Abstract
The YFL ribozyme is an artificial ligase ribozyme isolated by a ‘design and selection’ strategy, in which a modular catalytic unit was generated on a rationally designed modular scaffold RNA. This ligase ribozyme has a versatile catalytic unit that accepts not only β-nicotinamide mononucleotide (β-NMN) but also inorganic pyrophosphate as leaving groups for template-dependent RNA ligation. Although this property is interesting from an evolutionary viewpoint regarding primitive RNA ligation/polymerization systems in the RNA world, structural analysis of the YFL ribozyme has not been continued due to apparent structural nonuniformity of its folded state. To elucidate the active structure of the YFL ribozyme, we performed in vitro evolution experiments to improve its folding ability. Biochemical and phylogenetic analyses of evolved variants indicated that the catalytic unit of the YFL ribozyme is compact and the 3′ single-stranded region of the parent YFL-1 ribozyme contributes to mask an element that could form an inactive structure.
INTRODUCTION
Similar to a number of protein enzymes (1,2), naturally occurring RNA enzymes (ribozymes) often have modular architectures (3–5). Inspired by the modular architectures of natural ribozymes, we attempted to evolve artificial ribozymes by assembling structural modules. By employing an artificial self-folding RNA (Type B RNA) as a structural scaffold (Figure 1A, left), we have isolated two classes of ligase ribozyme that catalyze closely related but distinct reactions (6,7). Two ribozymes that share the scaffold structure have different catalytic modules adapted to different reactions in the evolutionary process. The first ribozyme, termed class DSL, utilizes inorganic pyrophosphate (PPi) as a leaving group for template-dependent RNA ligation (6). The remarkable catalytic ability, folding properties and explicit modular architecture of the DSL ribozyme are suitable for further in vitro evolution experiments, through which variants showing improved activity and/or having novel modular structures were generated (8).
Figure 1.
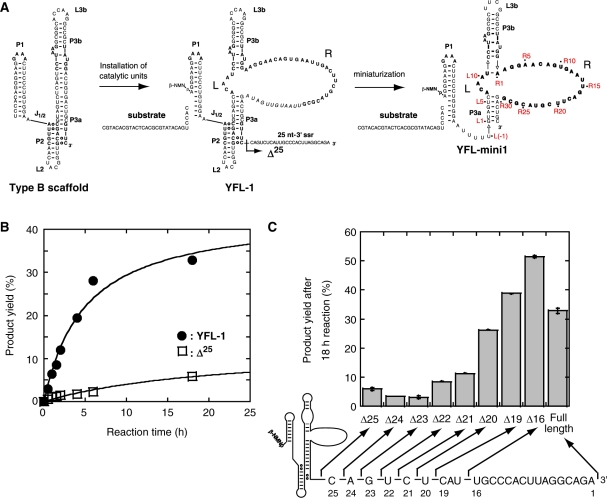
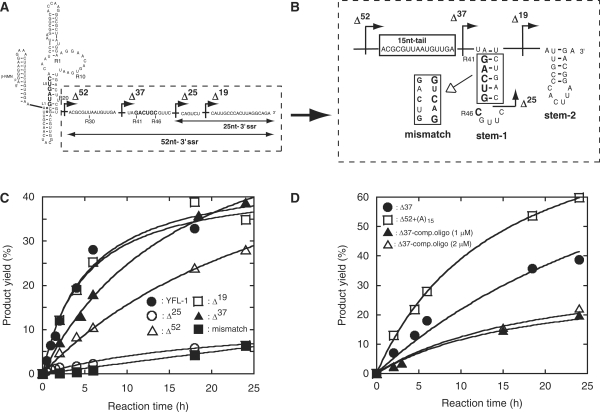
Construction and miniaturization of the class YFL ribozyme. (A) Generation and miniaturization of the class YFL ribozyme. The YFL-1 ribozyme (middle) was constructed by installing catalytic units into the Type B scaffold (left). YFL-mini1 (right) is a truncated variant lacking regions dispensable for minimal catalytic ability. In the structure of YFL-1 (middle), the arrow with a black arrowhead indicates the site downstream of which was removed in the Δ25 mutant. In the structure of YFL-mini1 (right), the nucleotide numbering for the catalytic unit and its surrounding elements is shown in red. (B) Time courses of the ligation reactions by the YFL-1 (filled circles) and its Δ25 mutant lacking the 25-nt-3′ssr (open squares). Reactions were performed in the presence of 30 mM Tris–HCl (pH 7.5), 50 mM MgCl2 and 200 mM KCl at 37°C. (C) Product yields of YFL-1 with the 25-nt-3′ssr, its variants lacking part of the 25-nt-3′ssr, and the Δ25 mutant lacking the whole 25-nt-3′ssr.
The second ribozyme, termed class YFL, was evolved to catalyze RNA ligation using the 5′-terminal phosphate activated by β-nicotinamide monophosphate (β-NMN) (Figure 1A, middle) (7). Biochemical analyses indicated that the YFL ribozyme has a versatile catalytic module that can utilize not only β-NMN but also PPi as a leaving group (7). The versatile catalytic properties of the artificial YFL ribozyme are interesting from the viewpoint of the molecular evolution of RNA ligase/polymerase ribozymes (9). According to one model of ribozyme evolution in the RNA world, primitive polymerase ribozymes may have utilized nucleotides as activated leaving groups because the evolution of RNA molecules capable of recognizing the nucleoside moiety would be easier than that capable of recognizing the anionic PPi moiety (9,10). As PPi is commonly used in modern RNA/DNA replication systems, primitive polymerase ribozymes utilizing nucleotide leaving groups had to evolve to an advanced form that utilized PPi leaving groups (7,9). From this viewpoint, the YFL ribozyme can be regarded as a model of intermediate species in the evolution of polymerase ribozymes, which should utilize both nucleotides and PPi as leaving groups.
