Abstract
Medical research in Africa has yet to benefit from the advent of genome-wide association (GWA) analysis, partly because the genotyping tools and statistical methods that have been developed for European and Asian populations struggle to deal with the high levels of genome diversity and population structure in Africa. However, the haplotypic diversity of African populations might help to overcome one of the major roadblocks in GWA research, the fine mapping of causal variants. We review the methodological challenges and consider how GWA studies in Africa will be transformed by new approaches in statistical imputation and large-scale genome sequencing.
Over the past 4 years, genome-wide association studies (GWA studies) have become a powerful tool for investigating the genetic basis of common diseases, and important new findings now emerge almost every week [1, 2, 3]. But so far there has been limited attention to health problems in Africa, such as the massive burden of infectious disease and the increasing prevalence of chronic diseases associated with changes in lifestyle [4, 5, 6]. By elucidating the molecular mechanisms that underlie resistance and susceptibility to disease, GWA studies in Africa might provide important insights into the development of more effective vaccines, therapeutics and public health interventions. As Africa is the ancestral home of all human populations, understanding the biology of disease in Africa could shed light on the genetic origins of common diseases worldwide [7, 8, 9].
African populations are genetically more diverse than European and Asian populations [10, 11, 12]. According to the out-of-Africa hypothesis of human origins, this is because groups migrating out of Africa experienced severe population bottlenecks, resulting in a reduction of genetic diversity in descendant populations [8, 13]. A reduction in nucleotide diversity outside Africa has been consistently observed in genotype and resequencing data8; similarly, levels of haplotype diversity tend to decrease and linkage disequilibrium (LD) tends to increase according to the geographic distance of a population from Africa [11, 12].
From a statistical genetic perspective, the high levels of haplotype diversity and low levels of LD in African populations have both advantages and disadvantages for genome-wide analysis. High levels of haplotype diversity are potentially a powerful tool for fine mapping the causal variants that underlie disease associations [14, 15, 16]. However, low levels of LD are disadvantageous when screening the genome for disease associations using current SNP-genotyping approaches, which essentially rely on the principle of LD mapping [17, 18]. The fundamental question underlying this Review is how to develop an appropriate methodology for GWA analysis in Africa that overcomes the difficulties of genome-wide screening for association and exploits the potential for fine mapping causal variants.
There is a growing body of data to address this problem. The first GWA study from Africa has recently been published [19], and others are close to completion. The International HapMap Project has generated a large catalogue of SNP allele frequencies and haplotypes in the Yoruba people of Nigeria [15, 20], and this effort has recently been expanded to include the Luhya and the Maasai people from Kenya. There has recently been remarkable progress in our understanding of genome sequence variation in Africa, starting with systematic resequencing of specific genomic regions [21, 22], followed by next-generation sequencing of the entire genome of an African individual [23], leading on to the 1000 Genomes Project. This large international endeavour will include whole-genome sequence data for several hundred people in different parts of Africa, and data have already emerged for the Yoruba group.
Here, we examine the implications of these and other recent findings for the design and analysis of GWA studies in Africa. We begin by outlining the practical importance of conducting GWA studies in Africa, and then examine in some detail the analytical problems that can arise at each stage of a conventional GWA study as a result of low LD and population structure (Box 1). We go on to consider how new approaches in statistical imputation and large-scale genome sequencing will help to overcome these problems and to accelerate the identification of causal variants. Finally, we briefly discuss the practical requirements for effective GWA studies in Africa, the need for attention to the ethical issues that arise in a resource-poor setting, and what is being done to build local research capacity in this area.
Box 1. Genome-wide association analysis by linkage disequilibrium mapping.
The table and diagrams describe the difference between African and European populations in the three stages of genome-wide association (GWA) analysis by linkage disequilibrium (LD) mapping.
| Stage of analysis | European populations | African populations |
|---|---|---|
| 1. Detecting genotype–phenotype associations by genome-wide SNP typing |
High levels of LD make it probable that many causal variants will be in sufficient correlation with nearby SNPs to show a significant genotype–phenotype association, even if the causal variant is not directly typed (see the figure, part a) |
Low levels of LD reduce the likelihood that a causal variant will have a sufficient level of correlation with nearby SNPs to show significant genotype–phenotype associations unless it is directly typed (part b) |
| 2. Replicating associations in multi-centre studies |
Different study sites tend to hove similar allele frequencies and patterns of LD. so there is a good chance of replicating associations even if the causal variant is not directly typed |
Different study sites may have different allele frequencies and/or patterns of LD. both of which reduce the likelihood of reproducing associations in multi-centre studies unless the causal variant is directly typed |
| 3. Localizing the causal variants through sequencing-and imputation-based strategies |
Localizing the causal variant can be difficult because it is in high LD with neighbouring SNPs, which give association signals of similar magnitude to the causal variant (part c) |
It might be relatively easy to localize the causal variant because it is in weak LD with neighbouring SNPs and will therefore stand out at the peak of the association signal (part d) |
Why GWA studies in Africa are needed
Most GWA studies are motivated by a desire to understand the underlying causes of disease at both the molecular and the environmental level. Although it is becoming apparent that the current generation of GWA studies will provide only a partial understanding of the genetic architecture of common diseases, they at least provide a foundation for systematic investigation of the problem, including the complex question of how disease risk is affected by gene–environment interactions [24, 25]. In high-income countries, GWA studies have provided many new leads for medical research on common diseases: for example, the discovery that common variants of the FTO gene are associated with risk of obesity – one of the first major findings from a GWA study – has led to new insights into the determinants of eating behaviour [26, 27, 28, 29]. It is important that Africa should not be excluded from this new research agenda. Here, we briefly outline two major public health problems that are of particular importance for GWA research in Africa: infectious diseases and the rising prevalence of chronic non-communicable diseases. In later sections we consider the potential importance of GWA studies in Africa for fine mapping the causal genetic variants that underlie common diseases found throughout the world.
