Abstract
Microbial infection of the intrauterine environment is a major cause of preterm birth. The current paradigm indicates that intrauterine infections predominantly originate from the vaginal tract, with the organisms ascending into the sterile uterus. With the improvements in technology, an increasing number of bacterial species have been identified in intrauterine infections that do not belong to the vaginal microflora. We have demonstrated previously that intrauterine infections can originate from the oral cavity following hematogenous transmission. In this study, we begin to systemically examine what proportion of the oral microbiome can translocate to the placenta. Pooled saliva and pooled subgingival plaque samples were injected into pregnant mice through tail veins to mimic bacteremia, which occurs frequently during periodontal infections. The microbial species colonizing the murine placenta were detected using 16S rRNA gene-based PCR and clone analysis. A diverse group of bacterial species were identified, many of which have been associated with adverse pregnancy outcomes in humans although their sources of infection were not determined. Interestingly, the majority of these species were oral commensal organisms. This may be due to a dose effect but may also indicate a unique role of commensal species in intrauterine infection. In addition, a number of species were selectively “enriched” during the translocation, with a higher prevalence in the placenta than in the pooled saliva or subgingival plaque samples. These observations indicate that the placental translocation was species specific. This study provides the first insight into the diversity of oral bacteria associated with intrauterine infection.
Preterm birth (PTB) is a significant public health concern, affecting 12.7% of births in the United States. This rate reflects a 36% increase in the past 25 years. Among these, the very preterm births, i.e., those occurring before 32 weeks of gestation, are the most costly and medically relevant group, accounting for approximately 2% of all deliveries. Intrauterine infection has been recognized as a major cause of PTB. The infection rate is inversely related to the gestational age and is highly prevalent among the very preterm deliveries (24). Intrauterine infection has also been implicated in other adverse pregnancy outcomes, such as late miscarriage and stillbirth (25). A wide variety of bacterial species have been identified in the infections, with Ureaplasma urealyticum, Mycoplasma hominis, Bacteroides spp., Gardnerella vaginalis, and Fusobacterium nucleatum (24, 31) as the most prevalent. It has been suggested that the majority of these bacterial species are of relatively low virulence. However, once inside the uterus, they stimulate the synthesis and release of proinflammatory cytokines, neutrophil infiltration and activation, and prostaglandin and metalloprotease synthesis and release, leading to cervical ripening, membrane weakening and rupture, uterine contractions, and PTB (24).
The current paradigm indicates that the majority of intrauterine infections originate in the lower genital tract, with the infectious agents ascending into the otherwise sterile womb. The bacteria can then infect the fetal membranes (causing chorioamnionitis), umbilical cord (funisitis), placenta (placental infection), amniotic fluid (amniotic fluid infection), and the fetus (sepsis) (24). However, an increasing number of studies report intrauterine infections caused by bacterial species not found in the urogenital tract, such as F. nucleatum and Bergeyella, Eikenella, and Capnocytophaga spp., all of which are commensal species in the oral cavity (28, 30, 31). Thus, a hematogenous transmission has been proposed as an alternative route of infection. The oral cavity houses more than 700 microbial taxa (1, 43) and hence is a potential reservoir for infection. During periodontal infection, oral bacterial titers increase dramatically, accompanied by gingival inflammation and bleeding. These conditions lead to increased bacteremia, thus enhancing the opportunities for hematogenous transmission.
We have previously demonstrated that hematogenous infection by oral F. nucleatum resulted in specific colonization in the mouse placenta, causing localized infection and inflammation, leading to preterm and term stillbirth. The pattern of infection mimicked those observed in ascending infections in humans, i.e., with the bacteria colonizing the decidua, followed by spread to fetal membranes, amniotic fluid, and fetus (29). This was the first experimental evidence that oral bacteria can cause adverse pregnancy outcomes following hematogenous transmission.