However, the YFL ribozyme has drawbacks regarding its folding properties. In the reaction of the most abundant isolate of the class YFL ribozyme (YFL-1), the yield of the ligated products did not exceed 45% (Figure 1B) (7). This makes it difficult to biochemically determine the active structure of the catalytic unit. To overcome this drawback, we performed in vitro evolution of the YFL ribozyme to isolate variants with improved folding ability. We also performed rational engineering of variants isolated by in vitro evolution, which enabled us to elucidate the active secondary structure of the YFL ribozyme.
MATERIALS AND METHODS
Oligonucleotides
Unmodified DNA oligonucleotides were purchased from Hokkaido System Science (Hokkaido, Japan) and Fasmac (Tokyo, Japan). 5′-Carboxyfluorescein (FAM)-labeled DNA/RNA chimeric oligonucleotide and IRD700-labeled DNA oligonucleotide were purchased from IDT (Coralville, IA, USA) and LI-COR Biosciences (Lincoln, NE), respectively. 5′-Biotin-modified RNA oligonucleotides were purchased from Dharmacon (Lafayette, CO, USA).
Oligonucleotides
The sequences of the synthetic oligonucleotides used in the experiments are listed in Supplementary Data.
Library construction
Template DNAs were constructed as follows. The 3′-half fragment of the library was prepared by polymerase chain reaction (PCR). The pool-R oligonucleotide, including 28 nt of 9% (Pool-R1) or 21% (Pool-R2) degenerate sequence, was amplified by PCR with ExTaq DNA polymerase (Takara, Tokyo, Japan) using a sense primer (Fw-R) and antisense primer (Rv-R). The 5′-half fragment of the library was also prepared in the same manner. The pool-L oligonucleotides, including 5 nt of random sequence (Pool-L), was amplified by PCR using a sense primer (Fw-L) and an antisense primer (Rv-L). Respective PCR products were digested with BsaI (New England Biolabs, Ipswich, MA, USA). The PCR R fragments were ligated with the L fragments using T4 DNA ligase (Takara).
The resulting ligated DNAs were used as templates for in vitro transcription with T7 RNA polymerase under standard reaction conditions except the nucleotide composition consisted of 1 mM GTP, UTP and CTP, 0.1 mM ATP and 50 mM β-NADH (Sigma, St Louis, MO, USA). The transcripts of the library were used for in vitro selection experiments.
In vitro selection
The purified RNAs were dissolved in distilled H2O, and then denatured at 80°C for 5 min, followed by preincubation at 37°C for 3 min. RNA folding was initiated by adding 5× reaction buffer. After incubation at 37°C for 10 min, substrate RNA was added to start the ligation reaction. The final concentrations of the ribozyme and substrate were both 1 μM. The reaction was carried out with 30 mM Tris–HCl (pH 7.5), 50 mM MgCl2 and 200 mM KCl at 37°C. Two 5′-biotinylated substrate RNAs (Sub-1 and Sub-2) were used (6,7); Sub-1 was used in rounds 1, 3 and 5, while Sub-2 was used in rounds 2 and 4.
The reaction was stopped by ethanol precipitation. The ligated RNAs were captured on streptavidin paramagnetic particles (Promega, Madison, WI, USA) and hybridized with a reverse transcription primer complementary to the 3′ region of the pooled RNAs. Reverse transcription was performed with ReverTra Ace MMLV RNaseH-minus reverse transcriptase (Toyobo, Osaka, Japan) with two primers: Rv-1 in rounds 1, 3 and 5; Rv-2 in rounds 2 and 4. The resulting cDNAs were eluted with 150 mM KOH, followed by neutralization with 150 mM HCl. The resulting cDNAs were subjected to selective PCR with the Rv primer and selective primers complementary to the sequences of the substrate RNAs (1st Fw-1 in rounds 1, 3 and 5; 1st Fw-2 in rounds 2 and 4). The PCR products were electrophoresed on 2% agarose gels and the desired products were isolated using Nucleospin ExtractII (Macherey-Nagel, Deuren, Switzerland). PCR to regenerate templates as well as replace the P1 sequence was carried out with the primer, including the sequence of the T7Ф2.5 promoter (11) followed by the sequence of one of the two P1 regions (2nd Fw-1 in rounds 1, 3 and 5; 2nd Fw-2 in rounds 2 and 4). The amplified DNAs were digested with BanI (New England Biolabs) to remove the 3′ region. Using T4 DNA ligase, the digested PCR product was then ligated with the 3′ region fragment (3SSR-1 in rounds 1, 3 and 5; 3SSR-2 in rounds 2 and 4). Additional PCR was performed with the primers 2nd Fw-1 and Rv-1 in rounds 1, 3 and 5, and 2nd Fw-2 and Rv-2 in rounds 2 and 4. The resulting DNAs were used as templates for the following round. The selective PCR products from the fifth round were cloned into the pGEM-T vector (Promega). Individual plasmid clones were isolated and sequenced.
Preparation of variants of the YFL-1 ribozyme
Plasmids encoding variants of the YFL-1 ribozyme were constructed by PCR-based mutagenesis with plasmid pYFL-1 carrying the parent YFL-1 sequence as a template. DNAs encoding variants with mutations at positions R4, R8, R20 and R22 were prepared directly by PCR with appropriate antisense primers possessing the mutations, the sense primer possessing the T7 promoter sequence, and pYFL-1. DNAs encoding variants with truncation in the 25-nt-3′ssr were also generated directly by PCR with appropriate antisense primers, the sense primer, and pYFL-1.
Ligation activity assays
The ribozymes were dissolved in H2O, then denatured by incubation at 80°C for 5 min, followed by additional incubation at 37°C for 3 min. RNA folding was initiated by adding 5× reaction buffer. After folding at 37°C for 10 min, the reaction was initiated by adding the 5′-FAM-labeled substrate. The reaction conditions were as follows. Final ribozyme and substrate concentrations were both 1 μM. The reactions were carried out at 37°C in the presence of 30 mM Tris–HCl (pH 7.5), 200 mM KCl, and MgCl2 at an appropriate concentration. For the standard reaction conditions, we employed 50 mM MgCl2. Aliquots were taken at various time points, and treated with a half volume of stop solution consisting of 80% formamide and 100 mM EDTA. Samples were separated on 12% polyacrylamide denaturing gels and quantified with a Pharos FX FluoroImager (Bio-Rad, Hercules, CA, USA). Rate constants were determined from at least two independent experiments.
