Abstract
Class I myosins, which link F-actin to membrane, are largely undefined in lymphocytes. Mass spectrometric analysis of lymphocytes identified two short tail forms: (Myo1G and Myo1C) and one long tail (Myo1F). We investigated Myo1G, the most abundant in T-lymphocytes, and compared key findings with Myo1C and Myo1F. Myo1G localizes to the plasma membrane and associates in an ATP-releasable manner to the actin-containing insoluble pellet. The IQ+tail region of Myo1G (Myo1C and Myo1F) is sufficient for membrane localization, but membrane localization is augmented by the motor domain. The minimal region lacks IQ motifs but includes: 1) a PH-like domain; 2) a “Pre-PH” region; and 3) a “Post-PH” region. The Pre-PH predicted α helices may contribute electrostatically, because two conserved basic residues on one face are required for optimal membrane localization. Our sequence analysis characterizes the divergent PH domain family, Myo1PH, present also in long tail myosins, in eukaryotic proteins unrelated to myosins, and in a probable ancestral protein in prokaryotes. The Myo1G Myo1PH domain utilizes the classic lipid binding site for membrane association, because mutating either of two basic residues in the “signature motif” destroys membrane localization. Mutation of each basic residue of the Myo1G Myo1PH domain reveals another critical basic residue in the β3 strand, which is shared only by Myo1D. Myo1G differs from Myo1C in its phosphatidylinositol 4,5-bisphosphate dependence for membrane association, because membrane localization of phosphoinositide 5-phosphatase releases Myo1C from the membrane but not Myo1G. Thus Myo1PH domains likely play universal roles in myosin I membrane association, but different isoforms have diverged in their binding specificity.
Keywords: Cytoskeleton, Evolution, Lymphocyte, Myosin, Plasma Membrane, Protein Domains, Class I Myosin, PH Domain
Introduction
Lymphocytes (and other cells of the hematopoietic system) are highly motile, and their motility is dependent on the well characterized class II conventional two-headed myosins (1). However, there are at least 36 other classes of myosins in eukaryotes (2), and in humans, there are 39 human myosin genes in 11 classes (3). The class I myosins in human are second only to class II myosins in terms of number of genes (8 class I genes and 14 class II genes in human). Class I myosin were first identified in amoeba and contribute to regulation of migration in amoeboid locomotion, which is in some respects reminiscent of hematopoietic cell migration. However, class I myosins in lymphocytes are largely unstudied.
The class I myosins are single-headed myosins (i.e. unconventional) whose function generally involves linking actin filaments to the membrane. This actin-membrane linking enables them to contribute to diverse functions, some of which involve linking actin to the plasma membrane (e.g. stabilization of microvilli, endocytosis, membrane ruffling, regulation of directional migration, and regulation of membrane tension) and others of which apparently involve vesicular transport (4–7). Class I myosins consist of an N-terminal actin-binding ATPase “head” domain, an α helical “neck” region that participates in calmodulin binding, and a C-terminal tail region. Class I myosins fall into “short” and “long” subsets differentiated by the length of their tail. Both subsets have a tail homology 1 (TH1)2 region of ∼200 amino acids identifiable by modest evolutionary conservation (8). In addition the long tail subset contains an additional Gly/Pro/Ala (GPA) region and an SH3 domain. Myosin I linking to actin occurs via the N-terminal head domain, a function that is common to all myosins and therefore widely investigated. The structural basis of myosin I binding to the membrane is much less well understood. It has been shown to be mediated by the C-terminal “tail” (9–12). However, some evidence also implicates the neck region in binding membrane lipid (13, 14). Coarse features of the tail domain have been elucidated by cryoelectron microscopy (15), but no high resolution structures are available.
Recently, we identified a PH-like domain embedded in the midst of the tail of Myo1C (14). PH-like domains comprise a diverse superfamily of domains (16). Although PH-like domains are very prevalent even in the earliest-branching eukaryotes, it is one of the relatively few domains for which a clear-cut ancestral version has not been identified in prokaryotes (17, 18). The conserved protein fold present in the PH-like domains is a composite fold consisting of two elements: 1) a four-stranded β-meander similar to a single blade of the β-propeller domains; and 2) an AP2-like three-stranded unit followed by a single helix (18). The sheets of the two elements are packed against each other with the single helix capping the orifice of the resultant partially open barrel (17, 18). This conserved scaffold has been widely utilized in diverse proteins to perform a range biochemical functions, because the overall β-barrel fold provides several distinct niches for potential interactions with substrates. The PH-like superfamily currently encompasses, in addition to the classic PH domain, several distinct families such as the phosphotyrosine binding, EVH1-DCP1, Ran-binding, GRAM, TFIIH TFB1 subunit, FERM-C-terminal, SSRP1, and VPS36-N-terminal domains (see, for examples, Structural Classification of Proteins, release 1.75 (June 2009) or family PF00169 from the Wellcome Trust Sanger Institute, both available on-line). The functions of these domains include membrane recognition, mRNA decapping, and peptide binding. The prototype of this superfamily, the classic PH domain family, is best known for the ability of some members to bind membrane phosphatidylinositols, especially PIP2 and/or PIP3 (19). The existence of a PH-like domain in class I myosins has previously been overlooked, because its sequence is quite divergent from other known PH-like domain families (see “Discussion”). We found that structure prediction algorithms predicted a PH-like domain in short tail class I myosins and confirmed that prediction by demonstrating that mutation of key basic residues within the predicted lipid binding pocket of that domain destroys the ability of Myo1C to bind membrane (14). Moreover, analysis of binding specificity indicates the domain to have relatively promiscuous binding to phosphoinositides. Subsequent studies recently identified a PH-like domain in Acanthamoeba long tail myosin (20).