Infectious disease
Over 10% of children in sub-Saharan Africa die before the age of 5 (compared with <1% in high-income countries), primarily due to infectious diseases, such as malaria, respiratory infections and diarrhoea [4]. AIDS is a major cause of death in young adults, and tuberculosis affects all age groups [5]. The difficulty of developing effective vaccines against malaria, AIDS and tuberculosis is a strong incentive for conducting GWA studies to discover natural mechanisms of resistance to infection, which has a significant genetic component [30, 31, 32, 33]. A classic example of how human genetic discoveries may translate into leads for vaccine development is given by a series of discoveries concerning Plasmodium vivax, a species of malaria parasite that causes much morbidity in the tropics. P. vivax infection is remarkably rare in sub-Saharan Africa, and over 30 years ago it was discovered that this is because most Africans lack the Duffy blood group, which is essential for erythrocyte invasion by P. vivax [34]. This lack is now known to be caused by a regulatory SNP in the Duffy blood group, chemokine receptor (DARC) gene; this discovery led to the molecular characterization of a crucial parasite protein that binds to the erythrocyte Duffy receptor, which in turn led to the development of a candidate vaccine against P. vivax [35]. Similarly useful genetic discoveries for Plasmodium falciparum, which is responsible for most malaria deaths, could revolutionize malaria vaccine development. Two further observations support this approach: malaria has been a strong force for selection on the human genome, and only a small fraction of host genetic resistance to malaria is explained by known factors, such as sickle haemoglobin, which implies that many genetic factors remain to be discovered [36]. A global partnership of malaria researchers, the Malaria Genomic Epidemiology Network (MalariaGEN), has been established to conduct multi-centre-scale genetic-association studies of resistance to malaria, and initial GWA data from The Gambia have been reported [19, 37]. An equally strong case can be made for genetic studies of tuberculosis, HIV/AIDS, invasive bacterial disease and other major infections [31, 32, 33]. GWA studies of tuberculosis have recently been completed in The Gambia and Ghana (A. V. Hill and R. D. Horstmann, personal communication), and a GWA study of bacteraemia in Kenya is currently being conducted by the Wellcome Trust Case Control Consortium.
Gene–environment interactions and chronic diseases
Changes in lifestyle in Africa are causing a rapidly rising prevalence of chronic non-communicable diseases, such as hypertension and diabetes [38, 39]. For example, the number of people in Africa with diabetes is estimated to rise from 10 million in 2006 to more than 18 million in 2025, and chronic diseases in general are expected to account for more than a quarter of deaths by 2015 (Ref. 39). The importance of genetic factors is highlighted by the observation that, among residents of high-income countries, there is higher prevalence of hypertension, diabetes and obesity in people of African ancestry than in those of European ancestry [40, 41]. Importantly, the prevalence of these diseases in people of African ancestry is much higher for those who reside in high-income countries [42, 43].
One approach being used to investigate the genetic basis of these differences is admixture mapping in groups of mixed ancestry, for example, African-Americans [44, 45, 46, 47, 48]. Admixture mapping – which has the advantage of requiring few genetic markers and the concomitant disadvantage that it localizes the genetic signal imprecisely – has led to the discovery of a number of important genetic loci [49, 50]. However, it is important that genetic studies are conducted in Africa itself, both to ensure relevance to the health problems that are encountered there and to take account of the great diversity of environments, which range from forest to urban, and of habitation, sanitation, diet, physical activity and other aspects of lifestyle. One of the most important environmental variables is the level of exposure to infection – for example, the prevalences of malaria, HIV/AIDS and helminthic infection vary widely across the continent.
One epidemiologically relevant change that is now occurring in Africa is a tendency for migration from rural to urban areas, which typically have a lower prevalence of parasitic diseases, such as malaria, but a higher prevalence of chronic non-communicable diseases, such as hypertension [51]. An important recent advance is a GWA study of hypertension in African-Americans, which identified several loci associated with systolic blood pressure that were replicated in a sample from West Africa [52]. There is a clear need to establish well-defined cohorts – such as the Africa America Diabetes Mellitus study, which involves five centres in Nigeria and Ghana – to investigate how genetic factors, and their interaction with the environment, contribute to the rapidly rising prevalence of hypertension, diabetes and other chronic diseases in Africa itself [53, 54, 55, 56].
GWA by LD mapping
Current methods for GWA analysis are based on the principle of LD mapping [17, 18]. This relies on a sufficiently high level of LD to screen for genetic associations across the whole genome by typing a subset of variants [15, 20, 23, 57]. GWA by LD mapping has three main stages of analysis, starting with genome-wide screening for associations, followed by replicating associations and then fine mapping of causal variants. In this section we summarize the different methodological issues that arise at each of these stages of GWA analysis in Africa.
Stage 1: Genome-wide screening for associations
At the first stage of GWA analysis, the aim is to screen the genome for regions that are associated with the disease or phenotype of interest. For example, a landmark study conducted by the Wellcome Trust Case Control Consortium in the UK population involved 17,000 individuals (2,000 cases for each of 7 common diseases and 3,000 population controls), and 0.5 million SNPs were genotyped in each individual [58]. To reduce the number of false-positive associations arising from multiple testing, it is necessary to impose a rigorous threshold for statistical significance that takes account of the large number of SNPs that have been genotyped. There are different ways of arriving at this genome-wide significance threshold [58, 59, 60, 61, 62], but typically it is set in the region of between p < 10−7 and p < 10−8. This threshold is more difficult to achieve in African populations than in European or Asian populations because of the lower levels of LD, and authentic loci with strong genetic effects may fail to reach genome-wide significance because of weak LD between causal variants and the SNPs that are genotyped [19]. In the following sections we consider how GWA signals might be boosted in African populations by improved SNP-genotyping platforms and by multipoint imputation from population-specific sequencing data.
Stage 2: Replicating associations
The second stage of GWA analysis aims to exclude false-positive associations due to systematic biases in genotyping and sampling by replicating associations in independent studies [59]. In European and Asian populations there has been considerable success in replicating GWA signals in large multi-centre studies across different locations [26, 63, 64], but in Africa there is a greater likelihood that authentic signals of association will fail to replicate across different locations because of high levels of population structure. A particular problem is that multi-centre replication is less likely to succeed when there is variation among locations in the level of LD between the causal variant and the SNPs that are genotyped [19, 65, 66].
Below, we discuss in more detail the problem that this LD variation and population structure creates for GWA studies in Africa, and how it might be addressed by using imputation to identify potential causal variants before attempting to replicate associations across different locations.
Stage 3: Fine mapping of causal variants
The final stage of GWA analysis is to make a high-resolution genetic map of those regions of the genome with replicable signals of association, with the aim of localizing the causal variants. How to achieve this remains open to debate, because so far there has been very limited success in identifying the causal variants responsible for GWA signals in European and Asian populations. Broadly speaking, fine mapping involves systematic resequencing of the genomic region of interest to identify all common variants, which are then tested for disease association using the largest possible sample size [67]. After the completion of the first tranche of large GWA studies in 2007 revealed novel genomic regions of association for several common diseases in European populations, there were initial hopes that this would shortly be followed by the identification of the causal variants. However, despite considerable efforts by several large research groups and consortia, progress has been slow, the fundamental problem being that high levels of LD make it difficult to distinguish causal variants from neighbouring non-functional variants. This has led to growing interest in trans-ethnic studies, which aim to increase the resolution of fine mapping by enlarging the haplotypic diversity of the sample. Studies in Africa could be of particular value in fine mapping because of the low levels of LD found in individual populations and because different populations in Africa have different patterns of LD [14, 15, 16, 68, 69].
In the following sections, we discuss in more detail the challenges of dealing with high levels of genome variation and population structure, and go on to consider how these challenges might be overcome as new GWA methodologies are developed, particularly those that are based on large-scale genome sequencing.