Apparent second-order rate constants for the bimolecular ligation reactions between 1 μM ribozyme and 1 μM substrate was calculated using the following equation (7):
where t is time, k is the apparent second-order rate constant (kapp), and Fa is the calculated final yield.
DMS modification
DMS modification was performed according to the published procedure (12). For the experiment, we first prepared a reaction mixture of the substrate RNA and a variant YFL-mini2SL ribozyme possessing a single-stranded region (5′-AGGUGUAUGCAUGGACAGUGCCAUGUGCUUCAGCUAAGUGUCAAAGUUUCCUGUGUGAAAUUGUUAUCCG-3′), which was added as a priming site for reverse transcription. The ligation reaction (250 μl) was carried out at 37°C for 18 h in the presence of the variant YFL-mini2SL ribozyme (1 μM), the substrate RNA (1 μM), 30 mM Tris–HCl (pH 7.5), 200 mM KCl and 150 mM MgCl2. One microliter of 10% DMS in ethanol was added to the resulting reaction mixture, followed by incubation for 5 min at 37°C. Reactions were stopped with 100 μl of 1 M DTT and RNAs were precipitated by adding 25 μl of 3 M NaOAc and 1 ml of EtOH. After treatment of the reaction mixture with DMS, the ligated RNA was isolated by denaturing 5% polyacrylamide gel electrophoresis. Aliquots of the ligated RNA (3.5 pmol) were subjected to reverse transcription with ReverTra Ace with the IRD700-labeled DNA primer complementary to the last 25 nt of the single-stranded region. The resulting cDNAs were electrophoresed and analyzed using a LI-COR DNA Analyzer Model 4300 (LI-COR Biosciences).
RESULTS
Effects of the 25-nt 3′ single-stranded region on YFL-1 ribozyme activity
Previously, we characterized the basic properties of the YFL-1 ribozyme (7). We have reduced the size of the YFL-1 ribozyme by deleting elements that are not essential for catalysis (Figure 1A). The resulting variant [designated as the quadruple mutant in the original report (7) and renamed YFL-mini1 in the present study, Figure 1A, right] retained its activity. However, the apparent second-order rate constant (kapp) and product yield (18 h) of YFL-mini1 were 5.0- and 2.2-fold lower than those of the parent YFL-1 ribozyme, respectively (7).
To identify the element responsible for the activity of the parent YFL-1, we carried out deletion analysis of the 3′ single-stranded region 25 nt in length (termed 25-nt-3′ssr) of the parent YFL-1. Deletion analysis indicated that the activity of the YFL-1 decreased markedly with removal of the last 21 nt or more of the 25-nt-3′ssr (Figure 1B and C). On the other hand, removal of the last 16 or 19 nt showed no negative effect on the activity (Figure 1C).
Optimizing the catalytic module of a small derivative of the YFL ribozyme
To simplify the structure–function relationship of the YFL ribozyme to facilitate its biochemical analysis, we carried out in vitro evolution of YFL-mini1 to obtain variants the activity of which would be not only higher than the original YFL-1 but that would also be independent of the particular nucleotide sequence of the 3′ single-stranded region.
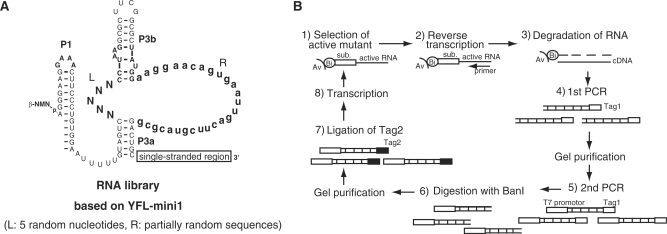
The starting pool was designed based on the YFL-mini1 ribozyme. A single-stranded region as a priming site for RT-PCR was attached to the 3′ end of the ribozyme (Figure 2A). To isolate the variants with activities that were independent of particular nucleotide sequence in the 3′ single-stranded region, two sequences were used as alternative RT-PCR priming sites. The resultant pool was composed of variants of the YFL-mini1, in which the L region (5 bases) was randomized, whereas the R region (28 bases) was partially mutagenized with 9% or 21% degeneracy. The pooled RNA was subjected to five rounds of in vitro evolution consisting of selection and amplification under the conditions listed in Table 1 (Figure 2B), and 21 clones were randomly isolated and sequenced (Figure 3A).
Figure 2.
In vitro evolution of the YFL-mini1 ribozyme. (A) Starting pool designed based on YFL-mini1. Five nucleotides in the L region were replaced with random nucleotides (N). The R region was partially mutagenized with 9% or 21% degeneracy (lowercase letters). (B) Scheme for in vitro evolution to identify variants, activity of which is independent of the 3′-end sequence. (1) The RNA pool was incubated with a biotinylated substrate and active variants were selectively retained on streptavidin magnetic beads. (2) Active variants were preferentially amplified by reverse transcription. (3) After degradation of RNA, (4 and 5) cDNAs complementary to active variants were amplified by PCR. (6) The 3′ Tag region of resulting DNAs, corresponding to the 3′ssr for RT-PCR in the RNA sequence, was digested with BanI. (7) An alternate Tag sequence was attached by T4 ligase. (8) The resulting DNAs were transcribed and used for the next round.
Table 1.
Selection conditions
| Round | RNA pool (μM) | Substrate (μM) | KCl (mM) | MgCl2 (mM) | Reaction time | Temp. (°C) | 3′ssr |
|---|---|---|---|---|---|---|---|
| 1 | 1.0 | 1.5 (S-1) | 200 | 50 | 18 h | 37 | 1 |
| 2 | 1.0 | 1.5 (S-2) | 200 | 50 | 2 h | 37 | 2 |
| 3 | 1.0 | 1.5 (S-1) | 50 | 25 | 10 min | 37 | 1 |
| 4 | 1.0 | 1.0 (S-2) | 50 | 25 | 5 min | 37 | 2 |
| 5 | 1.0 | 1.0 (S-1) | 0 | 5 | 5 min | 30 | 1 |
Figure 3.
