Abstract
Varicella-zoster virus (VZV), the cause of chickenpox and zoster, was the first human herpesvirus to be sequenced fully and the first for which vaccines have been licensed and widely used. Three groups have published genotyping schemes based on single nucleotide polymorphisms (SNPs) and, between them, have identified five distinct phylogenetic clades, with an additional two putative clades. Sequencing of over 23 whole VZV genomes from around the world further refined the phylogenetic distinctions between SNP genotypes. Widespread surveillance in countries in which the varicella vaccine is now in use and the difficulties posed by three unique genotyping approaches prompted an international meeting, at which a common nomenclature based on phylogenetic clades was agreed upon. In this paper, we review the original genotyping schemes and discuss the basis for a novel common nomenclature for VZV strains. We propose a minimum set of SNPs that we recommend should be used to genotype these viruses. Finally, we suggest criteria by which novel clades can be recognized.
Introduction
Varicella-zoster virus (VZV; Human herpesvirus 3) is a member of the genus Varicellovirus, subfamily Alphaherpesvirinae. VZV exclusively infects humans, causing two diseases: chickenpox (varicella) following primary infection and shingles (herpes zoster), which results from reactivation of latently persistent virus (Hope-Simpson, 1965). VZV is related closely to other members of the genus Varicellovirus, including suid herpesvirus 1 (which infects pigs) and equine herpesviruses 1, 3, 4, 8 and 9, which infect horses (McGeoch & Cook, 1994). The most closely related human herpesviruses are herpes simplex virus 1 and 2. Varicella, although a relatively benign childhood infection in temperate countries, is nonetheless a cause of medically and economically important morbidity and mortality (Meyer et al., 2000; Preblud, 1986). In most countries, between 2 and 4 % of children are hospitalized for complications of chickenpox, including secondary bacterial infection, pneumonia and encephalitis (Sengupta et al., 2008). Mortality in children is around 0–0.05 deaths per 100 000 cases of chickenpox, primarily among children who were previously healthy (Sengupta et al., 2008). Adult infection, which is more common in tropical countries, results in a 6-fold increase in hospitalizations and up to a 20-fold increase in deaths (Marin et al., 2008; Rawson et al., 2001). Herpes zoster is a major cause of morbidity in the elderly from persistent neuropathic pain (Dworkin et al., 2007) and mortality in the immunocompromised, particularly from dissemination of the acute rash. The medical and economic burden of VZV disease has led to the introduction of childhood vaccination programmes against varicella in Japan, North America, parts of Europe, other Asian countries and the Antipodes (Bonanni et al., 2009). More recently, a vaccine to prevent herpes zoster has been licensed and is being administered to older people in the USA (Oxman et al., 2005).
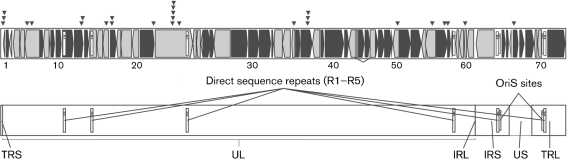
The VZV genome was one of the first human herpesvirus genomes to be sequenced fully; this was done at the MRC Virology Unit, Glasgow, UK (Davison & Scott, 1986). The Dumas strain, which was originally isolated from a patient with varicella in Holland, is approximately 125 kb in length. Seventy-two open reading frames (ORFs) numbered 0–71 have been identified, of which 69 are unique and three are duplicated (Fig. 1). Like other herpesviruses, VZV has unique long and short regions and two inverted-repeat regions, TR and IR, within which lie the duplicated genes (Fig. 1). VZV genomes differ in size by up to 500 bases and the differences are due to variation in the length of five tandem-repeat regions, R1–R5, and the viral origin of replication OriS (Tyler et al., 2007). R1, R2 and R3 lie with ORFs 11, 14 and 22, respectively, and are included in the coding regions of those genes (Davison & Scott, 1986). R4, R5 and the origin of replication OriS lie in non-coding regions. R4 and OriS are duplicated, occurring between ORFs 62 and 63 and ORFs 70 and 71. In recent years, 23 full-length sequences of 18 VZV genomes (Table 1) have been published, making the VZV genome the most-studied of the human herpesvirus genomes to date. The second virus to be sequenced fully was the parental vaccine strain pOka, as well as three preparations of the vaccine strain vOKa (Gomi et al., 2002). Two groups, one at the CDC and the other a collaboration between scientists at the University of Iowa and the Canadian National Public Health Laboratory, have published analyses of two and 11 novel genomes, respectively (Table 1) (Grose et al., 2004; Norberg et al., 2006; Peters et al., 2006).
Fig. 1.
