Abstract
Phagocytosis of invading microbes requires dynamic rearrangement of the plasma membrane and its associated cytoskeletal actin network. The polarization of Cdc42 and Rac1 Rho GTPases to the site of plasma membrane protrusion is responsible for the remodeling of actin structures. However, the mechanism of Rho GTPase recruitment to these sites and the identities of accessory molecules involved in this process are not well understood. In this study, we uncovered several new components involved in innate immunity in Drosophila melanogaster. Our data demonstrate that Rab35 is a regulator of vesicle transport required specifically for phagocytosis. Moreover, recruitment of Cdc42 and Rac1 to the sites of filopodium and lamellipodium formation is Rab35 dependent and occurs by way of microtubule tracks. These results implicate Rab35 as the immune cell-specific regulator of vesicle transport within the actin-remodeling complex.
The innate immune response is the first line of defense against invading microbes before activation of adaptive immunity in mammals (1, 13). In some eukaryotes, innate immunity triggers a diversity of humoral and cellular immune responses that provide sufficient protection against infection. In Drosophila melanogaster, microbial infection induces massive antimicrobial peptide synthesis via activation of the Toll and Imd pathways in the fat body and hemocytes (12, 14, 16). Moreover, hemocytic phagocytosis is activated to eliminate invading microbes. Several molecules have been reported to play important roles in recognizing invading pathogens and subsequently activating antimicrobial peptide synthesis (7, 10, 20). However, the mechanisms leading to activation of cellular immune responses are not fully understood. Therefore, significant effort has been focused on identifying the key regulatory components involved in these biological events (7, 17, 18, 37).
Activation of antimicrobial genes during infection is central to the innate immune response (9, 21). However, this response is also accompanied by signal-induced cell movement. Because an immune cell must migrate to the site of infection and engulf large particles via phagocytosis, participating cells undergo plasma membrane rearrangement along with actin-dependent cytoskeletal remodeling (26). Thus, tight regulation of peripheral actin filament assembly is necessary to generate proper cell protrusions. Several Rho GTPases have been suggested to play an important regulatory role in actin cytoskeleton rearrangement during immune cell migration (23). In particular, Cdc42 and Rac1 were shown to promote formation of actin-rich, finger-like membrane extensions (filopodia) and surface protrusions (lamellipodia), respectively. These Rho GTPases are recruited to the plasma membrane, where they contact foreign particles and induce actin assembly to generate the phagocytic cup (6). Cdc42 polarization is also an essential step during cell migration. Mammalian Scribble (Scrib) controls Cdc42 localization during astrocyte migration by interacting with the guanine nucleotide exchange factor βPIX (25). Because most studies on Cdc42 and Rac1 focused on their effects on downstream effectors, the mechanisms of their recruitment to the site of phagocytosis remain poorly understood.
In highly compartmentalized eukaryotic cells, Rab GTPases regulate vesicle trafficking between distinct cellular compartments (31, 36, 39). More than 60 mammalian Rab proteins have been identified. Each Rab GTPase regulates distinct steps in this process through temporal and spatial associations with various interacting proteins (27, 31). For example, Rab1 and Rab6 mediate endoplasmic reticulum (ER)-Golgi and intra-Golgi trafficking, respectively, while Rab8 is required for trafficking from the Golgi apparatus to the plasma membrane (31). Rab5, Rab11, and Rab35 regulate a distinct endocytic recycling step (31). Moreover, a recent study suggested a functional association between Rab35 and cell shape (5), suggesting that Rab35 may function as the regulator involved in cytoskeletal rearrangement.
Previously we identified genes required for the innate immune response against fungal infection (18). Among them, transcription factors involved in chromatin remodeling or lineage-specific differentiation, as well as genes involved in cytoskeletal remodeling, were revealed as major elements in this process. Studies of immune cell-specific transcription factors have identified the regulatory mechanisms governing the development of specific immune cell types and their expression of particular antimicrobial peptide genes. Despite this, the regulation of cell morphology and migration during the immune response remains unknown. To understand the mechanism regulating cytoskeletal rearrangement during phagocytosis and cell movement, we screened Drosophila mutants with the P element inserted for novel cytoskeletal regulators showing defects in defense against bacterial infection. In this study, we identified Rab35 as a specific regulator of the actin cytoskeleton in the plasma membrane and demonstrated its role in the formation of filopodia and lamellipodia. We show that Rab35 is required for Cdc42 and Rac1 localization at the activated plasma membrane through contact with foreign particles. Together, these results suggest that Rab35 plays a key regulatory role in phagocytosis of hemocytes by controlling actin rearrangement at the immune cell periphery.
MATERIALS AND METHODS
In vivo phagocytosis assay.
In vivo phagocytosis assays were carried out in flies as described previously (18, 22). Ten adult male flies (5 days old) were injected with fluorescein isothiocyanate (FITC)-conjugated Escherichia coli bioparticles (1 mg/ml, 50 nl) (Invitrogen) on the ventral side with a sharp capillary using a Picospritzer III injector (Parker Hannifin, Cleveland, OH). Injected flies were incubated for 1 h at 25°C to allow uptake of the injected fluorescent bioparticles by hemocytes. To quench the fluorescence signal of extracellular bioparticles, excess trypan blue solution (0.4%, 200 nl) (Sigma) was injected. Fluorescence was visualized using a Zeiss Axioplan 2 microscope (Carl Zeiss MicroImaging, Inc.) fitted with a digital camera (AxioCam) and the Axiovision 4.3 software (Carl Zeiss MicroImaging, Inc.). Fluorescence signals around the dorsal vessel were quantified using Image J software (NIH, Bethesda, MD). The phagocytic index was expressed as the area of the signal corresponding to the sum of the encircled areas.
In vitro phagocytosis assay.
In vitro phagocytosis assays were performed as described previously (28). SL2 cells (1 × 105) (CRL-1963; ATCC) were seeded in 12-well plates and incubated for 2 h at 25°C. The cells were chilled at 4°C for 30 min, and then 10 μg of FITC-labeled E. coli bioparticles (Invitrogen, Carlsbad, CA) in ice-cold Drosophila medium was added. After incubation for 30 min at 4°C to permit attachment between cells and FITC-E. coli particles, the cells were incubated for 15 min at 25°C to allow for E. coli particle uptake. After being washed with ice-cold phosphate-buffered saline (PBS), cells were analyzed by flow cytometry using CELLQuest software (BD Biosciences, San Jose, CA) to quantify the fluorescence signal. The fluorescence of extracellular particles was quenched by replacing the medium with 0.04% trypan blue (Sigma, St. Louis, MO) in 1× PBS immediately before measurement.
Real-time quantitative PCR.
Total RNA was extracted with Trizol (Invitrogen) and reverse transcribed using the Superscript II system (Invitrogen). The efficiency of knockdown of each Rab GTPase and the level of antimicrobial peptide mRNA were measured by real-time PCR using the LightCycler 480 (Roche, Basel, Switzerland). PCR was performed using a SYBR green mix (Applied Biosystems, Foster City, CA) and analyzed with LightCycler 480 software 4 (Roche). All results were normalized to the level of RpL32 mRNA in each sample. Primer sequences are listed in Tables 1 and 2.
TABLE 1.
