Abstract
γ-Glutamylamine cyclotransferase (GGACT) is an enzyme that converts γ-glutamylamines to free amines and 5-oxoproline. GGACT shows high activity toward γ-glutamyl-ϵ-lysine, derived from the breakdown of fibrin and other proteins cross-linked by transglutaminases. The enzyme adopts the newly identified cyclotransferase fold, observed in γ-glutamylcyclotransferase (GGCT), an enzyme with activity toward γ-glutamyl-α-amino acids (Oakley, A. J., Yamada, T., Liu, D., Coggan, M., Clark, A. G., and Board, P. G. (2008) J. Biol. Chem. 283, 22031–22042). Despite the absence of significant sequence identity, several residues are conserved in the active sites of GGCT and GGACT, including a putative catalytic acid/base residue (GGACT Glu82). The structure of GGACT in complex with the reaction product 5-oxoproline provides evidence for a common catalytic mechanism in both enzymes. The proposed mechanism, combined with the three-dimensional structures, also explains the different substrate specificities of these enzymes. Despite significant sequence divergence, there are at least three subfamilies in prokaryotes and eukaryotes that have conserved the GGCT fold and GGCT enzymatic activity.
Keywords: Crystal Structure, Enzyme Mechanisms, Enzyme Structure, Enzymes, Gene Structure, 5-Oxoproline, γ-Glutamylcyclotransferase Fold, γ-Glutamyl-ϵ-lysine Dipeptides, γ-Glutamylamine Cyclotransferase
Introduction
Proteins can be cross-linked via the side chains of glutamine and lysine by transglutaminases. This reaction results in the formation of ammonia and an l-γ-glutamyl-l-ϵ-lysine isopeptide bond linking the two polypeptide chains. The existence of such γ-glutamyl-ϵ-lysine links was unambiguously demonstrated in factor XIIIa-cross-linked fibrin: extensive proteolytic degradation of cross-linked fibrin resulted in the formation of l-γ-glutamyl-l-ϵ-lysine (1). It was concluded that l-γ-glutamyl-l-ϵ-lysine is not broken down by conventional proteolysis. The breakdown of the isodipeptide is instead catalyzed by γ-glutamylamine cyclotransferase (GGACT),2 first purified from rabbit kidney (2, 3). The partially purified enzyme was demonstrated to be active toward a range of l-γ-glutamyl conjugates with mono- and polyamines and amino acids. The action of GGACT in all cases is to cyclize the γ-glutamyl moiety, producing 5-oxo-l-proline and the free alkylamine. No activity was detected toward l-glutamine or l-β-aspartyl-l-ϵ-lysine. Furthermore, derivatives of l-γ-glutamyl-l-ϵ-lysine in which the α-amino or α-carboxyl functional group of the glutamyl moiety is blocked do not serve as substrates, nor do any of a range of l-γ-glutamyl-l-α-amino acids (3). Based on these results, it was proposed that GGACT functions in the latter stages of the catabolism of the products of transglutaminases (3).
We reported recently the identification, cloning, and three-dimensional structure of an enzyme with related catalytic activity to GGACT but with distinct specificity: γ-glutamylcyclotransferase (GGCT) (4). Unlike GGACT, this enzyme is active toward a range of l-γ-glutamyl-α-amino acids (5, 6). GGCT catalyzes the penultimate step in glutathione catabolism, whereby l-γ-glutamyl-l-α-cysteine and other l-γ-glutamyl-l-α-amino acid dipeptides formed by the γ-glutamyl cycle are catabolized to 5-oxo-l-proline and a free amino acid. (l-Glutamic acid is subsequently formed from 5-oxo-l-proline by the action of 5-oxoprolinase.) We demonstrated that GGCT is a homodimer of 20,994-Da subunits and has a distinctive mixed α/β-topology with six β-strands, five α-helices, and four short 310 helices, with strands β1–5 forming a barrel structure (4). This fold has also been observed in several proteins of unknown function from animals, plants, bacteria, and archaea. One such structure, solved by the Joint Center for Structural Genomics Consortium, is a mouse protein (AIG2-like domain 1; Protein Data Bank code 1VKB) of unknown function (7). The gene encoding this protein is distinct from the mouse gene encoding GGCT. Furthermore, a homolog of mouse AIG2-like domain 1 known as A2LD1 was predicted to exist in the human genome. The structure of the mouse protein possesses a cleft similar to the proposed active site of human GGCT. Tantalizingly, the proposed catalytic residue (Glu98 in human GGCT) and other active-site residues are structurally conserved in the mouse protein and in the amino acid sequence of its human homolog. These observations led us to the hypothesis that this mouse protein and its human homolog are also cyclotransferases, possibly the long sought after GGACT. Here, we describe the cloning, expression, purification, catalytic activity, and three-dimensional structure of human A2LD1 and positively identify it as GGACT. A common mechanism is proposed to act in both GGCT and GGACT. The proposed mechanism, combined with the three-dimensional structural data, also explains the different substrate specificities of these enzymes.
