Abstract
The concentration of the second messenger cAMP is tightly controlled in cells by the activity of phosphodiesterases. We have previously described how the protein kinase A-anchoring protein mAKAP serves as a scaffold for the cAMP-dependent protein kinase PKA and the cAMP-specific phosphodiesterase PDE4D3 in cardiac myocytes. PKA and PDE4D3 constitute a negative feedback loop whereby PKA-catalyzed phosphorylation and activation of PDE4D3 attenuate local cAMP levels. We now show that protein phosphatase 2A (PP2A) associated with mAKAP complexes is responsible for reversing the activation of PDE4D3 by catalyzing the dephosphorylation of PDE4D3 serine residue 54. Mapping studies reveal that a C-terminal mAKAP domain (residues 2085–2319) binds PP2A. Binding to mAKAP is required for PP2A function, such that deletion of the C-terminal domain enhances both base-line and forskolin-stimulated PDE4D3 activity. Interestingly, PP2A holoenzyme associated with mAKAP complexes in the heart contains the PP2A targeting subunit B56δ. Like PDE4D3, B56δ is a PKA substrate, and PKA phosphorylation of mAKAP-bound B56δ enhances phosphatase activity 2-fold in the complex. Accordingly, expression of a B56δ mutant that cannot be phosphorylated by PKA results in increased PDE4D3 phosphorylation. Taken together, our findings demonstrate that PP2A associated with mAKAP complexes promotes PDE4D3 dephosphorylation, serving both to inhibit PDE4D3 in unstimulated cells and also to mediate a cAMP-induced positive feedback loop following adenylyl cyclase activation and B56δ phosphorylation. In general, PKA·PP2A·mAKAP complexes exemplify how protein kinases and phosphatases may participate in molecular signaling complexes to dynamically regulate localized intracellular signaling.
Keywords: Phosphorylation, Protein/Protein-Protein Interactions, Signal Transduction/Adapter Proteins, Signal Transduction/Cyclic Nucleotides/Cyclic AMP, Signal Transduction/Phosphodiesterases, Signal Transduction/Phosphoprotein Phosphatases/PP1/PP2A, Signal Transduction/Protein Kinases/Cyclic Nucleotide, Tissue/Organ Systems/Muscle/Heart
Introduction
cAMP controls a plethora of processes in virtually every cell type, including gene expression, intermediary metabolism, and ion channel activity. In most cases, cAMP is produced through G protein-coupled receptor (GPCR)2 activation of the stimulatory Gα protein (Gαs), which in turn activates adenylyl cyclase that catalyzes the conversion of ATP to cAMP. In the heart, a variety of GPCRs promote the production of this small diffusible second messenger. However, a long standing finding is that the cellular response to cAMP signaling varies in the myocyte depending upon the upstream GPCR (1–4). This observation has led to the hypothesis that cAMP production in response to the stimulation of individual GPCRs is restricted to discrete subcellular domains, conferring spatiotemporal control of cAMP production as well as response specificity (5, 6). Understanding the mechanisms that confine cAMP to these microdomains remains of considerable interest.
Recent experimentation involving live cell imaging of cardiac myocytes has revealed that type 4 phosphodiesterases (PDE4) are important for the spatiotemporal control of cAMP following β-adrenergic receptor stimulation (7). Individual PDE4 isoforms may contribute to the local regulation of cAMP in different cellular compartments. For example, we have shown that the alternatively spliced PDE4 isoform D3 (PDE4D3) is bound by the scaffold protein mAKAP (muscle A kinase-anchoring protein) in cardiac myocytes (8–10). Because mAKAP is tethered to the nuclear envelope by nesprin-1α (11), PDE4D3-bound mAKAP likely controls perinuclear cAMP levels. Signaling through mAKAP multimolecular signaling complexes has been implicated in the regulation of myocyte hypertrophy and in the regulation of gene expression during hypoxia (9, 12–14). As its name implies, mAKAP binds the cAMP-dependent protein kinase PKA, and PKA binding to mAKAP is required for PDE4D3 phosphorylation. In response to elevated cAMP levels, mAKAP-bound PKA phosphorylates PDE4D3 on serine residues 13 and 54, resulting in 2–3-fold increased PDE4D3 binding to the complex and PDE activity (10, 15, 16). Because increased PDE4D3 activity accelerates cAMP degradation, PKA and PDE4D3 constitute a negative feedback loop that modulates both local cAMP levels and PKA activity (10). PDE4D3 bound to mAKAP serves not only as a PDE, but also as an adapter protein, recruiting the MAPKs MEK5 and ERK5 and the cAMP-dependent Rap1-guanine nucleotide exchange factor Epac1 to mAKAP complexes (9). Activation of MEK5 and ERK5 by upstream signals results in PDE4D3 phosphorylation on serine residue 579, inhibiting the PDE and promoting cAMP accumulation and PKA activation (9, 17, 18). Elevated cAMP levels will also activate mAKAP-associated Epac1. However, through Rap1, Epac1 inhibits ERK5 activity, preventing PDE4D3 inhibition (9). As a result, Epac1, ERK5, and PDE4D3 constitute a second negative feedback loop that will attenuate cAMP levels in the vicinity of mAKAP complexes. Importantly, both of these negative feedback loops intrinsic to the mAKAP signaling complex depend upon the regulation of PDE4D3 activity as the key element controlling local cAMP levels.
Although we have described how mAKAP-bound PDE4D3 may be regulated by PKA and ERK5 phosphorylation, the phosphatase(s) involved in PDE4D3 dephosphorylation have remained unknown. We now show that PP2A bound to the mAKAP complex catalyzes the dephosphorylation of PDE4D3 Ser-54, inhibiting the PDE. Although previously thought to be a constitutive, housekeeping enzyme, it has become apparent that PP2A contributes to the regulation of many phosphorylation events. For example, in the cardiac myocyte, PP2A is involved in the modulation of calcium and MAPK signaling (19–21). The current challenge in the study of this phosphatase is to understand its spatiotemporal regulation. PP2A is a serine/threonine phosphatase that exists as a heterotrimeric complex consisting of a stable, ubiquitously expressed catalytic (PP2A-C) and scaffolding (PP2A-A) subunit heterodimer and one of 21 known divergent B subunits (22, 23). PP2A-B subunits are grouped into three unrelated families termed B (or PR55), B′ (or B56) and B″ (or PR72) and are proposed to regulate both the catalytic activity and the intracellular targeting of the phosphatase (24). As now revealed, PP2A associated with mAKAP complexes contain B56δ B subunits. Recently published work demonstrates that B56δ is a PKA substrate, and its phosphorylation enhances PP2A catalytic activity (25). Accordingly, we show that phosphorylation of B56δ by mAKAP-bound PKA increases PDE4D3 dephosphorylation. Our results provide the mechanistic details of mAKAP-anchored PP2A regulation of PDE4D3, elucidating further how mAKAP signaling complexes may regulate a discrete intracellular cAMP signaling domain through a set of interlacing positive and negative feedback loops.
