This work demonstrates that region-specific interelement recombinational exchange, behind natural selection, plays a primary role in maintaining preexisting partnership and establishing new partnership between nonautonomous and autonomous long terminal repeat retrotransposons in soybean.
Abstract
Long terminal repeat (LTR) retrotransposons, the most abundant genomic components in flowering plants, are classifiable into autonomous and nonautonomous elements based on their structural completeness and transposition capacity. It has been proposed that selection is the major force for maintaining sequence (e.g., LTR) conservation between nonautonomous elements and their autonomous counterparts. Here, we report the structural, evolutionary, and expression characterization of a giant retrovirus-like soybean (Glycine max) LTR retrotransposon family, SNARE. This family contains two autonomous subfamilies, SAREA and SAREB, that appear to have evolved independently since the soybean genome tetraploidization event ∼13 million years ago, and a nonautonomous subfamily, SNRE, that originated from SAREA. Unexpectedly, a subset of the SNRE elements, which amplified from a single founding SNRE element within the last ∼3 million years, have been dramatically homogenized with either SAREA or SAREB primarily in the LTR regions and bifurcated into distinct subgroups corresponding to the two autonomous subfamilies. We uncovered evidence of region-specific swapping of nonautonomous elements with autonomous elements that primarily generated various nonautonomous recombinants with LTR sequences from autonomous elements of different evolutionary lineages, thus revealing a molecular mechanism for the enhancement of preexisting partnership and the establishment of new partnership between autonomous and nonautonomous elements.
INTRODUCTION
Retrotransposons are a class of mobile genetic elements that transpose via an RNA intermediate. Based on their structural features, retrotransposons are divided into a number of subclasses, including long terminal repeat (LTR) retrotransposons and non-LTR elements. The latter are the most abundant transposable elements in mammalian genomes, while the former account for a significant fraction of plant genomes (Kumar and Bennetzen, 1999). It has been documented that, in addition to polyploidization, the aggressive accumulation of LTR retrotransposons is the primary mechanism driving plant genome expansion (Bennetzen et al., 2005). In large-genome species, such as maize (Zea mays), barley (Hordeum vulgare), and wheat (Triticum aestivum), LTR retrotransposons make up >70 to 80% of their respective genomes, and the majority of the intact elements amplified within the last few million years (SanMiguel et al., 1996, 1998; Vicient et al., 1999; Wicker et al., 2001; Bruggmann et al., 2006). Myriad LTR retrotransposon families exist in plants, but, in general, only a few amplify to high copy numbers in a genome. For instance, >85% of LTR retrotransposons in the maize genome belong to the five largest families. A recent study shows that Oryza australiensis, a wild species of rice, has proliferated >90,000 copies of LTR retrotransposons belonging to only three families during the last three million years, leading to a twofold increase of its genome size in that time period (Piegu et al., 2006).
Despite extensive proliferation, LTR retrotransposons in plant genomes are nevertheless suppressed by a variety of mechanisms, including DNA methylation (Hamilton et al., 2002; Palmer et al., 2003; Emberton et al., 2005), conversion to heterochromatin (Lippman et al., 2004), formation of solo LTRs by unequal intraelement recombination (Devos et al., 2002; Ma et al., 2004), and accumulation of small deletions by illegitimate recombination (Devos et al., 2002; Ma et al., 2004). It was estimated that >190 Mb of retrotransposon DNA has been removed from the rice (Oryza sativa) genome by unequal recombination and illegitimate recombination within the past 4 million years, leaving a current genome of ∼400 Mb that contains <100 Mb of detectable retroelements or fragments (Ma et al., 2004). It can be predicted that the abundance of LTR retrotransposons in plant genomes is largely determined by the activities of competing mechanisms for the regulation of retrotransposon amplification and the generation of small deletions (Bennetzen et al., 2005).
In addition to two highly similar LTRs, an intact element, typically, contains gag, a gene that encodes a polyprotein comprising subcomponents of the virus-like particle (VLP) involved in the maturation and packaging of retrotransposon RNA, and pol gene products that encode protease, reverse transcriptase (RT), RNase H (RH), and integrase (IN) involved in the synthesis and integration of retrotransposon DNA into the host genome (Wicker et al., 2007). Based on the order of RT and IN in POL, LTR retrotransposons are classified into Gypsy (RT-RH-IN) and Copia (IN-RT-RH) types (Xiong and Eickbush, 1990). Regardless of its current transpositional activity, an intact element is generally defined as autonomous if it appears to encode all the protein-coding domains necessary for catalyzing its transposition. By contrast, an element that lacks some or all of the protein coding domains but appears to have or have had amplification capability is defined as nonautonomous (Wicker et al., 2007). A nonautonomous element is presumed to transpose by hijacking the transposition machinery of its autonomous partner. Several nonautonomous LTR retrotransposon families have been found in plants (Jin and Bennetzen, 1989; Hu et al., 1995; Lander et al., 2001; Witte et al., 2001; Jiang et al., 2002; Kalendar et al., 2004; Kejnovsky et al., 2006), but in most cases, their autonomous counterparts have not been discovered. Hence, little is known regarding their origins and the evolutionary processes shaping their structure. Nevertheless, putative autonomous and nonautonomous LTR retrotransposon partners, Dasheng and Rire2, were identified in rice, and these elements share substantial sequence identity in their termini, including LTRs, primer binding site (PBS), and polypurine tract (PPT) downstream of the PBS and upstream of the PPT (Jiang et al., 2002). These regions are assumed to be cis-sequences required for transpositions of both autonomous and nonautonomous elements (Jiang et al., 2002); thus, selection would favor their conservation. Despite this, nearly all the intact Dasheng and Rire2 elements were essentially grouped into two distinct families/phylogenetic clades based on their conserved sequences, such as LTRs (Jiang et al., 2002).
The availability of recently sequenced large genomes, such as that of soybean (Glycine max), provides unprecedented opportunities for investigation of transposable elements and their evolution in a complex plant genome. By screening the assembled soybean whole-genome sequence (http://www.phytozome.net; Schmutz et al., 2010), we identified 510 LTR retrotransposon families, and their copy numbers (intact elements and solo LTRs) vary from 1 to 5832 with a median value of 3. The family with the largest copy number, designated as SNARE, drew our attention because (1) it is the family that contains the largest size (up to 20 kb) LTR retrotransposons in the soybean genome; (2) it contains autonomous elements and nonautonomous elements; (3) the autonomous elements contain an env-like protein domain, a signature of putative endogenous plant retrovirus; (4) the autonomous elements can be clearly classified into two distinct lineages that roughly date the recent soybean genome allotetraploidization event; and (5) a subset of nonautonomous elements share an unique insertion of a piggybacking solo LTR of an unrelated retrotransposon family and thus can be used as a unique marker to track the evolutionary history of a specific lineage of SNARE elements. Here, we show evidence of extensive recombination between the autonomous and nonautonomous elements of SNARE family that is likely to have occurred during reverse transcription processes. These processes, followed by selection, have led to extensive region (particularly LTR)-specific sequence replacement of nonautonomous elements by their autonomous partners. This study provides a molecular description of the bifurcation and enhancement of autonomous and nonautonomous retrotransposon partnership by LTR swapping in any organisms.
RESULTS
Characterization of SNARE LTR Retrotransposon Family in the Soybean Genome
Using a combination of structural analysis and sequence homology comparison, as described earlier (Ma et al., 2004; Ma and Bennetzen, 2004), we mined 975 Mb of the assembled soybean genomic sequence for LTR retrotransposons. SNARE was the largest LTR retrotransposon family in the soybean genome. This family comprises 5832 copies, including 2851 intact elements flanked by target site duplications (TSDs) (referred to as intact elements), 45 intact elements without TSDs, 1463 solo LTRs with TSDs (referred to solo LTRs), 33 solo LTRs without TSDs, and 1440 truncated elements that contain at least one identified LTR (Table 1; see Supplemental Data Set 1 online). The elements with sequence gaps and severely degenerated fragments were not included in this analysis. Overall, SNARE makes up ∼113 Mb of DNA, accounting for ∼12% of assembled soybean genomic sequence.