The present studies emerged from a mass spectrometric analysis of proteins enriched in plasma membrane-containing preparations from lymphocytes in which we identified Myo1G as abundant in lymphocytes and apparently enriched at the plasma membrane (21). In investigating Myo1G, we sought a more general understanding of the structural basis of its membrane association, of the relationship of the myosin PH-like domain to the PH domain superfamily, and of potential functional differences between class I myosins in their membrane association. We find the Myo1PH domain to be a new family of the PH-like domain superfamily and identify an instance in prokaryotes. Unlike various classic PH domains (22), the Myo1PH domain is not sufficient for membrane localization but requires a Pre-PH and a Post-PH region, which together are likely to form a composite membrane-recognition module in these proteins. Moreover, Myo1G differs from Myo1C with respect to a basic residue (Lys-898) that is critical for its membrane association and in its dependence on PIP2 for membrane localization.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Jurkat Tag cells were provided by Dr. Gerald Crabtree (Stanford University), the mouse pre-B cell line 300.19 (provided by Dr. Geoffrey Kansas, Northwestern University Medical School), both were maintained in RPMI 1640. RBL cells and HeLa cells were obtained from the ATCC and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Both media were supplemented with 100 mm sodium pyruvate, nonessential amino acids, 2 mm l-glutamine, 100 μg/ml antibiotic mix (all from Invitrogen), and 50 mm β-mercaptoethanol (Sigma). Fresh human lymphocytes were isolated from the blood of healthy human volunteers by leukapheresis and elutriation (to prepare peripheral blood lymphocytes) and, when necessary, by immunomagnetic negative selection as previously described (23, 24). Mouse B- and T-lymphocytes were purified by negative selection following the manufacturer's protocol (Myltenyi Biotec). Rapamycin was purchased from Calbiochem. The following antibodies were used for Western blot (WB) and/or immunofluorescence staining: rabbit polyclonal antibodies raised against human Myo1G aa 2–14, human WIP (Dr. David Nelson, Metabolism Branch, NCI, National Institutes of Health), mouse monoclonal Ab to Myo1C (25), β-actin mAb (Sigma A1978), moesin mAb (Sigma M7060), goat anti-mouse IgG-Alexa 488 (Invitrogen), goat anti-mouse IRDye 680, goat anti-rabbit IRDye 800CW (LI-COR, Lincoln, NE). Mass spectrometric analysis was as previously described (21).
Western Blot
For WB analysis equal amounts of protein (60 μg) from each cell type were resolved by 4–12% SDS-NuPAGE gels, transferred to nitrocellulose membranes, and analyzed by WB with antibodies listed above using an Odyssey Infrared Imaging System (LI-COR Biosciences). To release Myo1G from cytoskeleton-membrane, 108 human PBT cells were lysed either in TNE buffer (25 mm Tris·Cl, pH 7.4, 150 mm NaCl, 5 mm EDTA, Triton X-100 1%), or the same buffer plus ATP 50 mm in the presence of protein inhibitors EDTA free (Roche Applied Science) for 1 h on ice. Then, samples were centrifuged at maximum velocity in a tabletop centrifuge for 15 min, and the supernatant was recovered to a new tube, the pellet was resuspended to the equivalent original volume in 1× sample buffer, and the same volume aliquots were run in NuPAGE 4–12 gradient gels and analyzed by Western blot.
Immunofluorescence Microscopy
PBT, Jurkat, and 300.19 cells (1–2 × 106 cells each) were dropped onto the glass bottom of 35-mm MatTek culture dishes (MatTek, Ashland, MA) pre-coated with poly-l-lysine (Sigma). Cells were allowed to settle for 10 min at room temperature and either analyzed alive or fixed (for endogenous Myo1G expression and 5-Ptase recruitment to the membrane) by the addition of 4% paraformaldehyde solution (Sigma). After 10 min at room temperature, samples were washed with PBS, permeabilized with 0.2% Triton X-100, and blocked for 1 h at room temperature in PBS with 3% BSA. Primary antibodies were added for 1 h at room temperature in PBS with 3% BSA. After washing, goat anti-rabbit Alexa-488-conjugated secondary antibodies (Molecular Probes) in PBS with 3% BSA were added for 1 h at room temperature. Samples were examined using a Zeiss LSM510 laser scanning confocal microscope using a 63× or 100× objectives (Carl Zeiss). Quantitative analysis was performed using the Imaging Examiner software (LSM, Carl Zeiss, Inc.). For each cell analyzed, a line was drawn manually at the plasma membrane, and another line was drawn just inside the plasma membrane in the cytosol. Average fluorescence intensity was determined for the set of pixels touching the plasma membrane line (plasma membrane mean intensity) and for those touching the cytosol line (cytosol mean intensity). Membrane enrichment for that cell is calculated as the (plasma membrane mean intensity)/(cytosol mean intensity). Results are summarized as the mean ± S.E. from at least eight representative cells from two to three independent experiments. Because pixels touching the line at the plasma membrane will often include a fraction of extracellular space, the average obtained for a construct that is not enriched at the plasma membrane is often somewhat less than 1.0.
Constructs
Human Myo1G was amplified from Human PBT cytoplasmic RNA (RNeasy Mini Kit, Qiagen) by reverse transcription-PCR (SuperScript One-Step RT-PCR for Long Templates, Invitrogen), and cloned into pENTR vector by recombination (pENTR/D-TOPO Cloning Kit, Invitrogen). Myo1F constructs were derived from an Open Biosystems clone (MHS1010-7507831), subcloned to pENTR. Myo1C construct was previously described (14). Desired subregions of class I myosin proteins were subcloned from the full-length constructs as follows: Myo1G (Motor-IQ, aa 1–779; IQ-Tail “IQ1,2 Ext TH1,” aa 702–1018; IQ2-Ext TH1, aa 735–1018; Extended-TH1, aa 750–1018; TH1, aa 763–1018; Myo1PH, aa 875–968; Myo1G-delta 967, aa 1–967; Myo1C FL, aa 1–1028), Myo1C (Motor-IQ, aa 1–765; Myo1C IQ-Tail, aa 691–1028), and Myo1F (FL, aa1–1098; Myo1F Motor-TH1, aa 1–922; and Myo1F IQ-Tail, aa 690–1098). Point mutants of Myo1G and Myo1F were generated using QuikChange site-directed mutagenesis (Stratagene); all the constructs and mutants were verified by sequencing. Inserts were transferred from pENTR into Gateway destination vectors from the Protein Expression Laboratory (NCI-Frederick, Frederick, MD) (pDest732 (N-terminal eGFP-tag) or pDest733 (N-terminal mRFP tag)), by an LR reaction according to the manufacturer's recommendations (Invitrogen) to construct mammalian expression vectors. CFP-tagged membrane-targeting construct containing FRB domain of human mTOR1 and mRFP-tagged human type IV 5-Ptase enzyme construct containing FKBP12 or mRFP-FKBP12 control construct without type IV 5-Ptase (26) were provided by Dr. Tamas Balla (NICHD, NIH, Bethesda, MD).
Transfection
Transfection was done using 10 μg of each plasmid in a BTX ECM 830 electroporator (Harvard Apparatus, 300 V for 10 ms). After electroporation, the cells were cultured for 16–24 h and used for immunofluorescence analysis as described above. For studies involving 5-Ptase recruitment to the plasma membrane Jurkat cells were transfected with equal amounts of the appropriate FRB and FKBP constructs and either Myo1G or Myo1C. After incubation at 37 °C cells were adjusted to 107cells/ml in Hanks' balanced salt solution containing 0.3% BSA, 200 μl were added onto the glass bottom of 35-mm culture dishes (MatTek) precoated with poly-l-lysine and allowed to settle for 10 min at 37 °C. 200 μl of a solution of 6 μm rapamycin (in Hanks' balanced salt solution 0.3% BSA) was then added to the cells. After 5 min of incubation at 37 °C, the cells were fixed by the addition of 1 ml of 4% paraformaldehyde solution. After 10 min at room temperature, the cells were washed four times with PBS and examined as above.
