Abstract
Alphaviruses are mosquito-borne viruses that cause serious human and animal diseases. Previous studies demonstrated that a determinant within the nsP1/nsP2 cleavage domain of the virulent Sindbis AR86 virus played a key role in regulating adult mouse virulence without adversely affecting viral replication. Additional characterization of this determinant demonstrated that a virus with the attenuating mutation induced more type I IFN production both in vivo and in vitro. Interestingly, this phenotype was not specific to the Sindbis AR86 virus, as a similar mutation in a distantly related alphavirus, Ross River Virus (RRV), also led to enhanced IFN induction. This effect was independent of virus-induced host shutoff, since IRF-3 phosphorylation, which occurs independently of de novo host transcription/translation, was induced more robustly in cells infected with the mutant viruses. Altogether, these results demonstrate that critical determinants within the nsP1/nsP2 cleavage domain play an important role in regulating alphavirus induced IFN responses.
Keywords: Alphaviruses, Sindbis, Ross River, nsP1 mutants, type I IFN induction
Introduction
Alphaviruses are mosquito-borne viruses capable of infecting humans and causing various diseases ranging from acute encephalitis to long-term virus-induced arthritis. The Sindbis-group viruses are among the most studied and well characterized of the alphaviruses, having been used to identify viral and host factors that contribute to virus-induced disease in mice (Morrison, Simmons, and Heise, 2008; Ryman et al., 2002; Suthar et al., 2005). The majority of the Sindbis virus strains are a virulent in adult mice, however, there are a few exceptions, including the neurovirulent neuroadapted Sindbis virus (NSV) (Tucker et al., 1993) and AR86 strains (Heise, Simpson, and Johnston, 2000; Simpson et al., 1996). Genetic studies have mapped virulence determinants of these neurovirulent viruses to both the structural and nonstructural proteins. Detailed genetic mapping studies using chimeric viruses that encode regions of the adult mouse virulent AR86 and the closely related a virulent Girdwood S.A identified four major neurovirulence determinants in the AR86 genome including, a Threonine at nsP1 position 538, an 18-amino acid deletion at nsP3 386, a Cysteine at nsP3 position 537, and a Serine at E2 position 243 (Suthar et al., 2005). While all four determinants were found to be essential for adult mouse virulence, the determinant within nsP1 was found to be critical; as a single coding change of this determinant from a Thr to Ile severely attenuated the virus, while introduction of a Thr at this position into non-virulent viruses partially rescued virulence (Heise, Simpson, and Johnston, 2000). Currently, the exact mechanism by which this determinant affects viral virulence is not understood.
The alphavirus nonstructural proteins form the viral replication complex that mediates cytoplasmic viral RNA synthesis within infected cells. Nonstructural polyprotein processing is intimately linked to viral RNA synthesis (Lemm and Rice, 1993; Shirako and Strauss, 1994). Upon viral entry, the virion-associated RNA is immediately translated to form the replication complex consisting of the nonstructural precursor, nsP123, and nsP4, which together mediate negative strand RNA synthesis (Strauss and Strauss, 1994). Later in the course of infection, the polyprotein precursor nsP123 is further processed by nsP2 to produce mature nsP1, nsP2, and nsP3. The mature nonstructural proteins, together with nsP4, form the replication complex to synthesize full length genomic RNA. The mature nsPs also bind to an internal subgenomic promoter to synthesize smaller subgenomic RNAs that encode the viral structural proteins. Previous studies using mutants that lack the ability to efficiently process the nonstructural polyprotein precursor nsP123 showed dysregulation of viral RNA synthesis (Lemm and Rice, 1993). Interestingly, the wild type AR86 Thr to Ile change at nsP1 position 538, is located in the P3 position of the conserved nsP1/2 cleavage recognition domain (Shirako and Strauss, 1994). The presence of an attenuating Ile at this position results in increased kinetics of nonstructural polyprotein processing as well as earlier induction of 26S subgenomic RNA synthesis. Furthermore, this mutation did not affect full length negative or positive strand RNA synthesis, though it did result in increased virus production (Heise et al., 2003). Whether differential nonstructural protein processing or the increased 26S RNA is important for the attenuated phenotype of this virus in vivo has yet to be determined.
In addition to mediating viral replication, the nonstructural proteins interact with host factors to alter environmental conditions that favor viral replication (Garmashova et al., 2006; Montgomery et al., 2006). The nonstructural proteins of Old World alphaviruses, such as Sindbis and Semliki Forest virus, have been shown to mediate shutoff of host transcription and translation (Garmashova et al., 2007). While the exact mechanism of virus-mediated host shutoff is not completely understood, it has been proposed that mature nsP2 is required (Gorchakov, Frolova, and Frolov, 2005; Gorchakov et al., 2008). Alphaviruses are thought to down regulate host mRNA and protein synthesis in order to limit competition for the cellular machinery to maximize viral replication while simultaneously limiting induction of type I IFN and related IFN stimulated genes (ISGs).
Sindbis viruses are highly sensitive to the effects of type I IFN (Frolova et al., 2002). Type I IFN receptor deficient mice exhibited enhanced susceptibility to Sindbis virus marked by increased virulence, broadened tissue tropism, and uncontrolled virus replication (Ryman et al., 2000), demonstrating that type I IFN plays an important role in controlling Sindbis virus infection. Therefore, it is likely that Sindbis virus utilizes several mechanisms to limit and/or suppress type I IFN induction. One likely mechanism involves generalized suppression of host macromolecular synthesis. In support of this, Sindbis virus mutants that are defective for host shutoff are more potent inducers of the type I IFN response (Gorchakov, Frolova, and Frolov, 2005; Gorchakov et al., 2008). However, host shutoff independent mechanisms likely exist, as a mutation in the nsP2 protein of Semliki Forest Virus, a related alphavirus, affects type I IFN induction independently of generalized host shutoff (Breakwell et al., 2007).
In this study, we demonstrate an important role for a determinant at the P3 cleavage position between nsP1 and nsP2 in regulating type I IFN induction. Mutation of this position in two distantly related alphaviruses, the AR86 strain of Sindbis and Ross River Virus, resulted in enhanced type I IFN induction and IRF-3 activation in comparison to the wild type viruses. Furthermore, the P3 mutation had little effect on kinetics of virus mediated host transcription and translation shutoff. Taken together, these results suggest that the P3 determinant, which regulates AR86 virulence, plays a major role in regulating type I IFN induction that is independent of virus mediated host shutoff.