Transmission of oral bacteria into the pregnant uterus has been observed in humans in two cases reported by our group. In one case, using 16S rRNA gene-based culture-independent technology, we identified uncultivated oral Bergeyella from the amniotic fluid complicated by PTB at 24 weeks. No Bergeyella cells were detected in the mother's vaginal tract, but interestingly, the same clonal type was identified in her subgingival plaque based on the 16S-23S rRNA gene sequences (28). In a second case, oral F. nucleatum was identified as the cause of a term stillbirth and was isolated from the lung and stomach of the stillborn infant. Using similar technology, the same clonal type was identified in the mother's subgingival plaque, but not in her supragingival plaque, or vaginal or rectal microflora (27). Such accumulating evidence underscores the importance of oral bacteria in intrauterine infection and adverse pregnancy outcome.
One important question that arises from these studies concerns what proportion of the complex oral microbiome is capable of oral-uterus transmission. With the majority of the bacteria in the oral cavity uncultivated, it is very likely that the involvement of oral bacteria in intrauterine infection has been significantly underestimated, because clinical microbiology laboratories still employ routine culturing methods for bacterial detection (30). As a result, clinical evidence involving oral bacteria in PTB is incomplete, and specifically, sufficient knowledge of the diversity of oral bacterial involved in PTB is lacking. In the current proof-of-concept study, we utilize the pregnant murine model established in our laboratory to identify species of the oral microbiome that can translocate to the placenta. We report that a diverse group of oral bacteria are capable of hematogenous transmission to colonize the murine placenta. Furthermore, most of these species have been reported to be associated with adverse pregnancy outcomes in humans.
MATERIALS AND METHODS
Oral sample preparation.
Our study was approved by the Case Western Reserve University Internal Review Board. Saliva samples were collected from five healthy volunteers, with each individual spitting nonstimulated saliva into a sterile tube, and pooled. Subgingival plaque was collected from deep periodontal pockets (pocket depth of >7 mm) of eight periodontitis patients, using sterile curettes, and pooled. The pooled subgingival plaque samples were centrifuged at 14,000 × g for 5 min, and the pellets were resuspended in 1 to 3 ml prereduced sterile phosphate-buffered saline (PBS). All samples were kept in a portable anaerobic jar containing ice and were injected into mice within an hour following collection.
Animal handling, infection, and placenta collection.
The animal protocol was approved by the Case Western Reserve University Institutional Animal Care and Use Committee. Ten-week-old outbred CF-1 mice (Charles River Laboratories, Wilmington, MA) were mated at a female/male ratio of 2:1. Positive mating was indicated by the presence of a white vaginal plug, and that day was termed day 1 of gestation. The normal length of gestation of CF-1 mice is 20 to 21 days. Injections were performed on days 15 to 17 of gestation. A total of 0.1 ml of undiluted saliva or 0.1 ml of the subgingival plaque suspension was injected into the tail vein of each pregnant mouse. At 24 h postinjection, placentas were harvested. For each mouse, placentas were pooled and stored at −80°C until use.
16S rRNA gene-based PCR and construction of the clone libraries.
The genomic DNA from pooled saliva, pooled subgingival plaque, and mouse placentas was extracted as previously described (28). The 16S rRNA gene was amplified from each pooled sample by PCR, using the universal primers A17F and 1512R as previously described (34). The PCR amplicons were cloned into the pCR8/GW/TOPO vector by using the TA cloning kit (Invitrogen, Carlsbad, CA) according to the supplier's instructions. Recombinant plasmids were extracted using the Wizard Plus SV Minipreps DNA purification system (Promega, Madison, WI). Plasmids harboring inserts with the expected size (∼1,500 bp) were selected, and the DNA sequences of the inserts were determined by the Genomics Core Facility of Case Western Reserve University (Cleveland, OH) using primers GW1 (GTTGCAACAAATTGATGAGCAATGC) and GW2 (GTTGCAACAAATTGATGAGCAATTA) (Invitrogen, Carlsbad, CA). Sequences were assembled using the VectorNTI program (Invitrogen, Carlsbad, CA).
Analysis of 16S rRNA gene.