EXPERIMENTAL PROCEDURES
Antibodies
The following primary antibodies were used for immunoblotting: mouse monoclonal anti-GFP (1:500; Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-VSV tag (1:1000; Sigma), mouse monoclonal anti-mAKAP (1:1000; Covance), 9E10 mouse anti-myc (1:500; Santa Cruz Biotechnology), polyclonal anti-PP2A-C (1:500; Santa Cruz Biotechnology), and polyclonal anti-PP1 catalytic subunit (1:500; Santa Cruz Biotechnology). A phospho-specific antibody for phospho-PDE4D3 Ser-54 was generated and affinity-purified using phosphorylated and nonphosphorylated human PDE4D3 peptides containing residues 70–81 (21st Century Biochemicals) and was used at a dilution of 1:500. Polyclonal B56δ antibodies, both non-phospho-specific and specific for phospho-Ser-566, are as described previously (25).
Expression Constructs
Expression vectors for FLAG-tagged B56δ, glutathione S-transferase (GST) PP2A-A fusion protein, and myc- and GFP-tagged rat and human mAKAP are as described previously (12, 25–27). The myc-tagged mAKAP construct deficient in PP2A binding was made by subcloning a cDNA fragment encoding rat mAKAP 1286–2083 generated by PCR into pCMV-Myc (Clontech). mAKAPα and mAKAPβ are two alternatively spliced isoforms of mAKAP expressed in the heart and brain, respectively (28). mAKAPβ is identical to mAKAPα residues 245–2314; all recombinant mAKAP proteins expressed in this paper are based on mAKAPα. The expression vector used for PDE4D3 throughout this paper was constructed by subcloning a cDNA encoding VSV-tagged PDE4D3 (10) into a GFP expression vector (Clontech), resulting in a double-tagged PDE4D3 protein. Further details of the expression vectors are available upon request.
Immunoprecipitation
HEK293 cells were used in this project as a heterologous system lacking mAKAP in which the various wild-type and mutant proteins could be easily expressed. Cells cultured on 60-mm plates were transfected at 50–70% confluence by the calcium phosphate method, using 6 μg of each DNA construct/plate. Cells were harvested 24 h after transfection in 0.5 ml of HSE buffer (HEPES, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, and protease inhibitors). Supernatants were incubated with 3 μg of antibody and 15 μl of prewashed protein A- or protein G-agarose beads. Following overnight incubation at 4 °C, the immunoprecipitates were washed three times with the same buffer. Bound proteins were analyzed by immunoblotting.
For immunoprecipitation of endogenous, native mAKAP complexes, adult rat hearts (Pel-Freez Biologicals) were homogenized in 10 ml of HSE buffer. After centrifugation at 15,000 × g for 25 min, clarified extracts were immunoprecipitated as above.
PDE Assay
PDE activity associated with immunoprecipitated protein complexes was assayed according to the method by Beavo et al. (29). Samples were assayed in 45 μl of PDE buffer A (100 mm MOPS, pH 7.5, 4 mm EGTA, 1.0 mg/ml bovine serum albumin) and 50 μl of PDE buffer B (100 mm MOPS, pH 7.5, 75 mm magnesium acetate, 1 μm cAMP, and 100,000 cpm of [3H]cAMP (PerkinElmer Life Sciences)). Inhibitors were included as indicated.
Phosphatase Assay
Phosphatase activity was measured according to the method of Ahn et al. (25) using 32P-labeled histone as substrate. Histone was radiolabeled in reactions containing 250 mm MOPS, pH 7.4, 2.5 mm magnesium acetate, 100 mm β-mercaptoethanol, purified PKA catalytic subunit, 1 μm ATP, 20 μm histone, and 1 mCi of [γ-32P]ATP (6000 Ci/mmol). The reaction was terminated by the addition of 50% trichloroacetic acid, and [32P]histone was purified from free radionucleotide by centrifugation. The [32P]histone pellet was washed with 1 ml of ether/ethanol/HCl (4:1:0.1) once and 1 ml of ether/ethanol (4:1) three times. The substrate was then suspended in 200 μl of PP2A assay buffer (25 mm Tris, pH 7.4, 1 mm dithiothreitol, and 10 mm MgCl2) before precipitation with 50% trichloroacetic acid. After repeated washing, the [32P]histone was suspended in 200 μl of PP2A buffer.
To measure phosphatase activity, immunoprecipitated protein complexes were washed twice in HSE buffer and once in PP2A reaction buffer. The immunoprecipitates were incubated for 30 min at 30 °C in 20 μl of PP2A assay buffer containing 100,000 cpm of [32P]histone in the presence and absence of inhibitors. The PP2A inhibitor (Calbiochem) was used at a concentration of 30 nm. Purified I-1 was phosphorylated by PKA before using as a specific PP1 inhibitor. Reactions were terminated by the addition of 100 μl of 20% trichloroacetic acid followed by a 10-min centrifugation. Trichloroacetic acid supernatants containing released 32PO4 were measured by scintillation counting.
GST Pulldowns
Glutathione resin adsorbed with PP2A-A subunit GST fusion protein or GST control protein were incubated with HEK293 cell extracts. After an overnight incubation, the beads were washed three times. Bound proteins were analyzed by immunoblotting.
Statistics
Each n refers to a completely independent experiment performed using separate cultures or heart preparations. All p values were calculated using Student's t test.