Schematic representation of the VZV genome showing the ORFs, numbered from left to right. ▾ represent the positions of single-nucleotide polymorphisms (SNPs) that have been identified as useful for strain genotyping. Black-filled shapes represent ORFs that are transcribed late in the VZV replication cycle. IR, Inverted repeat; TR, terminal repeat; UL, unique long region; US, unique short region.
Table 1.
VZV whole genomes, GenBank accession numbers and references
| GenBank accession no. | VZV strain | Clade | Reference |
|---|---|---|---|
| X04370 | Dumas | 1 | Davison & Scott (1986) |
| EU154348 | SVETA | Loparev et al. (2009) | |
| DQ674250 | NH29_3 | V. N. Loparev, direct submission to GenBank (5 June 2006)* | |
| DQ479963 | 32 passage 72 | 1 | Peters et al. (2006) |
| DQ479962 | 32 passage 22 | 1 | Peters et al. (2006) |
| DQ479961 | 32 passage 5 | 1 | Peters et al. (2006) |
| DQ479960 | 8 | 4 | Peters et al. (2006) |
| DQ479959 | 49 | 1 | Peters et al. (2006) |
| DQ479958 | 36 | 1 | Peters et al. (2006) |
| DQ479957 | 03-500 | 3 | Peters et al. (2006) |
| DQ479956 | 22 | 3 | Peters et al. (2006) |
| DQ479955 | 11 | 3 | Peters et al. (2006) |
| DQ479954 | Kel | 1 | Peters et al. (2006) |
| DQ479953 | SD | 1 | Peters et al. (2006) |
| DQ457052 | CA123 | 5 | Norberg et al. (2006) |
| DQ452050 | DR | 4 | Norberg et al. (2006) |
| AJ871403 | HJ0 | 3 | Norberg et al. (2006) |
| DQ008355 | VariVax | 2 | Tillieux et al. (2008) |
| DQ008354 | VarilRix | 2 | Tillieux et al. (2008) |
| AY548170 | MSP | 1 | Grose et al. (2004) |
| AY548171 | BC | 1 | Grose et al. (2004) |
| AB097933 | Oka, substrain pOka | 2 | Gomi et al. (2002) |
| AB097932 | Oka, substrain vOka | 2 | Gomi et al. (2002) |
*NCID, CDC, 1600 Clifton Rd, Atlanta, GA 30333, USA.
If the variable regions are excluded, there are between 30 and 200 single nucleotide polymorphism (SNP) differences between any two genomes, i.e. between 1 in 4000 and 1 in 600 substitutions (Tyler et al., 2007). Approximately 30 % of these result in amino acid changes, and substitutions are not distributed evenly. If mutations are standardized per 100 bases, ORFs 1 and 62 are the most variable, whereas ORFs 3, 25 and 49 had no substitutions in 11 genomes (Tyler et al., 2007). Considering the genome in its double-stranded, linear form, the most variation occurs in ORFs sited at either end of the genome, i.e. ORFs 1–3 and 57–68, as well as in ORFs 35–37 (Tyler et al., 2007). The numbers of substitutions observed in the unique short region appear to be higher than in the unique long region, suggesting a different evolutionary rate (McGeoch, 2009).
Although some VZV genotyping by restriction fragment length polymorphism (RFLP) analysis of the variable region was carried out in the 1980s and 1990s, it was the development of SNP-typing tools that led to a better understanding of VZV molecular epidemiology and evolution. From 2000 onwards, SNP-based methodologies for genotyping were published by two USA-based and one UK-based groups (Barrett-Muir et al., 2001; Faga et al., 2001; Loparev et al., 2004; Muir et al., 2002). SNPs are sites that contain single base-pair variations (Collins, 1997). These sites are generally considered biallelic, due to the limited number of transversions that have been found (Brookes, 1999; Kruglyak, 1997). Each allele must occur in >1 % of the population to be considered an SNP (Kruglyak, 1997; Wang et al., 1998). Individuals who share many of the same SNPs are likely to have arisen from a common ancestor and can therefore be grouped by inheritance (Collins, 1997; Faga et al., 2001).
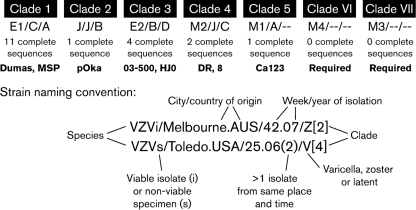
The three SNP-based genotyping methodologies differ in their approaches. The scattered SNP method, developed by the Breuer group in the UK, used heteroduplex-mobility assays to identify 92 polymorphisms in 37 ORFs spread evenly across the VZV genome (Barrett-Muir et al., 2001; Muir et al., 2002). This research group identified three distinct phylogenetic genotypes circulating in the UK and a fourth in Japan. The Iowa group, led by Grose, sequenced five glycoprotein genes and the ORF encoding the major immediate-early transactivating protein, IE62. Sixty-one polymorphisms, 21 in the glycoproteins and 40 in ORF 62, were identified (Faga et al., 2001). Phylogenetic analysis of these identified four clades (Peters et al., 2006). The third group, led by Schmid at the CDC, located a 447 bp fragment in ORF 22 upstream of the C-terminal coding region that was sufficiently polymorphic to allow identification of four viral clades (Table 2) (Loparev et al., 2004). Between them, the three groups identified five distinct clades, although no one group identified all five. The different nomenclatures used by each group are shown in Table 2 and the SNPs used for genotyping in Table 3.