Primer sequences used for formation of dsRNA
| Gene | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| Rab1 | CCACTGACAACGCTAGCAAA | CAAATCCCTTGTGTCACACG |
| Rab2 | GATTCGATTTCAGTGAGGTCC | ATGCCTCATTAAATTTGCCTT |
| Rab3 | TGTATATCCTTTGCGTAAGCTC | GCTGGTACTCTATGCGATACAT |
| Rab4 | CAGATATGGGACACAGCCG | GGGCTCTAGTGCTTCACGA |
| Rab5 | AAGAAATACCTAAAGCCGGCT | TGAGAACAACTGCAACTGGAA |
| Rab6 | GACAGCTCCATGTACTACGC | ACGAGGCATATTCCGGATGG |
| Rab7 | AACAATTTCAGCAAAACAGCA | CAAATTCACGCTCGAATGTT |
| Rab8 | TAAACATCAATCAGCAGCCG | GTGATATCTCGCAAATCTGCA |
| Rab9 | ATCGGACATCTGTCTGCTCT | ATCGGACATCTGTCTGCTCT |
| Rab10 | GTGGATCGGATAACGAGGAT | GTGCACCGTCCTTACATACAA |
| Rab11 | AAACAGATCAGAGATCCGCC | GTTGTTCTGCTCGCGAAAT |
| Rab14 | CATTGAGGTGGACGACAAAAA | CCTCCTGGATGTTCTGGTAAA |
| Rab18 | AAGCATTAAGCGGAGCTCAA | AAGTATTCGGAGGAGACGAT |
| Rab19 | TGGAGGGCAAACAAATCA | GCAAACTAAAGAGCGGATTT |
| Rab21 | ACCCCTTAATTGTAGGTTGTGT | TAAATGCCGCAACAGGAC |
| Rab23 | AGGCTTCCGTCCTGGTCTT | GCCCGAAAGATGGTGAAA |
| Rab26 | GACTATAACGATGGTTGGCGA | AGGACAGCTCCACATTGAGTC |
| Rab27 | GAATCGCAGATTCCTTCCAG | CCTAACAGTTGCGGCAGTT |
| Rab30 | CACGCAAAGCTACTATCGATC | GAAAGCGAAGAGAATTGCTC |
| Rab32 | TTCTAGTCATTGGTGAACTGGG | TGTTACGTTAAAACCCTAAGCA |
| Rab35 | TGAAATTAGGACCAGGACCTG | GCCAATAATTTGATCTCCCC |
| Rab39 | ATTTCGACTGATTTTAATCGGC | CGACGAGATTAAAGTCCAGACT |
| Rab40 | TGCAGGACAATTGTGCGA | GCTTCTCTTGCTGAATTCGC |
| RabX1 | GGTGGCGCCCATGTACTACA | CGGAGTTGGGTTGCTTGA |
| RabX2 | TGACAGCGGCGTGGGCAAAT | TGCTCAGATGGCATCGGGCT |
| RabX3 | ATCGGCGTGGGCAAATCCT | ACATTCGCGCCACTCTTGG |
| RabX4 | ACATCTCTACGATAGGCATTGA | TTAGCAGGTGCAGCGATT |
| RabX5 | GCACAGACTCCGCTGAGCAA | GGCAGCGATCCTTTGGAA |
| RabX6 | CTGGACCACATCGATAGGG | CCGAACTCAATGTTCCCC |
TABLE 2.
Primer sequences used for real-time PCR analyses
| Gene | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| RpL32 | CAGTCGGATCGATATGCTAAGCTGT | TAACCGATGTTGGGCATCAGATACT |
| Rab1 | CCACTGACAACGCTAGCAAA | AATTTAAATACGCACACGCA |
| Rab2 | CCAACACAATGCGGAATAA | CCGACATAGAAAGTGGAAATTG |
| Rab3 | TTGTCCTTATATTCGTGTGTGG | CGGGTATGCAGATGTGTGTT |
| Rab4 | TCCGTAACTTACAGACACGACA | TGTTCATCCTCTAGATCAGATCC |
| Rab5 | ACTAAACATGAGAAGAAAGCGC | TTGCAACTATCACACGTGTCC |
| Rab6 | GTGCATTTCCGATACCCC | GCCGCAAACAGAACAGAA |
| Rab7 | AGTCAAATTTTGCACGCAAC | CCAAAGAGCATGAGAATTCATAA |
| Rab8 | GCGTCCTTCTCGAAGAGGT | GTGTGCGGCAAAGAACAA |
| Rab9 | GATGTGGACCAGGACAAGTTT | CCGCATATATTCGATACGGTA |
| Rab10 | GAAAATCCGAGCTGCAGTG | CGGTAAATCATTTGGTGTGG |
| Rab11 | AACAACATCCGGCTGTAGTTA | GAAAATCCGAGCTGCAGTG |
| Rab14 | TTTTATCGCCCAAACAGTG | TTCATTTAGCACGAGCACTG |
| Rab18 | ACATCACCAGCAGGGACAG | AACACGTAGACGCGCTTG |
| Rab19 | CATAGAGCGACATGGGAACA | GGCTCGATTCGCTGATGT |
| Rab21 | GAAGGTTGTGTGGGCAAGA | CGCTCAGGAAGGGCAAGA |
| Rab23 | CCAACCGTTGGATGCAAC | AACTCCTATTCGGCTTACATG |
| Rab26 | CACAACGTGCCCTTCATG | CTTTCGAGTGCCCCACTT |
| Rab27 | TGCCGTACATCGAGACGA | AGCGAAACACACTTAGATTCG |
| Rab30 | TGACGTGATCGTTTGCCTT | CCAAAGATTGCACATTAATCG |
| Rab32 | TTCTAGTCATTGGTGAACTGGG | TGTTACGTTAAAACCCTAAGCA |
| Rab35 | TACAATAACAAGAGCGACCG | GCAGATCAGAGCTTTCGTTT |
| Rab39 | TCCAAGAGACAAATCCCTCG | ATCCATCCTCCGCCTTATAT |
| Rab40 | TTTACACAGTAAAGCATGCACC | TGCTTCTCTTGCTGAATTCG |
| RabX1 | CGAATATGGGCTCAAGGGCC | CAGCTGTCATTAAGAACTAA |
| RabX2 | TGTTTAAGGTTCTCGTCCTGG | GGCTGATCGGGTTGTGAA |
| RabX3 | TGCTCGTTTACGACATAACG | ACATTCGCGCCACTCTTGGC |
| RabX4 | TTAACCTCGATGGAGTACCCA | TCCCTTAACTGGTTTGCATC |
| RabX5 | AGGCCACTCAGGCATCTGTT | CTAGCAAGTACAGCCACTTT |
| RabX6 | TAATAGGAATTCCATCGATT | GGAGTATTACGTGCAATAAG |
| CecA2 | ATTAGATAGTCATCGTGGTT | GTGTTGGTCAGCACACT |
| Dpt | ATGCAGTTCACCATTGCCGTC | TCCAGCTCGGTTCTGAGTTG |
| Dfn | CGCTTTTGCTCTGCTTGCTTGC | TAGGTCGCATGTGGCTCGCTTC |
| Mtk | GCATCAATCAATTCCCGCCACC | CGGCCTCGTATCGAAAATGGG |
RNA interference (RNAi) and RNA analysis.
Double-stranded RNA (dsRNA) was prepared as described previously (19). SL2 cells (1×105) (CRL-1963, ATCC) were seeded in 12-well plates. After attachment, the cells were washed with serum-free Drosophila medium (Welgene, Daejeon, South Korea) and treated with dsRNAs (30 μg) in serum-free medium for 8 h. After this incubation, serum was added to a final concentration of 10% and incubated for an additional 72 h.
Drosophila stocks and P-element excision.
Strains of Drosophila melanogaster were cultured on a standard cornmeal-yeast medium at 25°C and 60% humidity. The EP lines were purchased from GenExel (Daejeon, South Korea). The Imd1, spzrm7, and RelE20 strains were kindly provided by Won-Jae Lee (Ewha Woman's University, Seoul, South Korea). W1118 was used as a wild-type (WT) stock, and P[ry+Δ2-3]sb/TM6B was used as a genomic transposase source. Rab35E17, the revertant for Rab35G11110, was generated through precise excision of the P element by crossing with P[ry+Δ2-3]sb/TM6B as described previously (29). The identity of the excision allele was confirmed by PCR and direct sequencing of the excision site.
Immunoblotting.
Whole-cell extracts of SL2 cells were prepared in lysis buffer containing 20 mM Tris (pH 7.6), 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM dithiothreitol (DTT), 2 mM EDTA, and protease inhibitors. For extracts prepared from whole Drosophila, 10 to 15 adult flies were homogenized with a small pestle prior to lysis. The extracts (30 μg) were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and then probed with anti-Rab35 (1:1,000) and anti-α-tubulin (1:1,000; Sigma) sera. The primary and secondary antibodies were diluted in blocking solution (5% skim milk in Tris-buffered saline-Tween 20 [TBST]) and in TBST (40 mM Tris [pH 7.4], 200 mM NaCl, 0.1% Tween 20), respectively. Proteins were detected using the ECL Plus detection system (GE Healthcare) after reaction with peroxidase-conjugated secondary antibody (1:5,000; Sigma).
Preparation of genomic DNA and PCR.
Approximately 10 to 15 adult flies were placed in a 1.5-ml centrifuge tube and frozen in liquid nitrogen for 5 min. The frozen flies were homogenized with a small pestle, and genomic DNA was isolated with a G-spin genomic DNA extraction kit (Intron, Gyeonggi-do, South Korea). The oligonucleotide primers used in PCR for confirmation of the revertant Rab35E17 were 5′-GCG AGG GAG TCG AGC TT-3′ (forward) and 5′-ATG TAA GTG TTG TTG CCG C-3′ (reverse). The standard thermal profile for PCR amplification was 30 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min.
Infection and survival experiments.