Results
An attenuating Isoleucine mutation at nsP1 position 538 (T538I) in the Sindbis virus AR86 backbone leads to increased type I IFN induction in vivo and in vitro
A virulence determinant located within the nsP1/nsP2 cleavage domain (nsP1 538) of the Sindbis virus, AR86, regulates viral nonstructural polyprotein processing and subgenomic RNA synthesis (Heise et al., 2003). Given that the presence of the attenuating Ile mutation led to earlier induction of viral 26S RNA synthesis, which could alter type I IFN induction, we assessed whether the attenuated mutant virus exhibited any differences in type I IFN induction from that of the wild type virus both in vivo and in vitro. To evaluate whether the viruses exhibited differential IFN induction in vivo, six-week old CD-1 mice were mock infected or infected with the wild type AR86 (previously referenced as s300 (Suthar et al., 2005)) or mutant T538I (nsP1 538 Ile previously referenced as s340) viruses by the intracranial route (i.c.) and bled between 9 and 18 hours post infection. Serum type I interferon responses were measured using an interferon bioassay on L929 cells. As shown in figure 1A, serum from mice infected with the mutant T538I virus exhibited robust type I interferon levels compared to mice infected with the wild type AR86 virus at both 12 and 18 hours post infection as detectable by bioassay. Similar results were observed in C57BL/6 mice (data not shown). Importantly, wild type AR86 did not induce detectable type I interferon up to 48 hours post-infection (data not shown), indicating that the wild type virus was not simply delayed in the induction of type I interferon. To further assess whether the mutant virus was a more potent inducer of type I IFN, L929 cells were infected with either the wild type AR86 virus or the T538I mutant and type I IFN levels in the supernatant were evaluated. As shown in figure 1B, similar to the in vivo results, the attenuated mutant was a more potent inducer of type I IFN than the wild type virus. These results were not restricted to L929 cells, since the mutant virus was also a more potent IFN inducer in human cell lines such as A549 cells (data not shown). Overall, these results suggest that the determinant at nsP1 position 538 plays a major role in regulating type I IFN induction by the virus.
Figure 1. The mutant SIN T538I virus induces more type I interferon.
A) Groups of six-week old (n=3) CD-1 mice were infected with diluent alone, wild type Sindbis AR86, or Sindbis T538I mutant at 1×103 pfu via the intracranial (i.c.) route. Serum was harvested at the indicated time points (hours) and diluted (1:10) into media. B) L929 cells were either mock infected or infected with wild type AR86 or T538I mutant at an MOI of 5.0 and supernatants were harvested at 18 hours post-infection. Serum and supernatants were subjected to an interferon bioassay on L929 cells. Each bar represents the average of triplicate samples and the p values were determined by ANOVA statistical analysis. Error bars represent the standard error of the mean. The limit of detection in each bioassay was 31IU/ml.
Previous studies with Sindbis viruses have suggested that virus-mediated host cellular macromolecular shutoff is linked to IFN induction. Some Sindbis virus mutants that are potent IFN inducers are also defective in their ability to shutoff host RNA synthesis (Gorchakov, Frolova, and Frolov, 2005), raising the possibility that the nsP1 mutation in AR86 affected type I IFN induction through effects on host macromolecular synthesis. Therefore, we assessed the kinetics of transcriptional and translational shutoff between the wild type and mutant AR86 viruses. These studies utilized L929 cells, since the T538I mutant exhibited enhanced type I IFN induction in this cell type (figure 1B). Unfortunately, we were unable to achieve 100% infection at low MOIs. Therefore, the MOI was increased to 50 to increase the percent of infected cells and to limit the confounding effect of nonproductively infected cells in the culture continuing transcription/translation and complicating the analysis of virus-induced shutoff of these processes. As shown in figure 2A, both the wild type and the mutant viruses exhibited similar kinetics of host transcription shutoff at 5 hours post-infection as observed by the loss of host mRNA, 28S rRNA, and 18S rRNA. By 8hpi, both viruses have efficiently turned off host transcription. Interestingly, in Neuro2A cells, the wild type Sindbis AR86 virus exhibited less efficient shutoff of host mRNA, 28S rRNA and 18S rRNA as compared to the mutant T538I virus suggesting that the T538I mutant may actually shutoff transcription more efficiently than the wild type virus (data not shown). Additionally, the wild type and mutant Sindbis viruses were equally efficient at inhibiting host protein translation (figures 2B and 2C). As early as 4 hours post-infection, both viruses exhibit greater than 25% shutoff of host protein synthesis and by 8 hours post-infection greater than 75% of host protein synthesis shutoff was observed (figure 2C). Most importantly, when comparing the β-actin band, the mutant T538I virus displayed similar kinetics of host protein shutoff as compared to wild type AR86 virus (figure 2B). Similar observations were also made in Neuro2A and BHK-21 cells (data not shown). These results suggest that the Sindbis virus mutant induced enhanced type I IFN production in the absence of a detectable defect in generalized host cell shutoff.
Figure 2. The Sindbis T538I mutant virus shuts off host RNA transcription and protein translation with similar kinetics to the wild type AR86.