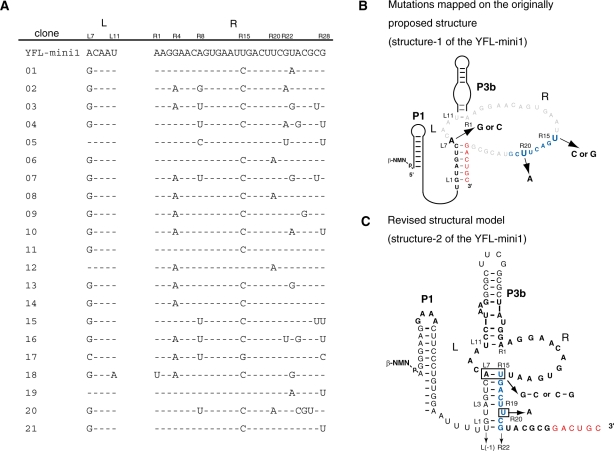
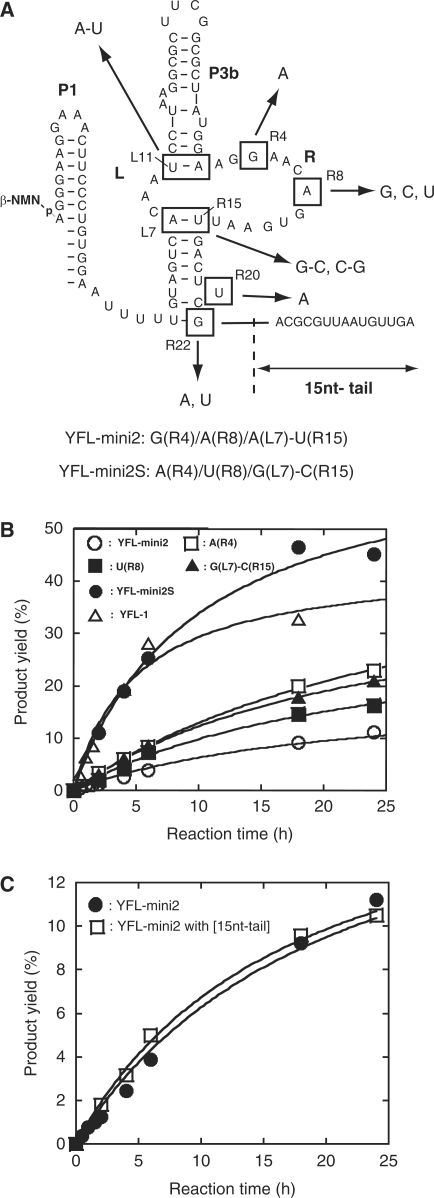
Sequence comparison of YFL-mini1 and its variants isolated by in vitro evolution. (A) Sequences of 21 clones from the pool after the 5th round. Dashes indicate the bases identical to those of the parent YFL-mini1. (B) Base substitutions found in the isolated clones were mapped on the originally proposed structure (structure-1) of YFL-mini1. (C) The revised structure model derived from the phylogenetic comparison of the isolated clones (structure-2).
Sequence comparison indicated that simultaneous substitutions occurred at A(L7) of the L region and U(R15) of the R region, which were changed to G(L7) and C(R15) in 17 clones and C(L7) and G(R15) in one clone (Figure 3B). These observations strongly suggested that position L7 forms a Watson–Crick base pair with position R15 (Figure 3C). This can be a part of five consecutive base pairs between L3-L7 and R15-R19 (Figure 3C). Moreover, U to A substitution at position R20, which was found in four clones, forms an additional base pair (Figure 3C), by which L(-1)-L7 and R15-R22 should form eight consecutive base pairs. These data strongly suggested that the catalytic module of the YFL-mini1 ribozyme is smaller than originally proposed (Figure 3C).
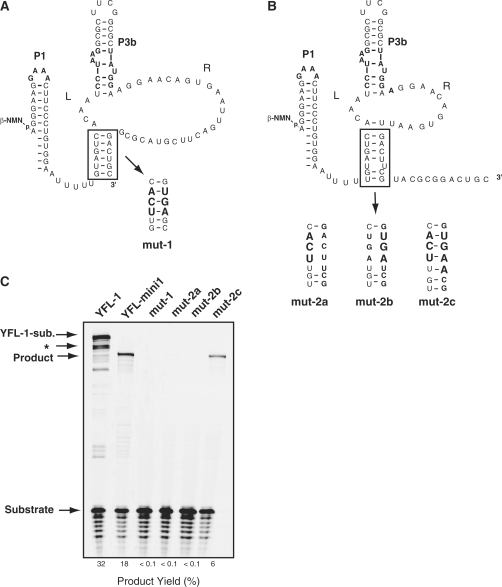
To address the actual secondary structure of YFL-mini1, we prepared four mutants based on the YFL-mini1 ribozyme (Figure 4). The mut-1 mutant was designed to selectively form the originally proposed structure (structure-1) (Figure 4A), whereas the mut-2c mutant was designed to selectively form the revised structure (structure-2) (Figure 4B). Both mut-2a and mut-2b were designed to disrupt the stem region of structure-2 (Figure 4B). The results of activity analyses indicated that mut-2c promotes ligation, whereas the other three mutants showed no detectable activity (Figure 4C). These results further support the suggestion that structure-2 corresponds to the active structure of the YFL-mini1 ribozyme. The mut-2c variant was less active than the parent YFL-mini1. Such incomplete complementation of P2 base pairs was also observed in analysis of the P2 regions of the DSL ribozyme (6) and the YFL-1 ribozyme (7). These observations suggest that the scaffold structure may prefer the original P2 sequence.
Figure 4.