Beauveria bassiana from 3-day cultures (containing, per 1 liter of distilled water, 10 g dextrose, 2.5 g peptone, and 5 g yeast extract; 25°C) and Erwinia carotovora subsp. carotovora 15 from overnight cultures (containing, per 1 liter distilled water, 3 g beef extract and 5 g peptone [pH 6.8]; 30°C) were recovered by centrifugation at 6,000 rpm for 10 min at 25°C. Supernatants were discarded, and pellets were resuspended in phosphate-buffered saline consisting of 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4. Septic injury was performed by either pricking the leg discs of adult flies with a tungsten needle dipped previously in concentrated B. bassiana or E. carotovora subsp. carotovora 15 or by injecting diluted bacteria (optical density [OD] of 0.1, 55 nl) into the ventral lateral side with a thin needle using a Picospritzer III injector (Parker Hannifin) (19). Survival rates for flies after pathogen infection were measured with groups of 30 adults (15 males and 15 females) aged 2 to 4 days. Septically injured flies were kept at 25°C and transferred to a fresh vial every day. Surviving flies were counted every day after E. carotovora subsp. carotovora 15 injection or on days 3 and 6 after B. bassiana injection. These experiments were repeated at least three times.
Antibody generation.
To generate an anti-Rab35 antibody, full-length rab35 from Drosophila was cloned into the BamHI/EcoRI-digested pRSET-A vector (Invitrogen). Prior to use in rat immunizations, recombinant Drosophila Rab35 was expressed and isolated as a His-tagged fusion protein using an Ni-nitrilotriacetic acid (NTA) protein purification system (Qiagen, Chatsworth, CA). Antiserum specificity was confirmed by Western blot analysis of extracts prepared from wild-type or Rab35G11110 fly- or Rab35 dsRNA-treated cells.
Plasmid construction.
To visualize Rab35 movement, a plasmid expressing enhanced green fluorescent protein (EGFP)-tagged Rab35 was constructed. Full-length Drosophila Rab35 was amplified by PCR from SL2 cells. The EGFP fragment was also prepared by PCR from the pEGFP-N1 plasmid (Clontech Laboratories, Inc., Mountain View, CA). Primers flanking EGFP were designed to introduce a KpnI site at the 5′ end and a BamHI site at the 3′ end. For Rab35, primers were designed to introduce a BamHI site at the 5′ end and a SalI site at the 3′ end. Each PCR product was then digested with restriction enzymes and cloned into KpnI/SalI-digested pRmHa3, an SL2 cell expression vector, through three-fragment ligation. The same strategy was used to construct the red fluorescent protein (RFP)-tagged Cdc42 expression plasmid. For Rab4, Rab5, and Rab8-RFP, EcoRI, KpnI, and BamHI restriction enzymes were used. For the actin promoter-driven expression plasmid, the RFP-Cdc42 fragment was subcloned into BamHI-digested Ract-HAdh, an SL2 expression vector containing the actin 5C promoter Drosophila Genomics Resource Center (DGRC) (https://dgrc.cgb.indiana.edu). To generate the constitutively active (CA) and dominant negative (DN) Rab35 mutant alleles, site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Primers used in the mutagenesis were as follows: CA forward, CTG GGA CAC GGC CGG GCT GGA GCG CTT CCG GAC; CA reverse, GTC CGG AAG CGC TCC AGC CCG GCC GTG TCC CAG; DN forward, GCG GCG TGG GCA AGA ACT CGC TGC TCA TCC GG; and DN reverse, CCG GAT GAG CAG CGA GTT CTT GCC CAC GCC GC.
Transfection and establishment of stable cell lines.
SL2 cells were maintained at 25°C in Schneider's medium (Welgene Inc., South Korea) containing 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen) plus 50 units/ml penicillin (Invitrogen). For establishment of stable cell lines, SL2 cells were cotransfected with the pCoHYGRO selection plasmid (Invitrogen) and pRmHa3-GFP-Rab35 WT, CA, or DN using Cellfectin (Invitrogen). Stable cell lines were obtained by hygromycin selection. For EGFP-Rab35 allelic expression, cells were treated with 500 μM CuSO4 for 8 h.
Imaging.
To stain filamentous actin, SL2 cells were rinsed in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) and then fixed for 20 min with 4% paraformaldehyde (Sigma) in PBS. The cells were then permeabilized with 0.3% Triton X-100 and 5% goat serum in PBS and stained with Alexa Fluor 568-conjugated or Alexa Fluor 647-conjugated phalloidin (1:200) (Molecular Probes, Invitrogen) and 0.5 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) (Sigma) in PBS. Fluorescent images from preparations mounted in Vectashield (Vector Laboratories) were generated using a confocal microscope (LSM510; Carl Zeiss MicroImaging, Inc.) equipped with 40×/1.4 and 100×/1.4 numerical aperture (NA) Plan-Apo objectives. For lamellocyte staining, hemocytes were prepared on polylysine-precoated glass coverslips (MatTek Corporation, Ashland, MA). Attached hemocytes were washed with PBS prior to permeabilization with 0.3% Triton X-100 in PBS and incubation with PBS containing 1% bovine serum albumin (BSA) for 20 min. The hemocytes were incubated overnight at 4°C with lamellocyte-specific L1 antibody diluted (1:30) in PBS containing 1% BSA. On the following day, the cells were washed, incubated with FITC-conjugated anti-mouse IgG antibody (Sigma, 1:250) at room temperature for 1 h, and then enclosed in Vectashield (Vector Laboratories). Lamellocyte-specific L1 antibody was kindly provided by Istvan Ando (Biological Research Center of the Hungarian Academy of Sciences, Szeged).
Live imaging.
To observe highly active actin filament formation, SL2 cells were plated onto concanavalin A (ConA)-treated glass-bottom microwell dishes (MatTek Corporation). For visualization of Cdc42-RFP and Rab35-GFP comigration, time lapse images were acquired every 30 s for up to 30 min. The same conditions were used in the Rab4-, Rab5-, or Rab8-RFP and Rab35-GFP experiments. To observe vesicular movement, SL2 cells were treated with Rab35 dsRNA and then with metal promoter-driven Rab35 CA-GFP and actin promoter-driven RFP-Cdc42 expression plasmids 72 h later. After the RFP signal was confirmed, SL2 cells were treated with CuSO4 to induce Rab35 CA-GFP expression and then tracked for 12 h, with images collected every 15 min. To observe Candida albicans phagocytosis by SL2 cells, time lapse images were acquired every 2.5 min for up to 90 min. C. albicans from 2-day cultures (containing, per 1 liter distilled water, 10 g Bacto yeast extract, 20 g Bacto peptone, 20 g glucose monohydrate, 40 mg adenine hemisulfate, and yeast-peptone-dextrose-adenine medium; 25°C) was recovered by centrifuging at 6,000 rpm for 10 min at 25°C. Supernatants were discarded, and pellets were resuspended in PBS. To isolate a single C. albicans cell, resuspended cells were sonicated briefly. All live images were taken and processed using a CoolSnap HQ-cooled charge-coupled device (CCD) camera on a DeltaVision Spectris Restoration microscope built around an Olympus IX70 stand with a 40×/0.75-numerical-aperture lens (AppliedPrecision).
Scanning electron microscopy (SEM).
Rab35 CA, WT, or DN-transfected SL2 cells were fixed in 2% glutaraldehyde in 25 mM phosphate buffer (pH 7.2) for 2 h at 4°C. The tissues were dehydrated in a graded ethanol series, sequentially dried with liquid CO2 to their critical point, and coated with gold in a sputter coater (Baltec, SCD 005). The specimens were observed with a LEO 1455VP ESEM microscope at 20 keV.
RESULTS
Rab35 mutant flies display an increased susceptibility to infection.
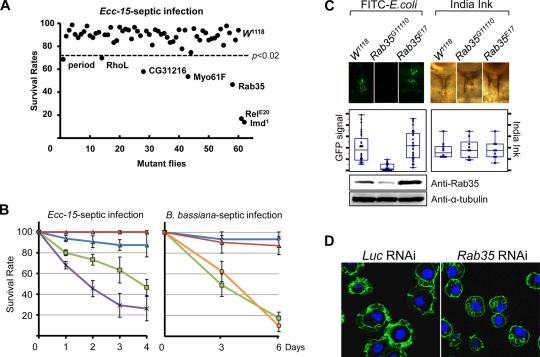
To identify EP lines with compromised defense against infection, 30 adult flies from each of the 61 lines were pricked on the leg disc with a needle covered with a concentrated solution of live pathogenic E. carotovora subsp. carotovora 15 bacteria. The EP lines were obtained from GenExel, and their P-element insertion sites were confirmed as described previously (18). Septic infection with E. carotovora subsp. carotovora 15 caused no major harm to wild-type flies; most survived the infection. Most mutant flies showed no defect in defense against the bacterial infection. However, the Imd1 and RelE20 mutants were severely affected by the E. carotovora subsp. carotovora 15 infection, as only 14 to 17% survived. In addition, five mutants, i.e., periodG10561, RhoLG15295, CG31216G18713, Myo61FG4411, and Rab35G11110, exhibited significantly higher lethality than wild-type flies after infection (Fig. 1A; Table 3). It is intriguing that mutation of genes involved in cytoskeletal remodeling correlated with defects in defense against bacterial infection. Among them, Rab GTPase mutation caused the most severe defect following infection. This result suggests a new link between the innate immune response and vesicle transport regulation. Thus, we examined further the immune defect caused by Rab35 mutation.