RESULTS
Regulation of mAKAP-bound PDE4D3 by an Okadaic Acid (OA)-sensitive Phosphatase
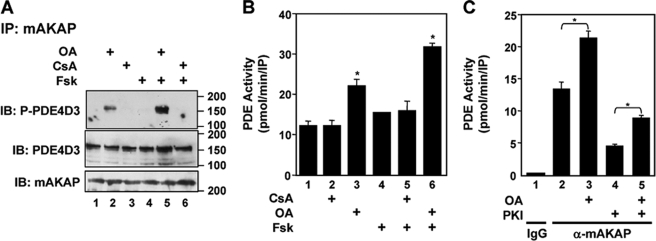
We have previously described a negative feedback loop intrinsic to mAKAP complexes that includes cAMP activation of PKA, PKA phosphorylation and activation of PDE4D3, and PDE4D3-catalyzed cAMP degradation (10). PDE4D3 phosphorylation was dependent on PKA binding to mAKAP. We speculated that, symmetrically, a mAKAP-bound phosphatase might be responsible for PDE4D3 dephosphorylation. We have found that both PP2A and the Ca2+/calmodulin-dependent protein phosphatase calcineurin (PP2B) associate with the mAKAP scaffold in cardiac myocytes (12, 14, 27). To begin this study, we tested in a heterologous system whether PP2A or PP2B might dephosphorylate PDE4D3 at Ser-54, the residue within the PDE4D3 upstream conserved region required for PKA activation (15). HEK293 cells overexpressing mAKAP and PDE4D3 were treated with 300 μm OA to inhibit PP2A (and PP1) activity or 500 μm cyclosporin A to inhibit PP2B activity (Fig. 1A). After immunoprecipitation of protein complexes using a mAKAP-specific antibody, PDE4D3 phosphorylation was assayed by immunoblotting with a phospho-specific antibody to residue Ser-54 that we had generated. OA treatment resulted in an increase in the base-line phosphorylation of PDE4D3 Ser-54, whereas inhibition of PP2B had no effect (Fig. 1A, top panel, lane 2). This increased phosphorylation was further enhanced 1.8-fold when PKA was activated by the addition of the adenylyl cyclase agonist forskolin (Fsk; Fig. 1A, top panel, lane 5). Notably, forskolin alone had no significant effect in the absence of phosphatase inhibition (Fig. 1A, lane 4). Immunoblotting using a non-phospho-specific antibody for PDE4D3 and an antibody for mAKAP demonstrated that two proteins were similarly precipitated under each condition (Fig. 1A, middle and bottom panels).
FIGURE 1.
An OA-sensitive phosphatase regulates mAKAP-associated PDE4D3. A, transfected HEK293 cells expressing both mAKAP and PDE4D3 were treated with either 300 μm OA or 500 μm cyclosporin A (CsA) for 30 min before stimulation with 5 μm Fsk for 10 min. The phosphorylation state of PDE4D3 present in mAKAP antibody immunoprecipitates (IP) was determined using a antibody specific for phosphorylated PDE4D3 Ser-54 (top panel). Total PDE4D3 (middle panel) and mAKAP (bottom panel) present in mAKAP antibody immunoprecipitates were detected using non-phospho-specific antibodies. Note that in these experiments mAKAP was GFP-tagged, and PDE4D3 was VSV and GFP-tagged, resulting in increased molecular weights. n = 3. IB, immunoblotted. B, PDE activity associated with mAKAP antibody immunoprecipitates prepared as in A was assayed using [3H]cAMP substrate. *, p < 0.05 compared with untreated cells (bar 1). C, endogenous protein complexes were isolated using control (IgG) or mAKAP-specific antibodies from clarified adult rat heart extracts (500 μg of total protein). PDE activity associated with the immunoprecipitates was assayed in the presence of 10 nm OA or 50 nm PKI. n = 3; *, p < 0.05.
Because phosphorylation of PDE4D3 Ser-54 increases PDE activity 2-fold (15), we tested whether OA treatment would also increase the activity of mAKAP-bound PDE4D3. mAKAP complexes were immunoprecipitated from transfected HEK293 cells and assayed for associated PDE activity (Fig. 1B). mAKAP-associated PDE activity in untreated cells was detected only when mAKAP was co-expressed with PDE4D3 (Fig. 1B, bar 1, and data not shown), consistent with our previous observation that PDE4D3 accounts for all of the PDE activity associated with mAKAP in cardiac myocytes (10). In agreement with the results obtained with the phospho-Ser-54 antibody, Fsk treatment alone was unable to stimulate mAKAP-bound PDE4D3 activity significantly in HEK293 cells, whereas Fsk and OA treatment together synergistically increased PDE4D3 activity (Fig. 1B, bars 3 and 6). Cyclosporin A had no effect on either basal or stimulated PDE4D3 activity, suggesting that PP2B does not regulate PDE4D3 bound to mAKAP in cells under these conditions. Together, these results show that in this heterologous system, an OA-sensitive phosphatase strongly inhibits both the base-line and Fsk-stimulated phosphorylation and activity of PDE4D3 bound to mAKAP.
The enhancement of PDE activity by OA was seen not only with expression of recombinant proteins in HEK293 cells, but also upon isolation of native mAKAP complexes from adult rat heart extracts (Fig. 1C). Both PDE4D3 and PKA are active in purified mAKAP complexes (10). PKA activity present in endogenous mAKAP complexes is responsible for increasing PDE activity 2-fold, as was evident upon inhibition of mAKAP-bound PKA with the specific PKA inhibitor PKI (Fig. 1C, bars 2 and 4). Importantly, OA inhibition increased mAKAP-associated PDE activity 30% (bars 2 and 3) and 60% when PKA was also inhibited (bars 4 and 5). Taken together, these data suggest that an OA-sensitive phosphatase associated with the mAKAP complex is responsible for the dephosphorylation of PDE4D3 and the regulation of PDE activity.
PP2A Associates with the mAKAP Scaffold in the Heart
Having established that an OA-sensitive phosphatase was associated with the mAKAP complex, we proceeded to identify the phosphatase by co-immunoprecipitation experiments. Phosphatase activity associated with mAKAP complexes isolated from heart cell extracts was measured using [32P]histone as a substrate. There was a 3-fold enrichment of phosphatase activity over control IgG immunoprecipitates (Fig. 2A, bars 1 and 2). The mAKAP-associated phosphatase responsible for the immunoprecipitated activity was identified as PP2A because the phosphatase activity was completely inhibited by 30 nm PP2A inhibitor 1 (30), but not by addition of 100 nm PKA-phosphorylated PP1 inhibitor 1 (31). As a positive control, the PKA-phosphorylated PP1 inhibitor 1 did inhibit PP1 isolated by immunoprecipitation with a PP1 antibody from HEK293 cell extracts (supplemental Fig. 1). The mAKAP-associated phosphatase activity was not due to mAKAP-bound PP2B because no Ca2+/calmodulin was included in the phosphatase assay buffer. Confirmation of these results was obtained by immunoblot analysis of mAKAP immunoprecipitates. PP2A-C subunit, but not PP1 catalytic subunit, was detected in mAKAP-specific immunoprecipitates (Fig. 2, B and C).