The 16S rRNA gene sequence analysis was performed by using the Human Oral Microbiome Database (HOMD; http://www.HOMD.org). Sequences displaying an identity percentage of at least 97% were retained for further analysis. Sequences with an identity percentage of <97% were reexamined by blasting each half part of the 16S rRNA gene sequence to identify chimeras. Sequences matching only with a genus but not a species or a defined taxon were discarded.
RESULTS
Infection of mice, and construction and analysis of 16S rRNA gene clone libraries.
The pooled saliva was injected into seven pregnant mice, and the pooled subgingival plaque was injected into 10 pregnant mice, all at gestation of 15 to 17 days. At 24 h postinfection, placentas were collected from each mouse. This time point was chosen because we have shown previously that bacterial colonization in the mouse was cleared by 24 h, except in the placenta, where it continued to proliferate (29). Thus, any bacteria detected at 24 h after injection was likely due to specific colonization, rather than hematogenous diffusion into the placenta.
A 16S rRNA gene clone library was constructed for each mouse. For each of these libraries, 8 to 21 independent clones were sequenced. When combined, a total of 59 clones were sequenced from the seven saliva-infected murine libraries, and a total of 106 clones sequenced from the 10 subgingival plaque-infected libraries (Table 1). Separate libraries were constructed for the pooled saliva and pooled subgingival plaque samples. A total of 64 and 105 clones were sequenced, respectively, from these two libraries (Table 1).
TABLE 1.
Clones analyzed for each 16S rRNA gene library
| 16S rRNA clone library | No. of clones sequenced | No. of clones retained for analysis |
|---|---|---|
| Pooled saliva | 64 | 41 |
| Pooled subgingival plaque | 105 | 81 |
| Placentas infected with saliva | 59 | 56 |
| Placentas infected with subgingival plaque | 106 | 104 |
The near full-length 16S rRNA gene sequences obtained from each sequenced clone were analyzed by BLAST against the HOMD. The bacterial identification was accepted if the sequence identity was ≥97%. For the clones with sequence identity of <97% (n = 52), the full-length 16S rRNA gene sequences were divided into halves, and each half was reanalyzed by BLAST against the HOMD to identify chimeras formed during PCR. The majority of these low-identity clones appeared to be chimeras (n = 49). The remaining three clones shared limited identities (94.6 to 96%) with Leptotrichia, Neisseria, and Veillonella, respectively. Only the sequences with ≥97% identity were retained for the analysis described below. The number of retained clones from each library is shown in Table 1.
Transmission of salivary bacteria into mouse placenta.
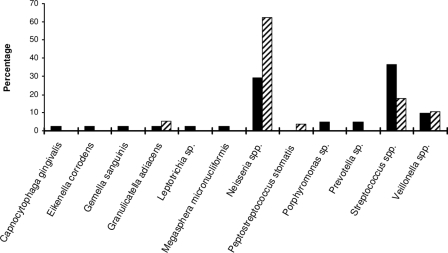
From the salivary 16S rRNA gene clone library, a total of 25 taxa belonging to 11 different genera and four different phyla, Firmicutes, Fusobacteria, Bacteroidetes, and Proteobacteria, were identified (Table 2). The most prevalent was Streptococcus spp. (37%), followed by Neisseria spp. (29%), Veillonella spp. (10%), Prevotella spp. (5%), Porphyromonas sp. (5%), and Capnocytophaga gingivalis, Eikenella corrodens, Gemella sanguinis, Granulicatella adiacens, Leptotrichia sp., and Megasphera micronucliformis, with each at 2% (Fig. 1).
TABLE 2.