FIG. 1.
Rab35 mutant flies show an increased susceptibility to infection. (A) Survival rates of 59 mutant flies at 4 days after E. carotovora subsp. carotovora 15 (Ecc-15) septic infection. Of the 59 lines, 5 lines containing mutations in Rab35 showed reduced resistance to E. carotovora subsp. carotovora 15 infection (P < 0.02). (B) Kinetics of survival rates from three independent experiments. The Imd1 and Spzrm7 mutants were used as lethal controls for E. carotovora subsp. carotovora 15 and B. bassiana infection, respectively. The P value for differences between W1118 and Rab35G11110 or against E. carotovora subsp. carotovora 15 was less than 0.02, and that against B. bassiana was less than 0.002. Error bars indicate standard deviations. Blue lines, W1118; red lines, Rab35E17; green lines, Rab35G11110; purple line, Imd1; orange line, Spzrm7. (C) India ink image of total hemocytes and fluorescent image of hemocyte clusters on the abdominal dorsal surface of a fly that internalized FITC-labeled E. coli extract. The box plot shows the amounts of phagocytosis and total hemocytes, which were calculated by multiplying the area and intensity of the fluorescent signal. In vivo phagocytosis assays were carried out with more than 30 adult flies. The P value for differences of fluorescent signals observed between W1118 and Rab35G11110 was below 0.001. Immunoblotting was performed to depict the expression level of Rab35 protein in Rab35G11110, Rab35E17, and W1118 flies. W1118 was used as a wild-type control. (D) Comparison between the actin structures from Luc RNAi- and Rab35 RNAi-treated cells. Actin structures were stained with Alexa Fluor 488-conjugated phalloidin.
TABLE 3.
P-element insertion mutant lines used and their survival rate after septic infection of E. carotovora subsp. carotovora 15
| Strain or mutant no. | Symbol | GenExel no. | Survival rate (%) on day: |
||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |||
| W1118 | Wild type | 100.00 | 98.47 | 96.71 | 94.86 | 93.99 | |
| Imd1 | 100.00 | 43.06 | 28.89 | 22.59 | 16.94 | ||
| RelE20 | 100.00 | 91.17 | 50.04 | 17.14 | 13.90 | ||
| 1 | Perioda | G10561 | 100.00 | 95.99 | 95.99 | 77.66 | 68.65 |
| 2 | Fimbrin (CG8649) | G10929 | 100.00 | 98.33 | 96.61 | 90.50 | 88.83 |
| 3 | CG3881 | G11876 | 100.00 | 97.33 | 96.67 | 94.67 | 94.00 |
| 4 | CycE | G12107 | 100.00 | 98.72 | 98.72 | 98.72 | 98.72 |
| 5 | Cdk4 | G12455 | 100.00 | 95.71 | 94.87 | 92.24 | 90.45 |
| 6 | CG6860 | G12512 | 100.00 | 95.83 | 90.83 | 88.33 | 87.50 |
| 7 | l(3)05822 | G13184 | 100.00 | 96.67 | 95.00 | 92.20 | 90.53 |
| 8 | G-ialpha65A | G13928 | 100.00 | 97.37 | 96.87 | 95.05 | 92.89 |
| 9 | AnnIX | G13932 | 100.00 | 97.70 | 97.70 | 97.70 | 97.70 |
| 10 | Puromycin-sensitive aminopeptidase | G13991 | 100.00 | 94.00 | 91.56 | 91.56 | 89.33 |
| 11 | Pak3 | G14159 | 100.00 | 96.11 | 92.70 | 90.95 | 87.58 |
| 12 | Non-claret disjunctional | G14231 | 100.00 | 98.64 | 94.62 | 93.29 | 91.26 |
| 13 | Mo25 | G14716 | 100.00 | 96.67 | 95.56 | 93.33 | 92.22 |
| 14 | RhoLa | G15295 | 100.00 | 93.83 | 91.84 | 81.20 | 69.75 |
| 15 | Ack | G16320 | 100.00 | 95.00 | 92.50 | 90.83 | 90.00 |
| 16 | EndoA | G16603 | 100.00 | 94.44 | 93.33 | 91.11 | 90.00 |
| 17 | NetB | G1622 | 100.00 | 99.00 | 97.33 | 97.33 | 96.50 |
| 18 | String | G16904 | 100.00 | 92.18 | 92.18 | 87.70 | 83.26 |
| 19 | CG12065 | G17117 | 100.00 | 98.33 | 95.34 | 91.12 | 84.98 |
| 20 | Inx2 | G17438 | 100.00 | 97.78 | 97.78 | 96.67 | 96.67 |
| 21 | CG9086 | G17767 | 100.00 | 97.78 | 95.55 | 93.33 | 93.33 |
| 22 | CG2803 | G18036 | 100.00 | 100.00 | 96.67 | 95.00 | 95.00 |
| 23 | CG10916 | G18115 | 100.00 | 99.17 | 96.67 | 95.83 | 94.17 |
| 24 | CG30015 | G18234 | 100.00 | 98.28 | 96.61 | 88.99 | 86.41 |
| 25 | CG10973 | G18338 | 100.00 | 98.00 | 97.31 | 96.62 | 96.62 |
| 26 | CG4080 | G18431 | 100.00 | 96.67 | 94.37 | 94.37 | 93.22 |
| 27 | Trxr-1 | G1853 | 100.00 | 95.56 | 92.96 | 89.26 | 89.26 |
| 28 | CG31216a | G18713 | 100.00 | 87.04 | 85.50 | 80.70 | 57.93 |
| 29 | CG8588 | G20048 | 100.00 | 97.78 | 96.67 | 94.44 | 93.33 |
| 30 | Dap160 | G2320 | 100.00 | 97.19 | 95.11 | 91.63 | 90.30 |
| 31 | Betaggt-II | G2472 | 100.00 | 94.45 | 90.00 | 85.56 | 81.11 |
| 32 | Wun | G2520 | 100.00 | 100.00 | 100.00 | 98.89 | 94.45 |
| 33 | CG11242 | G2655 | 100.00 | 99.33 | 96.55 | 96.55 | 93.14 |
| 34 | Dcp-1 (CG5370) | G2729 | 100.00 | 96.00 | 93.73 | 92.40 | 90.40 |
| 35 | CG10376 | G2887 | 100.00 | 98.81 | 96.35 | 95.12 | 91.32 |
| 36 | CG8445 | G3196 | 100.00 | 98.00 | 95.31 | 91.20 | 87.86 |
| 37 | Cysteine proteinase 1 | G3277 | 100.00 | 97.74 | 95.56 | 83.33 | 83.33 |
| 38 | CG17019 | G3302 | 100.00 | 94.14 | 90.80 | 86.58 | 84.05 |
| 39 | CG7816 | G3896 | 100.00 | 97.78 | 94.48 | 92.71 | 87.86 |
| 40 | Syx1A | G4126 | 100.00 | 98.89 | 98.89 | 97.78 | 94.44 |
| 41 | CG31547 | G4171 | 100.00 | 91.38 | 87.16 | 82.10 | 77.82 |
| 42 | CG17723 | G4288 | 100.00 | 97.78 | 95.38 | 95.38 | 94.10 |
| 43 | Myo61Fa | G4411 | 100.00 | 75.15 | 67.24 | 57.85 | 53.59 |
| 44 | CG9796 | G4657 | 100.00 | 92.84 | 82.73 | 74.66 | 74.66 |
| 45 | Best1 (CG6264) | G4725 | 100.00 | 97.79 | 97.79 | 95.39 | 95.39 |
| 46 | Darkener of apricot | G4792 | 100.00 | 92.59 | 90.00 | 88.59 | 88.59 |
| 47 | PGRP-LF | G4841 | 100.00 | 93.74 | 91.64 | 90.34 | 85.32 |
| 48 | CG9139 | G5077 | 100.00 | 97.34 | 97.34 | 93.60 | 93.60 |
| 49 | Syx13 | G5399 | 100.00 | 98.33 | 94.17 | 93.33 | 91.64 |
| 50 | Calpain-B | G5405 | 100.00 | 95.01 | 91.13 | 89.45 | 85.45 |
| 51 | CG16718 | G5419 | 100.00 | 90.83 | 90.83 | 87.50 | 86.67 |
| 52 | CG15234 | G5655 | 100.00 | 97.82 | 96.90 | 90.62 | 87.16 |
| 53 | a | G7875 | 100.00 | 97.61 | 94.27 | 90.60 | 89.49 |
| 54 | Ptp4E | G773 | 100.00 | 100.00 | 97.50 | 96.67 | 95.00 |
| 55 | Sktl | G8067 | 100.00 | 92.11 | 89.85 | 85.33 | 80.88 |
| 56 | Cher | G9093 | 100.00 | 98.48 | 98.15 | 97.78 | 97.78 |
| 57 | CG6454 | G9545 | 100.00 | 100.00 | 97.37 | 91.72 | 91.72 |
| 58 | Rab35 | G11110 | 100.00 | 80.00 | 73.33 | 63.33 | 46.67 |
| 59 | CG12004 | G4815 | 100.00 | 96.90 | 90.06 | 88.15 | 85.54 |
Mutants exhibited significantly higher lethality after infection compared to wild type flies. (P < 0.02).