Table 1.
The Structures of SNARE Elements
| Structurea | No. of SARE | No. of S | No. of O | No. of Other SNREb | Total |
| Intact elements with TSDs | 1244 | 1290 | 244 | 73 | 2851 |
| Solo LTRs with TSDs | NAc | NA | NA | NA | 1463 |
| Intact elements without TSDs | 22 | 21 | 1 | 1 | 45 |
| Solo LTRs without TSDs | NA | NA | NA | NA | 33 |
| Truncated elements | 683 | 411 | 47 | 299 | 1440 |
TSDs, target site duplications.
Cannot be determined whether these are SNREO or SNRES elements.
NA, not applicable.
SNARE Contains Two Autonomous Subfamilies and a Nonautonomous Subfamily
Although it was defined as a single family based on the high sequence similarity of LTRs, PBS, and PPT sites, SNARE exhibited extensive variation in size and sequence among elements.
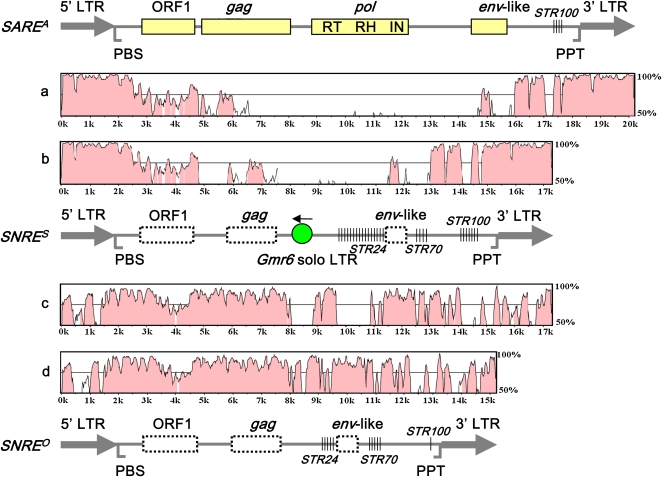
The majority of intact elements range from 13 to 20 kb with 1.6- to 2.2-kb LTRs. The size variation of LTRs is mainly due to tandem duplication and deletion. Annotation of the internal regions of intact elements uncovered two major distinct structural features that define them as gypsy-type soybean autonomous retroelements (SARE) and nonautonomous retroelements (SNRE) (Figure 1). In addition to the gag and pol genes that are considered to be necessary for its transposition, a typical SARE element contains a functionally unknown open reading frame (ORF) upstream of pol that matches the ORF1 found in the Ogre LTR retrotransposon of pea (Pisum sativum) (Neumann et al., 2003) (e-value = 4e−47) and another ORF upstream of PPT that shares 21% amino acid similarity to an env-like gene in Arabidopsis, the signature of putative endogenous retroviruses (Laten et al., 2003). By contrast, a typical SNRE element lacks pol genes and shows very low sequence similarity to the gag and env-like genes (Figure 1). SNRE also contains unique internal sequences that are absent in SARE, including two simple tandem repeat families (STR24 and STR70) and unknown sequences surrounding the degraded env-like gene remnant. Nevertheless, SARE and SNRE share identical PBS and PPT, and in most cases, display striking sequence similarity in LTRs, as well as regions ∼1 kb downstream of PBS and ∼2 kb upstream of PPT (Figure 1).
Figure 1.
Structure and Sequence Comparison of gypsy-Type SNARE Elements.
The protein coding domains gag, pol, and env-like in an autonomous element SAREA are represented by boxes with solid outlines, while their corresponding homologous sequences, if any, in nonautonomous elements SNRES and SNREO are represented by blank boxes with dashed outlines. The solid circle represents the copia-type piggybacking Gmr6 solo LTR, and the arrow above the circle indicates the proposed transcriptional orientation of the LTR in an intact Gmr6 element. Three different families of simple tandem repeats, STR100, STR70, and STR24, are indicated. The plots a and b show the relative nucleotide identity between the SAREA and SNRES elements, and the plots c and d show the relative nucleotide identity between the SNRES and SNREO elements. To reflect the sequence divergence level among three distinct subfamilies/subgroups, the three elements IN593 (SAREA), IN834 (SNRES), and IN9037 (SNREO), as indicated by arrows in Figure 3, were randomly chosen from the relatively young elements of the SNARE family.
[See online article for color version of this figure.]
Despite their structural conservation, SARE elements were consistently classified into two distinct subfamilies (dubbed SAREA and SAREB) by phylogenetic analysis of sequences from different regions, including 5 ′ LTRs, ORF1, gag, and RT sequences, and no recombination between SAREA and SAREB elements was detected (Figure 2B). Overall, the LTR sequences show a lower level of divergence between the two subfamilies than the ORF1, gag, and RT sequences. These two subfamilies were further differentiated by the presence of two different families of simple tandem repeats (STRs) upstream of their 3 ′ LTRs. The tandem repeat family STR100 is present only in SAREA, whereas STR62 is unique to SAREB (Figure 2A). STR100 is also present at the same positions of SNRE elements, but STR62 was not found in any SNRE element. This suggests that SNRE elements arose from the SAREA lineage after its divergence from the SAREB lineage. Based on the synonymous substitution of the consensus RT sequences between SAREA elements and SAREB elements from a random sample described below (see Methods), it was estimated that these two subfamilies diverged from each other ∼11 million years ago.
Figure 2.
Structural Comparison and Evolutionary Relationship of the SAREA and SAREB Subfamilies.
(A) Structural and sequence comparison of SAREA and SAREB elements. The protein coding domains in SAREA and their corresponding homologous sequences are represented with boxes. The plots a and b show the relative nucleotide identity between the SAREA and SAREB. The two elements IN593 and IN5410 were randomly chosen from relatively young elements in clade 3 (SAREA) and clade 4 (SAREB). Different sizes of simple tandem repeat STR100 and STR62 are also indicated. These two kinds of repeats share no sequence similarity, suggesting their independent origins.
(B) Evolutionary relationship and sequence divergence between SAREA and SAREB using 5 ′ LTR, conserved protein ORF1, gag, and RT, respectively. Clades 1, 3, and 4 were labeled corresponding to the autonomous element clades shown in Figure 3. The level of nucleotide sequence distance is indicated by the scales.
[See online article for color version of this figure.]
Proliferation of an Alien Solo LTR Mediated by Proliferation of a Single SNRE Element
Further examination of the internal regions of SNRE elements revealed an unexpected observation. Of 1556 intact elements of SNRE, 1311 (dubbed SNRES) harbor a foreign solo LTR at a single consensus site, unique to SNRE elements, and 244 (dubbed SNREO) do not have the solo LTR at this site (Table 1). The remaining 73 elements appear to have lost this segment due to deletions; thus, whether they once harbored this solo LTR cannot be determined (Table 1). The solo LTR is flanked by TSD and belongs to an unrelated copia-type LTR retrotransposon family, Gmr6 (see Supplemental Data Set 1 online). No intact Gmr6 elements, possessing both LTRs and internal domains, were found at this site. The proposed transcriptional orientation of this piggybacking Gmr6 solo LTR insertion is opposite to that of the SNRES elements in which it resides. In addition to the 1311 Gmr6 solo LTRs harbored in SNRES elements, the soybean genome contains 763 intact elements and 998 solo LTRs belonging to Gmr6 (see Supplemental Data Set 1 online), but none of these 1761 elements were found at the same site within any other transposable elements.