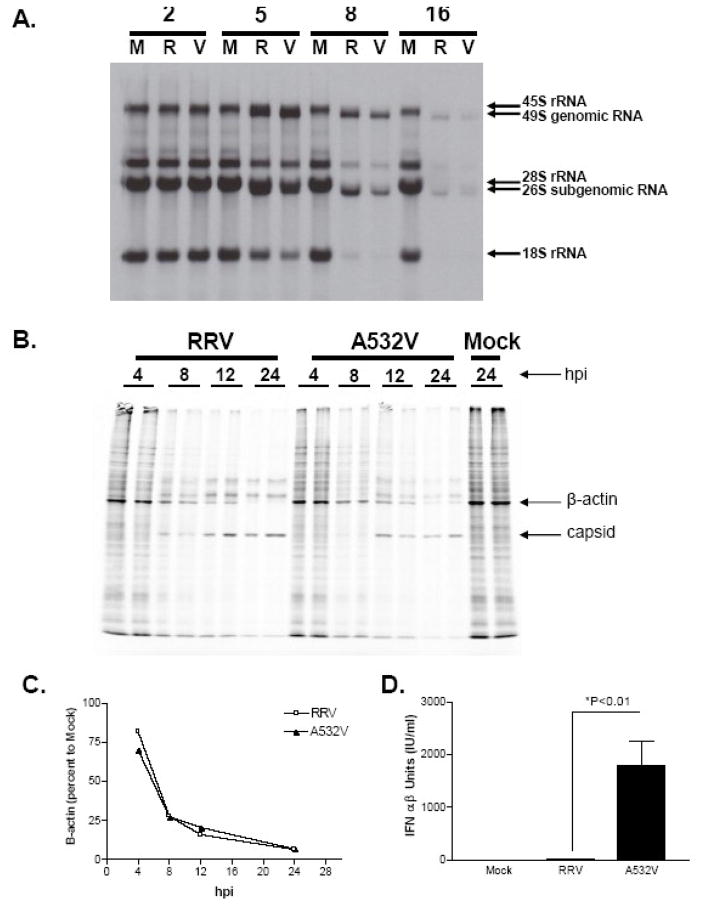
L929 cells were mock infected (M) or infected with wild type Sindbis AR86 (T) or mutant T538I (I) at an MOI of 50. A) To analyze host RNA synthesis, at various times post-infection (2, 5, 8, and 16 hours), the media was replaced with media containing 20μCi/ml of 3H-Uridine and cells were labeled for a total of 3 hours. Total RNA was harvested and analyzed by agarose gel electrophoresis as previously described. B) To analyze host protein synthesis, L929 cells were labeled with 35S Met/Cys for 1 hour at the hours indicated and cell lysates (in duplicates) were analyzed by SDS-PAGE. C) Residual host cell protein synthesis in figure 2B was evaluated by measuring the amount of radioactivity detected in the protein band corresponding to actin (as marked by the arrow) and normalized to the amount of radioactivity detected in the same protein band in mock infected cells (AR86-□, T538I-▲). The data shown are representative of two independent experiments.
Mutation of the nsP1 determinant in Ross River Virus results in enhanced type I IFN induction by the mutant virus
To determine if the effect of the amino acid within the nsP1/nsP2 cleavage domain on type I IFN induction was specific to AR86, similar mutations were introduced in the distantly related alphavirus, Ross River Virus (RRV) (figure 3A). RRV encodes an Alanine codon at the same position within the nsP1/2 cleavage site and we examined whether substitution with an Isoleucine (A532I) or Valine (A532V) would result in a viable virus with similar effects on type I IFN induction as seen with the Sindbis T538I mutant (figure 1). Substitution of an Isoleucine in RRV resulted in a significantly attenuated virus that grew poorly in BHK-21 cells (data not shown). In contrast, substitution of an Alanine to Valine resulted in a viable virus that grew to levels comparable to wild type RRV, though the Valine mutant displayed a smaller plaque phenotype on BHK-21 cells (data not shown). Furthermore, the RRV A532V mutant had a similar RNA specific infectivity and particle to PFU ratio to those observed with the wild type virus (data not shown). In both single (figure 3B) and multistep (figure 3C) growth curves in BHK-21 cells, the A532V mutant exhibited a significant and reproducible reduction in viral yield at intermediate times (7–16hpi), although the endpoint yield was identical to wild type RRV. Therefore, the A532V mutant is viable, though it may exhibit a slight replication defect as compared to the wild type virus.
Figure 3. Characterizations of the RRV A532V mutant virus.
A) Schematic diagram of the single amino acid substitutions in the Sindbis AR86 (T538I) and Ross River Virus (A532I and A532V) mutants. B) Single step growth curve. BHK cells were infected at an MOI of 5 with either RRV (■) or A532V (△) viruses. Supernatants at 1, 4, 7, 10, 13, and 25hpi were analyzed by plaque assay. C) Multi-step growth curve. BHK-21 cells were infected at an MOI of 0.01 with either RRV (■) or A532V (△) viruses. Supernatants at 4, 7, 10, 13, 16, and 25hpi were analyzed by plaque assays. Each data point represent the average of triplicate samples and significance was determined by a 2-factor ANOVA statistical analysis (*p<0.05, **p<0.01). Error bars represent the standard error of the mean.
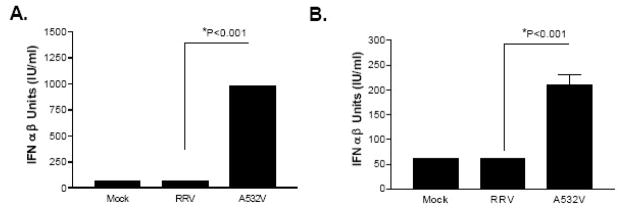
In order to assess the impact of the A532V mutation on type I IFN induction, L929 cells were infected with either the wild type RRV or the RRV A532V mutant, and type I IFN induction in the supernatant was measured by bioassay. As shown in figure 4A, the A532V mutant exhibited enhanced IFN induction (~90 fold) in comparison to wild type RRV at 24 hours post-infection. Similar results were found by qRT-PCR analysis for IFN-beta mRNA transcripts at early times post-infection (figure 4B and 4C). Altogether, these data, along with the earlier findings with the Sindbis AR86 viruses, highlight the importance of the determinant within the nsP1/nsP2 cleavage site in modulating type I IFN responses by at least two alphaviruses.
Figure 4. The RRV A532V mutant virus induces more type I IFN than the wild type.
L929 cells were infected with RRV and A532V viruses at an MOI of 5. A) Type I IFN bioassays were performed on the supernatants harvested at 24 hpi. The limit of detection for the bioassay is 7 IU/ml. B) and C) Total RNA was extracted at 6 hpi (B) or 12 hpi (C) and analyzed by quantitative real time PCR (Applied Biosystems) for IFN-beta message transcripts. The data are represented as the fold induction over Mock infected cells and have been normalized to 18S rRNA. Each bar above represents the average of triplicate samples and the p values were determined by ANOVA statistical analysis. Error bars represent the standard error of the mean.