Dealing with high levels of genome variation
How many SNPs should be genotyped?
What is the optimum number of SNPs to genotype at the first stage of a GWA study in Africa, and which are the best SNPs to include in this genotyping set? More specifically, what is the optimum genotyping set for a given population, and how much does this need to be enlarged to cover other ethnic groups and geographical locations, given the great genetic diversity of African populations [10, 11, 12]? Although there is no clear answer to these questions at present, data soon to emerge from the 1000 Genomes Project on different populations in Africa will greatly increase our understanding of this problem. Our current understanding relies primarily on HapMap data on 90 Yoruban individuals from Ibadan in Nigeria, who were genotyped for 3.4 million SNPs [15, 20]. A crude estimate from the initial HapMap publication, based on the concept of tagging SNPs, was that a GWA study of 1.5 million SNPs in an African population would have approximately the same statistical power as a study of 0.6 million SNPs in a European population [15]. In practical terms, the current commercial SNP-genotyping platforms, most of which have been designed based on HapMap data, provide considerably lower levels of genome coverage in Africa than in Europe or Asia, and this translates to a lower power to detect a GWA signal that achieves the genome-wide significance threshold [11, 70, 71, 72].
The problem of ascertainment bias
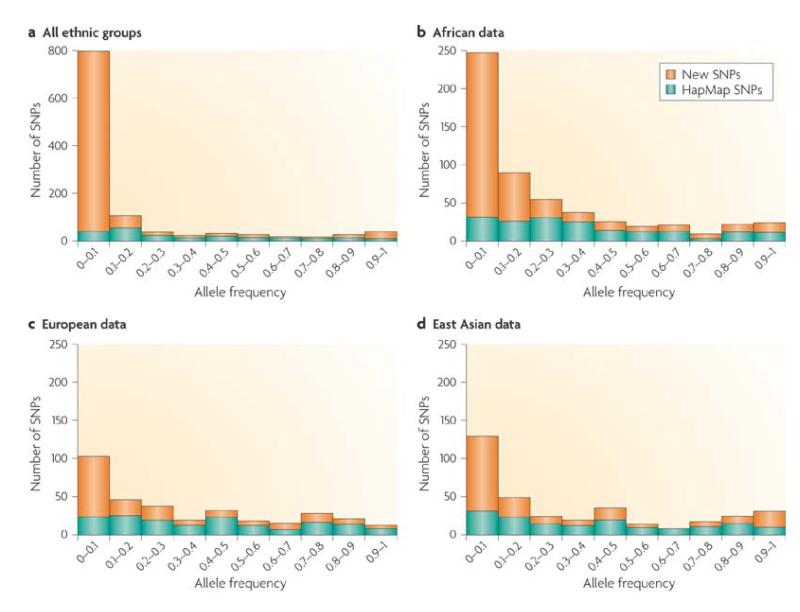
A fundamental limitation of using HapMap data for designing genotyping platforms for Africa is that the data focus on SNPs that were discovered in a relatively small number of individuals, predominantly of European descent [73]. This is particularly problematic because African populations have more private SNPs than European or Asian populations [74]. Resequencing of specific genome regions in the ENCODE Project revealed that data from Phase 2 of the HapMap Project provide 81% coverage of common SNPs in the Yoruba people compared with 94% in Europeans [75]. Because the HapMap Project prioritized SNPs that were common on all three continents, this leads to over-representation of high-frequency SNPs and under-representation of SNPs that are common in Africa but rare or absent elsewhere [22, 76] (Fig. 1). This was highlighted in a targeted resequencing study, which found that 91% of low-frequency SNPs discovered in the Yoruba people were missing from the HapMap data, compared with 86% in a comparable European sample [22].
Figure 1. African populations are subject to high levels of ascertainment bias in current SNP databases.
A study by Wall et al. [76] sequenced 40 intergenic regions in 90 individuals from 6 different ethnic groups. Within these regions, they observed almost all of the SNPs in the HapMap Phase 2 database, as well as discovering many new SNPs. The figure shows the number of SNPs in the HapMap data (green) compared with the number of SNPs that were discovered by resequencing and that were not present in the HapMap data (orange), categorized by derived allele frequency. a | Data from all ethnic groups combined. b | SNPs discovered in an African group (Mandinka) compared with African data (Yoruba people in Ibadan, Nigeria (YRI)) from the HapMap Project. c | SNPs discovered in a European group (Basque) compared with European data (Utah residents with Northern and Western European ancestry from the CEPH collection (CEU)) from the HapMap Project. d | SNPs discovered in an East Asian group (Han Chinese) compared with SNPs from a similar group (Han Chinese in Beijing (CHB)) in the HapMap Project. It can be seen that the HapMap data have greater SNP ascertainment bias for African than for European or Asian populations. In particular, African populations have many low-frequency alleles that are not well represented in current SNP databases. The figure is modified, with permission, from Ref. [76] © (2008) CSHL Press.
It remains uncertain to what extent data from the Yoruba people can be extrapolated to other populations in Africa, given the high level of haplotypic diversity and population structure [11]. The practical implication is that a SNP-genotyping platform based on HapMap data might have decreased coverage in regions of the genome in which LD differs between the population under study and the HapMap populations [65, 66]. Large-scale genotyping data sets are now being generated for other African populations: Phase 3 of the HapMap Project will include the Luhya and Maasai groups from Kenya in East Africa; the Human Genome Diversity Project includes individuals from eight African ethnic groups [12, 77]; and GWA studies of malaria and tuberculosis involving thousands of individuals are ongoing in The Gambia, Ghana and Malawi [19, 37]. Although these studies will provide valuable information, they all use commercial genotyping platforms for which SNP selection reflects the same biases as the HapMap data. Large-scale resequencing studies, such as the 1000 Genomes Project, are therefore required for the development of a comprehensive list of African variants and their LD structure across the continent.
Capturing structural variation
GWA studies must also take account of the importance of structural variation in the human genome. This includes copy-number variants (CNVs), such as insertions, deletions and duplications, as well as inversions and translocations. Structural variants seem to exhibit similar demographic patterns to SNPs (that is, a high proportion of common variants seem to be shared across continents), but there is evidence of greater structural variant diversity in Africa: among the HapMap populations, the Yoruba sample has more polymorphic CNVs than the European or Asian samples [78, 79, 80]. Structural variants can be genotyped with arrays that are specifically designed to interrogate known CNV regions, but there is potential ascertainment bias if these are based primarily on non-African reference data. Alternatively, they can be inferred from SNP-genotyping arrays, but the low levels of LD in Africa are an inherent limitation in LD-based strategies for CNV tagging. New approaches using next-generation sequencing technology will be particularly valuable in reducing ascertainment bias towards known, common variants.
Dealing with population structure
The potential confounding effect of population structure on genetic association studies in Africa is illustrated by the existence of more than 2,000 distinct language groups, most of which correspond to a specific ethnic group (see the Ethnologue website). There is growing evidence that these ethnic differences correlate with genetic differences and that levels of population structure are much greater within Africa than in other parts of the world [10, 19, 77].