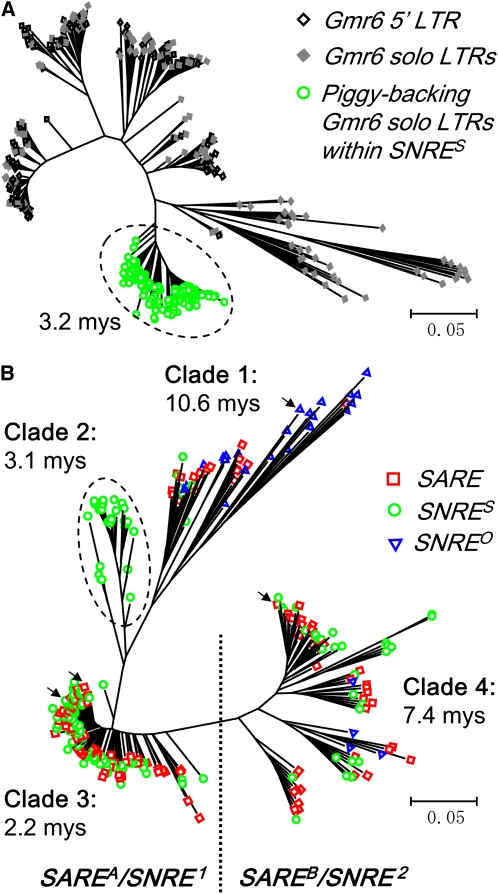
Phylogenetic analysis of a random set of Gmr6 LTRs was performed. These include 150 solo LTRs harbored in SNRES elements, 150 5 ′ LTRs from intact elements, and 150 solo LTRs outside of SNRES elements. The LTR sequences were aligned using MUSCLE (Edgar, 2004), and a neighbor-joining phylogenetic tree was generated using MEGA4 (Tamura et al., 2007). As shown in Figure 3A, all Gmr6 solo LTRs harbored in SNRES elements fell exclusively into a single clade and exhibited a high level of sequence identity within this clade but substantial sequence divergence from all other Gmr6 LTRs. This result, together with the observation that all Gmr6 solo LTRs in SNRES elements share the same insertion site, indicates that the Gmr6 solo LTRs harbored in the SNRES elements proliferated by the amplification of a single founding SNRE element after a Gmr6 element inserted in it. In addition, not a single intact Gmr6 was found at this insertion site, suggesting that the initial founding SNRE with the insertion of a Gmr6 did not amplify until the Gmr6 solo LTR was formed, most likely, by unequal intraelement recombination (Devos et al., 2002). Using a method described previously for determining the relative time of insertion of monophyletic groups of LTR retrotransposons in rice (Jiang et al., 2002), it was estimated that the ancestral Gmr6 solo LTR within a SNRES element was formed ∼3.2 million years ago (see Methods).
Figure 3.
Phylogenetic Analysis of LTR Sequences.
(A) Neighbor-joining tree of LTR sequences from random samples of Gmr6 solo LTRs harbored in SNRES elements (green circles enclosed in the dashed oval) and intact elements (open diamonds) and solo LTRs (filled diamonds) of Gmr6 outside of SNRES elements in the soybean genome. The level of nucleotide sequence distance is indicated by the scales.
(B) Neighbor-joining tree of LTR sequences from random samples of intact SARE (red rectangles), SNRES (green circles), and SNREO (blue triangles) elements. SNRE1 and SNRE2 indicate the two subgroups of SNRE elements formed by two independent subfamily-specific interelement recombination machineries. The green circles enclosed in the dashed oval indicate the lineage of ancestral SNRE elements. Representative elements used for comparisons of sequence divergence in Figure 1 are labeled as black arrows. The level of nucleotide sequence distance is indicated by the scales.
Extensive Interelement Recombination between SARE and SNRE Elements
Presuming that the amplification of each SNARE element was an independent event, one could propose that the recently amplified SNRES elements, which share the piggybacking Gmr6 solo LTR, must be distinguishable from the SAREA, SAREB, and SNREO subfamilies/subgroups based on their conserved regions. To test this assumption, we performed a phylogenetic analysis of 5 ′ LTR sequences of 300 SNARE elements randomly chosen, including 121 SARE elements, 150 SNRES elements, and 29 SNREO elements (see Supplemental Data Set 1 online). The neighbor-joining phylogenetic tree obtained exhibits four major monophyletic groups, clades 1, 2, 3, and 4, with different levels of sequence divergence and population structures that reflect different evolutionary time frames and lineages (Figure 3B). We estimated that the relative ages of monophyletic clades 1, 2, 3, and 4 are 10.6, 3.1, 2.2, and 7.4 million years, respectively (see Methods). As seen in Figure 3B, all SAREA elements were clustered into clades 1 and 3, while all SAREB elements were clustered into clade 4. Clade 1 is the oldest lineage that contains the SAREA elements and the majority of SNREO elements, and clade 3 is the youngest lineage that contains only SAREA elements and the majority of SNRES elements, further suggesting that SNRE was derived from SAREA.
Unexpectedly, the SNRES elements, derived from a single founding SNRE element ∼3 million years ago, were not clustered into a single clade. Instead, they were observed in all four monophyletic groups (Figure 3B). More intriguingly, many intersubfamily elements (e.g., between SAREA and SNRES elements or between SAREB and SNRES) show a substantially higher level of LTR sequence identities (e.g., >99%) than intrasubfamily elements (e.g., between SAREA elements, between SNRES elements, or between SAREB elements). In addition, of the 2457 intact elements of the SNARE family identified in this study, which contain complete LTRs, 920 (37.4%) SARE and SNRE elements were found to have LTR sequences best but cross-matching to those of SNRE and SARE elements, respectively (see Supplemental Table 1 online). Based on the pairwise comparison of these LTR sequences, 388 unique pairs of elements were identified, of which 121 (31.2%) are in the SARE and SNRE composition (Table 2). These observations suggest that extensive interelement recombination between SARE elements and SNRE elements occurred over the evolutionary time of this family, particularly after the formation of the first SNRES element, leading to subfamily-specific homogenization of LTR sequences and the bifurcation of SNRE elements (particularly SNRES elements) into two distinct subgroups (dubbed SNRE1 and SNRE2; i.e., SNRES1 versus SNRES2, and SNREO1 versus SNREO2) corresponding to their autonomous partners SAREA and SAREB (Figure 4A).
Table 2.
Best Matched SARE-SNRE Pairs Determined by Pairwise Comparison of LTR Sequences of All SNARE Elements
|
SARE-SNRE Pairsa |
||||||
| SARE | SNRES | SNREO | Total | No. | Percentage | |
| SARE | 123 | |||||
| SNRES | 107 | 121 | ||||
| SNREO | 14 | 4 | 19 | |||
| Subtotal | 244 | 125 | 19 | 388 | 121 | 31.2 |
Only the 5 ′ LTR of each element was used in this analysis.
Figure 4.
Evolutionary Model and Insertion Times of SNARE Elements.
(A) Evolutionary model of SNARE evolution. Letters a, b, and c indicate three evolutionary events that gave rise to the distinct structural features of SNARE elements: the divergence of SAREA and SAREB, the formation of SNREO, and the integration of Gmr6 solo LTR, respectively. I and II indicate the proposed two machineries for subfamily- and region-specific interelement recombination. Arrows indicates proposed replacement of LTR sequences of nonautonomous elements by two distinct lineages of autonomous partners during SNARE evolution. The numbers of intact elements within each category are indicated.
(B) Age distribution of intact elements. Although SNRE elements were derived from SARE elements, the existing oldest SNRE elements were dated to be older than the existing oldest SARE elements on the basis of their LTR sequence divergence. This may reflects different levels of selection for LTR sequence conservation.
[See online article for color version of this figure.]