The A532V mutation affects type I IFN induction
Recent work from our group and others has demonstrated that another alphavirus, Venezuelan equine encephalitis virus (VEEV) can antagonize STAT1 activation following treatment with type I IFN (Simmons et al., 2009; Yin et al., 2009). Although, to date we have found no evidence that wild type RRV can antangonize STAT1 activation (Simmons and Heise, unpublished), we were interested in determining whether the A532V mutation affected the early induction of type I IFN or the amplification phase of the type I IFN response, which is dependent upon type I IFN signaling and STAT1 activation. Thus, we compared the wild type and mutant A532V RRV viruses for their ability to induce type I IFN in primary MEFs derived from wild type mice or mice deficient in the type I IFN αβ receptor (IFNR), which are therefore unable to mount an amplified type I IFN response. As shown in figure 5A, similar to the results from the L929 cells, the mutant virus was a more potent type I IFN inducer than wild type RRV in wild type MEFs. In IFNR deficient MEFs, the overall type I IFN induction was reduced compared to wild type cells, however the mutant virus still induced significantly more type I IFN than the wild type virus (figure 5B). Therefore, though we cannot rule out an effect of the A532V mutation on type I IFN signaling and subsequent amplification of the IFN response, these results strongly suggest that the nsP1 mutation does have an effect on the inductive phase of the type I IFN response.
Figure 5. The RRV mutant virus induces more IFN than the wildtype RRV in the absence of the Type I IFN αβ Receptor.
A) Sv/129 MEFS and B) Sv/129 IFNR−/− MEFS were infected with RRV and A532V viruses at an MOI of 5. Type I IFN bioassays were performed on the supernatants harvested at 24 hpi. Each bar represents the average of triplicate samples and the p values were determined by ANOVA statistical analysis. Error bars represent the standard error of the mean and the limit of detection for each bioassay is 61 IU/ml).
The RRV A532V exhibits a mild defect in virus-induced shutoff of host RNA and protein synthesis
The Sindbis T538I mutant exhibited little defect in virus-induced shutoff, therefore we determined whether this was also the case with the RRV A532V mutant. RRV or A532V were evaluated for their ability to shut off host protein and RNA synthesis in a manner similar to that described above for AR86. L929 cells were infected with the RRV viruses at an MOI of 50 to enhance the percentage of infected cells in the culture. At this MOI, the percentage of infected cells was greater than 80% for both viruses as demonstrated by an Immunofluorescent Assay (IFA) (data not shown). Importantly, the RRV A532V mutant still induced an enhanced IFN response compared to the wild type virus at this MOI (Figure 6D). Both wild type RRV and the mutant A532V viruses inhibited cellular RNA transcription as early as 5 hours post-infection as indicated by the decline in 18S rRNAs as compared to mock infected cells (figure 6A). By 8 hours post-infection, both viruses efficiently turned off 18S rRNA synthesis. Similar results were also obtained in BHK cells though interestingly, in BHK cells, the A532V mutant displayed a slight delay in transcriptional shutoff at early times post-infection compared to the wild type RRV virus (data not shown). Therefore, though the A532V mutant is capable of shutting off host RNA synthesis we cannot rule out the possibility that the A532V mutant virus may be slightly delayed in its ability to turn off cellular RNA synthesis. Analysis of virus induced inhibition of host protein synthesis (figure 6B) demonstrated that both viruses inhibit cellular protein synthesis in L929 cells with similar kinetics as indicated by the loss of the β-actin band (figure 6C). Similarly to the transcriptional shutoff data, in BHK cells, the A532V mutant was slightly delayed in inhibiting protein translation in comparison to the wild type virus (data not shown). Therefore, though the mutant RRV is ultimately able to shut off host cell RNA and protein synthesis, based on these results, we cannot rule out the possibility that a slight defect in shutoff might contribute to the enhanced type I IFN induction by the A532V RRV mutant.
Figure 6. Characterization of virus-induced shutoff by the RRV A532V mutant.
L929 cells were either mock infected (M) or infected at an MOI of 50 with RRV (R) or A532V (V). A) At various times post-infection (2, 5, 8, and 16 hrs), cells were labeled with 20μCi/ml of 3H-Uridine (as previously described) for analysis of host RNA synthesis. B) L929 cells were labeled with 35S Met/Cys at the indicated times post-infection for 1 hour to monitor host protein synthesis. Total cell lysates (duplicates) were analyzed by SDS-PAGE as described in the materials and methods. C) The kinetics of cellular β-actin protein synthesis was evaluated by measuring the amount of radioactivity detected by densitometry in the β-actin protein band in the infected cells and normalized to the mock infected cells at 24hpi. D) L929 cells were either mock infected or infected with RRV or A532V mutant viruses at an MOI (50). Supernatants at 24hpi were analyzed for IFN by bioassay as previously described. The limit of detection in this assay is 2 IU/ml and each bar represents the average of triplicate samples. The p-values were determined by ANOVA statistical analysis and error bars represent the standard error of the mean
The A532V mutant virus is a strong inducer of IRF-3 phosphorylation than the wild type virus
The results with the Sindbis T538I mutant suggest that host shutoff defects do not contribute to the enhanced type I IFN induction by the mutant virus. Furthermore, though we could not detect a defect in the RRV A532V mutant’s ability to shutoff host transcription or translation in L929 cells, the delayed kinetics of shutoff by the A532V mutant in BHK cells left open the possibility of shutoff dependent effects for the RRV mutant. Therefore, to more clearly address this issue, we assessed the RRV viruses for their ability to activate IRF-3, a step that is independent of de novo host RNA and protein synthesis and therefore not susceptible to shutoff mediated effects (Preston, Harman, and Nicholl, 2001). IRF-3 is an essential transcription factor that is immediately activated by several pathogen-associated molecular pattern receptors to induce early transcription of IFN-β and IFN-α genes. Activation is initiated upon IRF-3 phosphorylation resulting in protein dimerization and nuclear translocation. Therefore, we assessed whether the mutant RRV virus exhibited differential IRF-3 activation compared to the wild type RRV in L929 cells, which exhibited high levels of type I IFN induction by the RRV A532V mutant (figure 4A). L929 cells were transfected with poly IC as a positive control (Hato et al., 2007), which resulted in robust IRF-3 phosphorylation as indicated by the shift in the IRF-3 protein band in comparison to the mock treated cells (Figure 7A). Additionally, phosphatase treatment of cell lysates confirmed that the upper band was indeed a phosphorylated protein (data not shown). Analysis of the virally infected cells demonstrated that the RRV A532V mutant virus induced IRF-3 phosphorylation at 10 hours post-infection; however, the wild type RRV virus did not display IRF-3 phosphorylation until as late as 12 hours post-infection. Also, there was substantially more IRF-3 phosphorylation at 12 hours post-infection in cells infected with the A532V mutant virus (figure 7B), consistent with our previous data that the RRV A532V mutant induces more IFN than wild type RRV. Similar to the results with RRV, the AR86 T538I mutant induced faster and more robust IRF-3 activation than the wild type virus in HEC-1B cells, human endometrial carcinoma cells, that have been well characterized in IRF-3 activation studies and do not respond to IFN treatment (Chen et al., 1981; Talon et al., 2000)(data not shown). Altogether, these results suggest that both the AR86 and RRV mutants affect IFN induction through a mechanism that is independent of virus-induced host shutoff. However, given that the RRV A532V mutant did exhibit delayed shutoff kinetics in BHK cells, we cannot rule out a synergistic role for shutoff dependent and independent effects in enhancing type I IFN induction by the RRV A532V mutant.