Examination of catalytic abilities of structures-1 and -2. (A, B) Secondary structures of YFL-mini1 and its mutants. mut-1 (A) was designed to form the originally proposed structure (structure-1). mut-2c (B) was designed to form the revised structure (structure-2). mut-2a and mut-2b (B) were designed to disrupt the stem region of structure-2. (C) Reactions of YFL-1, YFL-mini1 and the four mutants of YFL-mini1. Reactions were performed with 30 mM Tris–HCl (pH 7.5), 100 mM Mg2+, 200 mM K+ at 37°C. ‘YFL-1-sub’. indicates the ligated product of the YFL-1 ribozyme and the substrate oligonucleotide. ‘Product’ indicates the ligated product of the substrate and the YFL-mini1 or its mutant. The asterisk indicates the product presumably formed by specific hydrolytic cleavage of the ligated product of YFL-1, which should be promoted by the RNA tertiary structure (22).
Reinvestigation of 3′ssr of the parent YFL-1
Based on the possibility that structure-2 corresponds to the class YFL ribozyme active structure, we reinvestigated the role of the 3′ssr in the activity of the parent YFL-1 ribozyme (Figure 5). We predicted the secondary structure of the revised 3′ssr with 52 nt (termed 52-nt-3′ssr) using the mfold program (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) (13). The predicted structure showed that the region forms two hairpin stem–loops (Figure 5B). The first hairpin (stem-1) involves nucleotides (R41–R46) designed to form the original P3a base pairs (see Figure 1A). Reconsideration of the effect of the 3′ssr revealed that relatively active variants (full-length, Δ16, Δ19 and Δ20 in Figure 1C) seem to form a stable stem-1 structure (Figure 5B). On the other hand, in weakly active variants (Δ21–Δ25 in Figure 1C), the stem-1 structure should be unstable or missing (Figure 5B). Thus, comparison of the predicted structures of the 52-nt-3′ssr and the ligation activity of the corresponding variants suggested the importance of the stem-1 structure for YFL-1 ribozyme activity. If structure-2 corresponds to the active structure, a liberated 3′ strand of the original P3a (positions G(R41)-C(R46) in Figure 5A) in turn could act as an inhibitory element that can competitively regenerate the original but inactive P3a base pairs with G(L1)-C(L6). Formation of such an inactive structure, however, may be suppressed by the stem-1 structure in the 52-nt-3′ssr because it should mask the 3′ strand of the original P3a (Figure 5B). According to this model, we hypothesized that weakly active Δ25 mutant should be reactivated by removal of nucleotides R41–R46. We prepared and examined two mutants lacking the nucleotides R41–R46 (Figure 5B). Two mutants (Δ37 and Δ52) were distinctly more active than Δ25 mutant. Interestingly, Δ37 was more active than Δ52 (Figure 5C), suggesting that the 15-nt single-stranded region (15-nt-tail, see Figure 5B) plays a positive role. In addition, base substitutions in the stem region of stem-1 (mismatch variant in Figure 5B) resulted in severe reduction of activity (Figure 5C). This result also supports the model in which the stem-1 structure acts as a suppressor, which prevents formation of the inactive structure of YFL-1.
Figure 5.
Possible stem structure in the 52-nt-3′ssr. Sequence (A) and possible secondary structure (B) of 52-nt-3′ssr. The secondary structure was predicted using the mfold program. Arrows with black arrowheads indicate the truncation positions. (C) Activities of the parent YFL-1 and its mutants to examine the effects of 52-nt-3′ssr. (D) Activities of the parent YFL-1 and its mutants to examine the effects of the 15-nt-tail. Δ37–comp. oligo indicates the reaction of the Δ37 mutant in the presence of the indicated concentrations of a DNA oligonucleotide complementary to the 15-nt-tail region.
Additional experiments were carried out to clarify the role of the 15-nt-tail (Figure 5D). Addition of an oligonucleotide complementary to the 15-nt-tail decreased the activity of Δ37 (Figure 5D). On the other hand, substitution of the 15-nt-tail with poly (A)15 resulted in modest improvement of ribozyme activity (Figure 5D).
Effects of the mutations acquired by in vitro evolution
Sequence comparison and mutation analyses strongly suggested that the catalytic unit of the YFL-1 ribozyme involves an asymmetric loop composed of 3 and 13 nt (structure-2 in Figures 3C and 4B). Significant activity of the Δ52 mutant indicated that the intrinsic activity of the catalytic unit was independent of the 52-nt-3′ssr (Figure 5C).
Sequence comparison also revealed that mutations acquired by in vitro evolution could be categorized into 4 nt and two base pairs (Figure 6A). We constructed a series of variants of YFL-mini2 possessing one of these mutations, and analyzed their activities (Figure 6). A single mutation at position R20 (U to A) or R22 (G to A), or a base pair substitution of L11-R1 (U-A to A-U) showed almost no effect on the activity (data not shown). However, the mutants with a nucleotide substitution at positions R4 (G to A), R8 (A to U) or a base pair substitution at L7-R15 (A-U to G-C) were more active than the parent YFL-mini2, although the degrees of improvement in the activity by each mutation were modest (Figure 6B). However, the mutant with all of these mutations simultaneously (designated YFL-mini2S) exhibited significantly improved activity, which was comparable to that of YFL-1 (Figure 6B). The 15-nt-tail that improved the activity of Δ52 (Figure 5C) had no effect on the activity of YFL-mini2S (Figure 6C).
Figure 6.
Effects of mutations in the selected clones. (A) Mutations isolated by in vitro evolution. They were categorized into six positions in a shortened form of the YFL-1 (YFL-mini2). The YFL-mini2S mutant is the most active mutant of YFL-mini2, which possesses two nucleotide substitutions [G(R4)A and A(R8)U] and a base pair substitution [A(L7)G-U(R15)C]. (B) Activities of YFL-mini2 and its variants lacking the 15-nt tail. (C) Activities of the YFL-mini2 and its mutant with the 15-nt tail.