Table 2.
Genotyping nomenclatures in current use
| Nomenclature | Based on: | Prototypic strain | ||||
|---|---|---|---|---|---|---|
| Dumas | pOka | 11 | 8, DR | CA123 | ||
| Iowa–Canada | Glycoprotein | A | B | D | C | |
| UK | Scattered SNP | C | J | B | A2/J2 | A |
| CDC | ORF 22 | E1 | J | E2 | M2 | M1 |
| New | Whole genome | 1 | 2 | 3 | 4 | 5 |
Table 3.
Minimum complement of SNPs required to identify a putative clade
SNPs are given as the nucleotide position according to GenBank accession no. X04370 (Dumas). nd, Not determined.
| ORF | SNP | Clade | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | VI | VII | ||
| 1 | 508 | C | C | C | C | C | T | nd |
| 1 | 685 | G | A | G | A | G | G | nd |
| 1 | 790 | T | C | T | C | C | T | nd |
| 6 | 5827 | C | C | A | C | C | C | nd |
| 6 | 6850 | G | G | G | A | A | G | nd |
| 12 | 17834 | C | T | C | T | C | T | nd |
| 12 | 18082 | C | T | C | C | C | C | nd |
| 16 | 23294 | A | G | G | A | G | G | nd |
| 17 | 24533 | A | A | A | A | C | A | nd |
| 17 | 24578 | A | G | G | A | G | G | nd |
| 21 | 33725 | T | C | C | C | C | C | nd |
| 22 | 37902 | A | G | A | A | A | A | G |
| 22 | 38019 | G | G | G | A | G | G | G |
| 22 | 38055 | T | C | T | C | T | T | T |
| 22 | 38081 | A | C | A | C | C | C | C |
| 22 | 38177 | G | A | G | A | G | A | G |
| 22 | 39394 | G | A | A | G | A | A | nd |
| 35 | 64703 | G | G | G | A | A | G | nd |
| 37 | 68101 | T | T | C | T | T | T | nd |
| 37 | 68172 | G | A | A | A | A | A | nd |
| 37 | 68254 | C | C | T | C | C | T | nd |
| 50 | 87841 | C | T | T | T | T | T | nd |
| 54 | 95241 | T | C | T | C | C | T | nd |
| 55 | 98437 | T | C | T | C | T | C | nd |
| 56 | 98825 | T | T | T | C | C | C | nd |
| 60 | 101464 | C | C | C | C | A | C | nd |
| 66 | 113243 | A | A | C | C | A | C | nd |
Whole-genome sequencing
The phylogeny of VZV was further informed by whole-genome sequencing carried out by US–Swedish and US–Canadian collaborations (Norberg et al., 2006; Peters et al., 2006). Until this time, seven full-length sequences were available: the Dumas strain, the Biken, Merck and GSK strains of the Oka vaccine strain, parental Oka, and MSP and BC, in which the identical amino acid substitution D150N had been reported in glycoprotein E (Davison & Scott, 1986; Gomi et al., 2002; Grose et al., 2004). The US (Iowa)–Canadian collaboration reported an additional 11 strains, whilst the US–Swedish collaboration reported partial sequence from an additional two strains. In the latter, repeat and tandem-repeat regions, including the R1–R5 and OriS regions, were excluded (Norberg et al., 2006). In the first study, all 11 additional strains sequenced were collected from North America, whilst in the US–Swedish study, one strain originated from Morocco and the second from the USA (Norberg et al., 2006; Peters et al., 2006). At the same time, the Iowa–Canadian group carried out the definitive investigation of virus stability. Repeated passage of a virus in cell culture followed by whole-genome sequencing showed that, excluding unstable repeat regions, one substitution (8×10−6 substitutions per base) occurred following 20 passages and 28 substitutions (2.2×10−4 per base) after 72 passages. This suggests that, above 20 passages, the virus becomes unrepresentative of the original (Tyler et al., 2007). Table 1 shows the common clade nomenclature agreed upon at a meeting held in 2008, alongside each of the others; this will be used from this point forward. The power of full-length sequence to establish evolutionary history and shed light on pathogenesis was exemplified by work on the MSP and BC strains of VZV. In 2000, the Grose laboratory described the MSP virus, in which a mutation in a glycoprotein E (gE) B-cell epitope abrogated binding of an anti-gE antibody (Santos et al., 2000). Two years later, the same gE amino acid mutation was described by the Tipples group in a Canadian VZV community isolate, strain BC (Tipples et al., 2002). As the VZV genome is extremely stable, it was initially unclear whether the mutations in BC and MSP had arisen independently. The Iowa–Canadian collaboration hypothesized that MSP and BC might indeed represent a distinct subclade that was now spreading independently in North America. Sequencing of full-length genomes established that the viruses, although both belonging to clade 1, were not related more closely to each other than they were to other viruses within the same clade (Grose et al., 2004). Thus, the substitution in gE was a homoplasty, i.e. an identical mutation that had arisen in an evolutionarily independent manner in two unrelated viruses, most probably to escape neutralizing antibody. Subsequently, in an analysis of 634 VZV isolates collected at the Karolinska University Hospital, Sweden, two additional similar gE mutant viruses were discovered (Wirgart et al., 2006).