Figure 7. The RRV A532V mutant robustly induces IRF-3 phosphorylation compared to wild type virus.
A) L929 cells were either mock infected, infected with RRV and A532V viruses at an MOI of 5, or transfected with 1ug of poly I:C with Lipofectamine 2000 (Invitrogen). Cells were lysed at the indicated hours in NP40 lysis buffers containing protease and phosphatase inhibitors. 20ug of total protein was analyzed by SDS-PAGE and probed with an anti-IRF-3 antibody (Santa Cruz, C-20) for phosphorylated murine IRF-3 (p-IRF-3). The membranes were then re-probed with an anti-β-actin antibody (Sigma). B) The phosphorylated IRF-3 bands in (A) were quantified using ImageQuant 5.0 and represented as fold over mock. The data shown are representative of three independent experiments.
Discussion
Type I interferon is an essential component of the host response to viral infection, because it directly activates antiviral systems and modulates antiviral activities of other components of the host innate and adaptive immune systems. However, a number of viruses have evolved mechanisms to antagonize or evade type I IFN induction. These mechanisms range from nonspecific effects, such as rapid shutoff of host cell macromolecular synthesis (Ahmed et al., 2003; Gorchakov, Frolova, and Frolov, 2005), masking viral RNA from host cell sensory proteins (Cardenas et al., 2006; Mibayashi et al., 2007), to specific inhibition of host cell dsRNA sensors or signaling molecules that link these sensor molecules to transcription factors that regulate type I interferon transcription (Andrejeva et al., 2004; Basler et al., 2003; Li et al., 2005; Loo et al., 2006). Studies with Sindbis virus suggest that host cell shutoff plays a major role in regulating viral type I interferon induction (Gorchakov, Frolova, and Frolov, 2005), though studies with Semliki Forest virus (SFV) also indicate that alphaviruses can modulate type I IFN responses independently of host shutoff (Breakwell et al., 2007). In this report, we present evidence that an attenuating mutation at nsP1 position 538 (T538I) in the neurovirulent strain of Sindbis virus, AR86, modulates type I interferon induction without affecting this virus’s ability to shut off host cell macromolecular synthesis. Additionally, this determinant has been shown to play a key role in regulating viral neurovirulence (Heise, Simpson, and Johnston, 2000), and it is likely that the enhanced type I IFN induction by the nsP1 538 mutant virus in vivo (figure 1A) is at least partially responsible for its attenuating effect on AR86. Furthermore, the importance of this determinant in regulating type I IFN induction by alphaviruses is underscored by our finding that an analogous mutation in another alphavirus, Ross River Virus, exerts a similar effect on type I IFN induction (figure 4 and 5).
Using a genetically related Sindbis virus, Gorchakov et al. demonstrated that viruses with mutations resulting in defects in host translation or transcription shutoff induced more type I interferon (Gorchakov, Frolova, and Frolov, 2005). However, we found either no difference or a very mild defect in host shutoff with the mutant Sindbis and RRV viruses, as well as differences in the kinetics of virus induced IRF-3 activation, which is independent of host shutoff. This finding strongly suggests that these viruses can affect type I IFN induction independently of effects on host shutoff. It is also important to note that previous work with an SFV nsP mutant, which also demonstrated a role for the nonstructural proteins in modulating type I IFN induction independently of host shutoff effects, did not observe a differential effect on IRF-3 activation (Breakwell et al., 2007), which raises the possibility that the determinants in our AR86 and RRV mutants may affect interferon induction through different mechanisms than those observed with SFV. Furthermore, previous work from our lab and others have demonstrated a role for VEE to inhibit STAT-1 activation (Simmons et al., 2009; Yin et al., 2009), a necessary step in the IFN signaling pathway to amplify the type I IFN response indicating that these viruses may employ additional mechanisms to antagonize this pathway. However, our analysis in IFN αβ Receptor deficient MEFS, coupled with preliminary data that wild type RRV does not inhibit STAT-1 activation, further suggests that our determinant is affecting the initial IFN induction step.