Here, we discuss the analytical implications in two parts. First, we consider the consequences of local population structure – that is, variation in allele frequencies and LD between different ethnic groups who reside at a single location. Second, we discuss the effects of population structure on multi-centre studies – that is, variation in allele frequencies and patterns of LD at different geographical locations.
Local population structure
Failure to account for population structure in a community with multiple ethnic groups can result in a high false-discovery rate and reduce the power of the study [81, 82]. These confounding effects can be minimized by ethnic matching of cases and controls, but accurate matching can be difficult in communities in which there is substantial mixing between groups (Box 2). At the first stage of a GWA study, it is possible to correct for population structure using statistical approaches, such as genomic control and principal components analysis [83, 84]. In The Gambia, which is a community of considerable ethnic diversity, quantile–quantile plots of GWA data indicate that methods based on principal components analysis are highly effective in minimizing false-positive associations caused by population artefacts [19]. However, such statistical methods are more difficult to apply at the second stage of a GWA study because they require a substantial fraction of the assayed genetic markers to be independent of the phenotype being studied, and are therefore of limited value in replication studies in which only candidate SNPs are genotyped. In this situation it may be necessary to rely on surrogate markers, such as language or location of residence, to correct for population structure, and it has been shown that, at least in some populations, this can be reasonably effective19. An alternative is to replicate candidate signals with the use of family-based association studies – for example, with family trios – as such designs are generally more robust to the confounding effects of population structure85.
Box 2. Correcting for local population structure.
Population structure can affect genome-wide association (GWA) studies both at the local level and when combining data across multiple sites: here, we consider the effects of local population structure in The Gambia, West Africa. Jallow and colleagues [19] recruited case individuals (patients with severe malaria) at a large government hospital and control individuals from local birth clinics. The majority of cases and controls came from a relatively small geographical area of approximately 400 square miles. This community is made up of several ethnic groups, each with their own language, of which the most common are Mandinka, Fula, Wolof and Jola.
Part A of the figure illustrates the extent of population structure in The Gambia. It shows a principal components analysis of genome-wide SNP data from 2,500 individuals, revealing that the genetic population structure corresponds to self-reported ethnicity. Based on the genetic data, some individuals can be confidently assigned to a specific ethnic group, whereas others seem to have more complex ancestry.
Part B of the figure illustrates the effects of various statistical correction procedures in a case– control association study. It shows quantile–quantile plots of the trend test statistic for association with severe malaria. Part Ba shows that there are many false-positive associations in the raw data. Part Bb shows that the number of false-positive associations can be greatly reduced if the analysis is stratified by self-reported ethnicity. Part Bc shows a very low rate of false-positive associations after correction by principal components analysis.
These findings show that, in this particular community, the number of false-positive genetic associations in a case–control study can be reduced to an acceptable level by taking an individual’s self-reported ethnicity into account, despite high levels of population stratification and ethnic admixture. Such information can be valuable when conducting candidate gene studies in large population surveys.
However, there are several other ways in which population structure might confound GWA analysis that are not considered here. For example, variable patterns of linkage disequilibrium among different ethnic groups might act to reduce authentic GWA signals when the results from different groups are combined, as discussed in the main text and in Fig. 2. The figure is reproduced, with permission, from Nature Genetics Ref. [19] © (2009) Macmillan Publishers Ltd. All rights reserved.
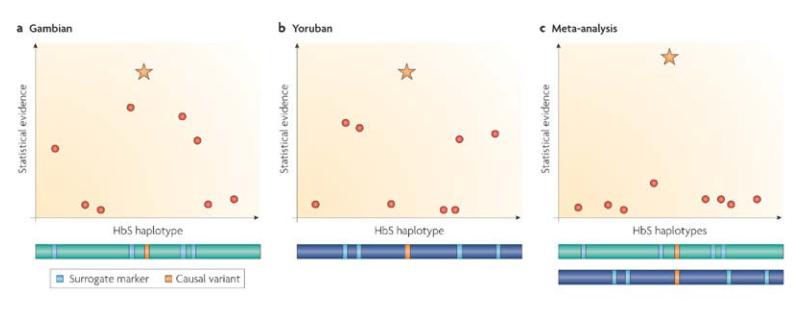
Population structure in multi-centre studies. A fundamental problem for multi-centre replication studies in Africa is that allele frequencies and patterns of LD may vary among the different study sites [8, 66, 86, 87]. Replication studies across European populations have been largely successful in reproducing the initial findings from GWA studies because the allele frequencies and patterns of LD are reasonably constant across Europe. This is not the case in Africa, and failure to replicate an association at different study sites may simply be due to varying patterns of LD between the causal variant and the SNPs that are genotyped. An analysis of the haemoglobin-β (HBB) gene region in different parts of West Africa provides a clear example of this: the SNP encoding ‘sickle cell’, haemoglobin (HbS), shows different patterns of LD in The Gambia compared with the HapMap Yoruba sample, and if data from both populations are combined using standard meta-analytic approaches, this tends to reduce, rather than improve, statistical power to detect signals of associations unless the causal SNP itself is genotyped [19, 66](Fig. 2).
Figure 2. Meta-analysis at a site with different associated haplotypes in two populations.
The ‘sickle cell’ variant of the haemoglobin-β (HBB) gene – encoding haemoglobin S (HbS) – is known to confer resistance to severe malaria. It is also known to exist on different haplotypes in different African populations. Here, we consider the major HbS haplotypes (green and blue horizontal bars) found in Gambia and in the Yoruba people of Nigeria: the HbS-encoding variant (orange strip) is in linkage disequilibrium with different SNPs (cyan strips) in the two populations. The graphs represent fictitious case–control studies of severe malaria in the Gambian (a) and Yoruban (b) populations, showing the strength of association signal expected from the causal variant (orange star) and other SNPs (red circles). Part c shows the results expected if data from a and b were combined in a standard meta-analysis: the association signal of the causal variant is boosted, but that of other SNPs is reduced.
This presents a quandary for the design and analysis of GWA studies in Africa. The standard approach in Europe aims to confirm initial GWA findings in multi-centre studies before attempting to identify the causal variants by regional sequencing and fine mapping. However, in Africa there is a lower probability that association signals will replicate in multi-centre studies unless the causal variants are assayed directly. It has been proposed that the term ‘transferability’ is more appropriate than ‘replication’ when testing SNP associations across genetically different populations [88].
In the next section we consider how new technologies for large-scale genome sequencing will help to overcome this problem in two ways: first, by starting to define the population genomic structure of African populations at the level of resolution needed to understand whether a particular multi-centre study is truly a test of replication as opposed to transferability; and second, by providing a method to refine the evidence of association in a GWA study, and to narrow this down to a shortlist of potential causal variants, before attempting to replicate putative causal variants across multiple populations.