FIGURE 2.
PP2A is associated with the mAKAP scaffold in adult rat heart. A, phosphatase activity associated with protein complexes immunoprecipitated using mAKAP antibody from clarified adult rat heart extracts (500 μg of total protein) was assayed using 32P-labeled histone substrate in the absence or presence of 30 nm PP2A inhibitor 1 (30) and 100 nm PKA-phosphorylated PP1 inhibitor 1 (31). n = 3; *, p < 0.05. B and C, protein complexes were isolated from adult rat heart extracts (2 mg of total protein) using control (IgG) or mAKAP-specific antibody. 80 μg of total control extracts and 25% of the total immunoprecipitates were loaded onto the gel. PP2A (B) and PP1 (C) catalytic subunits were detected by immunoblotting (IB). n = 3.
Like PKA, PP2A associates with many cellular substrates and is expected to be present in diverse intracellular compartments (24). Confocal fluorescent microscopy of cultured primary neonatal rat cardiomyocytes revealed that PP2A-C subunit is distributed throughout the cytoplasm in a fine punctuate pattern (supplemental Fig. 2, green). As found previously, mAKAP was localized primarily to the nuclear envelope (11). Consistent with the co-immunoprecipitation of mAKAP and PP2A from adult rat heart extracts, overlap of PP2A and mAKAP staining could be detected at the nuclear envelope (supplemental Fig. 2, composite image), supporting our model that a localized signaling complex consisting of discrete pools of PP2A, PKA, and PDE4D3 and the scaffold mAKAP is present in cardiac myocytes.
mAKAP Residues 2083–2319 Contain the PP2A Binding Domain
To map the PP2A binding site on mAKAP, we used a bacterially expressed PP2A-A subunit GST fusion protein to pull down GFP-tagged fragments of mAKAP expressed in HEK293 cells (Fig. 3, A and B). GST-PP2A-A consistently pulled down only fragments of mAKAP containing a domain C-terminal to residue 2085. Both human and rat mAKAP GFP fusion proteins bound GST-PP2A-A, including rat mAKAP 1835–2312 and human 2085–2319. As a negative control, the GFP-mAKAP fusion proteins did not bind PP1 in HEK293 cells, consistent with the lack of co-immunoprecipitation of PP1 and mAKAP from heart extracts (supplemental Fig. 3). To confirm the mapping of the PP2A binding site on mAKAP, myc-tagged mAKAP fragments expressed in HEK293 cells were immunoprecipitated with a myc tag antibody and assayed for associated PP2A activity (Fig. 3C). mAKAP 1286–2312, but not mAKAP 1286–2083, co-immunoprecipitated with OA-sensitive phosphatase activity. Together, these data show that PP2A binds a C-terminal site within mAKAP that is separate from the binding sites for PKA, PDE4D3, and other known mAKAP-binding proteins (Fig. 3A).
FIGURE 3.
PP2A binds a C-terminal mAKAP domain. A, schematic of mAKAP domains and GFP- and myc-tagged mAKAP proteins used in this paper. mAKAP fragments containing rat and human protein are drawn in black and gray, respectively. Hatched bars indicate the three spectrin repeat domains responsible for nuclear envelope targeting in myocytes (26). Binding sites are indicated for proteins known to bind mAKAP directly, including nesprin-1α (1074–1187) (11), ryanodine receptor (RyR2, 1217–1242) (39), PP2B (1286–1345) (14), PDE4D3 (1285–1833) (10), and PKA (2055–2072) (26). For reference, the binding site for 3-phosphoinositide-dependent kinase-1 (PDK1, mAKAP residues 227–232) (28) is indicated, although this protein is not discussed further in this paper. The stippled bar marks the PP2A binding site. The first and last residues of each fragment are indicated. B, purified GST-PP2A-A subunit fusion protein was incubated with extracts prepared from HEK293 cells expressing the indicated GFP-mAKAP fusion protein and pulled down using glutathione resin. GFP-mAKAP fragments were detected in the pulldowns (25% loaded, top panel) and the extracts (5% loaded, bottom panel) using a GFP antibody. n = 3. IB, immunoblotting. C, myc-tagged mAKAP fragments were expressed in HEK293 cells, and phosphatase binding was detected by immunoprecipitation using control (IgG) or myc-tag antibody followed by phosphatase assay using 32P-labeled histone substrate. n = 3. *, p < 0.05 compared with the other samples.
mAKAP-anchored PP2A Regulates PDE4D3 Phosphorylation in the Complex
Data obtained using mAKAP complexes isolated from rat heart extracts implied that mAKAP-bound PP2A regulated PDE4D3 in the complex (Fig. 1C). To test whether PP2A anchoring is required for PDE4D3 dephosphorylation, we expressed in HEK293 cells PDE4D3 and a mAKAP construct containing the binding sites for PDE4D3, PKA, and PP2A (myc-mAKAP 1286–2312), or a similar mAKAP construct lacking the PP2A binding site (myc-mAKAP 1286–2083). The cells were stimulated with Fsk and OA, and mAKAP complexes were subsequently isolated by immunoprecipitation. Phosphorylation of mAKAP-bound PDE4D3 was assayed by immunoblotting with the Ser-54 phospho-specific antibody. As was found upon expression of full-length mAKAP (Fig. 1A), phosphorylation of PDE4D3 bound to myc-mAKAP 1286–2312 was detected only when phosphatase activity was suppressed by OA (Fig. 4A, lane 3). Notably, upon expression of myc-mAKAP 1286–2083, which lacked significant PP2A binding (Fig. 4A, lanes 4–6), an increase in the base-line phosphorylation of mAKAP-bound PDE4D3 was detected (0.49 ± 0.19-fold of the level obtained with OA; Fig. 4A, lanes 4 versus 3). Moreover, upon deletion of the PP2A binding domain, Fsk alone increased phosphorylation of the PDE to levels equivalent to that associated with PP2A-containing complexes treated with both Fsk and OA (Fig. 4A, lanes 3, 5, and 6). The changes in PDE4D3 Ser-54 phosphorylation were mirrored by changes in PDE activity (Fig. 4B). PDE4D3 activity was 30% higher in myc-mAKAP 1286–2083 immunoprecipitates lacking PP2A than in complexes containing the phosphatase (bars 1 and 4). Importantly, no significant difference in PDE4D3 activity was seen between Fsk stimulation and Fsk stimulation in the presence of OA for the complexes lacking PP2A (bars 5 and 6). These data demonstrate the importance of PP2A anchoring for the regulation of PDE4D3 phosphorylation and activity. Furthermore, they suggest that PP2A serves not only to attenuate PKA-activated PDE activity, but also to maintain a low basal level of PDE4D3 activity in unstimulated cells.