Analysis of Recombinants and Their Parental Elements
To further illustrate the interelement recombination events and to shed light on the molecular mechanisms responsible for the recombination, we performed an in-depth analysis of the putative recombinants and their potential parental autonomous and nonautonomous forms. As shown in Figure 2B, the autonomous SAREA elements were consistently grouped into two clades, 1 and 3, using sequences from different regions of the elements, and clade 1 exhibits overall higher level of sequence identity than clade 3 for all the regions analyzed. These two clades, corresponding to clades 1 and 3 in Figure 3B, represent two SAREA lineages amplified within distinct evolutionary time frames. As supported by several lines of evidence that we have demonstrated earlier, the SNRE elements were originated from the older SAREA lineage (clade 1 in Figure 3B), and the SNRES elements were amplified from a single founding SNRE element within the last ∼3 million years. It is thus logical that the majority of recombination events occurred between SNRES and the younger SAREA lineage (clade 3) (Figure 3B).
Noticeably, clade 2 in Figure 3B comprised SNRES1 elements exclusively, all of which contain typical internal SNRE components that were clustered in the SNRES1-specific clades shown in Figures 5B to 5D, suggesting that these elements (dubbed ancestral SNRES elements) were amplified from the founding SNRES element without interelement recombination with SARE elements. The age of this clade was estimated to be 3.1 million years (Figure 3B), consistent to the formation time (3.2 million years ago) of the SNRES elements estimated based on the divergence of the piggybacking solo LTRs (Figure 3A). Similarly, the majority of SAREA elements clustered within clade 3 in Figure 3B contain the typical internal SARE components that were clustered in the SAREA-specific clades (Figures 5B to 5D), suggesting that these SAREA elements are not recombinants. This inference was further supported by the observation that the topology of the phylogenetic trees constructed from different regions of SAREA and SAREB elements are congruent (Figure 2B). Therefore, the SNRES1 elements mixed with autonomous elements would be inferred as nonautonomous recombinants that captured LTR sequences from SAREA elements by recombination. Similarly, the SNRES2 elements in clade 4 in Figure 3B would be inferred as nonautonomous recombinants whose original LTR sequences were replaced by the LTRs from SAREB elements.
Figure 5.
Evolutionary Relationship and Sequence Divergence between SAREA and SNRES1.
The bootstrap neighbor-joining trees were generated using 5 ′ LTR (A), ORF1 (B), gag (C), and env-like (D) homologous domains. Three clades defined in Figure 3B were labeled with each corresponding number. Clades 1 and 3 represent two SAREA lineages, while clade 2 represents the ancestral SNRES lineage. The level of nucleotide sequence distance is indicated by the scales
[See online article for color version of this figure.]
To further validate our proposition, we randomly chose four putative recombinants, two SNRES1 elements within clade 3 and two SNRES2 elements within clade 4 in Figure 3B, and identified their respective putative parental autonomous and nonautonomous elements by homology searches against all the SNARE elements in the whole soybean genome and subsequent sequence alignments. On the basis of LTR sequences, all of the four putative parental nonautonomous elements can be grouped into the ancestral SNRES clade as shown in Figure 3B, while the putative parental autonomous elements can be grouped into the same clades (i.e., clade 3 or clade 4 in Figure 3B) as their respective recombinants reside. In all cases analyzed, two recombination breakpoints in each of the recombinants can be roughly predicted: one is between PBS site and ORF1 and the other is between env-like gene and PPT site. The structural components of one (SNRES1 IN5127) of the four recombinants and its putative parental autonomous (SAREA IN4965) and nonautonomous (SNRES1 IN3618) elements were most clearly defined (see Supplemental Figure 1 online). According to sequence alignments (see Supplemental Figure 1E online), two recombination breakpoints in this putative recombinant element were identified, one is 3 ′ adjacent to the PBS site and the other is ∼900 bp upstream of the PPT site (see Supplemental Figures 1A and 1E online). Phylogenetic analysis of the predicted U3, R, and U5 subregions of LTRs (see Supplemental Figures 1C and 1D online) reveals that the nonautonomous recombinant shares much higher sequence identities with the parental autonomous element than with the parental nonautonomous element in all the three subregions (see Supplemental Figure 1B online), suggesting that the entire LTRs of the recombinant element were derived from its parental autonomous element. The exact sites for recombination in the other three recombinants cannot be precisely determined, probably due to the subsequent mutations (substitution, insertion, or deletion), new recombination between the initial recombinants and SARE elements, or on the other hand, the extremely high level of sequence conservation at the PBS and PPT sites and their adjacent downstream and upstream internal regions.
Following the analysis above, we characterized the structural components of all SAREA and SNRES1 elements listed in Figure 3. The results are summarized in Table 3 and exemplified in Supplemental Figure 2 online. Out of the 173 elements analyzed, 66 and 23 are autonomous and nonautonomous elements, respectively, without detected recombination, 58 are nonautonomous recombinants with two LTRs from SAREA elements, 23 are nonautonomous recombinants with two LTRs and ORF1 from SAREA elements, one is nonautonomous recombinant with two LTRs and the env-like gene from SAREA elements, and two are autonomous recombinants with the env-like gene from nonautonomous element. Overall, ∼78% SNRES1 elements are recombinants with at least LTRs from SAREA elements, while <3% SAREA elements carrying small internal fragments from nonautonomous elements. This difference in the proportions of chimeras within the autonomous and nonautonomous subfamilies may reflect the varying degree of functional constraints in recombinants and their parental elements for survival and subsequent proliferations.
Table 3.
Recombination between A and S1 Elements
| Conserved Protein Domain |
||||||
| Subfamily/Subgroup | 5 ′ LTR | ORF1 | gag | env | No. of Elementsa | Recombinant |
| SAREA | SAREA | SAREA | SAREA | SAREA | 66 | No |
| SNRES1 | SNRES1 | SNRES1 | SNRES1 | SNRES1 | 23 | No |
| SNRES1 | SAREA | SNRES1 | SNRES1 | SNRES1 | 58 | Yes |
| SNRES1 | SAREA | SAREA | SNRES1 | SNRES1 | 23 | Yes |
| SAREA | SAREA | SAREA | SAREA | SNRES1 | 2 | Yes |
| SNRES1 | SAREA | SNRES1 | SNRES1 | SAREA | 1 | Yes |
Intact elements chosen for the phylogenetic analysis are shown in Figure 3.
All the intact elements shown in Figure 3B are flanked by TSDs; thus, the interelement recombination events revealed above would have occurred prior to their integration into the soybean genome. It is possible that some of the detected recombinants flanked by TSDs were directly amplified from respective precursor recombinants generated by interelement recombination or from the recombinants amplified from the precursor recombinants. If one believes that only a few precursor recombinants were initially formed between SARE and the ancestral SNRES, it follows that extensive interelement recombination events must have taken place between SARE and the precursor recombinants or between SARE and the recombinants amplified from the precursor recombinants, given that SNRES elements appear in various clades or subclades that contain autonomous elements from different lineages or at different divergence levels (Figure 3B) and that 31.2% unique element pairs in SARE-SNRE composition were observed in the genome (Table 2).
Dating of Insertions of SNARE Elements and Divergence of SARE and SNRE
The insertion times of the SNARE intact elements were estimated based on sequence divergence of two LTRs from individual elements as described previously (Ma et al., 2004). As illustrated in Figure 4B, the majority (98.6%) of the intact elements were integrated into the soybean genome within the last 3 million years. The average ages of the SAREA elements and SNRES1 elements are 0.87 (0 to 3.75) and 0.48 (0 to 2.69) million years, with median ages of 0.64 and 0.35 million years, respectively, while the average ages of the SAREB elements and SNRES2 elements are 1.25 (0.09 to 3.03) and 1.26 (0.19 to 2.82) million years, with median ages of 1.11 and 1.19 million years, respectively. Within the last 1 million years, 576 SAREA and 826 SNRES1 elements were amplified, and of these elements, seven SAREA and six SNRES1 elements have identical LTRs. By contrast, 156 SAREB and 105 SNRES2 elements amplified within the last 1 million years, but none of these elements have identical LTRs. These data suggest that SAREA and SNRES1 elements were amplified within a similar and more recent time frame than SAREB and SNRES2 elements. The oldest SNRES element was dated to 2.82 million years, close to the estimated time for the birth of the ancestral SNRES lineage (3.1 million years ago) and for the initial integration of the Gmr6 solo LTR into SNRES (3.2 million years ago). Overall, SNREO elements are oldest, with the ages ranging from 0.66 to 4.39 million years. These observations are consistent with the evolutionary model of SNARE elements illustrated in Figure 4A.