The optimized catalytic unit was independent of the 3′ssr
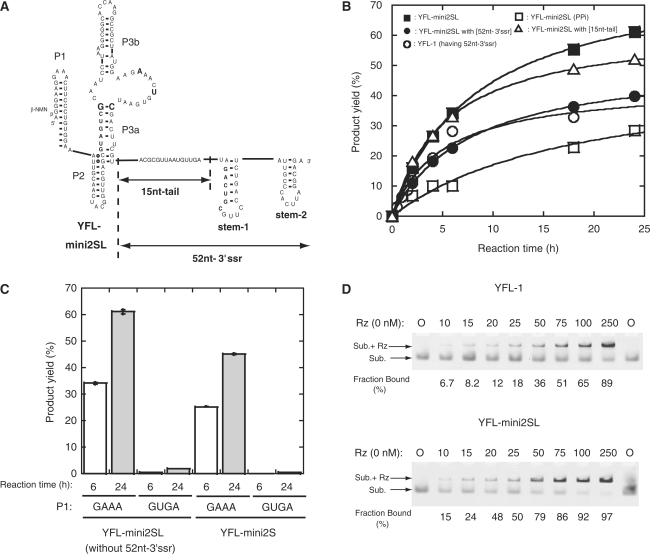
To examine whether the catalytic unit of YFL-mini2S can be installed in the original Type-B scaffold, we transplanted the catalytic unit of YFL-mini2S into the original scaffold structure of the YFL-1 ribozyme possessing the P2 region and L3b loop. The resulting variant (YFL-mini2SL, Figure 7) was more active than the YFL-mini2S with a 5′-AUUUU-3′ linker at the P2 region (Figure 6A). Moreover, the reaction time course of the YFL-mini2SL was comparable to that of cis-DSL-01, a modular ligase ribozyme shown to have excellent folding properties (6,7). We also investigated the effects of the 52-nt-3′ssr on the activity of YFL-mini2SL. The activity of YFL-mini2SL possessing the 52-nt-3′ssr was lower than that lacking the 3′ssr (Figure 7B). Specificity of the leaving group in the ligation reaction was also examined for YFL-mini2SL. A variant with PPi as a leaving group was catalytically active but its activity was substantially lower than that of the parent YFL-mini2SL using β-NMN (Figure 7B). We also examined the effects of the 15-nt-tail but addition of the tail had no positive effect on the activity of YFL-mini2SL (Figure 7B).
Figure 7.
Effects of 3′ssr on the YFL-mini2SL ribozyme. (A) Schematic representation of the secondary structure of YFL-mini2SL with or without the 52-nt-3′ssr. (B) Time courses of ligation reactions catalyzed by YFL-1, YFL-mini2SL and YFL-mini2SL variants. YFL-mini2SL (PPi) indicates a variant of YFL-mini2SL that uses inorganic pyrophosphate (PPi) as a leaving group. (C) Activities of the YFL-mini2SL and YFL-mini2S variants with mutations in the P1 loop. (D) Gel mobility shift assays of YFL-1 or YFL-mini2SL with the substrates in the presence of 50 mM Mg2+ ions at pH 7.5. The FAM-labeled substrate unit the 3′-end of which has deoxyribose (25 nM) was titrated with various concentrations of the ribozyme (0–250 nM). Rz and Sub. indicate the ribozyme and substrate, respectively. Upper and lower bands correspond to the substrate–ribozyme complex (Sub. + Rz) and free substrate (Sub.), respectively. Experiments were performed according to the published protocol (23).
To determine whether the scaffold structure was maintained in the revised structure (structure-2), derivatives lacking the interaction between GAAA at P1 and its receptor at P3b were constructed and their activities were examined (Figure 7C). The resulting variants (P1 GUGA) of YFL-mini2SL and YFL-mini2S both showed little activity (Figure 7C). Therefore, the tertiary structure of the scaffold, which was held by GAAA-11ntR interaction and P2 region (Figure 1A), supported the activity of the YFL-2SL ribozyme.
We investigated the physical affinities between the ribozyme and substrate by gel mobility shift assays with a modified substrate having a 3′-deoxy terminus (Figure 7D). In the presence of 50 mM Mg2+ ions, the concentrations of ribozyme where half of the fluorescently labeled substrate formed a complex with the ribozyme were ∼75 nM for YFL-1 (Figure 7D, top) and 25 nM for YFL-mini2SL (Figure 7D, bottom). Thus, the stability of the substrate–ribozyme complexes of YFL-mini2SL, which appears better than that of YFL-1, may be related to improvement of the product yield in the reaction with YFL-mini2SL.
DMS modification of the YFL-mini2SL
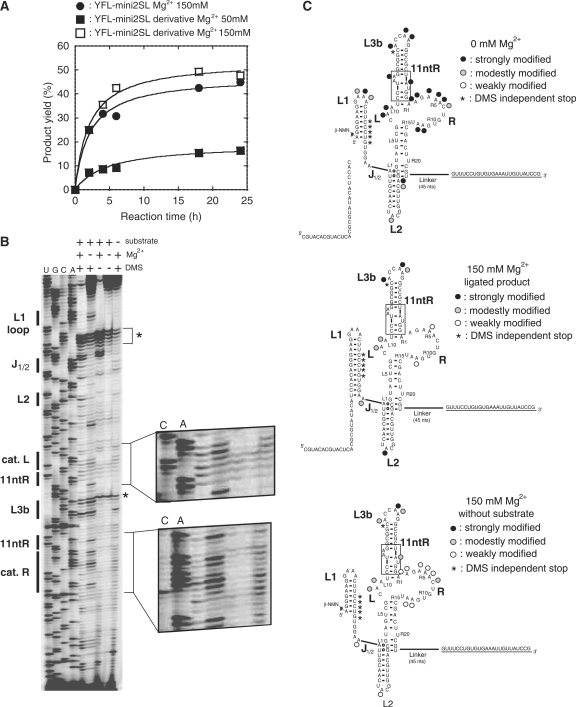
As the product yield of YFL-mini2SL was better than that of the parent YFL-1 and comparable to that of the class DSL ribozyme, YFL-mini2SL was subjected to DMS modification to analyze its secondary and tertiary structures (Figure 8A). We added a single-stranded sequence for primer extension using a dye-labeled M13 reverse primer (Figure 8C). Unfortunately, the activity of the resulting derivative was low in the presence of 50 mM MgCl2 (Figure 8A). In the presence of 150 mM MgCl2, however, the activity of the derivative became similar to that of the parent YFL-mini2SL (Figure 8A). The activity of the parent YFL-mini2SL with 150 mM MgCl2 was also similar to that with 50 mM MgCl2 (Figures 7B and 8A). In the absence of MgCl2, the overall pattern of DMS modification was consistent with structure-2 (Figure 8C, top). To obtain structural information regarding the active ribozyme, the YFL-mini2SL derivative was incubated under the conditions for ligation reaction in the presence of the substrate RNA and 150 mM MgCl2. The resulting reaction mixture was subjected directly to DMS modification to avoid perturbation of its active structure (14). After isolation of the DMS-modified ligated product by gel electrophoresis, reverse transcription of the ligated product was performed. The results of reverse transcription indicated that the positions of three adenines in the L1 loop and two adenines in the 11-ntR motif were protected from methylation (Figure 8). As protection of these adenines is regarded as a signature of GAAA-11-ntR interaction (15,16), this observation was consistent with the results of mutation analysis shown in Figure 7C. While DMS modification at L3b loop was comparable in the presence and absence of Mg2+ ions (Figure 8B), modification at the catalytic unit was distinctly weaker than that in the absence of MgCl2 (Figure 8B), suggesting that the catalytic unit forms an active tertiary structure that would be induced by Mg2+ ions. DMS modification pattern of the ribozyme without substrate was similar to that of the ligated product but accessibility of DMS to catalytic units in the absence of the substrate appeared higher than that of the ligated product (Figure 8C).