Sequence-Structure Analysis
Structure similarity searches were conducted using the DALIlite program, and structural alignments were made using the MUSTANG program. Protein structures were visualized and manipulated using the Swiss-PDB program. Sequence profile searches were performed against the NCBI non-redundant data base of protein sequences (National Center for Biotechnology Information, NIH, Bethesda, MD), and a locally compiled data base of proteins from eukaryotes with completely or near-completely sequenced genomes. PSI-BLAST searches were performed using an expectation value (E value) of 0.01 as the threshold for inclusion in the position-specific scoring matrix generated by the program. Hidden Markov Model (HMM) searches were performed using the newly released HMMER3 package (version β 2). Multiple alignments were constructed using the Kalign programs, followed by manual correction based on PSI-BLAST/HMMER3 high scoring pairs, secondary structure predictions, and information derived from existing structures. Protein secondary structure was predicted using a multiple alignment as the input for the JPRED2 program, which uses information extracted from a PSSM, HMM, and the seed alignment itself. Pairwise comparisons of HMMs, using a single sequence or multiple alignment as query, against profiles of proteins in the PDB data base were performed with the HHPRED program. Similarity-based clustering was performed using the BLASTCLUST program (ftp.ncbi.nih.gov/blast/documents/blastclust.html) with empirically determined length and score threshold parameters. Threading-based structural predictions were made using two publicly available software packages that have scored very highly in recent CASP (Critical Assessment of Techniques for Protein Structure Prediction) competitions: I-Tasser (27) and PHYRE (28).
RESULTS
Class I Myosins in Lymphocytes
We have previously performed mass spectrometric proteomic profiling of lymphocyte proteins (21) and here extend the data analysis to myosins. In those studies, analysis of post-nuclear lysate provided a global view of cytosolic proteins and of membrane/microvillus (MMV)-enriched fraction provided additional information on plasma-membrane proximal components. We identified 813 human proteins and 1361 mouse proteins in these fractions using criteria that required identification of at least two peptides for a protein (resulting in a calculated false discovery rate of <5%) (21). In resting human peripheral blood T cell post-nuclear lysate four myosins were detected: two conventional myosins, myosin IIA (a non-muscle myosin, MYH9) and smooth muscle myosin (MYH11); and two unconventional class I myosins, Myo1G and Myo1F (Fig. 1A, open bars). The number of peptides detected is a useful (albeit crude) estimator of protein abundance (especially when numerous peptides are detected for the proteins being compared and the number is normalized to protein length). Peptide detection data suggest that MyoIIA is the most abundant in post-nuclear lysate (8-fold more MyoIIA peptides in post-nuclear lysate than any other myosin but only 2-fold greater length). Because of the importance of myosins in events at the plasma membrane, particular importance was given to proteomic profiling data from microvilli/plasma membrane-enriched fraction from the same cells (Fig. 1A, closed bars). The same four myosins were detected, but their relative abundance was markedly different (again estimated by number of peptides, which in this context is a better estimate because the peptides come from the same protein). Of the four myosins, only one was enriched in the MMV fraction, namely Myo1G. In contrast, the other class I myosin, Myo1F, was decreased in the MMV preparation.
FIGURE 1.
Myosins identified in the fractions of human PBT and 300.19. Quantitation of the number of peptides identified from each myosin gene in mass spectrometric analysis of membrane/microvillus (MMV, filled bars) fraction versus post nuclear lysate (PNL, open bars) of two lymphoid cells: human PBT (A) and a mouse pre B cell line (300.19) (B). Conservative criteria were used for identifying each protein (namely detection of two distinct peptides from that protein using identification thresholds that gave a 5% false positive for any single peptide).
To assess whether the pattern of myosins observed in PBT was typical of other lymphoid cells, a similar analysis was performed on 300.19, a mouse pre-B cell line chosen because its morphology resembles primary lymphoid cells (spherical, and densely covered with microvilli), and it has been used as a model cell line for studies of lymphocyte dynamic adhesion (29, 30) (Fig. 1B). The pattern for mouse-pre B cells was similar in many respects to PBT, including Myo1G enrichment in MMV, and no enrichment of the abundant class II myosin in MMV. The notable differences are: 1) an additional class I myosin, myo1C, was present and resembled Myo1G in abundance and MMV enrichment; 2) presence of an additional isoform of myosin II (B versus A); and 3) absence of detectable Myo1F.
To better evaluate expression of Myo1G we developed a Myo1G-specific polyclonal rabbit antibody (supplemental Fig. S1). Western blotting for Myo1G and Myo1C confirmed the mass spectrometric findings that Myo1G was well expressed in PBT and 300.19, whereas Myo1C was abundant only in 300.19 (Fig. 2A). WB comparison of subsets purified from mouse spleen confirmed that Myo1G is comparable in expression in T- and B-cells while Myo1C is predominantly in B-cells (Fig. 2B). Our WB results (Fig. 2A) for another hematopoietic cell type (mast cells) and a non-hematopoietic (epithelial cells) are consistent with data available from other approaches (see “Discussion”), which indicates that Myo1G is hematopoietically restricted, whereas Myo1C is broadly expressed.
FIGURE 2.
Characterization of Myo1G expression by Western blot. A, WB of Myo1G in whole cell lysate (60 μg) from the indicated cell types with WB for actin as loading control; see supplemental Fig. S1 for entire molecular weight range of this WB; B, WB of Myo1G from purified mouse T- and B-lymphocytes with WB for moesin as loading control.
Localization of Myo1G to the Plasma Membrane
Myo1G distribution was analyzed by immunofluorescence in three different lymphoid cell populations: primary human T cells (PBT), the transformed human T-cell line Jurkat, and the murine pre-B cell line 300.19 (Fig. 3A). Two striking features were shared by images of all three cell types: 1) very strong localization at the plasma membrane with little or no detection in cytosol or nucleus; and 2) punctate distribution at the cell surface, especially evident in projection images, consistent with presence (and potentially enrichment) in peripheral processes such as microvilli or ruffles. Particulate localization in cytosol was not observed, providing no evidence for a major role in vesicular traffic.
FIGURE 3.
Localization of Myo1G at the plasma membrane. A, immunofluorescence analysis of fixed and permeabilized PBT (top), Jurkat (middle), and 300.19 (bottom) stained with rabbit serum anti Myo1G followed by secondary antibody conjugated to Alexa 488. A representative mid plane image (left panel) and projection image (right panel) are shown. Bars, 5 μm. B, WB of Myo1G in 1% Triton X-100 lysate, in the absence or presence of ATP. WB of cytoskeletal regulating cytosolic protein WIP is shown as control. C, mid plane immunofluorescence images of live cells transfected with full length GFP-Myo1G fusion protein: i, PBT; ii, 300.19; iii, Jurkat; and iv, HeLa. Bars, 5 μm.