The nsP1 virulence determinant described here might affect type I IFN induction through several different mechanisms. First, the nsP1 mutant may induce more type I IFN by producing earlier and/or increased quantities of a viral ligand, for efficient recognition by the host sensors. Previous studies examining very early times post-infection demonstrated that the AR86 mutant virus initiated 26S RNA synthesis more quickly than the wild type virus, though both viruses exhibited equivalent levels of 26S RNA synthesis by 4–6 hours post-infection (Heise et al., 2003). Although, we did not observe this kinetic difference in our L929 RNA labeling assays, this likely reflects the time-points analyzed in the current studies. Our earlier results raise the possibility that early or more abundant 26S RNA synthesis by the mutant virus might be linked to the difference in type I IFN induction. Preliminary analysis indicates that the RRV mutant may also produce more viral RNA than the wild type virus (data not shown). In this model, the wild type virus could delay 26S promoter induction until later times when virus-induced host shutoff efficiently antagonizes type I interferon induction. The attenuating mutation at nsP1, however, up-regulates 26S RNA synthesis prior to the time of effective virus-induced shutoff (Heise et al., 2003). This model suggests that the subgenomic RNA may be a potential target for host cell sensors, however; this does not rule out a role for multiple viral RNA species, such as the full length positive or negative sense RNA, or dsRNA complexes to activate IFN induction. Further investigations will be required to address the exact viral ligand required as well as the pathway that is mediating this response and whether this mechanism holds true for both viruses. Second, the nsP1 mutant may induce more type I IFN by altering the structure of one or more viral RNAs and thereby producing a stronger ligand. Gitlin et al. has reported that RIG-I plays an important role in responding to Sindbis virus infection (Gitlin et al., 2006), while PKR has also been shown to contribute to type I IFN induction during flavivirus infection (Gilfoy and Mason, 2007). Both RIG-I and PKR interact with 5-′triphosphates on uncapped RNAs (Hornung et al., 2006; Nallagatla et al., 2007; Pichlmair et al., 2006), though both proteins can also recognize other RNA ligands (Nallagatla and Bevilacqua, 2008; Saito and Gale, 2008). Both nsP1 and nsP2 proteins are involved in capping viral RNAs and though the C-terminus of nsP1 has not been implicated in capping activity, it is possible that the determinant at nsP1 is altering the efficiency of capping viral genomic or subgenomic RNAs (Ahola and Kaariainen, 1995; Ahola et al., 1997; Hornung et al., 2006; Mi et al., 1989; Rikkonen, Peranen, and Kaariainen, 1994; Vasiljeva et al., 2000). In this model, the nsP1 mutants are providing the host sensor, RIG-I, with viral ligands that contain free 5′-triphosphates that lead to stronger IFN-β induction. Finally, though there is no direct evidence to suggest that the wild type and T538I mutant Sindbis AR86 viruses or RRV and the A532V RRV mutant differ in their ability to actively suppress type I IFN induction, it is possible that the mutation is modulating type I IFN induction through different mechanisms in the context of each virus. Regardless of the ultimate mechanism responsible for the differential type I IFN induction phenotype, these results further support the idea that determinants in the viral nonstructural region of alphaviruses play a major role in regulating type I IFN induction, with a concomitant impact on viral virulence.
In summary, we have demonstrated that wild type AR86 virus was a poor inducer of type I interferon, while the mutant T538I virus containing an attenuating Ile codon at nsP1 position 538 induced a robust type I interferon response in vitro and in vivo. An analogous determinant exhibited a similar effect in another, distantly related alphavirus (RRV), suggesting that this determinant plays an important role in regulating type I IFN induction by multiple alphaviruses. The altered interferon induction was independent of virus-induced host translation or transcription shutoff, suggesting that the determinant within the nsP1/nsP2 cleavage site acts through a more specific mechanism to modulate the host type I interferon response.
Materials and Methods
Viruses and Cell Culture
The AR86 molecular clones pS300 (wild type nsP1 538 Thr) and pS340 (mutant nsP1 538 Ile) were described previously (Heise, Simpson, and Johnston, 2000). The wild type Ross River Virus molecular clone, pRR64 (Kuhn et al., 1991), was generously provided by Dr. Richard Kuhn (Purdue University). The Ross River Virus mutant, pRR64-A532V, was generated by introducing a single nucleotide change (thymine to guanine) at nucleotide position 1670, by PCR mutagenesis resulting in a single codon change from an Alanine (wild type) to Valine (A532V) at nsP1 position 532.
Viral stocks were generated by in vitro transcription as previously described (Heise et al., 2003; Morrison et al., 2006). Briefly, cDNA plasmids were linearized and used as templates for full length RNA transcripts generated by SP6-specific mMessage mMachine in vitro transcription kits (Ambion). Transcripts were electroporated into BHK-21 cells using a Bio-Rad electroporator. Supernatants were harvested 24 hours later, centrifuged for 20 min at 3,000 RPM, and frozen in 0.5 ml aliquots. Alternatively, electroporated supernatants were pelleted through a 20% (w/v) sucrose/phosphate buffered saline (PBS) cushion at 72,000 × g by ultracentrifugation (four hours) to concentrate viral stocks. The pelleted viruses were resuspended in PBS and 0.1ml aliquots were frozen at −80°C. Viral titers were determined by standard plaque assays on BHK-21 cells.
BHK-21 cells were maintained in alpha minimum essential medium (Gibco) supplemented with 10% bovine calf serum (BioWhittaker), 10% tryptose phosphate broth, and 0.29 mg/ml of L-glutamine. L929 mouse fibroblast cells were maintained in alpha minimum essential medium (Gibco) supplemented with 10% bovine calf serum, 10% tryptose phosphate broth, and 0.29 mg/ml of L-glutamine. Neuro2A mouse neuroblastoma cells (N2A) were grown in MEM containing nonessential amino acids and 10% fetal bovine serum (HyClone). HEC-1B cells (ATCC) were grown in MEM containing 10% fetal bovine serum, 0.29 mg of L-glutamine per ml, and penicillin/streptomycin. Primary Mouse Embryonic Fibroblast (MEFS) cells were generated from 13–15 day old Sv/129 wild type or IFN αβ receptor deficient (IFNR−/−) embryos and maintained in DMEM/F12 media containing 10% Fetal Bovine Serum, 10% tryptose phosphate broth, 0.29 mg/ml of L-glutamine, and 50ug/ml of gentamicin (Gibco).
Animal studies
Specific pathogen-free six-week old female CD-1 mice were obtained from Charles River Breeding Laboratories (Raleigh, NC), while C57Bl/6J mice were bred in house. Animal housing and care were in accordance with all University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee guidelines. In all studies, six- to eight-week-old mice (groups of 3 to 6 animals/study) were anesthetized with Ketamine supplemented with Xylazine (Barber Med.) prior to intracranial (i.c.) inoculation with a standard dose of 103 PFU of virus in diluent [phosphate-buffered saline (PBS, pH 7.4)], supplemented with 1% donor calf serum (DCS, Gibco)). Mock infected mice received diluent alone. At the indicated timepoints, mice were bled from the tail vein and sera were frozen at −80°C until analyzed for type I interferon by bioassay (see below).