FIGURE 4.
PP2A association with mAKAP·PDE4D3 complexes is required for inhibition of PDE4D3 phosphorylation. A, HEK293 cells expressing (VSV- and GFP-tagged) PDE4D3 and myc-tagged mAKAP 1286–2312 or 1286–2083 lacking the PP2A binding site were treated with 300 μm OA for 30 min before stimulation with 5 μm Fsk for 10 min. Protein complexes were immunoprecipitated (IP) using myc-tagged antibody in the presence of phosphatase inhibitors. The phosphorylation state of co-immunoprecipitated PDE4D3 was determined using an antibody specific for phosphorylated PDE4D3 Ser-54 (P-PDE4D3, top panel). Total PDE4D3, myc-mAKAP, and PP2A-C subunit present in the immunoprecipitates were detected using non-phospho-specific antibodies (bottom three panels). n = 3. IB, immunoblotting. B, PDE activity associated with myc-antibody immunoprecipitates isolated from additional cells treated as in A was assayed using [3H]cAMP. n = 3. *, p < 0.05 compared with bar 1.
mAKAP-bound PP2A Holoenzyme Containing B56δ Subunit Is Regulated by PKA
PP2A holoenzyme is composed of three subunits, including a core A and C subunit heterodimer and a B subunit that may target the holoenzyme to specific intracellular organelles (24). Three closely related B subunits have been identified that are expressed in the heart and are localized to the nucleus, B56δ, B56γ1, and B56γ3 (32, 33). Recent work demonstrated that PP2A holoenzyme containing B56δ is regulated by PKA phosphorylation (25). We, therefore, considered whether PP2A associated with mAKAP complexes might also be regulated by PKA activity. Native mAKAP complexes were immunoprecipitated from adult rat heart extracts and assayed for associated phosphatase activity (Fig. 5A). mAKAP-associated phosphatase activity was increased 2.5-fold by stimulation of bound PKA with the nonhydrolyzable cAMP analog CPT-cAMP (lanes 2 and 3). As controls, all immunoprecipitated phosphatase activity was inhibited by 10 nm OA (lane 4), and the CPT-cAMP-stimulated increase in phosphatase activity was blocked by the addition of the PKA inhibitor PKI (lane 5). Taken together, these data suggest that PP2A activity associated with mAKAP complexes in the heart is potentiated by PKA-dependent cAMP signaling.
FIGURE 5.
mAKAP-bound PP2A contains B56δ subunit and is cAMP-activated. A, protein complexes were immunoprecipitated from clarified adult rat heart extracts (500 μg of total protein) using control (IgG) or mAKAP-specific antibody as in Fig. 2B and assayed for associated phosphatase activity. As indicated, the immunoprecipitates were preincubated with no addition or with 50 μm CPT-cAMP, 10 nm OA, or 50 nm PKI for 5 min before the addition of [32P]histone substrate. n = 3; *, p < 0.05. B, endogenous protein complexes were immunoprecipitated from adult heart extract (2 mg of total protein) with B56δ and control (IgG) antibodies in 80 μg of extract, and 25% of the immunoprecipitates were loaded onto the gel. mAKAP was detected by immunoblotting (IB). n = 3. C, FLAG-tagged B56δ and/or GFP-tagged mAKAP were expressed in HEK293 cells. Protein complexes were immunoprecipitated (IP) using a mAKAP antibody. B56δ in the immunoprecipitates (25% loaded) and total extracts (5% loaded) was detected by immunoblotting with a FLAG antibody. n = 3. D, phosphatase activity associated with mAKAP-antibody immunoprecipitates prepared as in C was assayed using 32P-labeled histone substrate. n = 3. E, HEK293 cells expressing mAKAP and B56δ were treated with 5 μm Fsk and 10 μm IBMX (Fsk/IBMX) for 10 min before immunoprecipitation of protein complexes with mAKAP antibody. Phosphatase activity associated with the immunoprecipitates was assayed using [32P]histone substrate. n = 3. Note that PP2A B56δ and C subunit binding to mAKAP was not affected by Fsk/IBMX (see Fig. 6).
Because mAKAP-bound PP2A was regulated by PKA activity, we proceeded to test whether mAKAP-bound PP2A holoenzyme contained B56δ subunit. Protein complexes were immunoprecipitated from adult rat heart extracts using B56δ and control (IgG) antibody (Fig. 5B). mAKAP was consistently immunoprecipitated with the B56δ antibody. In addition, we expressed FLAG-tagged B56δ in HEK293 cells and showed that B56δ was immunoprecipitated with a mAKAP antibody only when co-expressed with (GFP-tagged) mAKAP (Fig. 5C). Finally, the binding of B56δ to mAKAP was shown to recruit PP2A-C subunit to the complex because mAKAP complexes immunoprecipitated from HEK293 cell extracts were associated with greater phosphatase activity when GFP-mAKAP was co-expressed with FLAG-B56δ (Fig. 5D, lanes 2 and 3). Based on these results, we propose that B56δ recruits the PP2A-A/C core heterodimer to mAKAP complexes in the heart, conferring cAMP-dependent phosphatase activity. Accordingly, elevation of intracellular cAMP with Fsk and the PDE inhibitor IBMX increased mAKAP-associated phosphatase activity in HEK293 cells, only when mAKAP was co-expressed with B56δ (Fig. 5E).
