPES pH 7.5 and 25%(w/v) PEG 8000. Crystals were flash-cooled in a liquid-nitrogen cryostream at 100 K and data were collected in-house on a Rigaku MicroMax-007 HF microfocus X-ray generator fitted with Varimax X-ray optics and a Saturn 944+ CCD detector (Rigaku, The Woodlands, USA).
3. Results and discussion
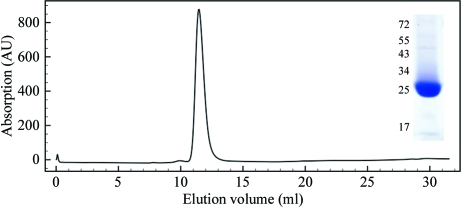
DAP5M was overexpressed in E. coli and purified to homogeneity, yielding approximately 8 mg protein per litre of bacterial culture. The protein was estimated to be greater than 95% pure by SDS–PAGE stained with Coomassie Brilliant Blue (Fig. 1 ▶). DAP5M migrated at an apparent molecular weight of approximately 26 kDa, although its calculated mass is 30.7 kDa. This anomalous migration could be explained by its relatively high theoretical pI of 8.79 as determined by the ProtParam tool on the ExPASy Proteomics Server (Gasteiger et al., 2003 ▶). Size-exclusion chromatography resulted in a single symmetrical peak at a mass corresponding to a monomer (Fig. 1 ▶). Electrospray mass spectrometry determined a mass of 30 688.8 Da, which corresponds well to the calculated theoretical weight of DAP5M of 30 689.7 Da (Fig. 2 ▶ a).
Figure 1.
Analytical size-exclusion chromatography profile and SDS–PAGE analysis (inset) of purified DAP5M. Size-exclusion chromatography was carried out on a Superdex 75 column (GE Healthcare, Little Chalfont, England). SDS–PAGE was carried out on a 12% polyacrylamide gel and the protein was visualized by Coomassie Brilliant Blue staining. Numbers indicate the migration of protein molecular-weight markers in kDa.
Figure 2.
Mass-spectrometric analysis of purified DAP5M: (a) native, (b) selenomethionine labelled.
Initial crystallization trials produced potential protein crystals in multiple conditions, although only two main crystal forms were observed (Fig. 3 ▶). Crystal form A was grown using an optimized reservoir solution consisting of 0.1 M HEPES pH 7.5, 0.2 M ammonium sulfate and 18–20%(w/v) PEG 5000 MME, while the optimized condition for crystal form B was 0.1 M HEPES pH 7.5 and 25%(w/v) PEG 8000. Even under optimized conditions, the crystals of form A were thin and fragile. Any attempt to cryoprotect the crystals resulted in breakage. Therefore, crystals were flash-cooled in a liquid-nitrogen cryostream without prior cryoprotection. For the second crystal form, large blades were found in 0.1 M HEPES pH 7.5 and PEGs of various molecular weights ranging from 3000 to 20 000 at 25–30%(w/v) concentration (crystal form B). The optimized condition for growth of crystal form B was determined to be 0.1 M HEPES pH 7.5 and 25%(w/v) PEG 8000.
Figure 3.
DAP5M crystal forms A (a) and B (b).
Crystal form A diffracted X-rays to 2.4 Å resolution using an in-house Cu Kα rotating-anode generator, whereas crystal form B diffracted to 2.75 Å resolution (Fig. 4 ▶). The data were processed and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). A summary of crystal parameters and diffraction data statistics is given in Table 1 ▶. The space group of crystal form A was determined to be C2, with unit-cell parameters a = 167.66, b = 56.69, c = 74.38 Å, β = 112.03°, while crystal form B was determined to belong to space group P43212, with unit-cell parameters a = b = 57.16, c = 169.08 Å. Matthews analysis revealed the presence of two molecules in the asymmetric unit for crystal form A and one molecule in the asymmetric unit for crystal form B, with solvent contents of 51.7 and 48.3% and Matthews coefficients (V M) of 2.54 and 2.30 Å3 Da−1 for crystal forms A and B, respectively (Matthews, 1968 ▶). Self-rotation analysis using the program GLRF (Tong & Rossmann, 1997 ▶) revealed the presence of a twofold noncrystallographic symmetry (NCS) axis in crystal form A (Fig. 5 ▶).
Figure 4.
Diffraction patterns of crystal forms A (a) and B (b) collected in-house on a Rigaku MicroMax-007 HF microfocus X-ray generator fitted with Varimax X-ray optics and a Saturn 944+ CCD detector. Resolution rings are shown in red. Resolutions are given in Å.