Early on, opportunistic genotyping by many groups identified differences in the global distribution of the five clades (Loparev et al., 2004; Muir et al., 2002; Wagenaar et al., 2003). Two clades (1 and 3) were found in European countries and areas of the world predominantly settled by Europeans, whilst clade 2 was identified as being predominantly present in Japan and surrounding countries (Loparev et al., 2004; Muir et al., 2002; Wagenaar et al., 2003). Because the varicella vaccine was developed in Japan, the majority of sequenced clade 2 strains originated from Japan. The two remaining clades, 4 and 5, are most prevalent in Asian and African countries (Loparev et al., 2004, 2007b; Muir et al., 2002; Wagenaar et al., 2003). The results obtained from genotyping of opportunistically collected samples were confirmed by analysis of zoster samples collected prospectively from European subjects living in the UK, the oldest of whom was over 100 years of age (Sengupta et al., 2007). Making the assumption that the virus reactivating as zoster and the virus responsible for primary infection were the same strain, it was confirmed that the European clades 1 and 3 were present in 90 % of Caucasians living in the UK and that this prevalence had not changed over the past 100 years. Reassuringly, this validated the conclusions about geographical distribution of strains and supported the value of opportunistic genotyping. These findings have enabled a number of groups to observe an increased circulation of clade 5 viruses in recent years (Hawrami et al., 1997; Quinlivan et al., 2002; Sauerbrei & Wutzler, 2007).
Following the initial publications, the CDC group added SNPs from ORFs 21 and 50 to those used originally in the ORF 22 genotyping scheme (Table 2) (Loparev et al., 2007b; Sergeev et al., 2006). Using these, they have suggested that, in addition to the five existing phylogenetic clades, there may be two additional stable clades, which they designated M3 and M4 (Loparev et al., 2007b, 2009; Sauerbrei et al., 2008; Sergeev et al., 2006) (Table 2; Fig. 2). M4 is negative for a BglI site in ORF 54, which is a characteristic feature of the ‘European origin’ clades 1 and 3 (Quinlivan & Breuer, 2006; Sengupta et al., 2007) and has in fact been shown by the CDC group to resemble European clades 1 and 3 (Loparev et al., 2009). It is possible that they may be the same as the viruses described as clade 1 and 3 ‘recombinants’ by the UK group (Muir et al., 2002). However, extensive SNP typing has shown M4 to have unique SNPs that are not shared with any other clade. The data suggest that, once full-length sequence has been obtained, the status of M4 as an independent clade is likely to be confirmed (Loparev et al., 2009). The status of M3 is less certain. The virus is positive for the ORF54 BglI site, in which respect it resembles the clade 2, 4 and 5 viruses that predominate in Japan, Asia and Africa, respectively. M3 has been described only a few times and more sequencing data are needed to confirm its existence as an independent clade (Loparev et al., 2007a; Sergeev et al., 2006).
Fig. 2.
Proposed new VZV nomenclature system, adopted 25 July 2008, Whitechapel, London, UK. Summary: major circulating clusters are designated as clades; there are no geographical implications in the naming system; established clades are designated by Arabic numerals; provisional clades are designated by Roman numerals. Reference strains for each clade are indicated in bold; additional reference strains are required for clades 2, 5, VI and VII.
The 2008 VZV nomenclature
To resolve the confusion associated with three different genotyping classifications, a meeting (see attendance list in Acknowledgements) to discuss a common nomenclature was held in London, UK, in 2008. Four principles were agreed upon: that the new nomenclature should be distinct from the three established nomenclatures, that it should reflect the phylogenetic structure of VZV strains, that it should reflect the order in which full-genome sequences were published or deposited in the databases, and that it should describe the provenance of the strain. To that end, the proposed nomenclature for VZV and the prototypic viruses for each clade are shown in Table 2 and Figs 2 and 3. A summary of the proposed nomenclature overall is shown in Fig. 2. We have used the word ‘clade’ to designate an evolutionarily distinct subspecies of VZV. The intention is that each clade should have at least two whole genomes. Clades 2 and 5 have only one genome and further work to generate a second prototype for each clade is ongoing. Several principles for naming strains have been agreed and are listed below.