PKA Binding Is Required for cAMP-dependent PP2A Activity in mAKAP Complexes
Previous work found that PKA phosphorylates B56δ on four serine residues (53, 68, 81, and 566), and Ser-566 is suggested to account for the induction of PP2A activity (25). Because mAKAP complexes include both PKA and PP2A, we considered that association of these molecules into a complex was important for PP2A phosphorylation, just as PP2A binding to mAKAP was required for PDE4D3 dephosphorylation (Fig. 4). To test this hypothesis, we expressed B56δ in HEK293 cells with wild-type full-length mAKAP or a full-length mAKAP mutant with an internal deletion of residues 2053–2073 comprising the PKA binding site (ΔPKA, Fig. 3A) (12). Following stimulation of the cells with Fsk/IBMX to elevate intracellular cAMP, mAKAP complexes were isolated by immunoprecipitation, and the phosphorylation state of B56δ was determined using a phospho-specific antibody to B56δ Ser-566 (Fig. 6A, top panel) (25). B56δ phosphorylation was detected only after FSK/IBMX treatment and only when B56δ was co-expressed with wild-type mAKAP and not the ΔPKA mutant (Fig. 6A, lanes 2 and 6). As a control, equivalent expression of mutant and wild-type mAKAP and B56δ proteins was demonstrated by immunoblotting with non-phospho-specific antibodies (Fig. 6A, middle and bottom panels). Additionally, wild-type mAKAP was co-expressed with a mutant B56δ form containing alanine residues at each of the four PKA substrate sites (S4A). As expected, Fsk/IBMX stimulation did not induce phosphorylation of B56δ S4A (Fig. 6A, lane 4). Because B56δ phosphorylation increases PP2A catalytic activity, we also assayed the mAKAP antibody immunoprecipitates for phosphatase activity (Fig. 6B). Consistent with the results obtained using the phospho-specific B56δ antibody, cAMP elevation increased phosphatase activity in mAKAP complexes 1.7-fold (Fig. 6B, lanes 2 and 3). This increase required phosphorylation of B56δ because complexes containing the S4A mutant showed no augmentation of PP2A activity by increased cAMP (lane 5). Likewise, PKA binding to mAKAP was required to induce PP2A activity because no increase was obtained when B56δ was co-expressed with the mAKAP ΔPKA mutant scaffold (lane 6). Interestingly, the Fsk/IBMX-induced increase in mAKAP-associated PP2A activity was not due to increased PP2A-C subunit binding to the mAKAP complexes (Fig. 6A, lanes 1 and 2). This result is in accord with our earlier suggestion that B56δ phosphorylation increases PP2A catalytic activity through conformational changes that do not affect holoenzyme formation (25).
FIGURE 6.
Phosphorylation of B56δ by PKA increases mAKAP-associated PP2A activity. A, B56δ is phosphorylated on serine residues 53, 68, 81, and 566 by PKA (25). B56δ wild-type or alanine-substituted at all four PKA sites (S4A) was co-expressed in HEK293 cells with wild-type (WT) mAKAP or a full-length mAKAP mutant lacking the PKA binding site (ΔPKA; cf. Fig. 3A). After stimulation with 5 μm Fsk and 50 μm IBMX, protein complexes were immunoprecipitated (IP) with mAKAP antibody, and associated proteins were detected by immunoblotting (IB) with B56δ, mAKAP, and PP2A-C antibodies (bottom three panels). PKA phosphorylation of B56δ was detected by immunoblotting with a B56δ phospho-Ser-566-specific antibody (P-B56δ, top panel). n = 3. B, immunoprecipitates prepared as in A were assayed for associated phosphatase activity. n = 3; * p < 0.05.
PP2A Regulates PDE4D3 Phosphorylation in a PKA-dependent Manner
The results described above imply that PP2A dephosphorylation of PDE4D3 in B56δ·mAKAP complexes should be enhanced by PKA-catalyzed phosphorylation of the phosphatase. To address the role of B56δ phosphorylation in the regulation of PDE4D3, we co-expressed PDE4D3 and mAKAP with either wild-type B56δ or the B56δ S4A mutant that is not responsive to PKA. Cells were stimulated with Fsk before isolation of mAKAP complexes. As detected by phospho-specific antibody immunoblot and enzymatic assay, Fsk stimulation of PDE4D3 Ser-54 phosphorylation and PDE activity was only observed for mAKAP complexes containing wild-type B56δ when PP2A was inhibited with OA (Fig. 7, lanes/bars 1–3), consistent with aforementioned data (Fig. 1). In contrast, expression of B56δ S4A resulted in detectable Fsk-stimulated PDE4D3 phosphorylation (0.39 ± 0.15-fold of Fsk/OA-stimulated cells; Fig. 7A, lane 5 versus 3) and a concomitant increase in PDE activity (Fig. 7B, bar 5), albeit not as strongly as when PP2A activity was directly inhibited by OA (Fig. 7, lanes/bars 3 and 6). Taken together with the results shown in Figs. 5 and 6, we conclude that the anchoring of a PKA-stimulated PP2A holoenzyme is responsible for the attenuation of both basal and PKA-stimulated PDE4D3 activity in the mAKAP signaling complex.
FIGURE 7.
Phosphorylation of B56δ by PKA enhances the dephosphorylation of mAKAP-associated PDE3D3. A, HEK293 cells expressing (GFP-tagged) mAKAP, (VSV- and GFP-tagged) PDE4D3 and either wild-type B56δ or B56δ S4A mutant at the PKA phosphorylation sites were treated as indicated with 300 μm OA for 30 min before stimulation for 10 min with 5 μm Fsk. Protein complexes were immunoprecipitated (IP) with mAKAP antibody in the presence of phosphatase inhibitors. The phosphorylation state of PDE4D3 present in the immunoprecipitates was determined using an antibody specific for phosphorylated PDE4D3 Ser-54 (top panel). Total PDE4D3, mAKAP, B56δ, and PP2A-C protein present in the immunoprecipitates were detected using non-phospho-specific antibodies (lower four panels). n = 3. IB, immunoblotting. B, PDE activity associated with protein complexes isolated from additional cells treated as in A was assayed using [3H]cAMP. n = 3; *, p < 0.05 compared with bar 1.
DISCUSSION
The results described in this paper define the biochemical mechanism for the dephosphorylation and inactivation of PKA-phosphorylated PDE4D3 bound by the scaffold protein mAKAP. We discovered that a PP2A heterotrimer composed of A, C, and B56δ subunits binds a C-terminal site on mAKAP distinct from the binding sites for other known mAKAP partners (Fig. 3). The association of PP2A with the mAKAP scaffold is of functional significance in two important and novel ways. First, by binding both PP2A and PDE4D3, mAKAP sequesters the phosphatase in close proximity to the PDE, allowing for efficient PDE4D3 dephosphorylation and down-regulation (Fig. 4). Second, by binding both PKA and PP2A, mAKAP promotes cAMP-dependent phosphorylation of the PP2A B56δ subunit and induction of PP2A activity (Fig. 6). The relevance of multimolecular signaling complex formation was evident upon expression of mAKAP mutants lacking binding sites for PP2A and PKA.