Transmission of salivary bacteria into murine placenta
| Bacterium identified in salivaa | No. of mice with placental infection of salivary speciesb |
|---|---|
| Capnocytophaga gingivalis oral taxon 337 | |
| Eikenella corrodens oral taxon 577 | |
| Gemella sanguinis oral taxon 757 | |
| Granulicatella adiacens oral taxon 534 | 2 |
| Leptotrichia sp. oral taxon 417 | |
| Megasphera micronucliformis oral taxon 122 | |
| Neisseria spp. | |
| N. flava oral taxon 609 | |
| N. flavescens oral taxon 610 | 7 |
| N. subflava oral taxon 476 | 1 |
| Peptostreptococcus stomatis oral taxon 112 | 1 |
| Porphyromonas spp. | |
| P. endodontalis oral taxon 273 | |
| Porphyromonas sp. oral taxon 284 | |
| Prevotella sp. oral taxon 308 | |
| Streptococcus spp. | |
| S. australis oral taxon 073 | |
| S. infantis oral taxon 638 | |
| S. mitis oral taxon 398 | 2 |
| S. mitis oral taxon 677 | 3 |
| S. oralis oral taxon 707 | |
| S. parasanguis II oral taxon 411 | 1 |
| S. pneumoniae oral taxon 734 | 1 |
| S. salivarius oral taxon 755 | 1 |
| Streptococcus sp. oral taxon 058 | |
| Veillonella spp. | |
| V. atypica oral taxon 524 | 2 |
| V. dispar oral taxon 160 | |
| V. parvula oral taxon 161 | 4 |
Species identified in the salivary clone library with sequence identities of ≥97% with those available in the HOMD, with the exception of P. stomatis oral taxon 112, which was present in the pooled saliva sample but not detected in the clone library. Taxa overlapping with those identified in subgingival plaque are in boldface type.
A total of seven mice were injected with the pooled saliva sample.
FIG. 1.
Transmission of salivary bacteria to murine placentas. The relative abundance of each bacterial genus/species in pooled saliva samples (solid bars) and placentas from all seven infected mice (striped bars) were expressed as the percentage of the species out of the total number of clones analyzed for the pooled saliva samples (n = 41) and the murine placentas (n = 56), respectively.
A different bacterial spectrum was detected in the mouse placenta (see Table S1 in the supplemental material for details). Except for one mouse, which was colonized only by Neisseria flavescens, all animals were infected by two or more different species in their placentas. A total of five different genera were detected (Table 2). Mice were preferentially infected by Neisseria spp. (in all seven mice tested), Streptococcus spp. (in five mice), Veillonella spp. (in four mice), G. adiacens (in two mice), and Peptostreptococcus stomatis (in one mouse). When each species was evaluated based on all 56 clones analyzed from placenta (Fig. 1), the order of prevalence remained unchanged, with 63% Neisseria spp., 18% Streptococcus spp., 11% Veillonella spp., 5% G. adiacens, and 4% P. stomatis. With the exception of P. stomatis, the species found in the placenta were also detected in the saliva. Using Peptostreptococcus-specific primers, the 16S rRNA gene of P. stomatis was amplified from the pooled saliva and verified by DNA sequencing (data not shown). Thus, P. stomatis is present in the pooled saliva but may exist in low quantities and hence was not detected by our random screening of the salivary clone library. These results suggest that P. stomatis was preferentially “enriched” in the mouse placenta. In addition, Neisseria, G. adiacens, and Veillonella were also enriched in the placenta, indicating the specificity of colonization (Fig. 1).
Transmission of subgingival bacteria into mouse placenta.
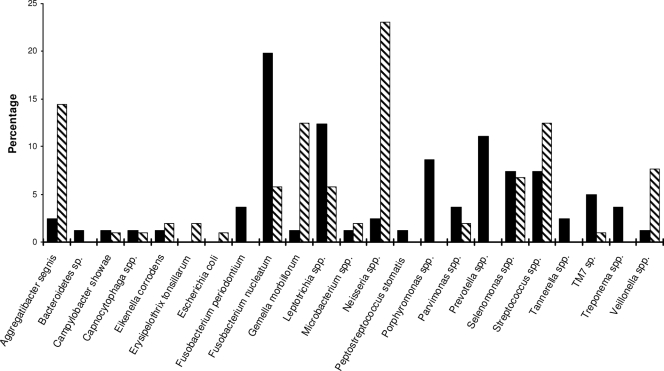
The bacterial composition in the pooled subgingival plaque sample was determined in a manner similar to that for the saliva. A total of 54 taxa belonging to more than 20 different genera/species were identified (Table 3). The seven most prevalent genera/species are listed below, in decreasing order: F. nucleatum (17%), Leptotrichia spp. (12%), Prevotella spp. (11%), Porphyromonas spp. (9%), Streptococcus spp. (7%), Selenomonas spp. (7%), and Neisseria spp. (5%). Ten of the 54 taxa were also identified in saliva (Table 3). All together, a total of 69 different taxa were identified in the saliva and subgingival plaque.