Strains of VZV will now be identified by the disease (varicella, zoster or latent).
Novel viruses will be designated a member of an existing clade, based on phylogenetic clustering or SNP profile as outlined in Table 3.
Strains will be identified as cultured isolates (i) or sequence (s).
The geographical and temporal origin of the virus will be identified.
Putatively novel clades will be identified by Roman numerals until confirmed as a clade.
Fig. 3.
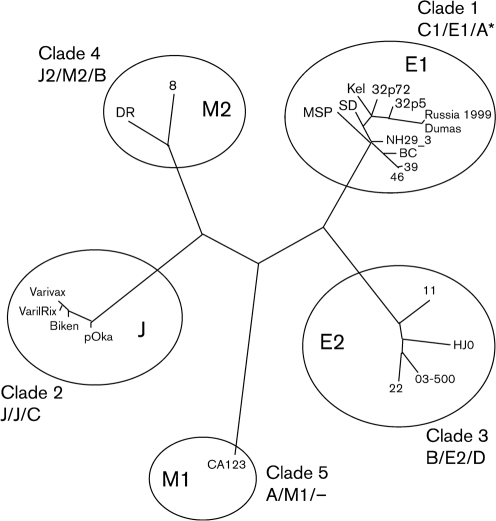
Phylogenetic tree showing five major VZV clades (adapted from Loparev, 2007b). Previous genotyping schemes are shown in the order Breuer/Schmid/Grose.
Some of the laboratories that can provide support for genome sequencing are listed in Table 4.
Table 4.
Location of laboratories providing reference VZV genotyping and genome sequencing
| Country | Institution | Contact | SNP genotyping | Whole-genome sequencing | Vaccine strain identification |
|---|---|---|---|---|---|
| USA | CDC | D. S. Schmid* | ORFs 21, 22, 38, 50, 54, 62 | 84 kb (targeted) by resequencing chip | Routinely perform four methods for SNPs in ORFs 38, 54 and 62 |
| UK | National VZV Reference Laboratory | Judy Breuer† | ORFs 21, 22, 38, 50, 54, 62 | 125 kb sequencing | Routinely perform four methods for SNPs in ORFs 38, 54 and 62 |
| France | Virology Department, Teacher Hospital, Limoges | Sophie Alain‡ | ORFs 38, 54, 62, to be completed soon by ORFs 21, 22, 50 | Not available | Routinely perform two methods for SNPs in ORFs 38, 54 and 62 (sequencing and RFLP) and RFLP in R5 region |
| Germany | National HSV, VZV Reference Laboratory | Andreas Sauerbrei§ | ORFs 1, 21, 22, 37, 38, 50, 54, 60, 62 | 125 kb sequencing | Routinely perform two methods for SNPs in ORFs 38, 54, and 62 |
| Australia | Institute of Clinical Pathology and Medical Research | Cheryl Toi|| | ORFs 21, 37, 60, 62 | Not available | Routinely perform two methods for SNPs in ORF 62 |
*Phone, +1-404-639-0066; fax, +1-404-639-4056; email, SSchmid@cdc.gov.
†Windeyer Building, 46 Cleveland St, London WC1 4JF, UK. Phone, +44 207 679 9559; email, j.breuer@ucl.ac.uk.
‡Laboratoire de Virologie CHU Dupuytren, 2 Avenue Martin Luther King, 87042 Limoges CEDEX, France. Phone, +33 5 55 05 67 28/67 24; email, sophie.alain@unilim.fr.
§Institute of Virology and Antiviral Therapy, University of Jena, Hans-Knoell-Strasse 2, 07745 Jena, Germany. Phone, +49-3641-9395700; fax, +49-3641-9395702; email, Andreas.Sauerbrei@med.uni-jena.de.
||Westmead Hospital, Westmead, NSW 2145, Australia. Email, cheryl.toi@swahs.health.nsw.gov.au.