The concept of phosphatase targeting to generate substrate specificity was first proposed in the mid-1980s with the identification of the glycogen particle-associated protein as the first PP1-targeting subunit (34). Because of this initial observation, several other phosphatase targeting motifs have been determined (24). AKAPs represent an important mechanism to link phosphatases with their appropriate substrates, and several AKAPs bind protein phosphatases. We have recently published that mAKAP binds PP2B and that this interaction is important for PP2B-dependent NFATc3 activation in myocytes (14). However, PP2B binding to mAKAP does not appear to regulate PDE4D3 because inhibition of PP2B did not affect PDE4D3 Ser-54 phosphorylation or PDE activity (Fig. 1). Our data support a unique role for PP2A bound to mAKAP in dephosphorylation of the PDE and, as a result, in the control of local cAMP levels.
The overall role of phosphatases in regulating cellular cAMP concentration has yet to be fully explored. In rat adipocytes, PP2A was found to regulate both PDE3B activity and phosphorylation (35). In addition to being phosphorylated by PKA on Ser-54, PDE4D3 is phosphorylated on Ser-579 by MAPKs, including by ERK5 present in mAKAP complexes (9, 18). Although PP1 does not appear to bind mAKAP (Fig. 2 and supplemental Fig. 3), PP1 may dephosphorylate PDE4D3 Ser-579 in other cellular domains because the addition of purified PP1 to isolated PDE4D3 decreased phosphorylation at this site. Further work will be required to identify the phosphatase(s) responsible for the dephosphorylation of mAKAP-bound PDE4D3 at Ser-579 as well as the second PKA site on PDE4D3, Ser-16 (16).
The anchoring hypothesis suggests that AKAPs function to target the actions of PKA toward specific substrates by localizing both proteins to the same signaling complex. In this report, we demonstrate a new target for PKA in the mAKAP complex, the PP2A B56δ subunit. Previous work found that phosphorylation of B56δ stimulated PP2A activity and enhanced dephosphorylation of DARPP-32 (25). In accordance with our results, stimulation of cardiac myocytes with β-adrenergic receptor agonists increases PP2A activity (36). We propose that the mAKAP scaffold may facilitate this event because the association of the anchoring protein with both PKA and PP2A is important for the cAMP-enhanced increase in phosphatase activity (Figs. 4 and 6). Hence, one can imagine a role for mAKAP in the regulation of phosphatase activity in the heart.
Based upon our results, we propose a model in which PP2A serves a dual role in regulating cAMP levels near mAKAP signaling complexes (Fig. 8). First, PP2A in mAKAP complexes should maintain PDE4D3 in a dephosphorylated, minimally active state in the absence of GPCR stimulation (Fig. 8A), presumably allowing for a more rapid rise in cAMP levels in response to agonist. Second, following induction of activating cAMP levels by GPCR stimulation, PKA will phosphorylate both PDE4D3 and PP2A (Fig. 8B). In contrast to the negative feedback on cAMP levels mediated by enhanced PDE4D3 phosphorylation, PKA phosphorylation of PP2A opposes PDE4D3 activation. By inhibiting PDE4D3 phosphorylation, PP2A presumably potentiates and prolongs the actions of local cAMP as part of a positive feedback loop. Thus, in conjunction with the potential inhibition of PDE4D3 by mAKAP-bound ERK5 that we have previously described (data not shown) (9), the mAKAP signaling complex is poised to finely regulate local cAMP levels both by multiple feedback loops intrinsic to the complex as well as by cross-talk with upstream MAPK signaling pathways. How these multiple signaling pathways that converge on PDE4D3 ultimately regulate the kinetics of cAMP metabolism in myocytes stimulated by different combinations of upstream signals, including, for example, adrenergic and cytokine receptors, will require further investigation. For example, it has been observed that PP2A expression and intracellular localization are altered in heart failure (37, 38). Whether PP2A-mediated positive feedback or PDE4D3-mediated negative feedback predominately controls cAMP levels local to mAKAP complexes may ultimately depend both on the stoichiometry of PP2A binding to mAKAP and the relative rates of PDE4D3 phosphorylation and dephosphorylation by PKA and PP2A in disease states.
FIGURE 8.
PKA and PP2A associated with mAKAP complexes coordinately regulate PDE4D3 activity and cAMP degradation. PKA is composed of two regulatory and two catalytic subunits. mAKAP-bound PP2A contains A, B56δ, and C (catalytic) subunits. A, in unstimulated cells, basal PP2A activity maintains PDE4D3 dephosphorylation, presumably allowing for a more rapid rise in cAMP levels in response to subsequent agonist than if PDE4D3 were phosphorylated and activated. At the same time, basal PDE4D3 activity should maintain low local levels of cAMP, preventing spurious signaling. B, Gs-coupled receptor stimulation induces cAMP synthesis, exceeding the rate of cAMP degradation by PDE4D3 and activating mAKAP-bound PKA. PKA phosphorylates and activates both PDE4D3 and PP2A. PDE4D3 activation should limit peak cAMP levels as well as accelerate the rate of cAMP clearance after GPCR down-regulation. In contrast, PP2A activation opposes PDE4D3 phosphorylation by PKA, attenuating cAMP degradation and contributing to greater, longer lasting cAMP signals.
In conclusion, we have discovered a novel mechanism by which the scaffold protein mAKAP maintains dynamic regulation of anchored PDE4D3 activity through the association with PDE4D3, PKA, and PP2A. Each of the three enzymes is likely to play an important role in the temporal control of cAMP concentration in the vicinity of perinuclear mAKAP complex. This intricate regulation of local cAMP by the mAKAP “signalosome” likely represents a broader role for AKAPs and phosphatase in the control of cAMP compartmentation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL82705 (to K. L. D.-K.), HL075398 (to M. S. K.), and DA10044 and MH074866 (to A. C. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- GPCR
- G protein-coupled receptor
- PDE4D3
- phosphodiesterase 4D3
- mAKAP
- muscle A kinase-anchoring protein
- PKA
- protein kinase A
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- PP1
- protein phosphatase 1
- PP2A
- protein phosphatase 2A
- PP2B
- protein phosphatase 2B
- GFP
- green fluorescent protein
- VSV
- vesicular stomatitis virus
- GST
- glutathione S-transferase
- MOPS
- 4-morpholinepropanesulfonic acid
- OA
- okadaic acid
- Fsk
- forskolin
- CPT-cAMP
- 8-(4-chlorophenylthio)adenosine 3′,5′-cyclic monophosphate
- PKI
- protein kinase inhibitor
- IBMX
- 3-isobutyl-1-methylxanthine.