TABLE 3.
Detection of subgingival plaque bacteria transmitted into murine placenta
| Bacterium identified in subgingival plaquea | No. of mice with placental infection of plaque speciesb |
|---|---|
| Aggregatibacter segnis oral taxon 762 | 3 |
| Bacteroidetes sp. oral taxon 511 | |
| Campylobacter showae oral taxon 763 | 1 |
| Capnocytophaga spp. | |
| C. granulosa oral taxon 325 | |
| Capnocytophaga sp. oral taxon 326 | 1 |
| Eikenella corrodens oral taxon 577 | 1 |
| Erysipelothrix tonsillarum oral taxon 484 | 1 |
| Escherichia coli oral taxon 574 | 1 |
| Fusobacterium nucleatum subspecies | |
| F. nucleatum subsp. animalis oral taxon 420 | |
| F. nucleatum subsp. nucleatum oral taxon 698 | 1 |
| F. nucleatum subsp. polymorphum oral taxon 202 | 1 |
| F. nucleatum subsp. vincentii oral taxon 200 | 2 |
| Fusobacterium periodontium oral taxon 201 | |
| Gemella morbillorum oral taxon 046 | 1 |
| Leptotrichia spp. | |
| L. buccalis oral taxon 563 | |
| L. goodfellowii oral taxon 845 | 1 |
| L. holfstadii oral taxon 224 | 2 |
| L. wadei oral taxon 222 | 1 |
| Leptotrichia sp. oral taxon 212 | |
| Leptotrichia sp. oral taxon 221 | 1 |
| Leptotrichia sp. oral taxon 225 | |
| Microbacterium sp. oral taxon 186 | 1 |
| Neisseria spp. | |
| N. elongata oral taxon 598 | 3 |
| N. sicca oral taxon 764 | |
| N. subflava oral taxon 476 | 5 |
| Parvimonas spp. | |
| Parvimonas sp. oral taxon 110 | 1 |
| Parvimonas sp. oral taxon 393 | 1 |
| Peptostreptococcus stomatis oral taxon 112 | |
| Porphyromonas spp. | |
| P. catoniae oral taxon 283 | |
| P. endodontalis oral taxon 273 | |
| P. gingivalis oral taxon 619 | |
| Porphyromonas sp. oral taxon 279 | |
| Prevotella spp. | |
| P. intermedia oral taxon 643 | |
| Prevotella sp. oral taxon 472 | |
| Selenomonas spp. | |
| S. noxia oral taxon 130 | 2 |
| Selenomonas sp. oral taxon 126 | 1 |
| Selenomonas sp. oral taxon 133 | |
| Streptococcus spp. | |
| S. cristatus oral taxon 578 | |
| S. mitis oral taxon 398 | 1 |
| S. mitis oral taxon 677 | 4 |
| S. pneumoniae oral taxon 734 | 1 |
| S. sanguinis oral taxon 758 | 2 |
| Streptococcus sp. oral taxon 058 | 1 |
| Streptococcus sp. oral taxon 064 | |
| Streptococcus sp. oral taxon 071 | 1 |
| Tannerella spp. | |
| Tannerella forsythia oral taxon 613 | |
| Tannerella sp. oral taxon 286 | |
| TM7 phylum | |
| Oral taxon 346 | 1 |
| Oral taxon 347 | |
| Oral taxon 349 | |
| Treponema spp. | |
| T. denticola oral taxon 584 | |
| T. medium oral taxon 667 | |
| Veillonella spp. | |
| V. dispar oral taxon 160 | 1 |
| V. parvula oral taxon 161 | 1 |
Species identified in the subgingival plaque clone library with sequence identities of ≥97% with those available in the Human Oral Microbiome Database, with the exception of E. tonsillarum oral taxon 484 and E. coli oral taxon 574, which were not detected in the clone library. Taxa overlapping with those identified in saliva are in boldface type.