This pattern of distribution is consistent with the localization of other well characterized mammalian class I myosins, for example Myo1A, which is enriched in intestinal brush border where it is bound to F-actin (31). To confirm Myo1G association with cytoskeleton, we investigated the association of Myo1G with the particulate fraction from Triton X-100 lysates of PBT (Fig. 3B). Myo1G is completely retained in the insoluble fraction under these conditions in which endogenous ATP is diluted by solubilization. In contrast, WIP (a cytoskeletal regulating protein used as control) is almost exclusively detected in the soluble fraction in the same conditions. Supplementation of the lysing buffer with 50 mm ATP results in release of ∼40% of the Myo1G from the particulate fraction, consistent with ATP preventing the stabilization of Myo1G binding to F-actin in an ATP-free rigor complex.
Myo1G constructs with the N-terminal GFP tag were generated to investigate whether they faithfully reproduce the pattern observed with endogenous Myo1G in lymphoid cells and whether plasma membrane localization required factors unique to hematopoietic cells (Fig. 3C). In hematopoietic cells, localization of the GFP-tagged construct mimicked that observed for endogenous Myo1G. Moreover, Myo1G is sharply localized to the plasma membrane in non-hematopoietic cells (e.g. HeLa cells, Fig. 3C) where it shows punctate localization resembling that observed in lymphoid cells. This evidence indicates that Myo1G does not require a hematopoietic specific factor for its membrane localization.
Identifying the Minimal Region Essential for Membrane Association
We investigated the role of the tail of lymphocyte class I myosins in their membrane localization (Fig. 4A). We compared Myo1G to Myo1C, whose requirements we have investigated previously (14), using N-terminally tagged constructs containing: full protein, motor-IQ, and IQ tail (Fig. 4B). Transfection into Jurkat cells demonstrated that the Myo1G IQ tail was sufficient for enrichment at the membrane (as it was for Myo1C). In contrast motor-IQ was not sufficient for enrichment at the membrane. The Myo1G IQ tail domain is sufficient to provide ∼2-fold enrichment at the plasma membrane (Fig. 4, B and C). The modest cytoplasmic signal from this construct appears to be diffuse without aggregates. The Myo1G IQ tail also was enriched in the nucleus, which we have not observed with immunofluorescence analysis of endogenous Myo1G. It is possible that deletion of the head domain unmasks a cryptic nuclear localization signal in this region rich with positive charge or that the relatively small construct enters the nucleus by diffusion and is retained by binding negatively charged nucleic acids (32). However, this feature would have to be unique to Myo1G but not Myo1C. Localization at the plasma membrane is markedly increased by inclusion of the motor domain (Fig. 4, B and C). This augmentation by the motor domain is not observed with Myo1C.
FIGURE 4.
IQ plus tail regions of Myo1G, Myo1C, and Myo1F are sufficient for membrane association. A, schematic representation of short tail (Myo1G and Myo1C) and long tail (Myo1F) class I myosin constructs studied. B, representative immunofluorescence images of Jurkat cells expressing constructs of Myo1G or Myo1C. Bar, 5 μm. C, quantitative analysis of membrane enrichment for the indicated mutants. Proteins not enriched at the plasma membrane typically have ratios of membrane/cytoplasmic localization in the range from 0.5 to 1.0 (dashed line). D, representative immunofluorescence images of Jurkat cells expressing Myo1F constructs. Bar, 5 μm. E, quantitative analysis of membrane enrichment for the indicated mutants. Proteins not enriched at the plasma membrane typically have ratios of membrane/cytoplasmic localization in the range from 0.5 to 1.0 (dashed line).
To lay the foundation for limited analysis of long tail myosins, we also assessed three Myo1F constructs, all of which demonstrated membrane localization (Fig. 4, D and E): Three additional features were notable: 1) full-length Myo1F was enriched at the plasma membrane, but much less strongly than Myo1G or Myo1C; 2) the GPA and SH3 regions (lacking in the motor-TH1 construct) are not necessary for this association; and 3) the motor domain appeared to impede membrane localization, because its removal augmented membrane association. The paradigm evident for all three isoforms (Myo1G, Myo1C, and Myo1F) was that the region C-terminal to the motor domain was sufficient for membrane localization.
It has not been established what are the minimal elements of the class I myosin tail that are necessary for membrane association. Since we have previously identified a PH-like domain (14) which is critical to membrane localization of Myo1C, one possibility was that the PH-like domain might be sufficient for membrane localization since various classical PH domains are sufficient for membrane localization (22). However, the isolated PH-like domain did not mediate membrane localization (Fig. 5B, C). To better understand the minimal region required for membrane association we prepared more inclusive constructs that contain N- and C-terminal extensions to the PH domain (Fig. 5A). We prepared four constructs, all of which included the additional 48aa found at the C terminus of the core PH domain. These constructs differed in having variable length N-terminal extensions: 1) the “IQ1, 2 Ext TH1” construct included both the predicted IQ motifs. 2) The “IQ2-Ext-TH1” construct included only the C-terminal IQ motif. 3) The “Extended TH1” construct left out both the IQ motifs but contained 115 residues N-terminal to the PH-like domain. 4) The “Tail-TH1” construct included 102 residues N-terminal to the PH-like domain (which encompasses the segment defined as the “TH1” region of class I myosins tails in the PFAM data base (PF06017)). 5) We additionally prepared a construct that deleted the 48 residues C-terminal to the PH domain from whole length Myo1G (Myo1G Δ967). Of these, the first three constructs localized to the membrane, whereas the Tail-TH1 and Myo1G Δ967 failed to do so (Fig. 5B).The shortest construct that was enriched at the membrane was the “Extended TH1” indicating that the minimal membrane-association segment consists of a “Pre-PH” region of ∼115 residues, the core PH-like domain and the C-terminal post-PH extension of 48 aa (Fig. 5, B and C). Of interest, addition of a single IQ motif augmented that membrane association, which was further enhanced by addition of the second IQ motif.
FIGURE 5.
Fine mapping of the “minimal” region of Myo1G tail for membrane localization. A, schematic representation of Myo1G fragments fused with GFP for membrane localization studies. The region shown is Myo1G C-terminal to the motor domain. The regions marked on the map include: IQ motifs, boundaries of the TH1 region identified by PFAM motif PF0617 (dashed line), the Myo1PH domain corresponding to PH-like domain previously described (14) and identified by our bioinformatic analysis, and the Pre-PH and Post-PH regions identified by our functional analysis. The boundaries of the constructs tested are shown relative to these regions. An arrowhead pointing to the left indicates that the construct includes the motor domain. B, representative mid plane images of Jurkat cells transfected with the indicated constructs. Bar, 5 μm. C, quantitative analysis of membrane enrichment for the indicated mutants. Proteins not enriched at the plasma membrane typically have ratios of membrane/cytoplasmic localization in the range from 0.5 to 1.0 (dashed line).