Future studies
Vaccination programmes to prevent varicella in children are now established in many resource-rich countries, and surveillance of viral genotypes has been set up by some to support these efforts (Table 4). Reference genotyping of varicella is provided by several countries and the establishment of a common nomenclature will facilitate interchange of information. In particular, the success of the vaccine programmes in preventing wild-type varicella and of genotyping the viruses causing outbreaks has been, and will continue to be, central to ongoing implementation and improvement. Most recently, the emergence of wild-type vaccine recombinant viruses provides a challenge for which the unified nomenclature and common genotyping will be important, not only for surveillance but also for pathogenesis (D. S. Schmid, unpublished data). Of the three such recombinants analysed so far, two are vaccine-clade 1 and one is vaccine-clade 3. This discovery was not totally unexpected, as recombination between two different live vaccine viruses has been documented previously with pseudorabies virus, an alphaherpesvirus that infects pigs (Dangler et al., 1993). A common nomenclature will also allow better understanding of VZV genome biology, including the emergence of new strains following biological and immune selection across functionally important regions. Finally, there is still work to be done on defining relatedness between strains of the same clade for investigation of outbreaks. The stability of the VZV genome and low numbers of substitutions between strains, particularly within the same clade, preclude the use of SNPs for transmission studies. However, signature sequences that are unique or uncommon may be useful for establishing chains of transmission, as was the case in an outbreak of varicella in a residential care home (Lopez et al., 2008). An alternative approach is to make use of repeat regions within the VZV genome, which evolve at a faster rate. Two such regions that have been used to investigate hospital transmissions are R1 in ORF 11 (Molyneaux et al., 2006; Tang et al., 2005) and OriS (J. Breuer, unpublished data). However, the precise parameters that will inform their use for investigation of outbreaks remain to be established. Ultimately, genome sequencing may prove to be the most useful tool for investigation of VZV transmission events.
Acknowledgments
We all acknowledge the importance of the initial VZV sequence analysis by A. Davison. Research by C. G. was supported by NIH grant AI22785. Research by G. T. was supported by a grant from the office of the Chief Scientist, Health Canada. Research by J. B. was supported by a grant from Barts and the London Special Trustees. J. B. is supported by UCLH Collaborative Biomedical Research Centre. Thanks also to the following who attended the 2008 VZV Nomenclature Meeting, Whitechapel, London, UK: Karin T. Averbeck, Inga Dry, Suzanna L. R. McDonald, Mark Quinlivan, Nitu Sengupta (Centre for Infectious Disease, Institute of Cell and Molecular Science, Barts, and the London School of Medicine and Dentistry, Queen Mary College, London E1 2AT, UK), Tomas Bergström (Department of Clinical Virology, Göteborg University, Guldhedsgatan 10B, S-413 46 Göteborg, Sweden), Vanda Bostik, Jane F. Seward (Division of Viral Diseases, NCID, CDC, Atlanta, GA 30333, USA), David Brown, Kevin Brown (Centre for Infection, Health Protection Agency, 61 Colindale Ave, London NW9 5EQ, UK), Randall J. Cohrs (Department of Neurology, University of Colorado Health Sciences Center, 4200 East 9th Avenue, Mail Stop B182, Denver, CO 80262, USA), Ruth Harbecke, Michael N. Oxman (VA San Diego Medical Center, 3350 La Jolla Village Drive, San Diego, CA 92161-0002, USA) and Richard A. Nichols (School of Biological and Chemical Sciences, Queen Mary University of London, London, UK). The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agencies.
References
- Barrett-Muir, W., Hawrami, K., Clarke, J. & Breuer, J. (2001). Investigation of varicella-zoster virus variation by heteroduplex mobility assay. Arch Virol Suppl 17, 17–25. [DOI] [PubMed] [Google Scholar]
- Bonanni, P., Breuer, J., Gershon, A., Gershon, M., Hryniewicz, W., Papaevangelou, V., Rentier, B., Rumke, H., Sadzot-Delvaux, C. & other authors (2009). Varicella vaccination in Europe – taking the practical approach. BMC Med 7, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes, A. J. (1999). The essence of SNPs. Gene 234, 177–186. [DOI] [PubMed] [Google Scholar]
- Collins, F. S. (1997). Sequencing the human genome. Hosp Pract (Minneap) 32, 35–54. [DOI] [PubMed] [Google Scholar]
- Dangler, C. A., Henderson, L. M., Bowman, L. A. & Deaver, R. E. (1993). Direct isolation and identification of recombinant pseudorabies virus strains from tissues of experimentally co-infected swine. Am J Vet Res 54, 540–545. [PubMed] [Google Scholar]