Table 1. Diffraction data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Crystal form A | Crystal form B | |
|---|---|---|
| Data collection | ||
| X-ray source | Rigaku MicroMax-007 HF | |
| Wavelength (Å) | 1.5418 | |
| Temperature (K) | 100 | |
| Detector | Saturn 944+ CCD | |
| No. of images | 360 | 360 |
| Exposure time (s) | 30 | 15 |
| Oscillation angle (°) | 1.0 | 1.5 |
| Crystal-to-detector distance (mm) | 55 | 70 |
| Data processing | ||
| No. of measured reflections | 176548 | 103149 |
| No. of unique reflections | 25004 | 7920 |
| Space group | C2 | P43212 |
| Unit-cell parameters (Å, °) | a = 167.66, b = 56.69, c = 74.38, β = 112.03 | a = b = 57.16, c = 169.08 |
| Resolution (Å) | 2.4 (2.49–2.40) | 2.75 (2.85–2.75) |
| Completeness (%) | 99.8 (99.4) | 99.8 (99.7) |
| Average redundancy | 7.1 (6.3) | 13.0 (11.1) |
| Mean I/σ(I) | 20.2 (3.7) | 25.3 (2.41) |
| Rmerge | 0.097 (0.498) | 0.108 (0.866) |
| No. of molecules per ASU | 2 | 1 |
| Matthews coefficient VM (Å3 Da−1) | 2.54 | 2.30 |
| Solvent content (%) | 51.7 | 46.6 |
Figure 5.
The κ = 180° section of the self-rotation function for DAP5M data using data in the 12–3 Å resolution range and a Patterson cutoff radius of 21 Å. The crystallographic twofold is found at the centre of the stereogram, whereas the twofold NCS axis is at ϕ = 64°, ψ = 65°, κ = 180°.
The data collected from crystal form A were analyzed first. Maximum-likelihood molecular replacement was carried out with the program Phaser (Read, 2001 ▶) using the structure of the MIF4G domain of human eIF4GII as a search model (PDB code 1hur; chain A; Marcotrigiano et al., 2001 ▶). This structure has 29% sequence identity to DAP5M. Nonconserved residues were replaced by alanines using the program CHAINSAW (Stein, 2008 ▶). Additionally, loops connecting the helices were removed manually. Phaser generated a single solution with acceptable Z scores and log-likelihood gain (RFZ = 5.3; TFZ = 11.3; LLG = 169). However, initial rounds of refinement including various combinations of rigid-body refinement with each of the two molecules in the asymmetric unit as one unit, simulated annealing, conjugate-gradient energy minimization and B-factor refinement using the CNS software package (Brünger et al., 1998 ▶) failed to reduce the R free and R factors below 55% and 50%, respectively.
We reasoned that given the relatively low sequence identity between DAP5M and the search model there might be slight differences in the positions and orientations of the individual helices within the DAP5M HEAT domain. Consequently, we broke up the search model into 20 separate rigid bodies, each corresponding to one helix of the two molecules in the asymmetric unit. Additionally, we trimmed all of the intervening loop regions. Using the rigid-body refinement option in REFMAC5 (Murshudov et al., 1997 ▶) with this strategy improved the R free and R factors to 50% and 47%, respectively. Inspection of the resulting structures revealed considerable displacements of the helices of up to approximately 3.0 Å. The N- and C-terminal helices shifted more significantly than the helices that were closer to the core of the molecules and the shifts were more pronounced in molecule A compared with molecule B. Subsequent simulated-annealing, energy-minimization and B-factor refinement in CNS further improved the R free and R factor to 44.4% and 38.0%, respectively. After density modification using DM (Cowtan & Main, 1993 ▶), including the application of twofold NCS averaging, the resulting maps showed reasonable electron density for the majority of the structure; however, considerable rebuilding and refinement will be required.
Molecular replacement using Phaser with data collected from crystal form B using the model produced from the data for crystal form A as described above generated one solution with lower Z scores and LLG values (RFZ = 4.4, TFZ = 7.3, LLG = 51) and an R factor of 59.9%. Given the solution of crystal form A and its higher diffraction limit, structure refinement will initially only be carried out with the data from crystal form A. The final model produced from this refinement will then be used for molecular replacement with the data from crystal form B.
To overcome potential model bias in the structure arising from the molecular-replacement technique, we have produced selenomethionine-labelled protein in order to determine experimental phases using the MAD/SAD technique at a synchrotron source (Hendrickson, 1991 ▶). DAP5M containing seven, six, five and four Se atoms was detected by mass-spectrometric analysis, with the largest fraction containing the maximum of seven Se atoms, which should provide an adequate signal for solution of the selenium substructure on a background of 261 residues (Fig. 2 ▶ b). Selenomethionine-labelled crystals of crystal from A were obtained by growth under the same conditions as the native crystals. Crystal form B has not been obtained with selenomethionine-labelled protein thus far. The crystal structure of DAP5M will be the first step towards determining the molecular basis of its regulation of the translation initiation complex and its role in the initiation of IRES-mediated translation.