Type I IFN bioassays
Total amounts of type I IFN were measured by using an interferon bioassay as previously described (Shabman et al., 2007). Briefly, L929 cells were seeded in 96 well plates. Samples, including the standards, were acidified to a pH ≤ 2.0 for 24 hours, then neutralized to pH of 7.4, UV treated for 10 minutes to inactivate residual virus, and added to cells in a titration of serial two-fold dilutions. After overnight incubation, 50ul of a 4×106 PFU/ml stock of encephalomyocarditis virus (EMCV) was added to each well. Twenty-four hours post-infection, cell viability was determined using 3-(4, 5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma) assay, and the absorbance was read on a microplate reader at 570nm. The IFN in each sample was compared to an IFN standard (from Chemicon or R&D Systems that has been normalized to the National Institutes of Health reference IFN) present in each plate and converted to international units (IU/ml).
Quantitative real-time PCR analysis
L929 cells were seeded in 6-well dishes and either mock infected or infected with RRV wild type and A532V RRV mutant viruses at an MOI of 5. At the indicated time points, total RNA was isolated using the Trizol (Invitrogen) and the PureLink RNA Mini Kit protocol (Invitrogen). Total RNA was reverse transcribed using random primers and SuperScript III Reverse Transcriptase Kit (Invitrogen). cDNAs were then probed for IFN-beta and 18S rRNA message using TaqMan real-time PCR with primer probe sets (Applied Biosystems) and analyzed on the Prism 7000 machine (Applied Biosystems).
Analysis of Protein Synthesis
L929, BHK-21, or Neuro2A cells were either mock infected or infected with the Sindbis AR86 or RRV viruses at an MOI of 50 and 10 (BHK and N2A). At various times post-infection, the media was removed and cells were starved with Minimum Essential Eagle Medium with Earle’s salts lacking methionine and cysteine (MP biomedicals) for 1 hour at 37°C. The starvation media was then replaced with media supplemented with 33μCi of 35S-methionine and 35S-cysteine (Amersham Pro-mix) and incubated at 37°C for one hour. Following each labeling period, cells were lysed in NP-40 Lysis buffer containing protease inhibitors (Roche). Equal cell lysates were analyzed on a 10% sodium dodecyl sulphate polyacrylamide gel (SDS-PAGE). Gels were fixed in buffer containing 10% acetic acid and 40% methanol. The gels were then dried, exposed to a phosphoimaging screen, and scanned using a Storm Phosphoimager (GE Healthcare). β-actin bands were quantified using ImageQuant software (GE Healthcare), and samples are represented as comparison to mock infected cells.
Analysis of Host RNA Synthesis
L929, BHK, or Neuro2A cells were either mock infected or infected with the Sindbis AR86 or the RRV viruses at an MOI of 50 and 10 (BHK and N2A). After 1 hour incubation, 2ml of media were added back to cells. At various times post-infection (2, 5, 8, and 16hpi), the media were replaced with media supplemented with 20uCi/ml [3H]-uridine (in the presence or absence of actinomycin D (1ug/ml)). The cells were labeled for a total of 3 hours, then washed with 1XPBS and lysed in Trizol (Invitrogen). RNA was extracted and equal volumes were denatured in glyoxal and dimethyl sulfoxide (DMSO) for 1 hour at 50°C. The RNA was analyzed on a 0.8% agarose NaPO4 gel. The agarose gel was washed twice in methanol followed by overnight incubation in 2.5% 2, 5-diphenyloxazole (PPO) in methanol. The gel was washed three times in water to precipitate the PPO, dried, and exposed to film.
SDS-PAGE and Western Blot Analysis
Protein extracts were resolved on 10% SDS-PAGE followed by transfer to a PVDF membrane. Anti-IRF-3 C-20 antibody (Santa Cruz) was used for detection of phosphorylated IRF-3 in L929 cells. For re-probing of the membranes, anti-β-actin (Sigma) antibodies were used. Membranes were washed and incubated in the appropriate anti-rabbit and anti-goat secondary antibodies.
Acknowledgments
This research was supported by NIH research grants R01AI 067641 (M.T.H.) and 1F31AI077324 (C.C.C.). We thank the members of the Carolina Vaccine Institute (CVI) and the Johnston and White laboratories for helpful scientific discussions. We thank Bianca Trollinger for providing excellent technical support with cell culture and Dr. Martin Ferris for helpful statistical analysis. We would also like to thank Dr. Charles Rice, Dr. Joe Marcotrigiano, Dr. Nancy Davis, and Eileen Hoyt for excellent scientific discussion and technical help with the host transcription experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed M, McKenzie MO, Puckett S, Hojnacki M, Poliquin L, Lyles DS. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J Virol. 2003;77(8):4646–57. doi: 10.1128/JVI.77.8.4646-4657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T, Kaariainen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci U S A. 1995;92(2):507–11. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T, Laakkonen P, Vihinen H, Kaariainen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71(1):392–7. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A. 2004;101(49):17264–9. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Muhlberger E, Bray M, Klenk HD, Palese P, Garcia-Sastre A. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol. 2003;77(14):7945–56. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakwell L, Dosenovic P, Karlsson Hedestam GB, D’Amato M, Liljestrom P, Fazakerley J, McInerney GM. Semliki Forest Virus Nonstructural Protein 2 Is Involved in Suppression of the Type I Interferon Response. J Virol. 2007;81 (16):8677–8684. doi: 10.1128/JVI.02411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80(11):5168–78. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Sato T, Fuse A, Kuwata T, Content J. Resistance to interferon of a human adenocarcinoma cell line, HEC-1, and its sensitivity to natural killer cell action. J Gen Virol. 1981;52(Pt 1):177–81. doi: 10.1099/0022-1317-52-1-177. [DOI] [PubMed] [Google Scholar]