- Davison, A. J. & Scott, J. E. (1986). The complete DNA sequence of varicella-zoster virus. J Gen Virol 67, 1759–1816. [DOI] [PubMed] [Google Scholar]
- Dworkin, R. H., Johnson, R. W., Breuer, J., Gnann, J. W., Levin, M. J., Backonja, M., Betts, R. F., Gershon, A. A., Haanpaa, M. L. & other authors (2007). Recommendations for the management of herpes zoster. Clin Infect Dis 44 (Suppl. 1), S1–S26. [DOI] [PubMed] [Google Scholar]
- Faga, B., Maury, W., Bruckner, D. A. & Grose, C. (2001). Identification and mapping of single nucleotide polymorphisms in the varicella-zoster virus genome. Virology 280, 1–6. [DOI] [PubMed] [Google Scholar]
- Gomi, Y., Sunamachi, H., Mori, Y., Nagaike, K., Takahashi, M. & Yamanishi, K. (2002). Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J Virol 76, 11447–11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose, C., Tyler, S., Peters, G., Hiebert, J., Stephens, G. M., Ruyechan, W. T., Jackson, W., Storlie, J. & Tipples, G. A. (2004). Complete DNA sequence analyses of the first two varicella-zoster virus glycoprotein E (D150N) mutant viruses found in North America: evolution of genotypes with an accelerated cell spread phenotype. J Virol 78, 6799–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrami, K., Hart, I. J., Pereira, F., Argent, S., Bannister, B., Bovill, B., Carrington, D., Ogilvie, M., Rawstorne, S. & other authors (1997). Molecular epidemiology of varicella-zoster virus in East London, England, between 1971 and 1995. J Clin Microbiol 35, 2807–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope-Simpson, R. E. (1965). The nature of herpes zoster: a long-term study and a new hypothesis. Proc R Soc Med 58, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak, L. (1997). The use of a genetic map of biallelic markers in linkage studies. Nat Genet 17, 21–24. [DOI] [PubMed] [Google Scholar]
- Loparev, V. N., Gonzalez, A., Deleon-Carnes, M., Tipples, G., Fickenscher, H., Torfason, E. G. & Schmid, D. S. (2004). Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J Virol 78, 8349–8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loparev, V., Martro, E., Rubtcova, E., Rodrigo, C., Piette, J. C., Caumes, E., Vernant, J. P., Schmid, D. S. & Fillet, A. M. (2007a). Toward universal varicella-zoster virus (VZV) genotyping: diversity of VZV strains from France and Spain. J Clin Microbiol 45, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loparev, V. N., Rubtcova, E. N., Bostik, V., Govil, D., Birch, C. J., Druce, J. D., Schmid, D. S. & Croxson, M. C. (2007b). Identification of five major and two minor genotypes of varicella-zoster virus strains: a practical two-amplicon approach used to genotype clinical isolates in Australia and New Zealand. J Virol 81, 12758–12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loparev, V. N., Rubtcova, E. N., Bostik, V., Tzaneva, V., Sauerbrei, A., Robo, A., Sattler-Dornbacher, E., Hanovcova, I., Stepanova, V. & other authors (2009). Distribution of varicella-zoster virus (VZV) wild-type genotypes in northern and southern Europe: evidence for high conservation of circulating genotypes. Virology 383, 216–225. [DOI] [PubMed] [Google Scholar]
- Lopez, A. S., Burnett-Hartman, A., Nambiar, R., Ritz, L., Owens, P., Loparev, V. N., Guris, D. & Schmid, D. S. (2008). Transmission of a newly characterized strain of varicella-zoster virus from a patient with herpes zoster in a long-term-care facility, West Virginia, 2004. J Infect Dis 197, 646–653. [DOI] [PubMed] [Google Scholar]
- Marin, M., Watson, T. L., Chaves, S. S., Civen, R., Watson, B. M., Zhang, J. X., Perella, D., Mascola, L. & Seward, J. F. (2008). Varicella among adults: data from an active surveillance project, 1995–2005. J Infect Dis 197 (Suppl. 2), S94–S100. [DOI] [PubMed] [Google Scholar]
- McGeoch, D. J. (2009). Lineages of varicella-zoster virus. J Gen Virol 90, 963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch, D. J. & Cook, S. (1994). Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Mol Biol 238, 9–22. [DOI] [PubMed] [Google Scholar]
- Meyer, P. A., Seward, J. F., Jumaan, A. O. & Wharton, M. (2000). Varicella mortality: trends before vaccine licensure in the United States, 1970–1994. J Infect Dis 182, 383–390. [DOI] [PubMed] [Google Scholar]
- Molyneaux, P. J., Parker, S., Khan, I. H., Millar, C. G. & Breuer, J. (2006). Use of genomic analysis of varicella-zoster virus to investigate suspected varicella-zoster transmission within a renal unit. J Clin Virol 36, 76–78. [DOI] [PubMed] [Google Scholar]