A total of 10 mice were injected with the pooled subgingival plaque sample.
The plaque bacteria translocated to the placenta were identified (see Table S2 in the supplemental material for details). Except for two mice which were infected only with Neisseria subflava, all mice had mixed infections with two or more different species. A total of 16 different genera/species were detected in the murine placenta (Table 3). Mice were preferentially infected by Neisseria spp. (in eight mice); Streptococcus spp. (in seven mice); Aggregatibacter segnis, Leptotrichia spp., and Selenomonas spp. (each in three mice); and Fusobacterium nucleatum and Eikenella corrodens (each in two mice). The remaining species, including Campylobacter showae, Capnocytophaga sp., Erysipelothrix tonsillarum, Gemella morbillorum, Microbacterium sp., Parvimonas sp., TM7 phylum sp., and Veillonella spp., were each identified in one mouse (Table 3). When each species was evaluated based on all 104 clones analyzed from the placenta, the order of prevalence remained the same, with 24% Neisseria spp., 14% Aggregatibacter segnis, 12% Streptococcus spp., and 12% Gemella morbillorum. The remaining species, C. showae, Capnocytophaga sp., E. corrodens, E. tonsillarum, Escherichia coli, F. nucleatum, Leptotrichia spp., Microbacterium sp., Parvimonas sp., Selenomonas spp., TM7 phylum sp., and Veillonella spp., showed prevalence of <10% (Fig. 2). With the exception of E. tonsillarum and E. coli, all species found in placenta were also detected in the dental plaque. Similar to P. stomatis discussed above, these two species may exist in low quantities in the subgingival plaque, undetected by random screening of the plaque clone library. Similarly, a few species were preferentially enriched in the placentas, including A. segnis, E. corrodens, E. tonsillarum, E. coli, G. morbillorum, Neisseria spp., Streptococcus spp., and Veillonella spp. (Fig. 2).
FIG. 2.
Transmission of bacteria from pooled subgingival plaque to murine placentas. The relative abundance of each bacterial genus/species in pooled subgingival plaque samples (solid bars) and placentas from all 10 infected mice (slashed bars) were expressed as the percentage of the species out of the total number of clones analyzed for the pooled subgingival plaque samples (n = 81) and the murine placentas (n = 104), respectively.
DISCUSSION
Utilizing the 16S rRNA gene-based PCR and clone analysis techniques, we provide initial insight into the diversity of oral bacteria that can translocate to the murine placenta following hematogenous infection. This approach mimics transient bacteremia, which can occur during periodontal infections and dental procedures.
Bacteremia caused by bacteria from both saliva and periodontal plaque has been documented, which was why both saliva and subgingival plaque samples were tested in our study (44, 46). These two samples represent two distinct yet closely related floras. The concordance between the two floras has been investigated in several studies, and the correlation was found to vary from species to species (12, 15). Our study, likewise, confirms these findings where the saliva and dental plaque microbiota share similarities and have differences. Out of 69 different taxa identified from both samples, 10 were shared by both. As a result, the species identified in the saliva-infected placenta only partially overlap with those from the plaque-infected placenta. Even for the species that do overlap, their prevalences differ. For example, Veillonella was one of the most prevalent bacteria in the saliva while it was not in the dental plaque. Consequently, Veillonella was detected in five out of seven mice infected with pooled saliva but only in 1 of 10 mice infected with pooled subgingival plaque. By testing both samples, our study provides a more complete spectrum of oral species capable of transmission to placenta.