REFERENCES
- 1.Hayes J. S., Brunton L. L., Mayer S. E. (1980) J. Biol. Chem. 255, 5113–5119 [PubMed] [Google Scholar]
- 2.Brunton L. L., Hayes J. S., Mayer S. E. (1979) Nature 280, 78–80 [DOI] [PubMed] [Google Scholar]
- 3.Keely S. L. (1977) Res. Commun. Chem. Pathol. Pharmacol. 18, 283–290 [PubMed] [Google Scholar]
- 4.Keely S. L. (1979) Mol. Pharmacol. 15, 235–245 [PubMed] [Google Scholar]
- 5.Steinberg S. F., Brunton L. L. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 751–773 [DOI] [PubMed] [Google Scholar]
- 6.Dodge-Kafka K. L., Langeberg L., Scott J. D. (2006) Circ. Res. 98, 993–1001 [DOI] [PubMed] [Google Scholar]
- 7.Fischmeister R., Castro L. R., Abi-Gerges A., Rochais F., Jurevicius J., Leroy J., Vandecasteele G. (2006) Circ. Res. 99, 816–828 [DOI] [PubMed] [Google Scholar]
- 8.Dodge-Kafka K. L., Kapiloff M. S. (2006) Eur. J. Cell Biol. 85, 593–602 [DOI] [PubMed] [Google Scholar]
- 9.Dodge-Kafka K. L., Soughayer J., Pare G. C., Carlisle Michel J. J., Langeberg L. K., Kapiloff M. S., Scott J. D. (2005) Nature 437, 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodge K. L., Khouangsathiene S., Kapiloff M. S., Mouton R., Hill E. V., Houslay M. D., Langeberg L. K., Scott J. D. (2001) EMBO J. 20, 1921–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pare G. C., Easlick J. L., Mislow J. M., McNally E. M., Kapiloff M. S. (2005) Exp. Cell Res. 303, 388–399 [DOI] [PubMed] [Google Scholar]
- 12.Pare G. C., Bauman A. L., McHenry M., Michel J. J., Dodge-Kafka K. L., Kapiloff M. S. (2005) J. Cell Sci. 118, 5637–5646 [DOI] [PubMed] [Google Scholar]
- 13.Wong W., Goehring A. S., Kapiloff M. S., Langeberg L. K., Scott J. D. (2008) Sci. Signal. 1, ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Negro A., Lopez J., Bauman A. L., Henson E., Dodge-Kafka K., Kapiloff M. S. (2010) J. Mol. Cell. Cardiol. 48, 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sette C., Conti M. (1996) J. Biol. Chem. 271, 16526–16534 [DOI] [PubMed] [Google Scholar]
- 16.Carlisle Michel J. J., Dodge K. L., Wong W., Mayer N. C., Langeberg L. K., Scott J. D. (2004) Biochem. J. 381, 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKenzie S. J., Baillie G. S., McPhee I., Bolger G. B., Houslay M. D. (2000) J. Biol. Chem. 275, 16609–16617 [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann R., Baillie G. S., MacKenzie S. J., Yarwood S. J., Houslay M. D. (1999) EMBO J. 18, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.duBell W. H., Lederer W. J., Rogers T. B. (1996) J. Physiol. 493, 793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.duBell W. H., Gigena M. S., Guatimosim S., Long X., Lederer W. J., Rogers T. B. (2002) Am. J. Physiol. Heart Circ. Physiol. 282, H38–H48 [DOI] [PubMed] [Google Scholar]
- 21.Liu Q., Hofmann P. A. (2004) Am. J. Physiol. Heart Circ. Physiol. 286, H2204–H2212 [DOI] [PubMed] [Google Scholar]
- 22.Lechward K., Awotunde O. S., Swiatek W., Muszyñska G. (2001) Acta Biochim. Pol. 48, 921–933 [PubMed] [Google Scholar]
- 23.Wera S., Hemmings B. A. (1995) Biochem. J. 311, 17–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virshup D. M. (2000) Curr. Opin. Cell Biol. 12, 180–185 [DOI] [PubMed] [Google Scholar]
- 25.Ahn J. H., McAvoy T., Rakhilin S. V., Nishi A., Greengard P., Nairn A. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapiloff M. S., Schillace R. V., Westphal A. M., Scott J. D. (1999) J. Cell Sci. 112, 2725–2736 [DOI] [PubMed] [Google Scholar]
- 27.Kapiloff M. S., Jackson N., Airhart N. (2001) J. Cell Sci. 114, 3167–3176 [DOI] [PubMed] [Google Scholar]
- 28.Michel J. J., Townley I. K., Dodge-Kafka K. L., Zhang F., Kapiloff M. S., Scott J. D. (2005) Mol. Cell 20, 661–672 [DOI] [PubMed] [Google Scholar]
- 29.Beavo J. A., Bechtel P. J., Krebs E. G. (1974) Methods Enzymol. 38, 299–308 [DOI] [PubMed] [Google Scholar]
- 30.Li M., Makkinje A., Damuni Z. (1996) Biochemistry 35, 6998–7002 [DOI] [PubMed] [Google Scholar]
- 31.Endo S., Zhou X., Connor J., Wang B., Shenolikar S. (1996) Biochemistry 35, 5220–5228 [DOI] [PubMed] [Google Scholar]
- 32.Gigena M. S., Ito A., Nojima H., Rogers T. B. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H285–H294 [DOI] [PubMed] [Google Scholar]
- 33.McCright B., Rivers A. M., Audlin S., Virshup D. M. (1996) J. Biol. Chem. 271, 22081–22089 [DOI] [PubMed] [Google Scholar]
- 34.Bauman A. L., Scott J. D. (2002) Nat. Cell Biol. 4, E203–E206 [DOI] [PubMed] [Google Scholar]
- 35.Resjo S., Oknianska A., Zolnierowicz S., Manganiello V., Degerman E. (1999) Biochem. J. 341, 839–845 [PMC free article] [PubMed] [Google Scholar]
- 36.De Arcangelis V., Soto D., Xiang Y. (2008) Mol. Pharmacol. 74, 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiken S., Gaburjakova M., Gaburjakova J., He Kl K. L., Prieto A., Becker E., Yi Gh G. H., Wang J., Burkhoff D., Marks A. R. (2001) Circulation 104, 2843–2848 [DOI] [PubMed] [Google Scholar]
- 38.Ai X., Pogwizd S. M. (2005) Circ. Res. 96, 54–63 [DOI] [PubMed] [Google Scholar]
- 39.Marx S. O., Reiken S., Hisamatsu Y., Jayaraman T., Burkhoff D., Rosemblit N., Marks A. R. (2000) Cell 101, 365–376 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.