Moving towards GWA by sequencing
GWA methodologies are about to be transformed by new sequencing technologies23, 89, 90, 91, 92, 93, 94. The cost of sequencing an individual human genome will probably fall to US$1000 in the next few years, and eventually it will be possible to conduct GWA analysis by genome sequencing of all of the cases and controls. This will be particularly beneficial for studies in Africa: it will increase the strength of GWA signals, because causal variants will be directly tested, and replication studies will be more likely to succeed because they will include the causal variant. In this situation, weak LD and variable LD between populations will become an advantage, as they will help to distinguish causal variants.
GWA by sequencing will greatly enhance our ability to detect associations with variants that are population-specific, and to dissect the problem of allelic heterogeneity. For example, there are two distinct variants of the HBB gene that confer resistance to malaria in West Africa: one encodes HbS (a valine substitution at codon 6) and the other encodes HbC (a lysine substitution, also at codon 6). HbS is relatively widespread, whereas HbC has a more localized distribution – for example, among the Dogon people of Mali, who have a low frequency of HbS [95, 96, 97]. This example is well understood because haemoglobin has been intensively studied by geneticists for many years, but allelic heterogeneity of this sort might be extremely difficult to dissect by GWA analysis, unless it is based on genome sequencing.
The 1000 Genomes Project will improve imputation accuracy
It will be some years before GWA analysis by sequencing becomes a practical proposition, and this raises the question of how to perform effective GWA studies in Africa using current genotyping resources. Within the next 2 years, the 1000 Genomes Project proposes to generate whole-genome sequence data on at least 60 individuals from each of 5 different African populations: data are currently being generated on two HapMap groups, the Yoruba of Nigeria and the Luhya of Kenya, and plans are under way to include groups from The Gambia, Ghana and Malawi. As well as enabling the optimization of new SNP-genotyping platforms, these data will increase the value of existing SNP-genotyping platforms by increasing the accuracy of multipoint imputation. Imputation is a method of statistically inferring an individual’s genotype at a variable position in the genome, based on that individual’s known genotypes at nearby variable positions combined with reference data on genome variation in the general population [98, 99, 100, 101]. The HapMap Project has provided an important reference panel for imputation in European populations, and it is now common for GWA studies to report association data at 3 million SNP positions, of which 1 million have been directly genotyped and the remainder imputed.
Accurate imputation requires the correct reference data
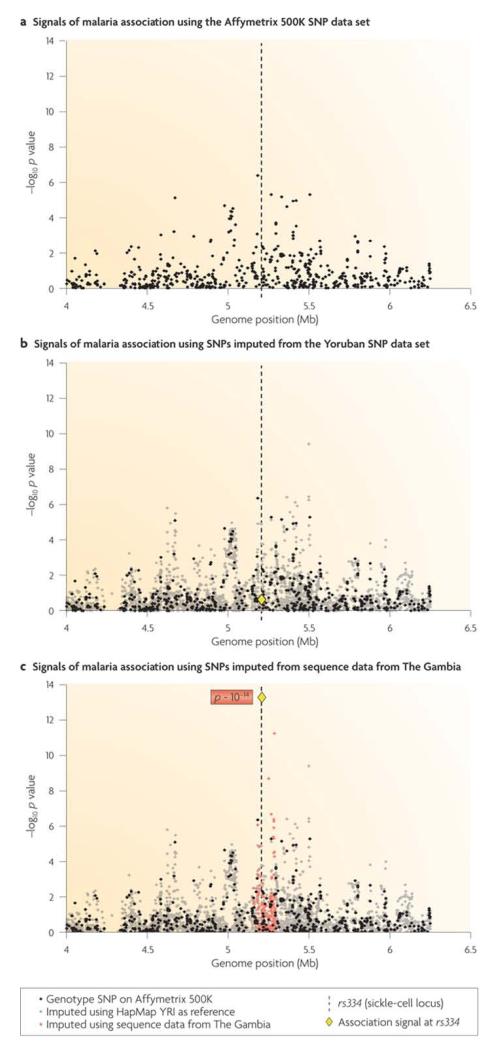
Imputation strategies are predicated on the assumption that the reference data accurately represent the haplotypes that exist in the GWA study population; if this is not the case, imputation can give misleading results. This is particularly problematic for African populations, in which lower imputation accuracy has been reported [65, 102]. The problem is well illustrated by a GWA study of severe malaria in Gambian children, in which a detailed analysis was undertaken of a 110-kb region of the genome containing the known malaria resistance variant HbS19. All common variants were imputed across this region using two different sets of reference data on genome variation. The first reference data set was obtained by resequencing 62 Gambian individuals across this region of the genome, and the second used HapMap data from the Yoruba people, who live in a different part of West Africa. Imputation based on the Gambian reference data accurately identified HbS as being strongly associated with resistance to malaria, whereas imputation based on the HapMap Yoruba reference data showed no association with HbS (Fig. 3). This finding is not entirely surprising given that the HbS-encoding allele occurs on different haplotypic backgrounds in different parts of Africa [103, 104, 105, 106].
Figure 3. Imputation and the choice of haplotype reference panel.
Imputation is a process of statistical inference that estimates the most likely genotype of an individual at a given position in the genome, based on what is known about the genotype of that individual at nearby positions and on a reference data set of genome variation in the general population. The accuracy of imputation depends on the appropriateness of the reference data set. The figure shows signals of association with severe malaria from SNPs distributed across a ~2.5-Mb region of chromosome 11 (Ref. 19). The vertical dashed line represents the position of rs334: this SNP is known to encode the haemoglobin S (HbS) variant of the haemoglobin-β (HBB) gene, which confers resistance to malaria. a | SNPs typed using the Affymetrix 500K genotyping platform (black circles). b | SNPs imputed using the HapMap Yoruba people in Ibadan, Nigeria (YRI) data as the reference (grey circles). The rs334 SNP is shown as a yellow diamond. c | SNPs imputed from regional sequencing data on 62 Gambian individuals (orange circles), including rs334 (yellow diamond). If we did not know that rs334 was the causal variant, imputation based on Gambian sequencing data would have been extremely useful, whereas imputation based on the HapMap YRI data would have been misleading. Parts a and c are modified, with permission, from Nature Genetics (Ref. 19) © (2009) Macmillan Publishers Ltd. All rights reserved.