- Frolova EI, Fayzulin RZ, Cook SH, Griffin DE, Rice CM, Frolov I. Roles of Nonstructural Protein nsP2 and Alpha/Beta Interferons in Determining the Outcome of Sindbis Virus Infection. J Virol. 2002;76(22):11254–11264. doi: 10.1128/JVI.76.22.11254-11264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmashova N, Gorchakov R, Frolova E, Frolov I. Sindbis Virus Nonstructural Protein nsP2 Is Cytotoxic and Inhibits Cellular Transcription. J Virol. 2006;80(12):5686–5696. doi: 10.1128/JVI.02739-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. The Old World and New World Alphaviruses Use Different Virus-Specific Proteins for Induction of Transcriptional Shutoff. J Virol. 2007;81(5):2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfoy FD, Mason PW. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81 (20):11148–58. doi: 10.1128/JVI.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103(22):8459–64. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, Frolov I. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol. 2005;79(15):9397–409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, Sawicki S, Atasheva S, Sawicki D, Frolov I. A New Role for ns Polyprotein Cleavage in Sindbis Virus Replication. J Virol. 2008;82 (13):6218–6231. doi: 10.1128/JVI.02624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hato SV, Ricour C, Schulte BM, Lanke KH, de Bruijni M, Zoll J, Melchers WJ, Michiels T, van Kuppeveld FJ. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell Microbiol. 2007;9(12):2921–30. doi: 10.1111/j.1462-5822.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- Heise MT, Simpson DA, Johnston RE. A single amino acid change in nsP1 attenuates neurovirulence of the Sindbis-group alphavirus S.A.AR86. J Virol. 2000;74(9):4207–13. doi: 10.1128/jvi.74.9.4207-4213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise MT, White LJ, Simpson DA, Leonard C, Bernard KA, Meeker RB, Johnston RE. An attenuating mutation in nsP1 of the Sindbis-group virus S.A.AR86 accelerates nonstructural protein processing and up-regulates viral 26S RNA synthesis. J Virol. 2003;77(2):1149–56. doi: 10.1128/JVI.77.2.1149-1156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Kuhn RJ, Niesters HG, Hong Z, Strauss JH. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991;182(2):430–41. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- Lemm JA, Rice CM. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J Virol. 1993;67 (4):1916–1926. doi: 10.1128/jvi.67.4.1916-1926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102(8):2992–7. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103(15):6001–6. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Durbin R, Huang HV, Rice CM, Stollar V. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology. 1989;170(2):385–91. doi: 10.1016/0042-6822(89)90429-7. [DOI] [PubMed] [Google Scholar]
- Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr, Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81(2):514–24. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SA, Berglund P, Beard CW, Johnston RE. Ribosomal Protein S6 Associates with Alphavirus Nonstructural Protein 2 and Mediates Expression from Alphavirus Messages. J Virol. 2006;80(15):7729–7739. doi: 10.1128/JVI.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TE, Simmons JD, Heise MT. Complement receptor 3 promotes severe ross river virus-induced disease. J Virol. 2008;82(22):11263–72. doi: 10.1128/JVI.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TE, Whitmore AC, Shabman RS, Lidbury BA, Mahalingam S, Heise MT. Characterization of Ross River virus tropism and virus-induced inflammation in a mouse model of viral arthritis and myositis. J Virol. 2006;80 (2):737–49. doi: 10.1128/JVI.80.2.737-749.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14 (6):1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-Triphosphate-Dependent Activation of PKR by RNAs with Short Stem-Loops. Science. 2007;318(5855):1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Preston CM, Harman AN, Nicholl MJ. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J Virol. 2001;75(19):8909–16. doi: 10.1128/JVI.75.19.8909-8916.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikkonen M, Peranen J, Kaariainen L. Nuclear targeting of Semliki Forest virus nsP2. Arch Virol Suppl. 1994;9:369–77. doi: 10.1007/978-3-7091-9326-6_37. [DOI] [PubMed] [Google Scholar]
- Ryman KD, Klimstra WB, Nguyen KB, Biron CA, Johnston RE. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J Virol. 2000;74(7):3366–78. doi: 10.1128/jvi.74.7.3366-3378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman KD, White LJ, Johnston RE, Klimstra WB. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 2002;15(1):53–76. doi: 10.1089/088282402317340233. [DOI] [PubMed] [Google Scholar]
- Saito T, Gale M., Jr Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J Exp Med. 2008;205(7):1523–1527. doi: 10.1084/jem.20081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabman RS, Morrison TE, Moore C, White L, Suthar MS, Hueston L, Rulli N, Lidbury B, Ting JP, Mahalingam S, Heise MT. Differential induction of type I interferon responses in myeloid dendritic cells by mosquito and mammalian-cell-derived alphaviruses. J Virol. 2007;81(1):237–47. doi: 10.1128/JVI.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirako Y, Strauss JH. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68(3):1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons JD, White LJ, Morrison TE, Montgomery SA, Whitmore AC, Johnston RE, Heise MT. Venezuelan equine encephalitis virus disrupts STAT1 signaling by distinct mechanisms independent of host shutoff. J Virol. 2009 doi: 10.1128/JVI.01041-09. JVI.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson DA, Davis NL, Lin SC, Russell D, Johnston RE. Complete Nucleotide Sequence and Full-Length cDNA Clone of S.A.AR86, a South African Alphavirus Related to Sindbis. Virology. 1996;222(2):464–469. doi: 10.1006/viro.1996.0445. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar MS, Shabman R, Madric K, Lambeth C, Heise MT. Identification of adult mouse neurovirulence determinants of the Sindbis virus strain AR86. J Virol. 2005;79(7):4219–28. doi: 10.1128/JVI.79.7.4219-4228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74(17):7989–96. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PC, Strauss EG, Kuhn RJ, Strauss JH, Griffin DE. Viral determinants of age-dependent virulence of Sindbis virus for mice. J Virol. 1993;67(8):4605–4610. doi: 10.1128/jvi.67.8.4605-4610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Merits A, Auvinen P, Kaariainen L. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J Biol Chem. 2000;275(23):17281–7. doi: 10.1074/jbc.M910340199. [DOI] [PubMed] [Google Scholar]
- Yin J, Gardner CL, Burke CW, Ryman KD, Klimstra WB. Similarities and differences in antagonism of the neuron interferon alpha/beta response by Venezuelan equine encephalitis and Sindbis alphaviruses. J Virol. 2009 doi: 10.1128/JVI.01209-09. JVI.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]