EXPERIMENTAL PROCEDURES
Cloning
A human expressed sequence tag clone (GenBankTM accession number BU156875) from a melanoma cell line was identified by sequence alignment with a mouse protein with a GGCT-like fold (Protein Data Bank code 1VKB). The clone was obtained from the I.M.A.G.E. Consortium, and the coding region of the cDNA was amplified with primers GGACT- F (5′-CTCCGCGGTGGAATGGCCCTAGTCTTCGTG-3′) and GGACT-R (5′-CTAAGCTTATCATCTGTTCTCCCGGGG-3′) and cloned into pGEM-T (Promega). The pGEM-T clone was sequenced by the Australian Cancer Research Foundation Biomedical Resource Facility of the John Curtin School of Medical Research to confirm the sequence. The cloned cDNA was excised from the pGEM-T vector with KspI and HindIII and then ligated into the same sites in the pHUE vector (8). The resulting clone, pHUE-GGACT, was transfected into Escherichia coli BL21(DE3) for protein expression. Production of E82Q and E82A mutants was undertaken using a QuikChange site-directed mutagenesis kit (Stratagene). The mutations were confirmed by DNA sequencing.
Expression and Purification
Expression of recombinant enzymes in pHUE allows the rapid purification of the expressed protein by nickel-agarose chromatography and the subsequent removal of all additional N-terminal residues by cleavage with the catalytic domain of mouse ubiquitin-specific protease 2 as described in detail previously (8). The recombinant enzyme was dialyzed in 20 mm Tris-HCl (pH 8.0) and 0.1 mm dithiothreitol and concentrated to ∼10 mg/ml. The purified enzyme was stored frozen until required.
Enzymatic Analysis
Activity with l-γ-glutamyl-l-α-alanine as a substrate was determined by a previously published spectrophotometric method that links the release of alanine to the oxidation of NADH (9, 10). Activity with l-γ-glutamyl-l-ϵ-lysine was determined by quantifying the release of 5-oxo-l-proline by a modification of a method described by Orlowski et al. (5). The reaction mixture contained 20 μl of 1 m Tris-HCl (pH 8.0), 20 μl of 0.15 m l-γ-glutamyl-l-ϵ-lysine (Bachem), 5 μl of enzyme, and 155 μl of water. The reaction was incubated at 37 °C for 15 min and stopped by heating at 90 °C for 5 min. The sample was then chilled on ice, and the inactivated protein was removed by centrifugation in a microcentrifuge. An aliquot of 100 μl of the supernatant was added to a 1-ml column of Dowex 50 equilibrated in water. The sample was then washed through the column with 0.9 ml of water, followed by a second wash with 1 ml of water. The unreacted l-γ-glutamyl-l-ϵ-lysine bound to the Dowex 50, and the 5-oxoproline was in the unbound fraction. The entire flow-through fraction was collected, and the concentration of 5-l-oxoproline was determined spectrophotometrically at 205 nm with an extinction coefficient of 2600 (5).
Crystallization and Data Collection
All crystals were grown at the Collaborative Crystallization Center using the vapor diffusion method. Initial screens were conducted using the PACT and JCSG screens. 50 μl of screening solution was dispensed into the reservoirs of Innovadyne plates, and equal volumes of human GGACT (6 mg/ml in 20 mm Tris-HCl (pH 8.0) and 0.1 mm dithiothreitol) were mixed with the precipitants (total volume of 0.3 μl). The trays were incubated in and drops imaged by Rigaku Minstrel systems at 8 or 20 °C. After the initial conditions were identified, crystals for x-ray analysis were grown by the vapor diffusion hanging drop method using Greiner® 24-well plates at 4 °C. 2 μl of protein was mixed with an equal volume of reservoir solution on a siliconized coverslip (Hampton Research) prior to suspension over reservoirs containing 1 ml of precipitant. A complex of wild-type GGACT with 5-oxo-l-proline was obtained by soaking crystals with artificial mother liquor containing this compound at 100 mm. Crystals of mutant E82Q (5.9 mg/ml in 20 mm Tris-HCl (pH 8.0) and 0.1 mm dithiothreitol) were grown under identical conditions.
X-ray data were collected at the Australian Synchrotron using Blu-Ice (11). Crystals were transferred to cryoprotectant prior to flash cooling using an Oxford Cryojet sample cooler at 100 K. Data were collected using an ADSC area detector. Data were processed with MOSFLM (12) and scaled with SCALA (13).
Structure Solution and Refinement
Automated molecular replacement in MOLREP (14) was used for the determination of initial structure factor phase angles. Model building and refinement were conducted using REFMAC5 (15) and COOT (16). Ramachandran plot statistics were derived from PROCHECK (17). Statistics for the final models are given in Table 1.
TABLE 1.