Because of extensive recombination between SARE and SNRE and varying selection pressures on different regions or different elements, the divergence time of SNRE from SARE cannot be precisely estimated based on the levels of LTR sequence divergence. However, a group of highly diverged SNREO elements were revealed by phylogenetic analysis of SNARE LTR sequences (clade 1, Figure 3B), and this group contains the oldest SAREA lineage that gave rise to SNRE. Therefore, we propose that SNRE was initially derived from SAREA ∼10.6 million years ago, shortly after the divergence of SAREA and SAREB lineages.
Similarity of SARE and SNRE Distributions and Insertion Sites
Chromosomal locations of the SARE and SNRE elements, including intact elements and solo LTRs, were investigated. As shown in Supplemental Figure 3 online, SARE and SNRE exhibit strikingly similar distribution patterns along each of the 20 soybean chromosomes, and the majority ( ∼98%) of these elements are clustered in heterochromatic regions where genetic recombination is nearly completely suppressed (Schmutz et al., 2009). The insertion sites of the SARE and SNRE elements were also investigated. We found that neither SARE elements nor SNRE elements inserted randomly into the host genome. Both SARE and SNRE elements show an overall bias for G and C content (50 and 49% respectively; see Supplemental Figure 4 online) within the TSD region. In addition to the 5-bp TSDs (coded as T1, T2, T3, T4, and T5), three nucleotides adjacent to T1 (coded as -1, -2, and -3) and three nucleotides adjacent to T5 (coded as 1, 2, and 3) are not random. Notably, these 11 nucleotides show almost identical consensus sequences between SARE and SNRE. Other sites show a level of G and C content similar to that of the whole genome ( ∼35%).
Expression of SARE and SNRE in Multiple Tissues
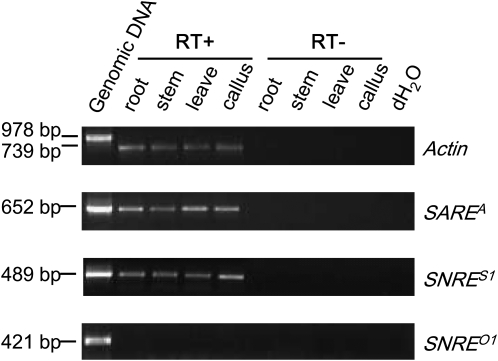
The transcriptional activity of SNARE elements was investigated by RT-PCR with total RNA extracted from roots, stems, leaves, and calli of the sequenced soybean cultivar William82. Primers (see Supplemental Table 2 online) were designed based on young elements and aimed to amplify the RT domain of SAREA, the 3 ′ junction region of the piggybacking Gmr6 solo LTR, and its downstream flanking sequence of SNRES1, and the region of SNREO that corresponds to the sequences flanking the Gmr6 solo LTR in SNRES1. As shown in Figure 6, fragments of expected sizes for SAREA and SNRES1 were amplified in all tissues examined, while the expected fragment for SNREO was not amplified in any of these tissues. In addition, we searched the SNARE elements with the ESTs deposited in the National Center for Biotechnology Information (NCBI) database and found 46, 71, 14, and 14 nonredundant soybean EST sequences that match ( ≥ 99% identity) SAREA, SNRES1, SAREB, and SNRES2, respectively, while no ESTs were found to match the unique region of SNREO1 (see Supplemental Table 3 online). It is likely that at least SAREA and SNRES1 are still transcriptionally and even transpositionally active, given that both subfamilies also contain young elements with two identical LTRs.
Figure 6.
Transcriptional Activity of SNARE Elements in Different Soybean Tissues.
The primers (see Supplemental Table 2 online) were designed based on relatively young elements to specifically amplify fragments unique to different subfamilies. RT domain from SAREA and an internal region between the gag and env-like gene remnants in SNREO and an internal region that covers 5 ′ upstream of Gmr6 solo LTR and part of the LTR in SNRES were amplified using these primers. RT-PCR reactions were performed parallel with total RNA (RT−) and with reverse transcribed RNA (RT+) into single strand cDNA. Primers amplifying the housekeeping actin gene fragment (spanning intron2), which is spliced, were used as a control.
DISCUSSION
The Hallmarks of the Autonomous-Nonautonomous Partnership
Several lines of evidence presented in this study indicate that SARE and SNRE are autonomous and nonautonomous partners in the soybean genome. In addition to the highly identical LTR sequences, identical PBS and PPT sites, conserved families of tandem repeats, similar chromosomal distribution patterns, and preferential integration sites, SARE and SNRE share detectable similarities between ORF1, gag, and env-like domains. Although not a single ORF can be predicted even in recently amplified SNRE elements, in which two LTRs remain identical, the highly degraded ORF remnants in SNRE show best matches to the corresponding ORFs predicted in the SARE elements in the soybean genome.
Substantial sequence similarity of LTRs and adjacent noncoding sequences are commonly observed between autonomous and nonautonomous retrotransposon partners (Jiang et al., 2002). In general, LTR sequences diverge faster than the protein-coding region of an element (Jordan and McDonald, 1998). Hence, the conservation of LTR sequences, which contain transcriptional regulatory sequences, between autonomous and nonautonomous partners would suggest the coevolution of the partners in a host genome. Despite their sequence conservation, the autonomous and nonautonomous retrotransposon partners previously identified were generally divided into two distinct monophyletic groups based on their conserved regions. For example, the putative autonomous and nonautonomous partners, Rire2 and Dasheng, were exclusively grouped into two highly diverged clades based on their LTR sequences, suggesting that few interelement recombinations occurred between Rire2 and Dasheng since the formation of Dasheng ∼10 million years ago (Jiang et al., 2002). In this study, we observed extensive subfamily- and region-specific sequence swapping within the recent evolutionary timeframe (e.g., 0 to 3 million years), which are primarily responsible for the observed LTR sequence conservation between autonomous elements and nonautonomous elements. These findings clearly indicate the partnership of SARE and SNRE elements
The Evolutionary History of the SNARE Family in the Context of the Host Genome Evolution
Because of the coevolution of autonomous and nonautonomous elements, it is difficult or impossible to precisely date the divergence of SAREA and SAREB lineages using LTR sequences. Based on the synonymous substitutions of RT sequences, it was estimated that SAREA and SAREB diverged ∼11 million years ago. Recent studies propose that the present soybean genome was evolved from an allotetroploid (Gill et al., 2009), which was formed by hybridization of two diploid progenitors that diverged from each other from a common diploid ancestor ∼13 million years ago (Shoemaker et al., 2006). If this is the case, then it follows that SAREA and SAREB lineages may be formed as a result of divergence and subsequent coalescence of the two subgenomes from the diploid progenitors.
For the same reason, the formation of SNRE cannot be dated precisely, but it is reasonable to deduce that it was derived from SAREA, given the facts that the majority of SNREO elements were clustered with the older SAREA elements in a monophyletic group, which does not contain SAREB, that the ancestral SNRES lineage was closely related to the SAREA monophyletic groups, and that the SNRE (both SNRE1 and SNRE2) and SAREA elements share the simple tandem repeat family STR100 at the same location, which was absent in all SAREB elements.