Previous studies have shown that when pregnant mice were injected with PBS, no bacteria were recovered from the placenta (29). Thus, all bacteria detected in the placenta originated from the starting material of pooled saliva or pooled subgingival plaque. This was further confirmed by sequencing of pooled saliva and pooled subgingival plaque sample libraries, which identified the same species isolated from the mouse placentas. Studies have shown that each individual harbors approximately 266 taxa in the oral microflora (55). Therefore, only a portion of the oral microbiome was identified, yet it appeared to be sufficient to confirm the origin of the placental infection in our study.
The bacterial prevalence in the placenta and that in the starting materials were compared. A few species that existed in high prevalence in the starting samples, such as Neisseria and Veillonella, continued to be detected in the placentas with a high prevalence, suggesting a potential dose-dependent effect. On the contrary, bacteria such as Leptotrichia colonized the placenta with decreased prevalence compared with the starting samples, suggesting a dose-independent effect. We also note the possibility of selective enrichment of several species, such as A. segnis and P. stomatis, where they may have existed in relatively low quantities in the starting pooled samples, but were identified in the placentas with increased prevalence. These observations suggest that bacteria utilize specific translocation mechanisms rather than random “diffusion” to colonize the placenta.
The number of clones analyzed from each mouse library was determined by the quality of the library and the bacterial taxa identified. For the majority of the mice, one or few predominant species were identified after sequencing just a few clones (see Tables S1 and S2 in the supplemental material). Analysis of more clones would not significantly alter the prevalence of these species. The prevalence may differ for the species that were identified only once or twice in the library. Yet, it would not affect our conclusion. In this proof-of-concept study, the answer we seek is “yes” or “no.” Even if the bacterium is identified only in the placental libraries once, it will suggest that this particular organism is capable of translocating to the placenta. Due to the intrinsic differences between humans and mice, the relative prevalence found in the mouse placenta may not be applicable to humans.
The majority of the species detected in the murine placenta have been associated with adverse pregnancy outcomes (Table 4), validating the relevance of our study. Among them, F. nucleatum is the best-recognized oral species. We have shown previously that F. nucleatum translocates hematogenously into mouse placenta when pure cultures were used (29). The current study demonstrates that such translocation also occurs in mixed species, better mimicking real-life situations. Furthermore, only F. nucleatum was detected in the placenta although additional fusobacteria, such as Fusobacterium periodontium, were present in the pooled plaque sample. This is again indicative of a species-specific translocation. This observation is consistent with those made in humans that only F. nucleatum, not other fusobacteria, is associated with intrauterine infections.
TABLE 4.
Bacteria identified in human intrauterine infections
| Bacterium | Association with adverse pregnancy outcome in humans | Reference(s) |
|---|---|---|
| Aggregatibacter | Preeclampsia | 5 |
| Campylobacter | Prenatal and neonatal sepsis; PLBW | 20, 21, 26, 39, 49 |
| Capnocytophaga | Chorioamnionitis, PTB | 18, 31 |
| Eikenella | PTB | 22, 30 |
| E. coli | PTB, chorioamniotis, stillbirth | 8, 24, 25 |
| F. nucleatum | PTB | 27, 30, 31 |
| Gemella | Amniotic fluid infection | 45 |
| Leptotrichia | PTB, abortion, fetal death | 9, 17, 30, 48 |
| Neisseria | PTB | 17 |
| Peptostreptococcus | PTB | 17, 30 |
| Streptococcus | PTB, urinary tract infection, neonatal sepsis | 23, 30 |
| Veillonella | PTB | 47 |
Some of the clones translocated to the murine placenta and those identified in intrauterine infection in humans were nonidentical but rather were closely related species of the same genus. They include Neisseria, Leptotrichia, Aggregatibacter, Campylobacter, Capnocytophaga, and Peptostreptococcus (Table 4). The discrepancy observed here may be due to the differences between humans and mice. It may also be due to the discrepancy in microbial nomenclature. It has been reported by our group and others that the nomenclature may differ based on the technology used (30). The fact that the species identified in our study, including Leptotrichia goodfellowii, N. flavescens, N. subflava, A. segnis, Campylobacter showae, and Capnocytophaga granulosa, have been associated with extraoral infections involving hematogenous transmission, such as endocarditis, meningitidis, and extraoral abscesses (2, 16, 19, 36, 50, 53), indicates their ability to translocate to different body parts and their virulence potential.