Crystallographic data
PDB, Protein Data Bank; r.m.s.d., root mean square deviation.
| Structure |
|||
|---|---|---|---|
| GGACT | GGACT/5-oxoproline | GGACT E82Q mutant | |
| PDB code | 3JUB | 3JUC | 3JUD |
| X-ray data | |||
| Beamline | PX-1 | PX-1 | PX-2 |
| Space group | P212121 | P212121 | P212121 |
| Unit cell parameters | a = 36.0, b = 41.9, c = 84.0 Å; α = β = γ = 90° | a = 36.2, b = 42.5, c = 84.8 Å; α = β = γ = 90° | a = 36.3, b = 42.4, c = 84.8 Å; α = β = γ = 90° |
| Resolution range (Å) | 30–1.2 (1.26–1.20)a | 43–1.2 (1.26–1.20) | 38–0.98 (1.03–0.98) |
| Total no. of observations | 277,499 (38,387) | 369,388 (39,839) | 494,078 (72,260) |
| No. of unique reflections | 40,593 (5782) | 40,928 (5805) | 74,200 (10,958) |
| 〈I/σI〉b | 22.2 (5.2) | 27.5 (10.7) | 10.4 (2.5) |
| Rmerge (%)c | 5.8 (32.3) | 5.5 (15.4) | 10.4 (68.8) |
| Completeness | 99.9 (99.7) | 98.5 (97.0) | 97.7 (100.0) |
| Multiplicity | 6.8 (6.6) | 9.0 (6.9) | 6.7 (6.8) |
| 〈B〉 from Wilson plot (Å2) | 8.7 | 8.7 | 5.0 |
| Refinement statistics | |||
| Resolution range (Å) | 42–1.20 (1.23–1.20) | 42–1.20 (1.23–1.20) | 42–0.98 (1.01–0.98) |
| No. of reflections used in refinement | 38,497 (2917) | 38,832 (2906) | 70,357 (5455) |
| No. of reflections (Rfree set) | 2032 | 2053 | 3736 |
| Rwork (%)d | 12.3 (14.9) | 11.7 (14.8) | 15.3 (23.9) |
| Rfree (%)d | 13.7 (17.0) | 9.5 (12.8) | 18.0 (26.0) |
| No. of atoms | 1433 | 1514 | 1450 |
| 〈B〉 of structure (Å2) | 9.0 | 9.8 | 8.4 |
| r.m.s.d. from ideal geometry | |||
| Bond lengths (Å) | 0.023 | 0.022 | 0.029 |
| Bond angles | 1.910° | 1.922° | 2.313° |
| Chiral centers (Å3) | 0.132 | 0.11 | 0.153 |
| General planes (Å) | 0.012 | 0.011 | 0.013 |
a Numbers in parentheses refer to the highest resolution bin.
b Angle brackets refer to mean values.
c Rmerge = ΣhΣi|Ihi − 〈Ih〉|/ΣhΣiIh.
d Rfactor = Σh‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree was calculated from 5% of the diffraction data not used in refinement.
Structure Analysis
The DALI server was used to find representatives of the cyclotransferase fold in the Protein Data Bank. STAMP (18) was used to superimpose the structures onto human GGACT and to create an initial structure-based sequence alignment, which was edited for clarity.
RESULTS
Gene, Protein, and Enzymatic Characterization
Examination of NCBI Human Genome Database Entrez Gene entries showed that the GGACT gene (A2LD1, Gene ID87769) is quite small (2.187 kb) and is located on chromosome 13q32.3. It is interesting to note that the gene contains only a single intron that falls within the 5′-noncoding region of the gene transcript. The human GGACT gene (A2LD1) encodes a protein of 153 amino acids with a predicted molecular mass of 17,327 Da. The human A2LD1 cDNA was expressed in E. coli as a 17,300-Da protein. SDS-PAGE of the purified recombinant enzyme under reducing conditions showed a protein that migrated within the expected size range. Gel filtration of the native protein revealed a molecular mass of 18 kDa, which is compatible with a monomeric structure (data not shown). Analysis of the potential γ-glutamylcyclotransferase activity of the recombinant enzyme revealed that it is inactive with l-γ-glutamyl-α-amino acid substrates such as l-γ-glutamyl-l-α-cysteine and l-γ-glutamyl-l-α-alanine but has distinct activity with l-γ-glutamyl-l-ϵ-lysine (2.7 ± 0.06 μmol/min/mg). In contrast, recombinant human GGCT is active with l-γ-glutamyl-l-α-alanine (50.3 ± 1.22 μmol/min/mg) but inactive with l-γ-glutamyl-l-ϵ-lysine. As a result of this clear specificity, we will refer to the product of the so-called A2LD1 gene as γ-glutamylamine cyclotransferase.