To confirm that this mutation results in a higher susceptibility to pathogen infection, we generated revertants in which the P element was excised precisely and compared their survival to that of the Rab35G11110 mutant after bacterial or fungal infection. As positive and negative controls for infection, Imd1 or Spzrm7 mutants and wild-type flies were also examined. Rab35G11110 mutants showed dramatically reduced resistance to B. bassiana and E. carotovora subsp. carotovora 15 infection, with survival rates of only 17.5% and 46.7%, respectively, which were comparable to those for Imd1 and Spzrm7 mutant flies (Fig. 1B). However, the revertants recovered their immune activity completely, showing a survival rate similar to that of wild-type flies. The higher lethality after infection resulted from loss of Rab35 protein. Homozygous mutants showed a significant reduction in Rab35 protein level compared to wild-type or P-element-excised flies. Despite this reduced level of Rab35, homozygote mutant flies were viable and showed no apparent developmental defect other than an increased susceptibility to infection. Therefore, the residual Rab35 activity supported development but did not support immune defense against infection.
Rab35 mutant flies show defects in phagocytosis in vivo.
It is not surprising that regulation of vesicular transport is required for cellular immunity, considering that cell movement is associated with phagocytosis and secretion of large amounts of antimicrobial peptides. To determine the defective immune responses responsible for reduced survival of the mutants, we examined the humoral and cellular immune responses in these flies after bacterial infection. Assessment of antimicrobial peptide mRNA levels revealed that the RelE20 mutant showed a severe defect in activation of most antimicrobial peptide genes tested (Fig. 2D). However, there was no apparent defect observed in the mutant flies. The apparent lack of antimicrobial peptide synthesis prompted us to examine the phagocytic activity of the mutant. Wild-type and Rab35 mutant adult male flies were injected with FITC-labeled E. coli, and the quantity of bacteria phagocytosed by fly hemocytes was measured after quenching excess fluorescent signal with trypan blue. A strong fluorescence signal from E. coli phagocytosed by hemocytes was visible in wild-type flies along the dorsal vessel of the abdomen. However, Rab35 mutant flies showed very weak signals in this in vivo phagocytosis assay (Fig. 1C). The numbers of hemocytes determined by India ink staining were comparable for the Rab35 mutant and wild-type flies, indicating that the reduced phagocytosis in the mutant resulted from defective hemocyte activity rather than a decrease in hemocyte number. This defect likely caused the increased lethality observed after bacterial infection in Rab35 mutant flies.
FIG. 2.
Rab35 is required for phagocytosis in Drosophila SL2 cells. (A) Percentage of cells that internalized FITC-labeled E. coli. RNAi was used to individually knock down 29 Rab GTPases (indicated on the horizontal axis). RNAi-treated cells were incubated with an extract of FITC-labeled E. coli for 30 min at 4°C and then incubated for 15 min at 25°C to allow internalization. To remove the fluorescence signal from the external bacterial extract, the cells were treated with 0.04% trypan blue immediately before fluorescence-activated cell sorter (FACS) analysis. As controls for genes involved in phagocytosis, Sr-C1 was tested along with Luc. The assay was performed three independent times. *, P < 0.01. Error bars indicate standard deviations. (B) Confirmation of RNAi-mediated Rab knockdown by PCR. The mRNA level of each Rab is shown relative to that from Luc RNAi-treated cells. (C) Confirmation of RNAi-mediated Rab35 knockdown by immunoblotting. Alpha-tubulin was assessed as a loading control. (D) Wild-type and mutant flies were injected with E. carotovora subsp. carotovora 15, and then total RNA was isolated after 6 h and analyzed by quantitative real-time PCR. Mtk, Metchnikowin; Dfn, Defensin; Dpt, Diptericin; CecA2, Cecropin A2.
Rab35 is required for phagocytosis in Drosophila SL2 cells.
To examine the specificity of Rab35 in phagocytosis, RNAi was used to eliminate individual Drosophila Rab GTPases in SL2 cells. The effect of this knockdown on phagocytosis was then examined. Once again, phagocytic activity was measured by monitoring the amount of fluorescent E. coli taken up by the cells (Fig. 2A; Table 4). As positive and negative controls, Scavenger Receptor class C type 1 (Sr-C1) and Luciferase (Luc) were tested for their effect on phagocytosis upon depletion. Quantitative reverse transcription-PCR (RT-PCR) was performed to demonstrate that double-stranded RNA treatment led to a severe reduction in the corresponding Rab RNA level (Fig. 2B; Table 5). However, with the exception of Rab35, knockdown of the Rab GTPases tested showed a negligible effect on phagocytic activity. Consistent with the in vivo results, Rab35 depletion decreased phagocytic activity significantly, to the level in Sr-C1 knockdown cells. Therefore, Rab35 appears to be the major regulator of vesicle transport specifically required for phagocytosis.
TABLE 4.
In vitro phagocytosis assay
| dsRNA | % of cells with internalized FITC-labeled E. coli extract |
|
|---|---|---|
| Mean | SD | |
| Luc | 53.02 | 5.42 |
| Sr-C1 | 27.07 | 1.74 |
| Rab1 | 45.75 | 2.82 |
| Rab2 | 47.75 | 3.56 |
| Rab3 | 55.35 | 5.91 |
| Rab4 | 52.92 | 7.13 |
| Rab5 | 50.26 | 5.44 |
| Rab6 | 43.57 | 4.05 |
| Rab7 | 45.91 | 12.26 |
| Rab8 | 43.79 | 2.92 |
| Rab9 | 45.72 | 4.16 |
| Rab10 | 49.36 | 4.80 |
| Rab11 | 54.13 | 5.98 |
| Rab14 | 43.47 | 4.25 |
| Rab18 | 55.89 | 7.97 |
| Rab19 | 52.53 | 5.25 |
| Rab21 | 46.78 | 4.57 |
| Rab23 | 49.23 | 3.32 |
| Rab26 | 47.01 | 2.65 |
| Rab27 | 51.08 | 3.98 |
| Rab30 | 48.67 | 3.51 |
| Rab32 | 55.63 | 7.02 |
| Rab35 | 31.63 | 3.38 |
| Rab39 | 55.20 | 8.44 |
| Rab40 | 45.04 | 3.47 |
| RabX1 | 46.44 | 3.58 |
| RabX2 | 54.75 | 13.48 |
| RabX3 | 52.62 | 4.76 |
| RabX4 | 51.00 | 6.20 |
| RabX5 | 42.82 | 6.74 |
| RabX6 | 44.73 | 6.02 |
TABLE 5.