The survival and subsequent proliferation of a SNRE element after the insertion of a Gmr6 element was an unexpected observation. Although nested LTR retrotransposons (SanMiguel et al., 1996) are ubiquitously seen in all plants investigated, amplification of one element after insertion of another had not been previously observed, even with the availability of nearly complete genomic sequences from several higher eukaryotes (Jiang and Wessler, 2001, Ma et al., 2004). In this study, 1722 SNRE elements were found to share the piggybacking solo LTR, indicating that neither transcriptional nor transpositional activities of SNRE elements were disrupted by the solo LTR. Nevertheless, SNRES elements did not proliferate until the formation of the Gmr6 solo LTR. The SNRES elements with the Gmr6 solo LTR also greatly outnumber the SNREO elements, suggesting that the Gmr6 solo LTR, upon its formation, may have facilitated the amplification of the SNRES elements. On the other hand, both transcription and age distribution analyses reveal paralleling transcription activities between SAREA and SNRE1 or between SAREB and SNRE2 elements; thus, it is likely that the activities of autonomous and nonautonomous elements are coregulated by the same factors.
The Biological Processes and Molecular Mechanisms
Our analysis provides evidence for extensive interelement recombination between SARE and SNRE elements. Because all SNRE elements included in this phylogenetic analysis share the structural features, such as the lack of pol genes, highly degraded gag and env-like genes, and a unique region, that distinguish them from SARE elements, and particularly, because all SNRES elements sharing the piggybacking solo LTR can be considered as a lineage arising from a single founding SNRE element within the past 3 million years, the mixture of the SARE and SNRE (mostly SNRES) LTR sequences within multiple distinct monophyletic groups as illustrated in Figure 3B and the considerably high proportion of SARE-SNRE pairs shown in Table 2 would be explained by frequent interelement recombination between SARE and SNRE elements. Our data also suggest that, except for the ancestral SNRES lineage (clade 2 in Figure 3), all other SNRES elements are essentially nonautonomous recombinants, which acquired LTRs (and adjacent regions in some cases) from autonomous elements. Therefore, different from Rire2 and Dasheng, SARE elements and the majority of SNRE elements cannot be distinguished as distinct groups based on their LTR sequences. Theoretically, SNRE elements lack the ability to transpose by themselves but are capable of hijacking the transposition machinery of their partners. Thus, natural selection must have played a central role in maintaining the structural completeness, sequence conservation, and independent transcriptional activities of autonomous elements. This deduction is supported by the observation that much fewer internal mosaic structures were detected in autonomous elements than in nonautonomous elements, regardless of the property of their LTR sequences (Table 3).
It is particularly interesting that, despite the lack of recombination between SAREA and SAREB, a subset of SNRES elements (dubbed SNRES2), originated from SAREA, were replaced by the LTRs of SAREB elements, leading to the divergence of the nonautonomous elements of the same origin into two distinct subgroups corresponding to the SAREA and SAREB lineages. A recent analysis of a family of centromere retrotransposons in maize, CRM1, revealed interelement recombination events between two different lineages (dubbed CRM1-A and CRM1-B, which were hypothesized to be derived from two diploid progenitors of the modern maize genome ∼12 million years ago [Swigonova et al., 2004]), which resulted in the creation of progressively more fit and complex CRM1 recombinants (Sharma et al., 2008). Unlike this observation, SAREA−SAREB recombinants were not identified in the soybean genome in our study (Figure 2B). A model that can explain our unique observation is that two distinct transpositional machineries existed in soybean cells and were responsible for the proliferation of SAREA and SAREB and their respective nonautonomous counterparts SNRE1 and SNRE2 (Figure 4A). The full details of the regeneration of a DNA retrotransposon from its genomic RNA, in any case, have not been carefully studied in plants. But the process is thought to proceed similarly to that of retroviruses. That is, the genomic RNA is transported to the cytoplasm, translated and bundled together with its gene products into virus-like particle, within which its full-length DNA form is recreated from the RNA (Wicker et al., 2007). We propose that the divergence and separation of the SAREA and SAREB in two diploid progenitors appear to have been long enough to compartmentalize the transposition machineries of the SAREA and SAREB subfamilies sufficiently that they were no longer copackaged in a single VLP during transposition. The SAREA and SNRE1 were copackaged in VLPs formed by active SAREA, whereas the SAREB and SNRE2 were copackaged in VLPs formed by active SAREB. Although the transposition processes have not been carefully investigated in most eukaryotes, a study of Ty1 LTR retrotransposons in yeast (Saccharomyces cerevisiae) indicated that multiple genomic RNAs are packaged during VLP formation (Feng et al., 2000).
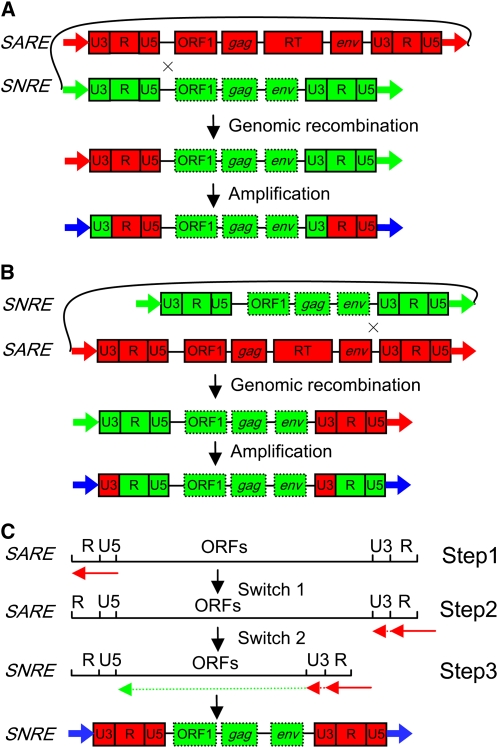
If this model above holds true, then the subfamily- and region-specific replacement of LTRs and their flanking regions can be well explained by RT-mediated recombination, such as interelement template switching during reverse transcription. Template switching initially referred to a part of the transposition process for regeneration of a new LTR retrotransposon from a single RNA template. The RNA template forms a loop using the homologous sequences (two ends of the RNA template) within the 5 ′ and 3 ′ LTRs. This allows the (−)-strand cDNA, which otherwise cannot proceed once it reaches the 5 ′ end of the RNA template, to switch to the 3 ′ end and continue the synthesis of cDNA (Sabot and Schulman, 2007). It has been proven or suggested that switches can occur between different RNA templates (intermolecularly) (Hu and Temin, 1990; Luo and Taylor, 1990) and in protein-coding regions (Archer et al., 2008), leading to retroviral recombination of genomic RNA. We propose that an intraelement switch and an interelement switch were involved in the formation of a nonautonomous recombinant, such as SNRES1 IN5127, with LTRs from an autonomous element; the first is the switch of the nascent cDNA strand from the 5 ′ end to the 3 ′ end of a SARE RNA, and the second is the switch of the synthesized cDNA strand that mainly covers the SARE LTR to an ancestral SNRE RNA copacked with the SARE RNA to synthesize the internal part of the SNRE element (as illustrated in Figure 7C). The locations of the recombination breakpoints identified in the SNRES1 IN5127 recombinant echo this model. After a nonautonomous recombinant was formed, it may amplify to generate additional copies of the recombinant or be involved in new recombinations with other autonomous partners to form new recombinants, in which the recombination breakpoints may not be precisely defined. It is also possible that more than two template switches or more than two elements are involved to form more complex mosaic structures.
Figure 7.
Models for Recombination between Autonomous and Nonautonomous Elements.
(A) and (B) Intrastrand unequal recombination between two SARE and SNRE elements to form chimeric structures of nonautonomous recombinants.
(C) Initiation of reverse transcription (step 1) from an autonomous element, intraelement template switch (step 2), followed by an interelement template switch (step 3) to form a nonautonomous recombinant with LTRs from the autonomous partner. The arrows underneath R, U5, and U3 represent the synthesized DNA fragments based on the SARE template, while the arrows underneath ORFs represent the synthesized DNA fragments based on the SNRE template.
[See online article for color version of this figure.]