Crystal Structure
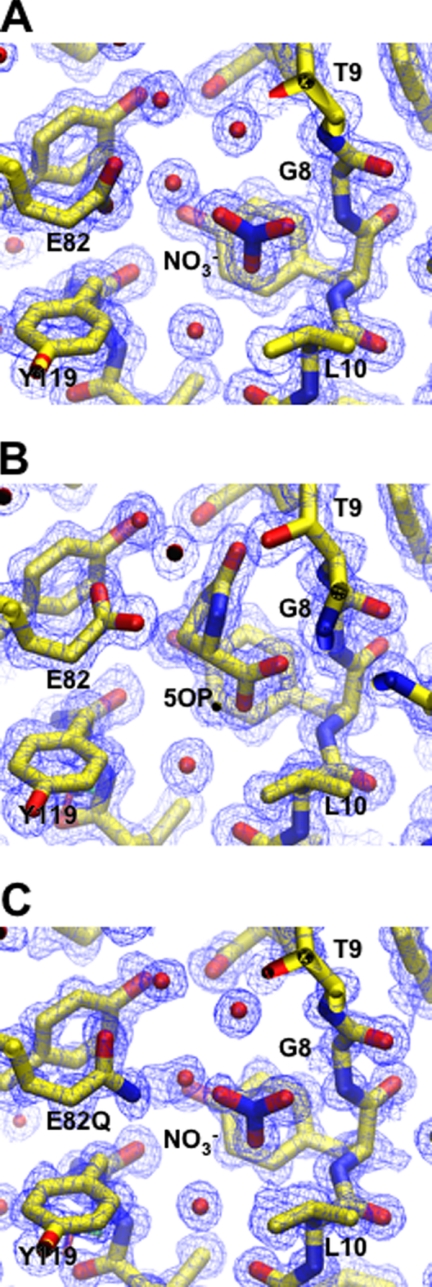
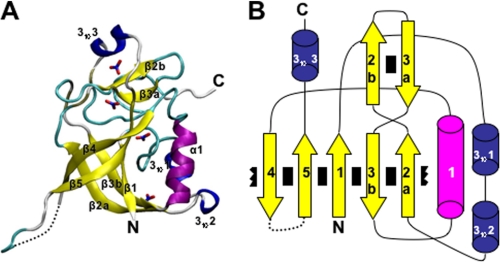
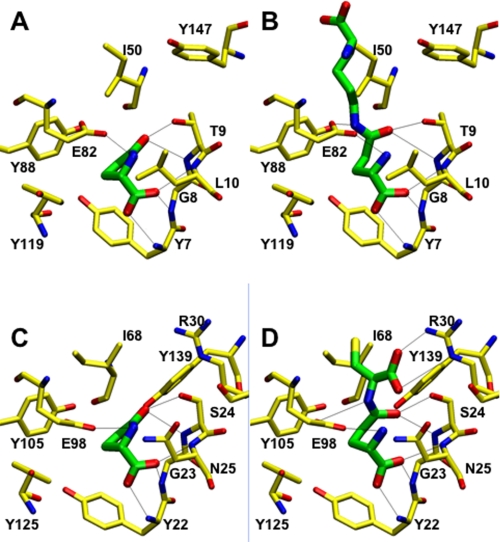
Crystals of human GGACT appeared in several polyethylene glycol-based conditions in the JCSG and PACT screens. The best crystals grew at 8 °C with JCSG condition C3 (20% (w/v) polyethylene glycol 3350 and 0.2 m ammonium nitrate) as the precipitant. Crystals grew as rods with dimensions of 220 × 100 × 80 μm. Crystals were cryoprotected by transfer to artificial mother liquor with the polyethylene glycol 3350 concentration increased to 25 or 30% (w/v). X-ray data collection statistics are given in Table 1. The structure was solved using the mouse GGACT homolog AIG2-like domain 1 (Protein Data Bank code 1VKB) as a search model. Several cycles of model building and refinement followed, during which the mouse sequence in the search model was mutated to match the human sequence. Water molecules and four NO3− groups, one of which binds in the active site, were also built into the model. The final model has excellent electron density (Fig. 1A) except for the surface loop linking strands β4 and β5 (residues 99–113), which appears disordered. The equivalent loop is also absent in the model of the mouse homolog. The final structure and its topology are illustrated in Fig. 2. The topology of GGACT is similar to that of GGCT, i.e. a five-strand β-barrel decorated with helices and connecting loops. In both structures, the active site is found in a cavity located on the side of the barrel adjacent to strands β1 and β5. The active site of GGACT is unambiguously identified by the binding of the reaction product 5-oxo-l-proline (Figs. 1B and 3A). In GGACT, strand β2a crosses over strand β3b so as to reverse topological order with respect to strands β2b and β3a (Fig. 2). This distinctive “crossover” feature is conserved in all structures with this fold. The cavity is formed by Tyr7, Gly8, and Thr9 in the loop following strand β1 (a conserved motif in the cyclotransferase family; see below), Leu10, Glu82, Tyr88, and Tyr119. The secondary amine group of oxoproline is 3.2 Å from the Glu82 carboxylate group. The loop containing residues 7–10 has a backbone conformation that causes the main chain amino groups to be oriented into the cavity. The oxoproline carboxylic acid moiety accepts hydrogen bonds from the main chain amino groups of Tyr7 and Gly8. The carbonyl oxygen of oxoproline accepts a hydrogen bond from the backbone amino and side chain O-γ moieties of Thr9. The aromatic face of the side chain of Tyr7 forms the bottom of the cavity. The ligand 5-oxo-l-proline sits lies ∼3.6 Å from this residue (Fig. 3). Using this structure as a template, the substrate l-γ-glutamyl-l-ϵ-lysine has been modeled into the active site (Fig. 3B). Similarly, substrate or oxoproline has been modeled in the active site of GGCT (Fig. 3, C and D). Patterns of hydrogen bonding are conserved in these structures.