Relative expression level after depletion of each Rab by RNAi
| dsRNA | % of expressed transcript relative to Luc RNAi as a negative control |
|
|---|---|---|
| Mean | SD | |
| Sr-C1 | 5.49 | 2.45 |
| Rab1 | 17.50 | 2.23 |
| Rab2 | 11.20 | 5.11 |
| Rab3 | 14.32 | 1.20 |
| Rab4 | 7.54 | 1.76 |
| Rab5 | 8.60 | 2.45 |
| Rab6 | 15.55 | 5.78 |
| Rab7 | 24.88 | 7.10 |
| Rab8 | 16.55 | 3.11 |
| Rab9 | 12.36 | 2.56 |
| Rab10 | 16.88 | 2.00 |
| Rab11 | 8.99 | 3.85 |
| Rab14 | 14.70 | 1.75 |
| Rab18 | 7.77 | 3.12 |
| Rab19 | 11.18 | 2.10 |
| Rab21 | 18.22 | 3.13 |
| Rab23 | 8.55 | 4.45 |
| Rab26 | 6.44 | 2.64 |
| Rab27 | 6.58 | 2.79 |
| Rab30 | 8.44 | 4.15 |
| Rab32 | 4.69 | 2.45 |
| Rab39 | 14.22 | 6.54 |
| Rab40 | 13.54 | 3.33 |
| RabX1 | 17.41 | 2.78 |
| RabX2 | 12.11 | 4.78 |
| RabX3 | 13.22 | 3.55 |
| RabX4 | 16.55 | 3.54 |
| RabX5 | 7.45 | 2.54 |
| RabX6 | 8.55 | 3.47 |
Rab35 is required for plasma membrane protrusion.
Phagocytosis requires proper regulation of cell protrusions such as lamellipodia and filopodia. To investigate how Rab35 affects phagocytosis in SL2 cells, we searched for structural abnormalities in cells treated with RNAi against Rab35. Phalloidin staining revealed large lamellipodia at the periphery of control cells (Luc RNAi) along with strong actin structures (Fig. 1D). However, RNAi-mediated depletion of Rab35 significantly reduced the formation of ruffles at the periphery. Instead, several large internal vesicles surrounded by actin filaments were generated, indicating that Rab35 is involved in plasma membrane rearrangement.
Rab35 shows a distinct localization pattern at the plasma membrane.
Plasma membrane rearrangement accompanying the synthesis of cell protrusions requires a diversity of Rab GTPases. Membrane and cytoskeletal components are transported to sites of new membrane protrusions, causing existing plasma membrane to be recycled through endocytic pathways. A distinct Rab GTPase regulates each of these transport processes, and its localization pattern usually reflects the specific function. To elucidate the function of Rab35, we compared its localization to those of other Rab GTPases (i.e., Rab1, Rab4, Rab5, Rab7, and Rab8) known for their involvement in a specific membrane vesicle transport process. GFP-tagged Rab35 was highly enriched at the plasma membrane, while protein in the cytoplasm appeared as punctuate areas (Fig. 3A). Unlike Rab35, the other Rab GTPases tested were not detected at the plasma membrane but were present mainly in the cytoplasm. Some of these cytoplasmic Rab GTPases appeared to colocalize with Rab35-GFP. In particular, Rab8, which is known to function in vesicle transport from the Golgi to the plasma membrane, appeared to colocalize with Rab35 in most cytoplasmic areas.
FIG. 3.
Rab35 shows a distinct localization pattern at the plasma membrane. (A) Rab35 colocalized with some Rab GTPases without stimulation of actin polymerization. Rab35-GFP colocalized with RFP labeled Rab1, Rab8, and Rab5 in the cytoplasm. (B) Rab35 showed a functionally distinct localization pattern on ConA-coated plates. The snap image was visualized every 30 s using a Delta Vision live imaging system. Unlike Rab35-GFP, RFP-labeled Rab4, Rab5, and Rab8 did not migrate to the plasma membrane with ConA treatment. The white bar represents 10 μm.
To test whether this colocalization is functionally relevant, we used live cell imaging to examine localization following concanavalin A (ConA) treatment, which induces dynamic plasma membrane protrusions. As reported previously, a strong ConA signal induces actin polymerization, leading to a significant increase in the number of filopodia and lamellipodia (Fig. 3B) (30). SL2 cells cultured on ConA-coated plates produce large, flat membrane protrusions at the periphery within 15 min after contact, as shown in Fig. 3B. Intriguingly, Rab35 migrated from cytoplasmic loci to the plasma membrane and gathered at the plasma membrane of newly developed cell protrusions. Unlike Rab35-GFP, RFP-labeled Rab4, Rab5, and Rab8 did not migrate to the plasma membrane with ConA treatment. These results suggest that Rab35 may be involved in vesicular transport from the cytoplasm to the plasma membrane during membrane protrusion induced by external stimuli.
Rab35 is required for cytoskeletal remodeling at the plasma membrane.
To study the role of Rab35 in plasma membrane organization, we examined the effect of overexpression of wild-type (WT), constitutively active (CA), and dominant negative (DN) Rab35 on cell morphology. Scanning electron microscopy analysis revealed that overexpression of CA Rab35 greatly induced filopodium and lamellipodium formation in most cells (Fig. 4A). In contrast, DN Rab35 caused a complete loss of filopodia and lamellipodia. The membranes of WT Rab35-transfected cells appeared to have many membrane ruffles and fine filopodia, indicating elevated actin polymerization activity at the plasma membrane. Therefore, Rab35 may act as a positive regulator of filopodium and lamellipodium formation from the plasma membrane.
FIG. 4.
Rab35 is required for cytoskeletal remodeling at the plasma membrane. (A) Morphology of Rab35 mutant cells visualized by SEM. Cells transfected with WT, CA, or DN Rab35 were visualized. The proportion of cells with filopodia is shown at the bottom. Error bars indicate standard deviations. (B) Changes in the morphology of Rab35 mutant cells resulting from abnormal actin structure. Cells transfected with GFP-tagged WT, CA, or DN Rab35 underwent phalloidin staining prior to image capture.
We further investigated the localization of these overexpressed proteins with actin structures within the cell. To accomplish this, WT, CA, and DN Rab35 were tagged with GFP to enable visualization. Overexpression of GFP-tagged CA Rab35 (GFP-Rab35CA) resulted in strong actin bundle development along the plasma membranes of the filopodia and lamellipodia (Fig. 4B). GFP-Rab35CA proteins were detectable throughout the cytoplasm but highly enriched on the extended regions of the plasma membrane, showing a significant overlap with the actin staining pattern. Reflecting the fine filopodia on the cell surface, GFP-tagged wild-type Rab35 (GFP-Rab35) localized to peripheral cell protrusions where strong actin staining was detected. Patches of GFP-Rab35 proteins were also present in the cytoplasm, but no obvious actin colocalization was observed. However, the overexpression of GFP-tagged DN Rab35 (GFP-Rab35DN) led to a complete loss of filopodium formation. The cells contained a large number of cytoplasmic vesicles surrounded by actin polymers, recapitulating the phenotype seen with RNAi-mediated Rab35 knockdown. Overall, the development of actin filaments was not affected by Rab35 activity. Instead, the localization of polymerized actin filaments and their association with Rab35 to form organized membrane structures such as filopodia and lamellipodia were affected dramatically by the mutations. Rab GTPases are mainly involved in vesicle transport and abnormalities of cell morphology. Therefore, our data suggest that Rab35 may be involved in transporting membrane-organizing materials between the plasma membrane and cytoplasmic vesicles rather than in actin polymerization itself.
Rab35 colocalizes with actin structure-remodeling Rho GTPases.
Previous reports have shown that Cdc42, a key regulator of actin polymerization, migrates to the plasma membrane during phagocytosis and cell migration (3). To determine whether Cdc42 is also involved in filopodium formation induced by hyperactive Rab35, we examined the localization of RFP-tagged Cdc42. Cdc42-RFP protein was localized at the plasma membrane and several actin bundles in the cytoplasm. Transfection of Cdc42-RFP and Rab35WT-GFP into SL2 cells revealed that the proteins colocalized. This colocalization pattern was maintained even under the severe morphological changes induced by the expression of the constitutively active or the dominant negative form of Rab35. The overexpression of Rab35WT-GFP did not cause a significant change in the location of Cdc42, indicating that Rba35 and Cdc42 may interact at the actin-remodeling loci (Fig. 5A). This colocalization was not limited to Cdc42. Rac1, the Rho GTPase involved in lamellipodium formation, also colocalized with Rab35 (Fig. 5B). Therefore, Rab35 is a good candidate as a general vesicle transport regulator of filopodium and lamellipodium formation.
FIG. 5.
Rab35 colocalizes with actin structure-remodeling Rho GTPases. Colocalization of Rab35 was demonstrated with Cdc42 (A) and Rac1 (B). Filamentous actin (F-actin) was visualized with Alexa Fluor 688-conjugated phalloidin staining using confocal microscopy.