Characterization of the Pre-PH Region
The Pre-PH region of Myo1G, shown above to be essential for membrane localization, has two striking sequence features: 1) it is predicted to be largely α helical, composed of four α helical regions with intervening less ordered regions (supplemental Fig. S2), and 2) it is highly basic, with a predicted pI of 10.3. This raised the possibility that electrostatic interactions of this region might contribute to association with the plasma membrane, which we explored by investigating sequence conservation among the six short tail class I myosins. Among its 18 Arg and Lys residues two in the third predicted α helix (Lys-815 and Arg-826) seemed promising based on each of two criteria. 1) They are the only two Arg and Lys in this region that are conserved among all six short-tail myosins and 2) they may be on the same face of a single α helix (Fig. 6A). Mutation of either of these residues individually in full-length Myo1G demonstrated that the mutations reduce membrane association (although not destroying it completely (Fig. 6, B and C)).
FIGURE 6.
Functional effect of mutating positively charged residues in the third predicted α helix in the Pre-PH region of Myo1G. A, helical wheel representation of the third predicted helix in the Pre-PH region of Myo1G. Arcs represent predicted hydrophobic surfaces. Positively charged residues are annotated with blue numbers indicating the number of human short tail myosins in which the residue is conserved. B, representative images of Jurkat cells transfected with the indicated point mutants in the Pre-PH of Myo1G; C, quantitative analysis of membrane enrichment for the indicated mutants. Proteins not enriched at the plasma membrane typically have ratios of membrane/cytoplasmic localization in the range from 0.5 to 1.0 (dashed line).
Characterization of the Myo1PH Domain
The relationship of the PH-like domain identified in Myo1C (14) to the superfamily of PH domains has not been clarified. We therefore undertook broad evolutionary analysis of the PH domain in class I myosins along with structure-guided sequence alignment. We first prepared a multiple alignment of the minimal region required in membrane-association (identified above) across all class I myosins and predicted secondary structure using the Jpred program. As a consequence we identified the 7-strand-helix core of the PH-like domain. We used this region to construct a PSSM and a HMM and used them in iterative search of the data base with the PSI-BLAST and HMMsearch programs. As a result we were able to detect homologous regions in short tail myosins and long tail myosins, including the distant long tail class I myosins from yeast, and the divergent kinetoplastid (e.g. Trypanosoma) myosin (Fig. 7 and supplemental Table S1). Additionally, these searches also recovered a series of lineage-specifically expanded proteins from ciliates, such as Paramecium and Tetrahymena, and the basal eukaryote Trichomonas with significant e-values (e < 10−5 upon first detection in the search). In these organisms the homologous segment was not fused to an N-terminal myosin region, but was combined to protein kinase domains or occurred as a standalone domain. Interestingly, these searches also recovered a divergent version of the domain in proteins from several green-sulfur bacteria such as Chlorobium chlorochromatii (Cag_0218, gi: 78188201; e = 10−3 in the 7th iteration with search initiated with the core PH-like domain profile). Prior to convergence, the searches also recovered certain classic PH domain (e-values ∼ 10−2). To confirm the relationship with the classic PH domains we seeded profile-profile comparisons with the HHpred program and obtained significant hits to structures in PDB of the PH domain (e.g. 2uzs and 2p0h p = 10−5-10−10). The results verify the previous analysis which focused on the β1/β2 region (14) and extend it to reveal a complete but divergent PH domain. These results also help clarify the boundaries of the PH-like domain in these proteins that have been previously subsumed in part under the PFAM TH1 module, which includes some non-PH elements (see above).
FIGURE 7.
Multiple sequence alignment of the Myo1PH domain of representative proteins identified by the Myo1PH sequence profile. A Myo1PH sequence profile was developed and used to screen the non-redundant (nr) sequence data base as described under “Experimental Procedures.” Shown here is the Myo1PH region of an informative panel of the protein hits, including all human class I myosins, a yeast long tail myosin, proteins from diverse eukaryotes, and the bacterial protein. For expansion of information on organism and listing of GI number see supplemental Table S1. In the histogram shown below the alignment represents ClustalX calculation of relative conservation at each position. The schematic above the alignment shows secondary structure predictions, which are concordant with those observed in the classic PH domain. The blue asterisks represent all basic residues in Myo1G, each of which was mutated to assess function. The red arrowheads indicate the three residues whose mutation impairs membrane localization. 1 (Lys-877) and 2 (Arg-887) are the signature residues in the β1/2 loop. 3 (Lys-898) in β3 strand was first identified as being important in our mutational screen.
A common structural feature of lipid binding PH domains is a “signature motif” of two basic residues conserved in the lipid-binding pocket, which bind to phosphate residues in acidic phospholipids (19). The multiple sequence alignment of the Myo1PH instances shows very strong conservation of these two residues (Fig. 7). The first is always lysine, and the second residue is arginine in almost all of the representative sequences shown (not a basic residue in only 2 of the 24, the bacterial and Trichomonas proteins). Our mutational analysis confirms the functional role of these residues in membrane localization of Myo1G and the more divergent long tail myosin Myo1F (Fig. 8, A and B). Note that the second signature motif residue in Myo1F, Arg-780, is less important functionally, consistent with its less consistent conservation during evolution.
FIGURE 8.
Identification of Arg and Lys residues in the Myo1PH that contribute to membrane localization. A, Jurkat cells were transfected with point mutants of the “signature residues” in the β1/β2 loop of the PH domain. B, quantitative analysis of membrane enrichment for the indicated mutants. Proteins not enriched at the plasma membrane typically have ratios of membrane/cytoplasmic localization in the range from 0.5 to 1.0 (dashed line). C, Jurkat cells transfected with point mutants in all the other Arg and Lys residues within “Myo1PH.” D, quantitative analysis of membrane enrichment for the indicated mutants. Proteins not enriched at the plasma membrane typically have ratios of membrane/cytoplasmic localization in the range from 0.5 to 1.0 (dashed line).