FIGURE 1.
Models of GGACT in the vicinity of the active site shown in stick form with 2mFo − DFc electron density, contoured at 1σ shown in chicken wire representation. A and B, GGACT and GGACT, respectively, in complex with 5-oxo-l-proline; C, GGACT E82Q mutant.
FIGURE 2.
A, schematic representation of GGACT, with bound nitrate groups represented in stick form. The N and C termini are labeled. B, topology diagram of GGACT. Interactions between β-strands are indicated by black rectangles. The broken rectangles adjacent to strands β2a and β4 indicate the wrapping of the β-sheet to form a barrel.
FIGURE 3.
A, structure of the active site of GGACT in complex with 5-oxo-l-proline; B, model of GGACT in complex with l-γ-glutamyl-l-ϵ-lysine; C, model of GGCT in complex with 5-oxo-l-proline; D, model of GGCT in complex with l-γ-glutamyl-l-α-cysteine. In all cases, carbon atoms of the protein and ligand are yellow and green, respectively. Potential hydrogen bonds are shown as thin black lines.
Site-directed Mutagenesis
We demonstrated previously that Glu98 is crucial for catalytic activity in GGCT (4). To determine the importance of the equivalent residue in GGACT (Glu82), we mutated this residue to glutamine or alanine. The E82Q and E82A mutants had no catalytic activity, and the E82A enzyme was unstable and readily precipitated. We crystallized the E82Q mutant under conditions identical to those used for the wild-type enzyme (Fig. 1C). The E82Q mutant and wild-type structures superimpose with a root mean square deviation of 0.352 Å over 144 C-α atoms. The only difference in the active site is a small rotation of Gln82 about the C-γ–C-δ bond with respect to Glu82. The binding patterns of the water molecule and an NO3− group binding in the active site are identical in the mutant and wild-type structures.
Comparison of GGACT with Related Structures
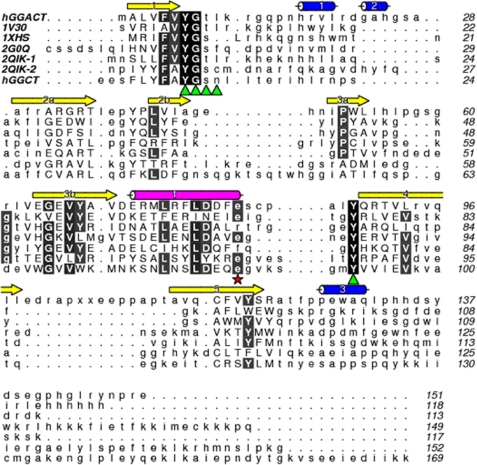
A DALI search for structural homologs revealed a limited number of proteins with this fold. These were from both prokaryotic and eukaryotic species and included Pyrococcus horikoshii (Protein Data Bank code 1V30), E. coli (code 1XHS), Arabidopsis thaliana (code 2G0Q), and Bacillus subtilis (code 2QIK), as well as the previously identified human GGCT and mouse GGACT homologs (codes 2PN7 and 1VKB, respectively). The superposed structures and structure-based sequence alignment (Fig. 4) show several conserved features: β-barrel topology with two strands “crossing over” as described above, a highly conserved helix (in GGACT numbering, helix α1), a binding cavity formed on one side of the β-barrel, a loop following strand β1 containing a conserved (V/A)YG(S/T) motif, a conserved tyrosine in strand β4, and an aromatic residue in strand β5.
FIGURE 4.
Sequence alignment of Protein Data Bank structures adopting the cyclotransferase fold. Sequences are labeled by Protein Data Bank codes, except for human GGACT (hGGACT) and GGCT (hGGCT). Protein Data Bank code 2QIK contains two cyclotransferase domains that have been split into 2QIK-1 and 2QIK-2. Secondary structure elements of human GGACT are denoted above the sequences. Where significant structural homology exists, the residues are shown in uppercase letters. Residues lining the active-site cavity are indicated with green triangles. Possible catalytic glutamate residues are highlighted with a red star. Completely conserved (black) and highly conserved (gray) residues are highlighted.