To verify Cdc42 and Rab35 colocalization during the dynamic process of cytoskeletal remodeling, live imaging of cells expressing both Rab35-GFP and Cdc42-RFP was performed following ConA treatment. During ConA induction of large membrane protrusions, Rab35 colocalized and comigrated with Cdc42, especially at the plasma membrane ruffles of newly formed lamellipodia (Fig. 6A). This result demonstrates that under conditions of enhanced actin polymerization, these molecules comigrate to the plasma membrane. Intriguingly, during phagocytosis of Candida albicans, Cdc42 and Rab35 localization was linked to new membrane protrusion development (Fig. 6B). Once SL2 cells became attached to C. albicans, the cells formed actin-rich protrusions such as filopodia toward the pathogen. Cdc42 and Rab35 colocalized strongly at the periphery of the newly formed protrusions. Therefore, Rab35 may participate in actin structure remodeling at the tips of membrane protrusions by transferring Cdc42 and Rac1 to the plasma membrane, where these events occur.
FIG. 6.
Rab35 colocalizes with Rho GTPases during actin cytoskeleton remodeling. (A) Rab35 colocalized and comigrated with Cdc42 following ConA treatment. Cells transfected with Rab35-GFP and Cdc42-RFP were imaged immediately after plating onto ConA-treated dishes. The snap image was visualized using a Delta Vision live imaging system. The arrowheads indicate a representative colocalization point in newly forming lamellipodium ruffles. (B) RFP-Cdc42 and GFP-Rab35 were recruited to phagocytic protrusions during phagocytosis of Candida albicans.
Localization of Cdc42 is dependent on Rab35 activity.
Next we proceeded to determine whether Rab35 is required for Rho GTPase activity or only for its transport to the plasma membrane. Therefore, the effect of Rab35 mutation on the distribution of constitutively active Rho GTPases was examined (Fig. 7A). RFP-tagged constitutively active Cdc42 or Rac1 mutants were cotransfected with GFP-fused WT, CA, or DN Rab35. Phalloidin staining showed increased filamentous actin caused by expression of constitutively active Cdc42 or Rac1 with Rab35. However, neither filopodia nor lamellipodia were observed in cells cotransfected with DN Rab35 and either constitutively active Cdc42 or Rac1. Although the filopodium actin structures disappeared from the cell surface, internal vesicles were surrounded by high-actin polymers that likely resulted from enhanced Cdc42 or Rac1 localization. These data indicate that inappropriate Cdc42 and Rac1 localization prevents filopodium and lamellipodium formation despite strong actin polymerization regulator activities. Moreover, Rab35 may be a key determinant in Cdc42 and Rac1 localization.
FIG. 7.
Localization of Cdc42 is dependent on Rab35 activity and microtubule tracks. (A) Changes in morphology and distribution of Cdc42 in cells cotransfected with RFP-tagged constitutively active Cdc42 and GFP-tagged WT, CA, or DN Rab35. Neither filopodia nor lamellipodia were observed in cells cotransfected with DN Rab35 despite transfection with constitutively active Cdc42. (B) Translocation of Cdc42 from the internal vesicles to the plasma membrane upon transition of Rab35 activity from depletion of Rab35 to CA. An actin promoter-regulated Cdc42-RFP construct and an inducible metal promoter-regulated GFP-Rab35 CA construct were transfected in Rab35-depleted cells. Continuous expression of RFP-Cdc42 regulated by an actin promoter made it possible to monitor translocation of Cdc42. Confirming that the Cdc42-RFP signal located on internal vesicles was caused by depletion of Rab35, we overexpressed GFP-Rab35 CA by treatment with Cu2+ to transit Rab35 activity from depletion of Rab35 to CA. DMSO, dimethyl sulfoxide. (C) Rab35-mediated transport of Cdc42/Rac1-containing vesicles is dependent on a microtubule track. A number of filopodia and lamellipodia induced by expression of the Rab35 CA proteins were eliminated by treatment with 0.5 mM colchicine, an inhibitor of microtubule polymerization. (D) Numerous vesicles but not lamellipodia were observed inside a Rab35G11110 lamellocyte. Lamellocytes from third-instar larvae were stained with L1 antibody, a lamellocyte-specific marker, and Alexa Fluor 568-conjugated phalloidin.
To verify that Rab35 is the only determinant of actin polymerization regulator transport, we monitored Cdc42 translocation from internal vesicles to plasma membrane protrusions upon induction of Rab35 activity. Rab35-depleted cells were introduced to an actin promoter-regulated Cdc42-RFP construct and an inducible metal promoter-regulated GFP-Rab35 CA construct. In contrast to continuous expression of Cdc42-RFP by the actin promoter, GFP-Rab35 CA expression is restricted because the metal promoter is operated only by Cu2+ treatment. Continuously expressed Cdc42-RFP proteins located on internal vesicles were caused by depletion of Rab35. We then treated cells with CuSO4 to induce expression of the GFP-Rab35 CA proteins regulated by an inducible metal promoter. Induction of CA Rab35 via a metal-inducible promoter in Rab35-depleted cells led to the production of large protrusions and Cdc42 migration from internal vesicles to the plasma membrane of newly developed protrusions (Fig. 7B). Therefore, Rab35 appears to be the key molecular switch of Cdc42 and Rac1 translocation to the plasma membrane.
Transport of Cdc42/Rac1-containing vesicles by Rab35 is dependent on a microtubule track.
To determine the microtubule tracks used by Rab35 to transport Cdc42 or Rac1 to the plasma membrane, we disrupted actin or microtubule tracks with specific drugs and examined their effect on Rab35-induced formation of plasma membrane protrusions. When cells expressing CA Rab35 were treated with colchicine, an inhibitor of microtubule polymerization, the phenotype became similar to that of DN Rab35-expressing cells (Fig. 7C). In particular, the cells contained many enlarged vesicles with high-actin structures. Inhibition of actin polymerization with cytochalasin D also caused a complete loss of plasma membrane protrusions in CA Rab35-expressing cells due to a lack of actin bundle formation at the plasma membrane. However, CA Rab35 proteins were highly localized at the plasma membrane along with Cdc42, suggesting that Rab35-mediated transport of actin polymerization regulators to the plasma membrane depends on microtubule tracks.
Hemocytes of Rab35 mutant flies show defects in lamellipodium production.
To verify that the defect in phagocytosis observed in mutant flies arose due to defective actin filament formation, as shown above in vitro, we examined the actin structures of Drosophila hemocytes. Because lamellocytes are relatively large, ranging from 15 to 40 mm, and produce dynamic lamellipodia upon immune challenge, we isolated lamellocytes from larvae and observed their actin structure by phalloidin staining (Fig. 7D). Wild-type lamellocytes developed strong lamellipodia and showed high actin staining at the cellular periphery. However, Rab35 mutant lamellocytes showed no obvious lamellipodium formation, and the periphery was devoid of actin structures. Instead, numerous internal vesicle structures developed, which was similar to the case for SL2 cells transfected with DN Rab35. Consistent with the in vitro results, these data demonstrate that Rab35 is a key regulator of Cdc42/Rac1 transport to the plasma membrane during phagocytosis in Drosophila hemocytes.
DISCUSSION
Phagocytes, including macrophages and monocytes, play a critical role in host defense mechanisms (11, 32, 33). They migrate to sites of infection and eliminate invading microbes by using a number of different methods, including phagocytosis (26). These processes require morphological changes that are typically mediated by changes in actin structure. Therefore, actin filament rearrangement must be tightly regulated during this process. Previous reports have demonstrated that Rho GTPases such as Cdc42 and Rac1 are essential to actin filament rearrangement during the formation of lamellipodial and filopodial membrane protrusions, respectively (6, 24). These Rho GTPases are recruited to specific areas where actin rearrangement will occur. For example, Cdc42 and Rac1 are recruited to the phagocytic cup during phagocytosis (15). The location and distribution of these Rho GTPases are critical for cell migration and morphology. As such, how these proteins polarize within the cell is an important topic under research by many laboratories. A positive feedback loop involving vesicular active transport has been suggested to promote Cdc42 polarization (2, 35). Alternatively, a combination of transport and lateral diffusion in the plasma membrane of budding yeast appears to be responsible for Cdc42 accumulation at sites adjacent to the bud scar (4, 8). However, how these Rho GTPases are involved in the regulation of plasma membrane protrusion during phagocytosis is not well understood.
Here, we demonstrate that Cdc42 and Rac1 polarity on the plasma membrane is established through microtubule-based vesicle transport rather than lateral diffusion toward the locus of actin filament rearrangement in Drosophila. The translocation of Cdc42 and Rac1 to the site of dynamic actin rearrangement was completely dependent on Rab35 activity. It is intriguing that Rab35 activity was manifested mainly in Drosophila hemocytes, resulting in specific defects in the immune response against infection. Several reports suggested the possibility that polar distribution of Cdc42 is established through actomyosin-directed vesicle transport via actin cables (34, 35). However, blockade of Cdc42 polarization by inhibition of microtubule polymerization, as well as proper Cdc42 localization to the plasma membrane despite the absence of actin polymerization, suggests that translocation of this Rho GTPase occurs through the microtubule network.