Imputation based on population-specific sequencing as an interim strategy
The above result represents a proof of principle that causal genetic variants can be fine mapped in Africa by imputation based on population-specific genome variation data. Fig. 3 illustrates three important points. First, imputation should be based on population-specific reference data. Second, multipoint imputation can be highly effective in boosting GWA signals when the genotyping array contains no individual SNP that is in strong LD with the causal variant; in the case of HbS, the imputed GWA signal was several orders of magnitude greater than that obtained by direct genotyping. Third, low levels of LD can be valuable in localizing a causal variant by fine mapping: in the case of HbS, after imputation had been performed with the appropriate reference panel, the causal SNP (rs334) could be clearly identified at the peak of the association signal. It remains to be determined whether other causal variants will be as amenable to fine mapping by imputation as the HbS variant. The success of this approach will be affected by different patterns of genome variation, and the HbS-encoding locus has several features that may be particularly favourable, namely a strong phenotypic effect, an extended ancestral haplotype owing to recent positive selection, and a genomic region with generally weak LD19. But until it becomes possible to conduct GWA by sequencing all cases and controls, imputation based on population-specific reference data provides an interim strategy to boost GWA signals of association in Africa and to enable multi-centre studies, the results of which can be usefully combined because all common (including causal) variants have been imputed with a reasonable level of accuracy at each centre.
Conclusions
GWA research in Africa presents a wide range of practical, scientific and ethical challenges, which we have not attempted to cover comprehensively here. For example, it can be a huge undertaking to establish the clinical research infrastructure needed to recruit large numbers of patients and to ensure accurate phenotypic data when working in a resource-poor setting. There is a need for robust epidemiological platforms to investigate how variations in environment and lifestyle affect the results of genetic-association studies in different populations, and for statistical methodologies that can account for such interactions when combining data from different locations in multi-centre GWA studies. GWA studies in Africa require the building of local resources and the development of data-sharing networks that meet the needs of the African research community, and it is of crucial importance to pay attention to the ethical issues that arise in medical and genetic research in resource-poor settings (Box 3).
Box 3. Developing capacity for genome-wide association studies in Africa.
Building local resources
Obtaining well-characterized phenotypic data from thousands of individuals in poor communities with no systematic medical records requires investment to strengthen infrastructure for clinical research and data management. There also needs to be a commitment to build the resources for genetic and genomic research in Africa, and to foster a cadre of African scientists with the expertise needed to lead this research area. The African Society of Human Genetics provides an important forum for knowledge-sharing, networking and interactive training programs, with an annual conference held in a different African region each year [107, 108]. The Society also recognizes the need to communicate with policymakers and to attract global attention to the efforts of African scientists, which will be crucial in persuading African institutions and governments to engage with large-scale projects in human genetics and genomics. Another example of capacity building is the Malaria Genomic Epidemiology Network (MalariaGEN) scheme of data fellowships and data bursaries, which provides training and support in statistics and informatics for researchers in 15 malaria-endemic countries [37]. Institutions also need to develop infrastructure to manage genetic samples and data within an appropriate regulatory and ethical framework, an example being the Gambian National DNA Bank [109].
Data-sharing networks
Data-sharing networks, such as the Wellcome Trust Case Control Consortium and the Genetic Association Information Network, have been instrumental in driving forward genome-wide association (GWA) research [58, 110]. Bringing together African and non-African partners will require work at many different levels – for example, reaching decisions about the ownership and permitted uses of shared data and samples, intellectual property and data release, and developing web information systems to integrate data from different research groups and to make these data widely accessible to partners in Africa [111, 112]. The MalariaGEN network has developed policies for data sharing that take account of the disparities of research in high- and low-income countries [37]. For example, MalariaGEN’s policy for GWA data release promotes data access while guarding against uses that might lead to ethnic stigmatization, in addition to setting timelines for data release that are fair to contributing investigators in malaria-endemic countries, who might not be in a position to analyse their data as rapidly as researchers in high-income countries [37, 113].
Ethical issues
Genetic research in developing countries raises a wide range of ethical and social issues [114], and considering them all is beyond the scope of this Review. An example is the problem of obtaining valid consent from study participants for whom terms such as ‘genetics’ and ‘database’ might be meaningless unless carefully explained, drawing from local experience [115, 116, 117]. In this situation, it is important to take account of local language and cultural practices and it may be appropriate to seek consent from community leaders as well as from individuals. Another sensitive issue is the fear that the use of information about ethnicity in genetic studies might lead to ethnic stigmatization [118].
Towards an African Genome Project
Plans to study African genome variation within the 1000 Genomes Project are currently limited to populations that speak Niger-Kordofanian languages, and will therefore encompass only a fraction of African genetic diversity [10]. The African Society of Human Genetics has put forward a strong case for an African Genome Project [108], one component of which would involve systematic sampling of at least 100 ethnic groups across the continent, including minority populations that will be poorly represented in the 1000 Genomes Project. The fundamental aim is to develop population-based resources to investigate genetic and environmental determinants of disease. As discussed above, there is also a need for investments in training and infrastructure. Although these plans remain embryonic, they demonstrate that the time is ripe for an expansion of genome research in Africa, led by African investigators.
Our main purpose in this Review has been to consider the specific methodological roadblocks to GWA analysis that arise in Africa owing to the high levels of genome diversity and population structure. Many of these roadblocks will be removed when new sequencing technologies make it possible to conduct GWA analysis by genome sequencing. Because all variants will be directly observed, including causal variants, this will increase the strength of GWA signals and make it easier to perform meta-analyses across multiple study sites. An interim strategy is to conduct imputation of all common variants, and for GWA studies in Africa it is particularly important that this should be based on population-specific genome variation data. By providing a framework for accurate imputation in a number of different African populations, the 1000 Genomes Project will be an important first step towards reliable multi-centre GWA studies in Africa and the fine mapping of causal variants.
Acknowledgements
We thank our colleagues in Africa and at the MalariaGEN resource centre for their many contributions to the ideas developed here. We are also grateful for helpful suggestions by two anonymous reviewers. This work was funded by the Wellcome Trust, the Medical Research Council and the Bill and Melinda Gates Foundation through the Foundation for the US National Institutes of Health.
Footnotes
Links
DATABASES
Competing interests statement The authors declare no competing financial interests.
References
- 1.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 3.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 4.Black RE, Morris SS, Bryce J. Where and why are 10 million children dying every year? Lancet. 2003;361:2226–2234. doi: 10.1016/S0140-6736(03)13779-8. [DOI] [PubMed] [Google Scholar]
- 5.Mathers CD, Boerma T, Ma Fat D. Global and regional causes of death. Br. Med. Bull. 2009;92:7–32. doi: 10.1093/bmb/ldp028. [DOI] [PubMed] [Google Scholar]
- 6.Mayosi BM, et al. The burden of non-communicable diseases in South Africa. Lancet. 2009;374:934–947. doi: 10.1016/S0140-6736(09)61087-4. [DOI] [PubMed] [Google Scholar]
- 7.Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nature Rev. Genet. 2002;3:611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- 8.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genomics Hum. Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. This paper provides a comprehensive discussion on the implications of genetic diversity in Africa for complex disease mapping and understanding the origins of modern humans.
- 9.Sirugo G, et al. Genetic studies of African populations: an overview on disease susceptibility and response to vaccines and therapeutics. Hum. Genet. 2008;123:557–598. doi: 10.1007/s00439-008-0511-y. [DOI] [PubMed] [Google Scholar]
- 10.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. The most detailed genetic survey of Africans and African-Americans to date.