Phylogenetic Relationships
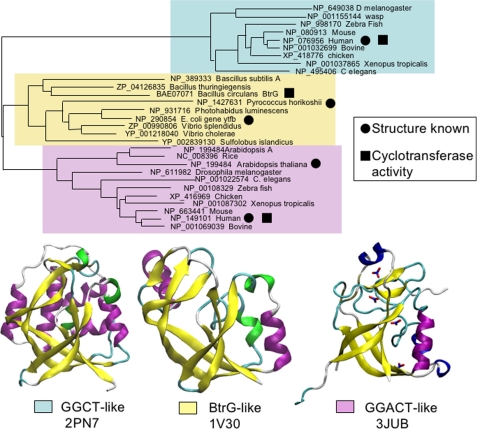
Despite the conservation of the overall fold and the active-site architecture, there is considerable sequence divergence between the different proteins exhibiting the GGCT fold. A direct alignment of the sequence of human GGACT with that of human GGCT showed <10% amino acid identity (Fig. 4). Similarly, pairwise alignment of the GGCT and GGACT sequences with the prokaryotic members of this structural family revealed a similar low level of sequence identity. To gain some insight into the evolution of this fold and the diversity within this broad protein family, we undertook BLAST searches to identify homologous proteins in different species. Multiple sequence alignments using the COBALT alignment tool (NCBI) (19) were then used to build a phylogenetic tree. Although there are many sequences available for this type of analysis, the sequences used here were selected to provide a broad evolutionary perspective. A neighbor joining tree (Fig. 5) indicated that there are three main families of proteins with the γ-glutamylcyclotransferase fold (GGCT-like, GGACT-like, and BtrG-like), and there is evidence that each group has retained γ-glutamylcyclotransferase activity. It is evident from the lack of sequence identity that these families have evolved separately for a considerable period. The GGCT-like family appears to be restricted to animals with sequences identified in a range of species from Caenorhabditis elegans to humans. Our previous studies showed that the human GGCT enzyme (Protein Data Bank code 2PN7) has γ-glutamylcyclotransferase activity (4). The GGACT-like family is also restricted to eukaryotes but clearly includes plants, as confirmed by the structure from A. thaliana (Protein Data Bank code 2G0Q). In this study, we have shown that members of the GGACT-like family have γ-glutamylcyclotransferase activity. In contrast, the BtrG-like family appears to be restricted to prokaryotes. The BtrG protein from Bacillus circulans has been shown to have γ-glutamylcyclotransferase activity in an antibiotic synthesis pathway (20), and the structures of the sequence-related proteins from P. horikoshii (Protein Data Bank code 1V30) and E. coli (code 1XHS) confirm the association between this fold and γ-glutamylcyclotransferase enzymatic activity in prokaryotes. Although we did not find evidence for GGCT fold proteins in yeast or fungal species, it is possible that they have diverged beyond the level of sequence identity that can be identified by sequence similarity searches, and these await identification via structural studies.
FIGURE 5.
Neighbor joining tree depicting the probable phylogenetic relationships between selected proteins with the γ-glutamylcyclotransferase fold. Representative structures from each subfamily are shown.
DISCUSSION
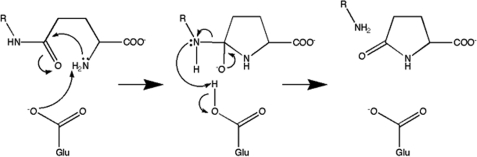
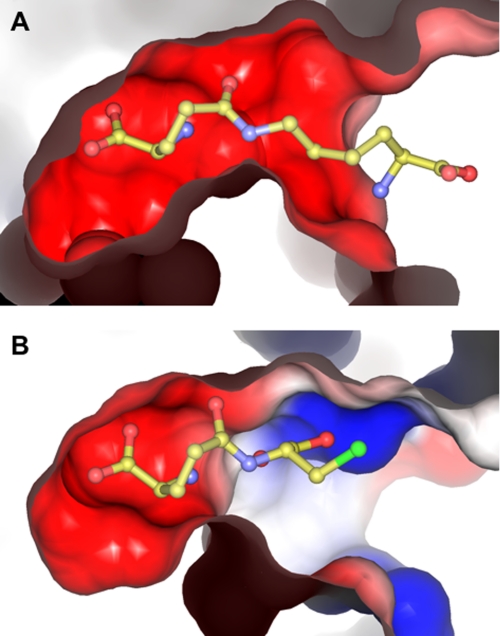
The enzymatic activity known as γ-glutamylamine cyclotransferase has been unambiguously assigned to the human gene known as A2LD1. The human GGACT protein is clearly similar to the mouse protein AIG2-like domain 1, and the human and mouse genes are on syntenic chromosomes. The two proteins can be superimposed with a root mean square deviation of 0.68 Å over 138 C-α atoms, and the putative active-site residues are conserved. It is therefore likely that the mouse protein has GGACT activity. The structure of GGACT bound to the reaction product 5-oxo-l-proline (Figs. 1B and 3A) unambiguously identifies the active site. The GGACT residues in proximity to this entity, Tyr7, Gly8, Thr9, Glu82, and Tyr88, are structurally equivalent to Tyr22, Gly23, Ser24, Glu98, and Tyr105 in GGCT, respectively, strongly suggesting that this compound binds in an analogous fashion in both enzymes. A model of the 5-oxo-l-proline-GGCT complex was therefore produced (Fig. 3C). We propose that these residues form the binding site for the l-γ-glutamyl portion of the substrates of both enzymes (Fig. 3, B and D). In both models, the α- and γ-amino groups of the glutaminyl moieties of the substrates are in close proximity to Glu82 (GGACT) and Glu98 (GGCT). We propose that Glu82 (GGACT) and Glu98 (GGCT) share the same function, i.e. a general acid/base, and propose the following reaction mechanism: the α-amino group of the l-γ-glutamyl moiety is deprotonated by Glu82/Glu98, with the concomitant nucleophilic attack of this amine onto the side chain amide carbon atom. The resulting