The RNA-mediated recombination would also explain the initial establishment of the parasitic affiliation between SNRE and SAREB and the subsequent recombination between the new autonomous and nonautonomous partners. Theoretically, recombination can occur in any homologous regions shared by autonomous and nonautonomous elements, but 3 ′ downstream of PBS sites and 5 ′ upstream of PPT sites appear to be recombination hotspots (Table 3). As illustrated in Figure 7C, recombination between autonomous and nonautonomous elements in these regions would generate nonautonomous recombinants with LTRs from the parental autonomous elements. Autonomous and nonautonomous partners are believed to share the same transposition machineries. It thus is reasonable to deduce that the recombinants, which captured the entire LTR sequences from autonomous partners, are likely to be more successful. Theoretically, nonautonomous elements should diverge from their autonomous partners far enough that they will eventually not be copackaged, and they would not necessarily be transcribed at the same time due to accumulated differences in their promoter regions in their LTR sequences. Thus, this swapping of the LTRs seems to be a mechanism that can resurrect dying nonautonomous elements by autonomous element sequences, ensuring that they get transcribed at the same time as younger autonomous elements. By contrast, genomic recombination, as illustrated in Figures 7A and 7B, only exchanges partial LTR sequences between two parental elements to form mosaic structures.
Unequal intrastrand genomic recombination is considered to be the primary mechanism for generating solo LTRs and other LTR retrotransposon recombinants in plants (Devos et al., 2002; Ma et al., 2004). Indeed, the initial recombination events either between SAREA and the ancestral SNRES elements or between SAREB and the ancestral SNRES elements could have occurred at the genomic level. However, genomic recombination is less likely to be the major process for the subsequent extensive subfamily-specific LTR swapping between autonomous and nonautonomous elements. On the other hand, the frequency of interelement recombination within the host genome reflected by the proportion of intact elements without TSDs in the soybean genome is very low ( ∼1.5%). In general, interelement recombination eliminates DNA between two elements involved and thus would have deleterious effects if the interelement space contains functional genes. Even if all these elements without TSDs are assumed to be the products of genomic interelement recombination (not the outcome of sequence variations in the region surrounding an intact element (e.g., nucleotide substitutions, deletions, duplications, and other sorts of DNA rearrangements), such a low proportion may still not be able to explain the high frequency of interelement recombination events reflected by the high proportion of SARE-SNRE pairs detected by pairwise distance comparison. Therefore, the explanation of genomic recombination alone for the generation of many nonautonomous recombinants is rendered less tenable.
Previously, recombination between two closely related families of yeast LTR retrotransposons, Ty1 and Ty2, were identified by analysis of 45 elements in the entire yeast genome (Jordan and McDonald, 1998). The interelement recombination generated a subclass of hybrid Ty1 elements (recombinants). The phylogenies of LTR sequences showed that the Ty1 recombinants were more closely related to Ty2 elements than they were to other (nonrecombinant) Ty1 elements and that all the recombinants were distinguished from either nonrecombinant Ty1 or Ty2 lineages. Further analysis revealed that each of the Ty1 recombinants contains Ty2-like U3 in both LTRs and defined one of the recombination breakpoints approximately at the beginning of the R subregion of the LTR. On the basis of these observations, it was proposed that two interelement template switches be involved in reverse transcription process to generate the Ty1 recombinants (Jordan and McDonald, 1998). Following this study, the authors observed a small piece of Ty2-like ORF domain adjacent to the 3 ′ LTR in the Ty1 recombinants (Jordan and McDonald, 1998, suggesting a recombination breakpoint within the ORF region. However, it would be equally possible that, similar to the model illustrated in Figure 7B, the Ty1 mosaic structure was formed by a genomic crossing over between the Ty1 and Ty2 elements at this breakpoint, followed by subsequent amplification of the initial recombinant. Hence, the exact mechanism responsible for the formation of the Ty1 recombinants in yeast remains to be determined.
We want to point out that the components and processes involved in LTR retrotransposon amplification in plant cells, as described in all literature to date, are still widely believed but never proven hypotheses. Thus, the evidence for template switching that we provided in this study may not be further experimentally proven. However, when considering the whole set of observations that we garnered, together with the discoveries gained from retroviruses about the reverse transcription mechanism and retroviral recombination, and with the proposition regarding the biological properties and affiliation between autonomous and nonautonomous partners, RNA-mediated recombination through reverse transcription appears to be an convincing explanation for the generation of the nonautonomous SNRES recombinants in two independent machineries and for the evolutionary model of SNARE family described in this study. We would also like to point out that genomic recombination may be also involved, particularly, in the initial formation of autonomous and nonautonomous recombinants.
Concluding Remarks
The findings obtained from this study provide new insights into the timing, nature, dynamics, and mechanisms of autonomous and nonautonomous LTR retrotransposon coevolution in the context of the soybean genome evolution. The majority of recombination events described in this study are likely to have occurred between autonomous and nonautonomous partners during the transposition processes, which led to the bifurcation of the parasitic affiliation between the autonomous and nonautonomous subfamilies/subgroups and the enhancement of the preexisting and newly established partnerships. It is likely that interelement recombination is a primary mechanism, behind natural selection, that drives the homogenization and/or divergence of autonomous and nonautonomous retrotransposon partners and their coevolution within host genomes. This study provides several lines of evidence in support of an assumption that nonautonomous elements share the same machineries with their autonomous partners for transposition. It is likely that extensive recombination by the same mechanisms may have also occurred, though not detected, between autonomous elements sharing certain levels of sequence similarity. If this is the case, diversified patterns for LTR retrotransposon evolution among plant species at different levels and status of ploidy would be expected. Careful identification and characterization of additional autonomous and nonautonomous partners in different plants and analysis of their evolutionary patterns and transpositional activities will deepen our understanding about how autonomous and nonautonomous elements communicate and interact with each other to sustain their transpositional activities and to drive their host genome evolution.
METHODS
Identification of LTR Retrotransposons
A combination of structural analyses and sequence similarity comparisons, as previously described (Ma et al., 2004; Ma and Bennetzen, 2004), were used to identify LTR retrotransposons in the soybean (Glycine max) genome (http://www.phytozome.net/). The structures and boundaries of all of the identified LTR retrotransposons were confirmed by manual inspection. The elements were classified into different structural categories as previously described (Ma et al., 2004). The SNARE LTR retrotransposon family was defined based on the homology of LTR sequences consistent with the criteria previously described (Wicker et al., 2007). The subfamilies were classified based on the sequence divergence and unique features of the internal regions of SNARE elements. The protein coding domains were predicted using ORF finder in NCBI and defined by searching Conserved Domains database (http://www.ncbi.nlm.nih.gov/structure/cdd/cdd.shtml). The boundaries of the U3, R, and U5 subregions of LTRs were defined based on the alignment of LTR sequences from relatively young autonomous elements with soybean EST sequences (accession numbers are listed in Supplemental Figure 1D online and are available in GenBank) matching the LTR regions, and the putative regulatory signals (e.g., TATA box), predicted by SoftBerry (http://www.softberry.ru), and ProScan (http://www-bimas.cit.nih.gov/molbio/proscan/).
Phylogenetic Analysis and Pairwise Sequence Comparison
Sequence alignments were performed using MUSCLE (Edgar, 2004) and edited manually when misalignments were seen. The neighbor-joining trees were built using the Kimura two-parameter method integrated in the MEGA4 program (Tamura et al., 2007). The pairwise comparisons of 5 ′ LTR sequences extracted from all the SNARE elements were performed using MEGA4.
Dating of LTR Retrotransposon Insertions and Subfamily Divergence
The insertion times of LTR retrotransposons with relatively complete LTRs (>60% of the full-length LTR) were determined in a manner described previously (Ma et al., 2004). The mutation rate of 1.3 × 10−8 substitutions per base per year proposed for intergenic sequences of rice (Oryza sativa; Ma and Bennetzen, 2004) was employed to convert sequence divergence into dates of insertion. The phylogenetic groups were dated by the methods previously described (Jiang et al., 2002), except that the mutation rate of 1.3 × 10−8 substitutions per base per year was employed. The divergence of two autonomous subfamilies was dated based on the synonymous substitution between the consensus RT sequences from individual subfamilies and mutation rate of 6.5 × 10−9 substitutions per base per year proposed for the adh1 and adh2 loci of grasses (Gaut et al., 1996).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the accession number J01298 for soybean housekeeping gene actin 1 and the accession number ACUP00000000 for the soybean pseudomolecule. The accession numbers for the EST sequences are listed in Supplemental Figure 1 and Supplemental Data Set 1 online, and the genes used for phylogenetic analysis can be found in Supplemental Data Sets 2 to 5 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. A Nonautonomous Recombinant and Its Putative Parental Forms.