- 11.Conrad DF, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nature Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. This article reports the extent of haplotype diversity in humans and the applicability of genome-wide studies across many populations.
- 12.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. This paper provides a detailed exposition of genetic variation across the populations of the Human Genome Diversity Project.
- 13.DeGiorgio M, Jakobsson M, Rosenberg NA. Out of Africa: modern human origins special feature: explaining worldwide patterns of human genetic variation using a coalescent-based serial founder model of migration outward from Africa. Proc. Natl Acad. Sci. USA. 2009;106:16057–16062. doi: 10.1073/pnas.0903341106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todd JA, et al. Identification of susceptibility loci for insulin-dependent diabetes mellitus by trans-racial gene mapping. Nature. 1989;338:587–589. doi: 10.1038/338587a0. An insightful study from 20 years ago that illustrates the problem of identifying causal genetic variants and the value of examining African haplotypes.
- 15.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgason A, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nature Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 17.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 18.Clark AG. Finding genes underlying risk of complex disease by linkage disequilibrium mapping. Curr. Opin. Genet. Dev. 2003;13:296–302. doi: 10.1016/s0959-437x(03)00056-x. [DOI] [PubMed] [Google Scholar]
- 19.Jallow M, et al. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nature Genet. 2009;41:657–665. doi: 10.1038/ng.388. The first report of a genome-wide study performed in Africa, describing population structure and imputation from population-specific sequencing data.
- 20.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford DC, et al. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am. J. Hum. Genet. 2004;74:610–622. doi: 10.1086/382227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhangale TR, Rieder MJ, Nickerson DA. Estimating coverage and power for genetic association studies using near-complete variation data. Nature Genet. 2008;40:841–843. doi: 10.1038/ng.180. A well-conducted resequencing study that highlights the level of ascertainment bias in existing databases for African populations in particular.
- 23.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. The first whole-genome sequence of an individual of African ancestry.
- 24.Goldstein DB. Common genetic variation and human traits. N. Engl. J. Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 25.Hirschhorn JN. Genomewide association studies – illuminating biologic pathways. N. Engl. J. Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 26.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dina C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 28.Wardle J, Llewellyn C, Sanderson S, Plomin R. The FTO gene and measured food intake in children. Int. J. Obes. (Lond.) 2009;33:42–45. doi: 10.1038/ijo.2008.174. [DOI] [PubMed] [Google Scholar]
- 29.Tanofsky-Kraff M, et al. The FTO gene rs9939609 obesity-risk allele and loss of control over eating. Am. J. Clin. Nutr. 2009;90:1483–1488. doi: 10.3945/ajcn.2009.28439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 2005;77:171–190. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill AV. Aspects of genetic susceptibility to human infectious diseases. Annu. Rev. Genet. 2006;40:469–486. doi: 10.1146/annurev.genet.40.110405.090546. [DOI] [PubMed] [Google Scholar]
- 32.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldstein DB. Genomics and biology come together to fight HIV. PLoS Biol. 2008;6:e76. doi: 10.1371/journal.pbio.0060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 35.Moreno A, et al. Preclinical assessment of the receptor-binding domain of Plasmodium vivax Duffy-binding protein as a vaccine candidate in rhesus macaques. Vaccine. 2008;26:4338–4344. doi: 10.1016/j.vaccine.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackinnon MJ, Mwangi TW, Snow RW, Marsh K, Williams TN. Heritability of malaria in Africa. PLoS Med. 2005;2:e340. doi: 10.1371/journal.pmed.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malaria Genomic Epidemiology Network A global network for investigating the genomic epidemiology of malaria. Nature. 2008;456:732–737. doi: 10.1038/nature07632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daar AS, et al. Grand challenges in chronic non-communicable diseases. Nature. 2007;450:494–496. doi: 10.1038/450494a. [DOI] [PubMed] [Google Scholar]
- 39.World Health Organization . Preventing Chronic Diseases: A Vital Investment. World Health Organization; Geneva: 2005. [Google Scholar]
- 40.Cooper RS, Rotimi CN, Ward R. The puzzle of hypertension in African-Americans. Sci. Am. 1999;280:56–63. doi: 10.1038/scientificamerican0299-56. [DOI] [PubMed] [Google Scholar]
- 41.Smith MW, O’Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nature Rev. Genet. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 42.Cooper R, et al. The prevalence of hypertension in seven populations of West African origin. Am. J. Public Health. 1997;87:160–168. doi: 10.2105/ajph.87.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper RS, et al. Prevalence of NIDDM among populations of the African diaspora. Diabetes Care. 1997;20:343–348. doi: 10.2337/diacare.20.3.343. [DOI] [PubMed] [Google Scholar]
- 44.Smith MW, et al. A high-density admixture map for disease gene discovery in African Americans. Am. J. Hum. Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKeigue PM. Prospects for admixture mapping of complex traits. Am. J. Hum. Genet. 2005;76:1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu X, et al. Admixture mapping for hypertension loci with genome-scan markers. Nature Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 47.Patterson N, et al. Genetic structure of a unique admixed population: implications for medical research. Hum. Mol. Genet. 2009 Nov 18; doi: 10.1093/hmg/ddp505. doi: 10.1093/hmg/ddp505. [DOI] [PubMed] [Google Scholar]
- 48.Price AL, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng CY, et al. Admixture mapping of 15,280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLoS Genet. 2009;5:e1000490. doi: 10.1371/journal.pgen.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reich D, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. A detailed investigation that used admixture mapping to identify a genomic region of interest for a common phenotype and then used association fine mapping to find a plausible causal variant.
- 51.Kaufman JS, Owoaje EE, Rotimi CN, Cooper RS. Blood pressure change in Africa: case study from Nigeria. Hum. Biol. 1999;71:641–657. [PubMed] [Google Scholar]
- 52.Adeyemo A, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. An important GWA study in African-Americans with replication studies in West Africa. This work sets the scene for African GWA studies of hypertension and other chronic diseases.
- 53.Rotimi CN, et al. A genome-wide search for type 2 diabetes susceptibility genes in West Africans: the Africa America Diabetes Mellitus (AADM) study. Diabetes. 2004;53:838–841. doi: 10.2337/diabetes.53.3.838. [DOI] [PubMed] [Google Scholar]
- 54.Rotimi CN, et al. In search of susceptibility genes for type 2 diabetes in West Africa: the design and results of the first phase of the AADM study. Ann. Epidemiol. 2001;11:51–58. doi: 10.1016/s1047-2797(00)00180-0. [DOI] [PubMed] [Google Scholar]
- 55.Rotimi C, et al. Prevalence and determinants of diabetic retinopathy and cataracts in West African type 2 diabetes patients. Ethn. Dis. 2003;13:S110–S117. [PubMed] [Google Scholar]
- 56.Steinthorsdottir V, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nature Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 57.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nature Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 58.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chanock SJ, et al. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 60.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 62.Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet. Epidemiol. 2008;32:179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 63.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teo YY, et al. Genome-wide comparisons of variation in linkage disequilibrium. Genome Res. 2009;19:1849–1860. doi: 10.1101/gr.092189.109. Provides a quantitative metric for assessing the extent of variation in patterns of LD between two populations.