Acknowledgments
This work was supported by a Career Development Award to BN from the Human Frontiers Science Program (CDA 0018/2006-C/1) and an operating grant from the Canadian Institutes of Health Research (CIHR grant MOP-82929). FF was supported by a Boehringer Ingelheim Fonds PhD Fellowship. We thank Yazan Abbas, Alexei Gorelik, Ahmad Kanaan and Marc Fabian for helpful discussions and Rose Szittner and Katalin Illes for technical assistance.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Cowtan, K. D. & Main, P. (1993). Acta Cryst. D49, 148–157. [DOI] [PubMed]
- De Gregorio, E., Preiss, T. & Hentze, M. W. (1998). RNA, 4, 828–836. [DOI] [PMC free article] [PubMed]
- De Gregorio, E., Preiss, T. & Hentze, M. W. (1999). EMBO J.18, 4865–4874. [DOI] [PMC free article] [PubMed]
- Derry, M. C., Yanagiya, A., Martineau, Y. & Sonenberg, N. (2006). Cold Spring Harb. Symp. Quant. Biol.71, 537–543. [DOI] [PubMed]
- Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D. & Bairoch, A. (2003). Nucleic Acids Res.31, 3784–3788. [DOI] [PMC free article] [PubMed]
- Hendrickson, W. A. (1991). Science, 254, 51–58. [DOI] [PubMed]
- Henis-Korenblit, S., Shani, G., Sines, T., Marash, L., Shohat, G. & Kimchi, A. (2002). Proc. Natl Acad. Sci. USA, 99, 5400–5405. [DOI] [PMC free article] [PubMed]
- Holcik, M. & Sonenberg, N. (2005). Nature Rev. Mol. Cell Biol.6, 318–327. [DOI] [PubMed]
- Hundsdoerfer, P., Thoma, C. & Hentze, M. W. (2005). Proc. Natl Acad. Sci. USA, 102, 13421–13426. [DOI] [PMC free article] [PubMed]
- Imataka, H., Olsen, H. S. & Sonenberg, N. (1997). EMBO J.16, 817–825. [DOI] [PMC free article] [PubMed]
- Levy-Strumpf, N., Deiss, L. P., Berissi, H. & Kimchi, A. (1997). Mol. Cell. Biol.17, 1615–1625. [DOI] [PMC free article] [PubMed]
- Lewis, S. M., Cerquozzi, S., Graber, T. E., Ungureanu, N. H., Andrews, M. & Holcik, M. (2008). Nucleic Acids Res.36, 168–178. [DOI] [PMC free article] [PubMed]
- Liberman, N., Dym, O., Unger, T., Albeck, S., Peleg, Y., Jacobovitch, Y., Branzburg, A., Eisenstein, M., Marash, L. & Kimchi, A. (2008). J. Mol. Biol.383, 539–548. [DOI] [PubMed]
- Lomakin, I. B., Hellen, C. U. & Pestova, T. V. (2000). Mol. Cell. Biol.20, 6019–6029. [DOI] [PMC free article] [PubMed]
- Marash, L., Liberman, N., Henis-Korenblit, S., Sivan, G., Reem, E., Elroy-Stein, O. & Kimchi, A. (2008). Mol. Cell, 30, 447–459. [DOI] [PubMed]
- Marcotrigiano, J., Gingras, A. C., Sonenberg, N. & Burley, S. K. (1999). Mol. Cell, 3, 707–716. [DOI] [PubMed]
- Marcotrigiano, J., Lomakin, I. B., Sonenberg, N., Pestova, T. V., Hellen, C. U. & Burley, S. K. (2001). Mol. Cell, 7, 193–203. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed]
- Rost, B., Yachdav, G. & Liu, J. (2004). Nucleic Acids Res.32, W321–W326. [DOI] [PMC free article] [PubMed]
- Schütz, P., Bumann, M., Oberholzer, A. E., Bieniossek, C., Trachsel, H., Altmann, M. & Baumann, U. (2008). Proc. Natl Acad. Sci. USA, 105, 9564–9569. [DOI] [PMC free article] [PubMed]
- Shaughnessy, J. D., Jenkins, N. A. & Copeland, N. G. (1997). Genomics, 39, 192–197. [DOI] [PubMed]
- Stein, N. (2008). J. Appl. Cryst.41, 641–643.
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed]
- Warnakulasuriyarachchi, D., Cerquozzi, S., Cheung, H. H. & Holcík, M. (2004). J. Biol. Chem.279, 17148–17157. [DOI] [PubMed]
- Wells, S. E., Hillner, P. E., Vale, R. D. & Sachs, A. B. (1998). Mol. Cell, 2, 135–140. [DOI] [PubMed]
- Yamanaka, S., Poksay, K. S., Arnold, K. S. & Innerarity, T. L. (1997). Genes Dev.11, 321–333. [DOI] [PubMed]