oxyanion intermediate collapses to form 5-oxo-l-proline, with the now protonated Glu82/Glu98 residue donating a hydrogen ion to the amine of the α- or ϵ-linked amino portion of the substrate (Fig. 6). We have demonstrated by site-directed mutagenesis and kinetics experiments that Glu82 (GGACT) and Glu98 (GGCT) (4) are essential for activity. The structural integrity of E82Q indicates that a glutamyl residue at this position is not essential for folding. Our model of substrate binding in GGACT and GGCT can further explain the substrate specificities of both enzymes. In our models, the groups to which the glutamyl moieties are linked are orientated out of the active site and toward the surface, and the residues of GGACT and GGCT share little structural similarity in this vicinity. In our GGCT model with substrate, Arg30 is involved in a salt bridge with the α-carboxylic acid of the cysteine residue, compatible with the known preference of GGCT for l-γ-glutamyl-l-α-amino acid substrates. GGACT lacks an arginine residue at the equivalent location and does not act on l-γ-glutamyl-l-α-amino acids. The active site is narrower in GGACT compared with GGCT, partly due to the presence of Phe81 (Fig. 7). This is compatible with the preference of GGACT for substrates with extended aliphatic amines (lysine, spermidine, etc.) conjugated to the γ-glutamyl group.
FIGURE 6.
Proposed reaction mechanism for GGACT and GGCT. Glu represents Glu82 (GGACT) or Glu98 (GGCT).
FIGURE 7.
Surface of GGACT (A) and GGCT (B) with substrate modeled in the active site. Electrostatic potential at the surface is represented as blue (positive potential) and red (negative potential).
The B. subtilis homolog (named YkqA, of unknown function; Protein Data Bank code 2QIK) has two cyclotransferase domains contained within one polypeptide chain. Only the second has a glutamyl residue equivalent to Glu82 in GGACT. This protein possibly has cyclotransferase-like activity in its C-terminal domain, with a regulatory function in the N-terminal domain. The E. coli homolog (named YtfP, of unknown function; Protein Data Bank code 1XHS) has an arginine residue in place of GGACT residue Glu82, suggesting that this protein is not a cyclotransferase but might bind a similar substrate. The A. thaliana protein (known as At5g39720.1, of unknown function; Protein Data Bank code 2G0Q) is clearly a member of the GGCT structural family and has a conserved active-site glutamyl residue, suggesting that it may have cyclotransferase activity. This is consistent with the previous detection of γ-glutamylcyclotransferase activity in tobacco suspension cultures (21).
At this juncture, the lack of structural similarity of the GGCT family of cyclotransferases and the glutaminyl cyclases (QCs) (EC 2.3.2.5) should be noted. The QCs catalyze the formation of N-terminal 5-oxoproline from its glutaminyl (or glutamyl) precursor on peptides. This modification is required by certain proteins for biological function in either mediating interaction with receptors or stabilizing proteins against N-terminal degradation. QCs are classified as belonging to two groups: mammalian QCs and a second group containing enzymes from bacteria, plants, and parasites. The structure of human QC, representative of the first group, adopts an α/β-open sandwich fold similar to the two-zinc exopeptidases (22) and requires zinc as a cofactor. Although human QC is unrelated to the GGCT family in either sequence or three-dimensional structure, it does appear to contain a catalytic acid/base residue (Glu201) analogous in function to Glu82 in GGACT. In human QC, a Zn2+ ion is proposed to stabilize an oxyanion intermediate during catalysis. In GGACT, the Thr9 amino and side chain hydroxyl groups appear to perform this function. As with GGCT and GGACT, mutation of the proposed acid/base residue in human QC (E201Q) inactivates the enzyme. Papaya QC, representative of the second group, adopts a five-blade β-propeller (23). Although this enzyme binds a Zn2+ ion, this does not appear to be involved in catalysis. Nevertheless, the proposed mechanism for papaya QC involves the use of an acid/base residue (Glu69) in an analogous fashion to Glu82 in GGACT, Glu98 in GGCT, and Glu201 in human QC. In the proposed mechanism, papaya QC stabilizes the oxyanion using the side chain amino group of Lys225. Taken together, the structural and functional data for QC, GGACT, and GGCT enzymes suggest convergent evolution in the catalytic mechanism.