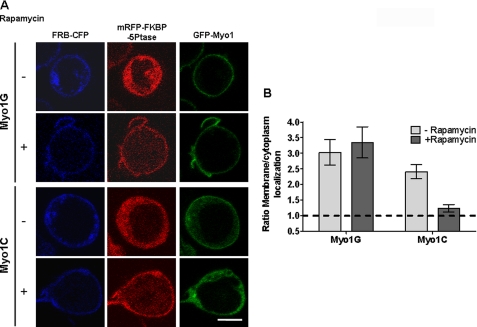
Previous structural and mutational analyses of other PH-like domains have implicated basic residues beyond the signature motif in mediating ligand binding. Because myosin I tail domains (and their divergent Myo1PH domains) have so far not been successfully crystallized, we used mutational analysis to identify other functionally important basic residues in this Myo1PH. We mutated all 12 Arg or Lys residues (distinct from the signature motif) to assure that unexpected contributions would not be missed. The results (Fig. 8, C and D) show that only one of those residues in the β3 strand, Lys-898, is critical to membrane localization. Notably, unlike the signature motif, which is conserved among all class I myosins, Lys-898 is conserved only among the Myo1G/Myo1D subfamily (Fig. 7). This suggested that the binding specificity of Myo1G might be significantly different from the previously characterized Myo1C binding to PIP2. To test this, we exploited an in-cell assay that acutely reduces membrane PIP2. Specifically, drug-inducible reduction in PIP2 can be achieved by transfection with a pair of constructs (26). One construct localizes to the plasma membrane. The other is a cytosolic phosphoinositide 5-phosphatase, which inducibly translocates to the membrane by rapamycin-induced association of the two constructs. Analysis of Myo1G and Myo1C localization at the plasma membrane before and after such inducible PIP2 hydrolysis demonstrates a marked difference (Fig. 9). As expected by previously demonstrated Myo1C dependence on PIP2, Myo1C was released from the membrane by addition of rapamycin. In contrast, Myo1G remained at the membrane. Thus, Myo1G localization at the membrane is not highly dependent on PIP2 levels.
FIGURE 9.
Effect of acute PIP2 reduction at the plasma membrane on localization of Myo1G. Jurkat cells were transfected with a rapamycin-inducible system to acutely reduce PIP2 levels in the plasma membrane (see “Experimental Procedures”). A, representative single-color images of individual cells transfected with the membrane-targeting construct (FRB-CFP, blue); the 5-Ptase construct (mRFP-FKBP-5-Ptase, red), and either Myo1G or Myo1C (green) in the presence or absence of Rapamycin. B, quantitative analysis of membrane enrichment for the indicated conditions. Proteins not enriched at the plasma membrane typically have ratios of membrane/cytoplasmic localization in the range from 0.5 to 1.0 (dashed line).
DISCUSSION
The foregoing results characterize Myo1G as a plasma membrane-associated class I myosin that is abundant in lymphocytes. Myo1G is abundant in several of the hematopoietic cells we studied (T-lymphocytes and B-lymphocytes as well as mast cells) and has previously been identified by Luna and colleagues in neutrophils (33). The tissue distribution of Myo1G has also been previously characterized as hematopoietic cell-restricted at a time when it was known only as the molecule carrying a minor histocompatibility HA-2 (34); HA-2 was subsequently shown to be a polymorphism of Myo1G (35). Broad evidence for the hematopoietic-specific expression of Myo1G is also provided from data in SymAtlas (36). Our further studies of Myo1G focus primarily on extending current understanding of the structural basis of its association with the membrane. The topics for discussion include: 1) optimal localization of Myo1G at the membrane is cooperative, involving head, IQ motifs, and Myo1PH region; 2) Myo1PH is an ancient divergent PH domain; 3) Myo1PH is not sufficient for membrane association but requires additional N- and C-terminal sequence; and 4) Class I myosins differ in their mechanisms of membrane association, because Myo1G does not share the PIP2 dependence of Myo1C and differs with respect to a critical basic residue in the Myo1PH domain.
First, efficient Myo1G localization at the membrane is a co-operative process involving multiple elements of the protein: head, IQ motifs, and Myo1PH-containing region. Although the Myo1G IQ tail region is sufficient for membrane localization (and the motor domain is not), the membrane/cytoplasm intensity ratio is >4-fold greater when Myo1G constructs contain both the motor and tail domains (Fig. 4, B and C). This result is similar to previous studies with Myo1B and Myo1C in epithelial cells, which showed that correct subcellular localization requires both the tail and motor domain (11, 37). This motor-dependent localization likely shows the importance of the actomyosin interaction in contributing to myosin localization with these proteins in these particular cells. Interestingly, the localization of Myo1C to the membrane in lymphoid cells does not appear to be dependent on the presence of the motor domain (Fig. 4, B and C), which may indicate that the Myo1C IQ tail region has a higher affinity for the plasma membrane than Myo1G in this cell type. In addition, the region containing IQ motifs also contributes in Myo1G, as evidenced by their augmentation of binding by the extended tail (Fig. 5), which previous reports suggest may entail binding to membrane lipid (13, 14).
Second, these studies identify the Myo1PH domain as an ancient, divergent family of PH-like domains. As outlined in the introduction, PH-like domains are a diverse superfamily with at least eight distinct families with strikingly different biochemical activities. Operationally, sequence profiles created to detect one family (e.g. PF00169 for classic PH) generally often do not detect other families (e.g. the third domain of FERM identified by PF09380 and vice versa). Thus, until recently, the presence of a PH-like domain in the tail of class I myosins had been missed: the existing PH-like domain family profiles were not able to detect it, because of sequence divergence. It is notable that the existence of a PH-like domain in class I myosins was first suggested by a threading-based algorithm for structure prediction (PHYRE) applied to mouse short tail Myo1C (14) and subsequently to Acanthamoeba long tail myosin (20). We have found that, in addition to PHYRE (28), another prediction server that scored very highly in recent CASP (Critical Assessment of Techniques for Protein Structure Prediction) competitions, the I-Tasser server (27) provides excellent PH-like models of the Myo1PH domains of all human class I myosins. As an example, an overlaid collection of the five best scoring I-Tasser models for Myo1G Myo1PH domain are included as a supplementary file (supplemental File 1).3
We sought a more general understanding beyond the two previously important instances (Myo1C and Acanthamoeba long tail myosin (14, 20)) and therefore undertook a more robust sequence analysis. Our objective was to assess what proteins across the evolutionary spectrum contain PH domains like those in class I myosins and how they relate to other PH domains. To do so we developed a sequence profile that is sensitive to the sequence elements found in the class I myosin PH-like domains, and we refer to the PH-like domains found by this profile as “Myo1PH.” Myo1PH domains constitute a distinct family of the diverse superfamily family of PH domains. Among myosins, the only myosins containing Myo1PH are class I myosins (short tail and long tail) and no others. However, Myo1PH are also found in other eukaryotes as diverse as green algae, Trichomonas and Trypanosoma, in which the Myo1PH occurs as a standalone domain with an N-terminal coiled-coil extension or combined to a protein kinase and classic PH domains (Fig. 7 and supplemental Table 1). The presence of a Myo1PH domain is predicted in the ancestral eukaryote on account of its presence in early-branching lineages such as Trichomonas and Trypanosoma.