Figure 8.
DMS modification of a derivative of YFL-mini2SL. (A) Time courses of the ligation reactions of YFS-mini2SL and its variant for DMS analysis. (B) DMS modification of a derivative of YFL-mini2SL. Asterisks indicate the stops of reverse transcription that occur independently from DMS modification. (C) Positions and extents of DMS modification in the absence (top) or presence (middle and bottom) of 150 mM MgCl2. In the absence of MgCl2, DMS modification was performed with the ribozyme in the presence of the substrate. In the presence of 150 mM MgCl2, DMS modification was performed with either the ligated product produced in situ (middle) or with the ribozyme in the absence of the substrate (bottom).
DISCUSSION
In vitro evolution of the YFL-1 ribozyme with improved folding ability
In this study, we performed in vitro evolution of the class YFL ribozyme to facilitate elucidation of its structure–function relationship and identified YFL-mini2S as a small but active variant. YFL-mini2S was active without the 3′ssr, which was important for optimal activity of the parent YFL-1. Phylogenetic comparison together with biochemical analyses of the YFL-1, YFL-mini2S, and other variants revealed that the catalytic module forms an asymmetric internal loop with 3 and 13 nt.
The reaction of YFL-mini2S lacking the P2 stem and 3′ssr was comparable to that of the parent YFL-1 (Figure 6B). The product yield of YFL-mini2SL possessing the P2 stem was better than that of the parent YFL-1 (Figure 7B) and comparable to that of the cisDSL-01 ribozyme with excellent folding properties (6,7). The improved product yield of YFL-mini2SL was partly due to the enhanced substrate binding activity (Figure 7D). As DMS was more accessible to the catalytic unit in the absence of the substrate than in the ligated product form (Figure 8), tertiary folding of the active catalytic unit may act in concert with the substrate binding in YFL-mini2SL. In contrast to the significant improvement of folding properties, the kapp values of YFL-mini2S (0.10 μM–1 h–1 calculated from Figure 6B) and YFL-mini2SL (0.12 μM–1 h–1 calculated from Figure 7B) were similar to that of the parent YFL-1 (0.20 μM–1 h–1) (7). These values were still 50–160-fold smaller than the typical reaction rates of artificial RNA ligase ribozymes (0.12–0.37 min–1) with PPi leaving groups (17). On the other hand, the moderate catalytic ability of the YFL ribozyme may be comparable with that of a shortened (and presumably ancestral) form of the group I intron ribozyme possessing P3–P7 helices as a minimal catalytic domain and lacking the P4–P6 helices (18). The reaction rate of the corresponding mutant of the T4td group I ribozyme (0.10 h–1), which was comparable to that of the YFL ribozyme (0.14 h–1), was 103-fold smaller than that of the full-length T4td group I ribozyme (18).
Structure of the catalytic unit and modular organization of the YFL ribozyme
Phylogenetic comparison and biochemical analysis based on in vitro evolution revealed the actual secondary structure of the catalytic unit of the YFL ribozyme as well as an important role of its 3′ssr. The catalytic unit is composed of an asymmetric internal loop with 3 and 13 nt in the L and R regions, respectively (Figures 4B and 6A). This structure was much shorter than the initial design of the RNA library, which had 5 and 40 randomized nucleotides in the L and R regions, respectively (see YFL-1 in Figure 1A) (7). Significant truncation of the R region was due to restructuring of P3a base pairs, in which the original 3′ strand was replaced with the sequence within the original R region consisting of 40 nt (see YFL-1 in Figure 1A). The liberated 3′ strand of the original P3a in turn acts as an inhibitory element that can competitively regenerate the original but inactive base pairs. Formation of such an inactive structure, however, was suppressed by a secondary structure in the 3′ssr masking the 3′ strand of the original P3a (Figure 5B).
The activity of the Δ52 mutant was improved by attaching a 15-nt-tail (the resulting variant was Δ37, Figure 5D). This tail, however, had no effect on YFL-mini2SL, the catalytic unit of which was evolutionarily optimized on the scaffold structure with no P2 stem (Figure 2B). As the tail also had no effect on the activity of YFL-mini2 with the 5′-AUUUU-3′ linker instead of the P2 region (Figure 6C), activation by the 15-nt-tail appears to require the P2 region. The activation was also canceled by addition of an oligonucleotide complementary to the 15-nt-tail, suggesting that the tail is involved in an interaction. However, no particular sequence of the 15-nt-tail seems to be required for activation because the activity of the Δ52 mutant was also improved by adding the (A)15 tail (Figure 5D). Taken together, these observations suggest that the 15-nt-tail may stabilize the structural scaffold by forming a specific kind of interaction, such as a triple helix with the P2 stem. The triple helix motif contributes the original Type B scaffold (Figure 1A-left), but whether a similar structure exists in the revised structure is unclear (Figure 5A). In the revised structure of the YFL-1, the 15-nt-tail in Δ37 and (A)15 tail in Δ52 + (A)15 interacted with the P2 stem, but the Δ52 mutant never showed such an interaction due to complete removal of the tail.