The wide distribution of proteins adopting the cyclotransferase fold across Eukarya, Archaea, and Bacteria may indicate an ancient evolutionary origin of the cyclotransferase fold. The structural features most strongly conserved are the β-barrel and helix α1 (GGACT numbering). The STAMP alignment also shows key conserved sequence motifs. Despite the lack of significant sequence identity between GGCT and GGACT, the conservation of topology (including the unusual β-strand crossover described above) and active-site residues leads us to propose that these enzymes are derived from a common ancestral gene and functionally specialized after a gene duplication event, with GGACT optimized for catalysis on substrates with a γ-l-glutamyl moiety linked to extended alkylamines and GGCT optimized for catalysis on substrates with a l-γ-glutamyl moiety linked to l-α-amino acids. There is recent evidence that some prokaryotic members of this structural family also have γ-glutamylcyclotransferase activity, as the BtrG protein from B. circulans catalyzes the formation of 5-oxoproline from an intermediate in the synthesis of the antibiotic butirosin (20). Thus, although the primary amino acid sequences of this structural family are incredibly diverse, the conserved fold is very strongly associated with conserved γ-glutamylcyclotransferase activity. Because there is such great sequence diversity between the three groups within this structural family, there is a strong possibility that additional subfamilies with sequence diversity beyond the reach of BLAST alignments may be identified by structural studies in the future.
Acknowledgments
We thank the technical staff of the Bio21-C3 Center for help with crystallization. This work was undertaken on the PX-1 and PX-2 beamlines at the Australian Synchrotron (Victoria, Australia).
This work was supported by Australian Research Council Discovery Grant DP0880027 and National Health and Medical Research Council Project Grant 525458.
The atomic coordinates and structure factors (codes 3JUB, 3JUC, and 3JUD) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GGACT
- γ-glutamylamine cyclotransferase
- GGCT
- γ-glutamylcyclotransferase
- QC
- glutaminyl cyclase.
REFERENCES
- 1.Pisano J. J., Finlayson J. S., Peyton M. P. (1968) Science 160, 892–893 [DOI] [PubMed] [Google Scholar]
- 2.Fink M. L., Chung S. I., Folk J. E. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 4564–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fink M. L., Folk J. E. (1981) Mol. Cell. Biochem. 38, 59–67 [DOI] [PubMed] [Google Scholar]
- 4.Oakley A. J., Yamada T., Liu D., Coggan M., Clark A. G., Board P. G. (2008) J. Biol. Chem. 283, 22031–22042 [DOI] [PubMed] [Google Scholar]
- 5.Orlowski M., Richman P. G., Meister A. (1969) Biochemistry 8, 1048–1055 [DOI] [PubMed] [Google Scholar]
- 6.Orlowski M., Meister A. (1973) J. Biol. Chem. 248, 2836–2844 [PubMed] [Google Scholar]
- 7.Klock H. E., Schwarzenbacher R., Xu Q., McMullan D., Abdubek P., Ambing E., Axelrod H., Biorac T., Canaves J. M., Chiu H. J., Deacon A. M., DiDonato M., Elsliger M. A., Godzik A., Grittini C., Grzechnik S. K., Hale J., Hampton E., Han G. W., Haugen J., Hornsby M., Jaroszewski L., Koesema E., Kreusch A., Kuhn P., Miller M. D., Moy K., Nigoghossian E., Paulsen J., Quijano K., Reyes R., Rife C., Sims E., Spraggon G., Stevens R. C., van den Bedem H., Velasquez J., Vincent J., White A., Wolf G., Hodgson K. O., Wooley J., Lesley S. A., Wilson I. A. (2005) Proteins 61, 1132–1136 [DOI] [PubMed] [Google Scholar]
- 8.Catanzariti A. M., Soboleva T. A., Jans D. A., Board P. G., Baker R. T. (2004) Protein Sci. 13, 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Board P. G., Moore K. A., Smith J. E. (1978) Biochem. J. 173, 427–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Board P. G., Smith J. E., Moore K. (1978) J. Lab. Clin. Med. 91, 127–131 [PubMed] [Google Scholar]
- 11.McPhillips T. M., McPhillips S. E., Chiu H. J., Cohen A. E., Deacon A. M., Ellis P. J., Garman E., Gonzalez A., Sauter N. K., Phizackerley R. P., Soltis S. M., Kuhn P. (2002) J. Synchrotron Radiat. 9, 401–406 [DOI] [PubMed] [Google Scholar]
- 12.Leslie A. G. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 13.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 14.Lebedev A. A., Vagin A. A., Murshudov G. N. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 16.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 17.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 18.Russell R. B., Barton G. J. (1992) Proteins 14, 309–323 [DOI] [PubMed] [Google Scholar]
- 19.Papadopoulos J. S., Agarwala R. (2007) Bioinformatics 23, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 20.Llewellyn N. M., Li Y., Spencer J. B. (2007) Chem. Biol. 14, 379–386 [DOI] [PubMed] [Google Scholar]
- 21.Steinkamp R., Schweinhofen B., Rennenberg H. (1987) Physiol. Plant. 69, 499–503 [Google Scholar]
- 22.Huang K. F., Liu Y. L., Cheng W. J., Ko T. P., Wang A. H. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13117–13122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wintjens R., Belrhali H., Clantin B., Azarkan M., Bompard C., Baeyens-Volant D., Looze Y., Villeret V. (2006) J. Mol. Biol. 357, 457–470 [DOI] [PubMed] [Google Scholar]