It is notable that the Myo1PH profile identifies clear-cut sequence relatives in bacteria such as those from Chlorobium, Pelodictyon, and Prosthecochloris among other members of the green sulfur bacteria. Previously, domains with a PH-like fold in bacteria proteins had been recognized based on structural similarities (e.g. Shew0819, pdb code: 3dcx), but no evidence of an evolutionary relationship to eukaryotic PH domains has been reported for these bacterial proteins based on sequence analysis. As a confirmatory approach, we investigated whether threading-based structural prediction algorithms could also identify this sequence as a PH-like domain. Indeed it can. I-Tasser predicted PH-like domain organization for each of its five best models for the bacterial sequence. Notably this prediction occurs despite only 12% sequence identity between the query sequence and the PH domain structures on which it based its model. It is interesting that the PH-like structures on which the bacterial Myo1PH domain is modeled come from three different families (classic, phosphotyrosine binding, and FERM-like), emphasizing its possible ancestral relationship to multiple PH-like domain families. It is also interesting that the bacterial Myo1PH domain lacks the characteristic excess of basic residues found in all the eukaryotic Myo1PH domains around the lipid binding pocket. Perhaps its function was not lipid binding, or maybe this reflects the predominance of phosphatidylethanolamine in bacterial membranes (38) whose binding may not be dependent on patches of positively charged residues (see PDB 2IQX).
The third point of discussion is that the Myo1PH domain is necessary for membrane localization, but, unlike various classic PH domains, the Myo1PH domain alone is not sufficient for membrane localization. Our experiments were designed to test this possibility raised by bioinformatic analysis. The hint from bioinformatic analysis is that the region conserved during evolution is not restricted to the Myo1PH itself but rather extends both N-terminally and C-terminally from it. For example, a sequence profile called “TH1” (PF06017) exists in the automatically generated PFAM-B catalogue of conserved regions. This TH1 motif identifies a region of sequence conservation in class I myosin tails, which includes the Myo1PH in Myo1G but extends 51 aa N-terminally and 48 aa to the C terminus. Based on such information we initiated our search for a minimal Myo1G tail construct with one that included the entire TH1 profile-defined region plus 51 residues more at the N terminus. This construct (“tail”) was not sufficient for membrane localization, but a slightly longer construct (extended tail that included 64 residues N-terminal to PFAM TH1) was sufficient for membrane localization. There are a variety of possible reasons why the surrounding region may be required, but one plausible model is that Myo1PH is one lobe of a complex TH1 “domain,” which also includes a pre-Myo1PH lobe and a post-Myo1PH lobe. This would be similar in concept, for example, to the FERM module, in which one of three lobes is a PH domain (39). However, because the strength of the sequence conservation is much less than the FERM, such a TH1 domain must be more loosely organized than the FERM domain. Cryoelectron microscopic analysis suggest that the LB (lipid binding) region is not highly compact and, rather, includes two extensively interacting globular elements (15). Secondary structure predictions indicate that post-Myo1PH region is composed of β strand elements, but threading predictions do not succeed in modeling them on known folds. We have functionally characterized the post-PH region only by demonstrating that its deletion from the intact Myo1G destroys membrane localization. It is likely to form an additional C-terminal four-stranded sheet that could potentially provide a membrane-interaction interface via the basic residues found in it.
The 116-aa Pre-PH region is predicted to consist of multiple α helices. These helices could be in: 1) extended conformation (like the IQ motifs), 2) in a loosely organized helical bundle, or 3) in a compact helical domain-like lobe B of the FERM domain. The lack of strong sequence conservation of this region across evolution argues against a compact helical domain. Previous cryoelectron microscopic analysis suggests it may not be in extended conformation (15). Instead we favor the model of a loosely organized helical bundle. Such bundles can evolve much more rapidly than compact helical domains, which would explain limited conservation of this region and the failure of the PFAM TH1 profile to detect the critical functional boundary. How does this region contribute to membrane localization? Our mutational analysis of two basic residues on the same face of a predicted α helix suggests electrostatic interactions with membrane lipids may contribute. However, those results do no exclude other possibilities for the function of this α helical Pre-PH such as stabilization of conformation of the Myo1PH, or facilitating membrane association by binding to other elements, including possibly small G-proteins (40). One possible model for this membrane interaction is suggested by the α-helical coiled-coil domain found in the p85α subunit of the phosphatidylinositol 3-kinase α. This region (also called iSH2, because it occurs as a linker between two SH2 domains) lies against the lipid heads and cooperates with another lipid-binding domain, the C2 domain, from the p110α subunit of phosphatidylinositol 3-kinase α to localize the enzyme to the membrane (41). In a similar fashion it is conceivable the helical N-terminal extension of the Myo1PH domains might form an accessory membrane-contacting interface.
The fourth point of discussion is that our studies of Myo1G establish that the PH-dependent mechanism of membrane association is not uniform between class I myosins but instead differs substantially between Myo1G and Myo1C. Our functional characterization demonstrates that Myo1G association with the plasma membrane is strikingly different from Myo1C with respect to its apparent independence from plasma membrane PIP2. Inspection of the multiple sequence alignment of the Myo1PH domains (Fig. 7) demonstrates that each subfamily of class I myosins have different loop insertions, which is potentially interesting because there are three “specificity-determining regions” (SDRs), also known as “variable loops” (VLs), that have been shown to contribute to lipid binding specificity (42–44). Myo1A/B/C/H have insertions in the β6/7 loop (called SDR3 or VL3), and the long tail myosins have an insert in the β3/4 loop (SDR2 or VL2). However, the insertion in Myo1G/D is in the β7/α loop, which has not been reported to contribute to phospholipid binding specificity. Our systematic mutational analysis of basic residues in the Myo1PH domain of Myo1G highlights the functional importance of a basic residue Lys-898 in the β3 strand that is conserved in Myo1G/D but absent in all other Myo1PH domains. That position in classic PH domains is typically occupied by a tyrosine, which can be seen in solved structures to coordinate via hydrogen bonding to the P4 position of PIP2. Thus the exceptional Lys-898 is in the right area of the binding pocket to contribute to phosphate binding. The lipid binding specificities of Myo1G remain an important question to be addressed by future studies.
Supplementary Material
Acknowledgments
We thank Drs. T. Balla, G. R. Crabtree, G. S. Kansas, D. L. Nelson, and L. M. Wahl for reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant GM57247 and the Intramural Research Program of the National Institutes of Health, NCI. This work was also supported by the National Library of Medicine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2, Table S1, and File 1.
Supplemental File 1 contains the sequence of the Myo1G Myo1PH domain that was submitted to I-Tasser. All of the five top models returned were aligned using the iterative magic fit algorithm of Deepview. The results illustrate consistency of the models to overall PH domain architecture but modest variability among the models in the exact conformation of the β5/6 loop through the β7 strand.
- TH1
- tail homology 1
- 5-Ptase
- 5-phosphatase
- FERM
- 4.1 ezrin radixin moesin
- Myo1C
- myosin 1C
- Myo1F
- myosin 1F
- Myo1G
- myosin 1G
- PH
- pleckstrin homology
- Myo1PH
- myosin I PH-like domain
- PBT
- peripheral blood T cells
- RBL
- rat basophilic leukemia
- SDR
- specificity determining region
- VL
- variable loop
- WB
- Western blot
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PIP3
- phosphatidylinositol 1,4,5-trisphosphate
- aa
- amino acid(s)
- mAb
- monoclonal antibody
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- eGFP
- enhanced green fluorescent protein
- HMM
- Hidden Markov Model
- MMV
- membrane/microvillus
- mRFP
- monomeric red fluorescent protein
- PFAM
- Protein Family data base.