- 66.Teo YY, et al. Power consequences of linkage disequilibrium variation between populations. Genet. Epidemiol. 2008;33:128–135. doi: 10.1002/gepi.20366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lowe CE, et al. Large-scale genetic fine mapping and genotype–phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nature Genet. 2007;39:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- 68.McKenzie CA, et al. Trans-ethnic fine mapping of a quantitative trait locus for circulating angiotensin I-converting enzyme (ACE) Hum. Mol. Genet. 2001;10:1077–1084. doi: 10.1093/hmg/10.10.1077. [DOI] [PubMed] [Google Scholar]
- 69.Sanna S, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nature Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Bakker PI, et al. Efficiency and power in genetic association studies. Nature Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 71.Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nature Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 72.Pe’er I, et al. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nature Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 73.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller RD, et al. High-density single-nucleotide polymorphism maps of the human genome. Genomics. 2005;86:117–126. doi: 10.1016/j.ygeno.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wall JD, et al. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 78.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nature Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 79.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 Oct 7; doi: 10.1038/nature08516. doi:10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nature Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 82.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case–control association study. Nature Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 83.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 84.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet. 2006;38:904–909. doi: 10.1038/ng1847. An invaluable approach for dealing with genetic association artefacts caused by ethnic admixture.
- 85.Ewens WJ, Spielman RS. The transmission/disequilibrium test: history, subdivision, and admixture. Am. J. Hum. Genet. 1995;57:455–464. [PMC free article] [PubMed] [Google Scholar]
- 86.Tishkoff SA, et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 87.Tarazona-Santos E, Tishkoff SA. Divergent patterns of linkage disequilibrium and haplotype structure across global populations at the interleukin-13 (IL13) locus. Genes Immun. 2005;6:53–65. doi: 10.1038/sj.gene.6364149. [DOI] [PubMed] [Google Scholar]
- 88.Shriner D, et al. Transferability and fine-mapping of genome-wide associated loci for adult height across human populations. PLoS ONE. 2009;4:e8398. doi: 10.1371/journal.pone.0008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 91.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 92.Branton D, et al. The potential and challenges of nanopore sequencing. Nature Biotech. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nature Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 94.Shendure J, Ji H. Next-generation DNA sequencing. Nature Biotech. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 95.Agarwal A, et al. Hemoglobin C associated with protection from severe malaria in the Dogon of Mali, a West African population with a low prevalence of hemoglobin S. Blood. 2000;96:2358–2363. [PubMed] [Google Scholar]
- 96.Modiano D, et al. Haemoglobin S and haemoglobin C: ‘quick but costly’ versus ‘slow but gratis’ genetic adaptations to Plasmodium falciparum malaria. Hum. Mol. Genet. 2008;17:789–799. doi: 10.1093/hmg/ddm350. [DOI] [PubMed] [Google Scholar]
- 97.Modiano D, et al. Haemoglobin C protects against clinical Plasmodium falciparum malaria. Nature. 2001;414:305–308. doi: 10.1038/35104556. [DOI] [PubMed] [Google Scholar]
- 98.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 99.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. A state-of-the-art imputation method that is particularly relevant to the availability of whole-genome sequence data.
- 102.Huang L, et al. Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hanchard N, et al. Classical sickle β-globin haplotypes exhibit a high degree of long-range haplotype similarity in African and Afro-Caribbean populations. BMC Genet. 2007;8:52. doi: 10.1186/1471-2156-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chakravarti A, et al. Nonuniform recombination within the human β-globin gene cluster. Am. J. Hum. Genet. 1984;36:1239–1258. [PMC free article] [PubMed] [Google Scholar]
- 105.Pagnier J, et al. Evidence for the multicentric origin of the sickle cell hemoglobin gene in Africa. Proc. Natl Acad. Sci. USA. 1984;81:1771–1773. doi: 10.1073/pnas.81.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chebloune Y, et al. Structural analysis of the 5′ flanking region of the β-globin gene in African sickle cell anemia patients: further evidence for three origins of the sickle cell mutation in Africa. Proc. Natl Acad. Sci. USA. 1988;85:4431–4435. doi: 10.1073/pnas.85.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rotimi CN. Inauguration of the African Society of Human Genetics. Nature Genet. 2004;36:544. doi: 10.1038/ng0604-544. [DOI] [PubMed] [Google Scholar]
- 108.Newport MJ, Rotimi CN. Reducing the global genomic inequity gap: development of an African genome project. Public Health Genomics. 2009;12:251–252. doi: 10.1159/000197973. [DOI] [PubMed] [Google Scholar]
- 109.Sirugo G, et al. A national DNA bank in The Gambia, West Africa, and genomic research in developing countries. Nature Genet. 2004;36:785–786. doi: 10.1038/ng0804-785. [DOI] [PubMed] [Google Scholar]
- 110.Manolio TA, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nature Genet. 2007;39:1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 111.Chokshi DA, Parker M, Kwiatkowski DP. Data sharing and intellectual property in a genomic epidemiology network: policies for large-scale research collaboration. Bull. World Health Organ. 2006;84:382–387. doi: 10.2471/blt.06.029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kaye J, Heeney C, Hawkins N, de Vries J, Boddington P. Data sharing in genomics – re-shaping scientific practice. Nature Rev. Genet. 2009;10:331–335. doi: 10.1038/nrg2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parker M, et al. Ethical data-release in genome-wide association studies in developing countries. PLoS Med. 2009;6:e1000143. doi: 10.1371/journal.pmed.1000143. This article discusses the ethical implications of data sharing and data release in large-scale genetic studies conducted in Africa.
- 114.Chokshi D, Kwiatkowski D. Ethical challenges of genomic epidemiology in developing countries. Genomics Soc. Policy. 2005;1:1–15. [Google Scholar]
- 115.Chokshi DA, et al. Valid consent for genomic epidemiology in developing countries. PLoS Med. 2007;4:e95. doi: 10.1371/journal.pmed.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marshall PA, et al. Voluntary participation and informed consent to international genetic research. Am. J. Public Health. 2006;96:1989–1995. doi: 10.2105/AJPH.2005.076232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tekola F, et al. Tailoring consent to context: designing an appropriate consent process for a biomedical study in a low income setting. PLoS Negl. Trop. Dis. 2009;3:e482. doi: 10.1371/journal.pntd.0000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Caulfield T, et al. Race and ancestry in biomedical research: exploring the challenges. Genome Med. 2009;1:8. doi: 10.1186/gm8. [DOI] [PMC free article] [PubMed] [Google Scholar]