The Δ16 mutant also gave a better product yield than the parent YFL-1 (Figure 1C), suggesting that the stem-2 region plays a negative role or the last 16 nt induces an alternative structure that should cancel the positive effects of the 15-nt-tail (see predicted secondary structure in Supplementary Figure S1).
Modular in vitro selection using the type B self-folding RNA as a structural scaffold
Structurally defined RNAs bearing a tertiary interaction between the GAAA loop and its receptor have been designed and employed as structural scaffolds to generate novel ribozymes, aptamers and self-assembling RNAs (6,7,19–21). By inserting random nucleotides at the same position of the P3a–P3b helical domain of the type B self-folding RNA, closely related RNA libraries were designed, from which two classes of modular ribozyme and one class of modular aptamer were isolated (6,7,19). While successful isolation of three classes of functional RNAs suggested the utility of the type B-based libraries, structural scaffolds of two classes of RNA were reconstructed during in vitro evolution by employing particular portions of random sequences (7,9). This observation suggested that the degrees of sequence and structural diversity of the current type B-based libraries are still limited, presumably because the structural constraints are too stringent. Further improvements in the current design of type B-based libraries are required to maximize sequence and structural diversity and also to minimize unexpected structural rearrangements during in vitro evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aids on Innovative Areas ‘Emergence in Chemistry’ (No.21111518 to Y.I.); for Exploratory Research (No.19657071 to Y.I.); for JSPS Fellows (No. 08J02792 to Y.F.); and for the Global COE Program ‘Science for Future Molecular Systems’ (to H.F.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan. Funding for open access charge: PRESTO, Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency (JST).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Campbell ID, Baron M. The structure and function of protein modules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1991;332:165–170. doi: 10.1098/rstb.1991.0045. [DOI] [PubMed] [Google Scholar]
- 2.Campbell ID, Downing AK. Building protein structure and function from modular units. Trends Biotechnol. 1994;12:168–172. doi: 10.1016/0167-7799(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 3.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 4.Jaeger L. The new world of ribozymes. Curr. Opin. Struct. Biol. 1997;7:324–335. doi: 10.1016/s0959-440x(97)80047-4. [DOI] [PubMed] [Google Scholar]
- 5.Westhof E, Masquida B, Jaeger L. RNA tectonics: towards RNA design. Fold. Des. 1996;1:R78–R88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 6.Ikawa Y, Tsuda K, Matsumura S, Inoue T. De novo synthesis and development of an RNA enzyme. Proc. Natl Acad. Sci. USA. 2004;101:13750–13755. doi: 10.1073/pnas.0405886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita Y, Furuta H, Ikawa Y. Tailoring RNA modular units on a common scaffold: a modular ribozyme with a catalytic unit for β-nicotinamide mononucleotide-activated RNA ligation. RNA. 2009;15:877–888. doi: 10.1261/rna.1461309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voytek SB, Joyce GF. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. Proc. Natl Acad. Sci. USA. 2007;104:15288–15293. doi: 10.1073/pnas.0707490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellington AD, Chen X, Robertson M, Syrett A. Evolutionary origins and directed evolution of RNA. Int. J. Biochem. Cell Biol. 2009;41:254–265. doi: 10.1016/j.biocel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Hager AJ, Szostak JW. Isolation of novel ribozymes that ligate AMP-activated RNA substrates. Chem. Biol. 1997;4:607–617. doi: 10.1016/s1074-5521(97)90246-5. [DOI] [PubMed] [Google Scholar]
- 11.Huang F. Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res. 2003;31:e8. doi: 10.1093/nar/gng008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumura S, Ikawa Y, Inoue T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 2003;31:5544–5551. doi: 10.1093/nar/gkg760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic. Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue T, Cech TR. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl Acad. Sci. USA. 1985;82:648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy FL, Cech TR. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993;32:5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- 16.Ikawa Y, Fukada K, Watanabe S, Shiraishi H, Inoue T. Design, construction, and analysis of a novel class of self-folding RNA. Structure. 2002;10:527–534. doi: 10.1016/s0969-2126(02)00739-6. [DOI] [PubMed] [Google Scholar]
- 17.Joyce GF. Forty years of in vitro evolution. Angew. Chem. Int. Ed. Engl. 2007;46:6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- 18.Ikawa Y, Shiraishi H, Inoue T. Minimal catalytic domain of a group I self-splicing intron RNA. Nat. Struct. Biol. 2000;7:1032–1035. doi: 10.1038/80947. [DOI] [PubMed] [Google Scholar]
- 19.Shiohara T, Saito H, Inoue T. A designed RNA selection: establishment of a stable complex between a target and selectant RNA via two coordinated interactions. Nucleic Acids Res. 2009;37:e23. doi: 10.1093/nar/gkn1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeger L, Leontis NB. Tecto-RNA: one-dimensional self-assembly through tertiary interactions. Angew. Chem. Int. Ed. Engl. 2000;39:2521–2524. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 21.Yoshioka W, Ikawa Y, Jaeger L, Shiraishi H, Inoue T. Generation of a catalytic module on a self-folding RNA. RNA. 2004;10:1900–1906. doi: 10.1261/rna.7170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa J, Matsumura S, Jaeger L, Inoue T, Furuta H, Ikawa Y. Rational optimization of the DSL ligase ribozyme with GNRA/receptor interacting modules. Arch. Biochem. Biophys. 2009;490:163–170. doi: 10.1016/j.abb.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.