Consistent with our observation, several studies recently reported that Rab35 appears to regulate cell shape and actin cytoskeletal rearrangement during neurite outgrowth and bristle development (5, 38). Based on the effect of Rab35 on cell shape, Chevallier et al. (5) suggested that that Rab35 may activate Cdc42 during actin remodeling. However, our data indicate that Rab35 does not regulate the activity of Rho GTPase directly. Instead, it appears to control the recruitment of the Rho GTPases to the plasma membrane during cytoskeletal rearrangement. These results indicate that Rab35 plays an important role in actin filament rearrangement in diverse developmental processes.
Examination of whether a physical interaction between Rab35 and Cdc42 or Rac1 exists by using immunoprecipitation demonstrated a lack of direct physical association among these molecules. Thus, how the vesicles regulated by Rab35 specifically load Cdc42 and Rac1 as cargo remains unknown. Additional studies are required to further determine how vesicles select Rho GTPases as well as how Rab35 functions during the loading and selection steps of vesicle transport.
Acknowledgments
We thank Jeongsil Kim-Ha for helping with excision of mutants and Li Hua Jin for screening mutants.
This research was supported by the International Research & Development Program and the WCU (World Class University) program of the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (MEST) of Korea (grants K20704000006-09A0500-00610 and R31-2008-000-10086-0).
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 2.Altschuler, S. J., S. B. Angenent, Y. Wang, and L. F. Wu. 2008. On the spontaneous emergence of cell polarity. Nature 454:886-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bokoch, G. M. 2005. Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 15:163-171. [DOI] [PubMed] [Google Scholar]
- 4.Casamayor, A., and M. Snyder. 2002. Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 5:179-186. [DOI] [PubMed] [Google Scholar]
- 5.Chevallier, J., C. Koop, A. Srivastava, R. J. Petrie, N. Lamarche-Vane, and J. F. Presley. 2009. Rab35 regulates neurite outgrowth and cell shape. FEBS Lett. 583:1096-1101. [DOI] [PubMed] [Google Scholar]
- 6.Cox, D., P. Chang, Q. Zhang, P. G. Reddy, G. M. Bokoch, and S. Greenberg. 1997. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J. Exp. Med. 186:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elrod-Erickson, M., S. Mishra, and D. Schneider. 2000. Interactions between the cellular and humoral immune responses in Drosophila. Curr. Biol. 10:781-784. [DOI] [PubMed] [Google Scholar]
- 8.Etienne-Manneville, S. 2004. Cdc42—the centre of polarity. J. Cell Sci. 117:1291-1300. [DOI] [PubMed] [Google Scholar]
- 9.Ferrandon, D., J. L. Imler, C. Hetru, and J. A. Hoffmann. 2007. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7:862-874. [DOI] [PubMed] [Google Scholar]
- 10.Franc, N. C., J. L. Dimarcq, M. Lagueux, J. Hoffmann, and R. A. B. Ezekowitz. 1996. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4:431-443. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg, S., and S. Grinstein. 2002. Phagocytosis and innate immunity. Curr. Opin. Immunol. 14:136-145. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann, J. A. 2003. The immune response of Drosophila. Nature 426: 33-38. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, Jr., and R. A. B. Ezekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 284:1313-1318. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, J. A., and J. M. Reichhart. 2002. Drosophila innate immunity: an evolutionary perspective. Nat. Immunol. 3:121-126. [DOI] [PubMed] [Google Scholar]
- 15.Hoppe, A. D., and J. A. Swanson. 2004. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol. Biol. Cell 15:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hultmark, D. 2003. Drosophila immunity: paths and patterns. Curr. Opin. Immunol. 15:12-19. [DOI] [PubMed] [Google Scholar]
- 17.Irving, P., J. M. Ubeda, D. Doucet, L. Troxler, M. Lagueux, D. Zachary, J. A. Hoffmann, C. Hetru, and M. Meister. 2005. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell. Microbiol. 7:335-350. [DOI] [PubMed] [Google Scholar]
- 18.Jin, L. H., J. Shim, J. S. Yoon, B. Kim, J. Kim, J. Kim-Ha, and Y. J. Kim. 2008. Identification and functional analysis of antifungal immune response genes in Drosophila. PLoS Pathog. 4:e1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, L. K., U. Y. Choi, H. S. Cho, J. S. Lee, W. B. Lee, J. Kim, K. Jeong, J. Shim, J. Kim-Ha, and Y. J. Kim. 2007. Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila. PLoS Biol. 5:e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kocks, C., J. H. Cho, N. Nehme, J. Ulvila, A. M. Pearson, M. Meister, C. Strom, S. L. Conto, C. Hetru, L. M. Stuart, T. Stehle, J. A. Hoffmann, J. M. Reichhart, D. Ferrandon, M. Rämet, and R. A. B. Ezekowitz. 2005. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123:335-346. [DOI] [PubMed] [Google Scholar]
- 21.Lemaitre, B., and J. Hoffmann. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25:697-743. [DOI] [PubMed] [Google Scholar]
- 22.Manaka, J., T. Kuraishi, A. Shiratsuchi, Y. Nakai, H. Higashida, P. Henson, and Y. Nakanishi. 2004. Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J. Biol. Chem. 279:48466-48476. [DOI] [PubMed] [Google Scholar]
- 23.Massol, P., P. Montcourrier, J. C. Guillemot, and P. Chavrier. 1998. Fc receptor-mediated phagocytosis requires CDC42 and Rac1. EMBO J. 17:6219-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niedergang, F., and P. Chavrier. 2005. Regulation of phagocytosis by Rho GTPases, Curr. Top. Microbiol. Immunol. 291:43-60. [DOI] [PubMed] [Google Scholar]
- 25.Osmani, N., N. Vitale, J. P. Borg, and S. Etienne-Manneville. 2006. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr. Biol. 16:2395-2405. [DOI] [PubMed] [Google Scholar]
- 26.Paladi, M., and U. Tepass. 2004. Function of Rho GTPases in embryonic blood cell migration in Drosophila. J. Cell Sci. 117:6313-6326. [DOI] [PubMed] [Google Scholar]
- 27.Pfeffer, S., and D. Aivazian. 2004. Targeting Rab GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 5:886-896. [DOI] [PubMed] [Google Scholar]
- 28.Rämet, M., P. Manfruelli, A. Pearson, B. Mathey-Prevot, and R. A. Ezekowitz. 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416:644-648. [DOI] [PubMed] [Google Scholar]
- 29.Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Johnson-Schlitz, W. K. Benz, and W. R. Engels. 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118:461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers, S. L., U. Wiedemann, N. Stuurman, and R. D. Vale. 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells The J. Cell Biol. 162:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz, S. L., C. Cao, O. Pylypenko, A. Rak, and A. Wandinger-Ness. 2007. Rab GTPases at a glance. J. Cell Sci. 120:3905-3910. [DOI] [PubMed] [Google Scholar]
- 32.Stuart, L. M., and R. A. B. Ezekowitz. 2005. Phagocytosis: elegant complexity. Immunity 22:539-550. [DOI] [PubMed] [Google Scholar]
- 33.Underhill, D. M., and A. Ozinsky. 2002. Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 20:825-852. [DOI] [PubMed] [Google Scholar]
- 34.Wedlich-Soldner, R., S. Altschuter, L. Wu, and R. Li. 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299:1231-1235. [DOI] [PubMed] [Google Scholar]
- 35.Wedlich-Soldner, R., S. C. Wai, T. Schmidt, and R. Li. 2004. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 166:889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zerial, M., and H. McBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107-117. [DOI] [PubMed] [Google Scholar]
- 37.Zettervall, C. J., I. Anderl, M. J. Williams, R. Palmer, E. Kurucz, I. Ando, and D. Hultmark. 2004. A directed screen for genes involved in Drosophila blood cell activation. Proc. Natl. Acad. Sci. U. S. A. 101:14192-14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, J., M. Fonovic, K. Suyama, M. Bogyo, and M. P. Scott. 2009. Rab35 controls actin bundling by recruiting fascin as an effector protein. Science 325:1250-1254. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, J., K. L. Schulze, P. R. Hiesinger, K. Suyama, S. Wang, M. Fish, M. Acar, R. A. Hoskins, H. J. Bellen, and M. P. Scott. 2007. Thirty-one flavors of Drosophila rab proteins. Genetics 176:1307-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]