- Muir, W. B., Nichols, R. & Breuer, J. (2002). Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J Virol 76, 1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg, P., Liljeqvist, J. A., Bergstrom, T., Sammons, S., Schmid, D. S. & Loparev, V. N. (2006). Complete-genome phylogenetic approach to varicella-zoster virus evolution: genetic divergence and evidence for recombination. J Virol 80, 9569–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxman, M. N., Levin, M. J., Johnson, G. R., Schmader, K. E., Straus, S. E., Gelb, L. D., Arbeit, R. D., Simberkoff, M. S., Gershon, A. A. & other authors (2005). A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N Engl J Med 352, 2271–2284. [DOI] [PubMed] [Google Scholar]
- Peters, G. A., Tyler, S. D., Grose, C., Severini, A., Gray, M. J., Upton, C. & Tipples, G. A. (2006). A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating clades. J Virol 80, 9850–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preblud, S. R. (1986). Varicella: complications and costs. Pediatrics 78, 728–735. [PubMed] [Google Scholar]
- Quinlivan, M. & Breuer, J. (2006). Molecular studies of varicella zoster virus. Rev Med Virol 16, 225–250. [DOI] [PubMed] [Google Scholar]
- Quinlivan, M., Hawrami, K., Barrett-Muir, W., Aaby, P., Arvin, A., Chow, V. T., John, T. J., Matondo, P., Peiris, M. & other authors (2002). The molecular epidemiology of varicella-zoster virus: evidence for geographic segregation. J Infect Dis 186, 888–894. [DOI] [PubMed] [Google Scholar]
- Rawson, H., Crampin, A. & Noah, N. (2001). Deaths from chickenpox in England and Wales 1995–7: analysis of routine mortality data. BMJ 323, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, R. A., Hatfield, C. C., Cole, N. L., Padilla, J. A., Moffat, J. F., Arvin, A. M., Ruyechan, W. T., Hay, J. & Grose, C. (2000). Varicella-zoster virus gE escape mutant VZV-MSP exhibits an accelerated cell-to-cell spread phenotype in both infected cell cultures and SCID-hu mice. Virology 275, 306–317. [DOI] [PubMed] [Google Scholar]
- Sauerbrei, A. & Wutzler, P. (2007). Different genotype pattern of varicella-zoster virus obtained from patients with varicella and zoster in Germany. J Med Virol 79, 1025–1031. [DOI] [PubMed] [Google Scholar]
- Sauerbrei, A., Zell, R., Philipps, A. & Wutzler, P. (2008). Genotypes of varicella-zoster virus wild-type strains in Germany. J Med Virol 80, 1123–1130. [DOI] [PubMed] [Google Scholar]
- Sengupta, N., Taha, Y., Scott, F. T., Leedham-Green, M. E., Quinlivan, M. & Breuer, J. (2007). Varicella-zoster-virus genotypes in East London: a prospective study in patients with herpes zoster. J Infect Dis 196, 1014–1020. [DOI] [PubMed] [Google Scholar]
- Sengupta, N., Booy, R., Schmitt, H. J., Peltola, H., Van-Damme, P., Schumacher, R. F., Campins, M., Rodrigo, C., Heikkinen, T. & other authors (2008). Varicella vaccination in Europe: are we ready for a universal childhood programme? Eur J Pediatr 167, 47–55. [DOI] [PubMed] [Google Scholar]
- Sergeev, N., Rubtcova, E., Chizikov, V., Schmid, D. S. & Loparev, V. N. (2006). New mosaic subgenotype of varicella-zoster virus in the USA: VZV detection and genotyping by oligonucleotide-microarray. J Virol Methods 136, 8–16. [DOI] [PubMed] [Google Scholar]
- Tang, J. W., Eames, I., Li, Y., Taha, Y. A., Wilson, P., Bellingan, G., Ward, K. N. & Breuer, J. (2005). Door-opening motion can potentially lead to a transient breakdown in negative-pressure isolation conditions: the importance of vorticity and buoyancy airflows. J Hosp Infect 61, 283–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillieux, S. L., Halsey, W. S., Thomas, E. S., Voycik, J. J., Sathe, G. M. & Vassilev, V. (2008). Complete DNA sequences of two Oka strain varicella-zoster virus genomes. J Virol 82, 11023–11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipples, G. A., Stephens, G. M., Sherlock, C., Bowler, M., Hoy, B., Cook, D. & Grose, C. (2002). New variant of varicella-zoster virus. Emerg Infect Dis 8, 1504–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, S. D., Peters, G. A., Grose, C., Severini, A., Gray, M. J., Upton, C. & Tipples, G. A. (2007). Genomic cartography of varicella-zoster virus: a complete genome-based analysis of strain variability with implications for attenuation and phenotypic differences. Virology 359, 447–458. [DOI] [PubMed] [Google Scholar]
- Wagenaar, T. R., Chow, V. T., Buranathai, C., Thawatsupha, P. & Grose, C. (2003). The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21, 1072–1081. [DOI] [PubMed] [Google Scholar]
- Wang, D. G., Fan, J. B., Siao, C. J., Berno, A., Young, P., Sapolsky, R., Ghandour, G., Perkins, N., Winchester, E. & other authors (1998). Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- Wirgart, B. Z., Estrada, V., Jackson, W., Linde, A. & Grose, C. (2006). A novel varicella-zoster virus gE mutation discovered in two Swedish isolates. J Clin Virol 37, 134–136. [DOI] [PubMed] [Google Scholar]