Supplemental Figure 2. Three Nonautonomous Recombinants and Their Putative Parental Forms.
Supplemental Figure 3. Distribution of SNARE Elements along the Soybean Chromosomes.
Supplemental Figure 4. Insertion Site Bias of SARE and SNRE Elements.
Supplemental Table 1. Categories of the Query Elements and Their Best Matches.
Supplemental Table 2. Primers Used for the Expression Analysis of SNARE Elements.
Supplemental Table 3. ESTs Matching to SNARE Elements.
Supplemental Data Set 1. Chromosomal Locations of SNARE Elements.
Supplemental Data Set 2. Text File of Alignment Corresponding to the Phylogenetic Tree in Figure 2B.
Supplemental Data Set 3. Text File of Alignment Corresponding to the Phylogenetic Trees in Figures 3A and 3B.
Supplemental Data Set 4. Text File of Alignment Corresponding to the Phylogenetic Trees in Figures 5A to 5D.
Supplemental Data Set 5. Text File of Alignment Corresponding to the Phylogenetic Tree in Supplemental Figure 1B.
Supplementary Material
Acknowledgments
We thank Phillip SanMiguel, David Sanders, and Jeff Bennetzen for insightful comments of this manuscript and the anonymous reviewers for constructive suggestions. This work is supported by USDA–Agricultural Research Service Specific Cooperative Agreement to R.C.S. and J.M., Purdue University faculty startup funds to J.M., and the National Science Foundation Plant Genome Research Program (DBI-0822258) to J.M.
References
- Archer J., Pinney J.W., Fan J., Simon-Loriere E., Arts E.J., Negroni M., Robertson D.L. (2008). Identifying the important HIV-1 recombination breakpoints. PLOS Comput. Biol. 4: e1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L., Ma J., Devos K.M. (2005). Mechanisms of recent genome size variation in flowering plants. Ann. Bot. (Lond.) 95: 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggmann R., et al. (2006). Uneven chromosome contraction and expansion in the maize genome. Genome Res. 16: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M., Brown J.K., Bennetzen J.L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberton J., Ma J., Yuan Y., SanMiguel P., Bennetzen J.L. (2005). Gene enrichment in maize with hypomethylated partial restriction (HMPR) libraries. Genome Res. 15: 1441–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.X., Moore S.P., Garfinkel D.J., Rein A. (2000). The genomic RNA in Ty1 virus-like particles is dimeric. J. Virol. 74: 10819–10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B.S., Morton B.R., McCaig B.C., Clegg M.T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93: 10274–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill N., Findley S., Walling J.S., Hans C., Ma J., Doyle J.J., Stacey G., Jackson S.A. (2009). Molecular and chromosomal evidence for allopolyploidy in soybean, Glycine max (L.) Merr. Plant Physiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A., Voinnet O., Chappell L., Baulcombe D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Das O.P., Messing J. (1995). Zeon-1, a member of a new maize retrotransposon family. Mol. Gen. Genet. 248: 471–480 [DOI] [PubMed] [Google Scholar]
- Hu W.S., Temin H.M. (1990). Retroviral recombination and reverse transcription. Science 250: 1227–1233 [DOI] [PubMed] [Google Scholar]
- Jiang N., Jordan I.K., Wessler S.R. (2002). Dasheng and RIRE2. A nonautonomous long terminal repeat element and its putative autonomous partner in the rice genome. Plant Physiol. 130: 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Wessler S.R. (2001). Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13: 2553–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.K., Bennetzen J.L. (1989). Structure and coding properties of Bs1, a maize retrovirus-like transposon. Proc. Natl. Acad. Sci. USA 86: 6235–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I.K., McDonald J.F. (1998). Evidence for the role of recombination in the regulatory evolution of Saccharomyces cerevisiae Ty elements. J. Mol. Evol. 47: 14–20 [DOI] [PubMed] [Google Scholar]
- Kalendar R., Vicient C.M., Peleg O., Anamthawat-Jonsson K., Bolshoy A., Schulman A.H. (2004). Large retrotransposon derivatives: Abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166: 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky E., Kubat Z., Macas J., Hobza R., Mracek J., Vyskot B. (2006). Retand: A novel family of gypsy-like retrotransposons harboring an amplified tandem repeat. Mol. Genet. Genomics 276: 254–263 [DOI] [PubMed] [Google Scholar]
- Kumar A., Bennetzen J.L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Lander E.S., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Laten H.M., Havecker E.R., Farmer L.M., Voytas D.F. (2003). SIRE1, an endogenous retrovirus family from Glycine max, is highly homogeneous and evolutionarily young. Mol. Biol. Evol. 20: 1222–1230 [DOI] [PubMed] [Google Scholar]
- Lippman Z., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476 [DOI] [PubMed] [Google Scholar]
- Luo G.X., Taylor J. (1990). Template switching by reverse transcriptase during DNA synthesis. J. Virol. 64: 4321–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Devos K.M., Bennetzen J.L. (2004). Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14: 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P., Pozarkova D., Macas J. (2003). Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol. Biol. 53: 399–410 [DOI] [PubMed] [Google Scholar]
- Palmer L.E., Rabinowicz P.D., O'Shaughnessy A.L., Balija V.S., Nascimento L.U., Dike S., de la Bastide M., Martienssen R.A., McCombie W.R. (2003). Maize genome sequencing by methylation filtration. Science 302: 2115–2117 [DOI] [PubMed] [Google Scholar]
- Piegu B., Guyot R., Picault N., Roulin A., Saniyal A., Kim H., Collura K., Brar D.S., Jackson S., Wing R.A., Panaud O. (2006). Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16: 1262–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabot F., Schulman A.H. (2007). Template switching can create complex LTR retrotransposon insertions in Triticeae genomes. BMC Genomics 8: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P., Gaut B.S., Tikhonov A., Nakajima Y., Bennetzen J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43–45 [DOI] [PubMed] [Google Scholar]
- SanMiguel P., Tikhonov A., Jin Y.K., Motchoulskaia N., Zakharov D., Melake-Berhan A., Springer P.S., Edwards K.J., Lee M., Avramova Z., Bennetzen J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 [DOI] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature http://dx.doi.org/10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Sharma A., Schneider K.L., Presting G.G. (2008). Sustained retrotransposition is mediated by nucleotide deletions and interelement recombinations. Proc. Natl. Acad. Sci. USA 105: 15470–15474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R.C., Schlueter J., Doyle J.J. (2006). Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 9: 104–109 [DOI] [PubMed] [Google Scholar]
- Swigonova Z., Lai J., Ma J., Ramakrishna W., Llaca V., Bennetzen J.L., Messing J. (2004). On the tetraploid origin of the maize genome. Comp. Funct. Genomics 5: 281–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Vicient C.M., Suoniemi A., Anamthawat-Jonsson K., Tanskanen J., Beharav A., Nevo E., Schulman A.H. (1999). Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11: 1769–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8: 973–982 [DOI] [PubMed] [Google Scholar]
- Wicker T., Stein N., Albar L., Feuillet C., Schlagenhauf E., Keller B. (2001). Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 26: 307–316 [DOI] [PubMed] [Google Scholar]
- Witte C.P., Le Q.H., Bureau T., Kumar A. (2001). Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc. Natl. Acad. Sci. USA 98: 13778–13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T.H. (1990). Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9: 3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.