REFERENCES
- 1.Jacobelli J., Chmura S. A., Buxton D. B., Davis M. M., Krummel M. F. (2004) Nat. Immunol. 5, 531–538 [DOI] [PubMed] [Google Scholar]
- 2.Richards T. A., Cavalier-Smith T. (2005) Nature 436, 1113–1118 [DOI] [PubMed] [Google Scholar]
- 3.Foth B. J., Goedecke M. C., Soldati D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalhammer G., Bähler M. (2000) Essays Biochem. 35, 33–42 [DOI] [PubMed] [Google Scholar]
- 5.Engqvist-Goldstein A. E., Drubin D. G. (2003) Annu. Rev. Cell Dev. Biol. 19, 287–332 [DOI] [PubMed] [Google Scholar]
- 6.Kim S. V., Flavell R. A. (2008) Cell. Mol. Life Sci. 65, 2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nambiar R., McConnell R. E., Tyska M. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11972–11977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammer J. A. (1991) Trends Cell Biol. 1, 50–56 [DOI] [PubMed] [Google Scholar]
- 9.Adams R. J., Pollard T. D. (1989) Nature 340, 565–568 [DOI] [PubMed] [Google Scholar]
- 10.Doberstein S. K., Pollard T. D. (1992) J. Cell Biol. 117, 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruppert C., Godel J., Müller R. T., Kroschewski R., Reinhard J., Bähler M. (1995) J. Cell Sci. 108, 3775–3786 [DOI] [PubMed] [Google Scholar]
- 12.Tyska M. J., Mooseker M. S. (2002) Biophys. J. 82, 1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirono M., Denis C. S., Richardson G. P., Gillespie P. G. (2004) Neuron 44, 309–320 [DOI] [PubMed] [Google Scholar]
- 14.Hokanson D. E., Laakso J. M., Lin T., Sept D., Ostap E. M. (2006) Mol. Biol. Cell 17, 4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jontes J. D., Milligan R. A. (1997) J. Mol. Biol. 266, 331–342 [DOI] [PubMed] [Google Scholar]
- 16.Lemmon M. A., Ferguson K. M. (2000) Biochem. J. 350, 1–18 [PMC free article] [PubMed] [Google Scholar]
- 17.Ingley E., Hemmings B. A. (1994) J. Cell. Biochem. 56, 436–443 [DOI] [PubMed] [Google Scholar]
- 18.Balaji S., Babu M. M., Iyer L. M., Aravind L. (2005) Nucleic Acids Res. 33, 3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemmon M. A., Ferguson K. M. (2001) Biochem. Soc. Transact. 29, 377–384 [DOI] [PubMed] [Google Scholar]
- 20.Hwang K. J., Mahmoodian F., Ferretti J. A., Korn E. D., Gruschus J. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao J. J., Wang G., Pisitkun T., Patino-Lopez G., Nagashima K., Knepper M. A., Shen R. F., Shaw S. (2008) J. Proteome Res. 7, 2911–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cozier G. E., Carlton J., Bouyoucef D., Cullen P. J. (2004) Curr. Top. Microbiol. Immunol. 282, 49–88 [DOI] [PubMed] [Google Scholar]
- 23.Brown M. J., Nijhara R., Hallam J. A., Gignac M., Yamada K. M., Erlandsen S. L., Delon J., Kruhlak M., Shaw S. (2003) Blood 102, 3890–3899 [DOI] [PubMed] [Google Scholar]
- 24.Wahl S. M., Katona I. M., Stadler B. M., Wilder R. L., Helsel W. E., Wahl L. M. (1984) Cell. Immunol. 85, 384–395 [DOI] [PubMed] [Google Scholar]
- 25.Wagner M. C., Barylko B., Albanesi J. P. (1992) J. Cell Biol. 119, 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varnai P., Thyagarajan B., Rohacs T., Balla T. (2006) J. Cell Biol. 175, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S., Skolnick J., Zhang Y. (2007) BMC Biol. 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 29.Ley K., Tedder T. F., Kansas G. S. (1993) Blood 82, 1632–1638 [PubMed] [Google Scholar]
- 30.Dwir O., Kansas G. S., Alon R. (2001) J. Cell Biol. 155, 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyska M. J., Mackey A. T., Huang J. D., Copeland N. G., Jenkins N. A., Mooseker M. S. (2005) Mol. Biol. Cell 16, 2443–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee W. L., Ostap E. M., Zot H. G., Pollard T. D. (1999) J. Biol. Chem. 274, 35159–35171 [DOI] [PubMed] [Google Scholar]
- 33.Nebl T., Pestonjamasp K. N., Leszyk J. D., Crowley J. L., Oh S. W., Luna E. J. (2002) J. Biol. Chem. 277, 43399–43409 [DOI] [PubMed] [Google Scholar]
- 34.de Bueger M., Bakker A., Van Rood J. J., Van der Woude F., Goulmy E. (1992) J. Immunol. 149, 1788–1794 [PubMed] [Google Scholar]
- 35.Pierce R. A., Field E. D., Mutis T., Golovina T. N., Von Kap-Herr C., Wilke M., Pool J., Shabanowitz J., Pettenati M. J., Eisenlohr L. C., Hunt D. F., Goulmy E., Engelhard V. H. (2001) J. Immunol. 167, 3223–3230 [DOI] [PubMed] [Google Scholar]
- 36.Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang N., Ostap E. M. (2001) Curr. Biol. 11, 1131–1135 [DOI] [PubMed] [Google Scholar]
- 38.Cronan J. E. (2003) Annu. Rev. Microbiol. 57, 203–224 [DOI] [PubMed] [Google Scholar]
- 39.Pearson M. A., Reczek D., Bretscher A., Karplus P. A. (2000) Cell 101, 259–270 [DOI] [PubMed] [Google Scholar]
- 40.Lemmon M. A. (2004) Biochem. Soc. Trans. 32, 707–711 [DOI] [PubMed] [Google Scholar]
- 41.Mandelker D., Gabelli S. B., Schmidt-Kittler O., Zhu J., Cheong I., Huang C. H., Kinzler K. W., Vogelstein B., Amzel L. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16996–17001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baraldi E., Djinovic Carugo K., Hyvönen M., Surdo P. L., Riley A. M., Potter B. V., O'Brien R., Ladbury J. E., Saraste M. (1999) Structure 7, 449–460 [DOI] [PubMed] [Google Scholar]
- 43.Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., Lemmon M. A. (2000) Mol. Cell 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 44.Lietzke S. E., Bose S., Cronin T., Klarlund J., Chawla A., Czech M. P., Lambright D. G. (2000) Mol. Cell 6, 385–394 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.