Some species identified in the placenta exist in multiple maternal microfloras, including Campylobacter, E. coli, Leptotrichia, Neisseria, Peptostreptococcus, and Streptococcus (Table 4). For example, E. coli is widely known as an enteric pathogen, but it has been also associated with stillbirth (25, 41). It has been considered that intra-amniotic infection with E. coli results from the lower genital tract through an ascending route (25). However, E. coli can be isolated from the oral cavity even if it is not one of the most prevalent bacteria (43). Our results indicate that the maternal-fetal infection by E. coli and the above-mentioned species can also originate from the oral cavity as a result of bacteremia.
Some bacteria detected in the murine placenta have not been found in intrauterine infections in humans, including Erysipelothrix, Granulicatella, Microbacterium, Parvimonas (formerly known as Micromonas), Selenomonas, and the TM7 phylum. Again, this could be due to the difference between humans and mice. Interestingly, this group includes uncultivated and relatively new species, such as TM7. Thus, it is also possible that they are implicated in human intrauterine infections but have not been identified because the clinical laboratories still use routine culturing to detect microbial infections.
One very interesting observation is that many species that translocate to the murine placenta and are associated with human intrauterine infections are commensal organisms (Tables 2 to 4). For example, we identified S. mitis, a ubiquitous oral species, in the mouse placenta. This organism, associated with various infections such as endocarditis and meningitis (14, 35), was recently identified in amniotic fluid from three cases of PTB (17). Similarly, Veillonella is also a well-recognized oral commensal species. Several reports described its association with human extraoral infections, including bacteremia, endocarditis, osteomyelitis, and meningitides, confirming its status as an opportunistic pathogen (7, 10, 11, 13, 51, 54). It has also been isolated from amniotic fluid accompanying PTB (47). Therefore, our results indicate the importance of commensal oral species in intrauterine infection.
In contrast, several well-recognized periodontal pathogens, such as Porphyromonas, Tannerella, and Treponema, i.e., the “red-complex” organisms (52), although present in the plaque sample, were not detected in the placenta. This could be due to several reasons. First, these species existed in low quantities in the plaque, placing them in a disadvantaged position in this “numbers game.” Second, the clones in the tested samples may not be the most “transmissive” isolates. Previous studies have shown that placental translocation of P. gingivalis is strain dependent (6).
How do these different bacteria colonize the mouse placenta? We have shown previously that F. nucleatum colonizes the murine placenta by invading and crossing the endothelial lining (29). This process requires the FadA adhesin of F. nucleatum (32). Similarly, we speculate that the placental species identified in our study may possess invasive properties. Our study does not address the question as what happens after these different bacteria colonize in the mouse placenta. We have, however, demonstrated previously that after F. nucleatum colonized the placenta, it proliferated quickly to reach a titer of 107 CFU per gram of tissue in 72 h and spread to the fetus and amniotic fluid, causing fetal death (29). We have shown that fetal death was the result of localized TLR4-mediated inflammation following the bacterial infection. When TLR4 activation was blocked by its antagonist, F. nucleatum still colonized the placenta to the same extent without causing fetal death (37). It will be interesting to see if similar or different events occur with different species in the placenta.
In summary, we identified a diverse group of oral bacteria that can translocate to the mouse placenta as a result of bacteremia. These findings are consistent with the observation made in humans that various microbial species have been detected in intrauterine infection. With the exception of the two cases reported from our laboratory, the origins of most intrauterine infections were not determined. Results from the current study suggest that the oral cavity may be a previously overlooked source of intrauterine infection. Furthermore, for the first time, our study indicates the potential significance of commensal oral species in intrauterine infection. This is consistent with the previous report that many intrauterine species are of low virulence. Most of the species reported in this study have been detected in transient bacteremia in humans (4). Previous studies have identified periodontal disease as a potential risk factor for PTB. However, intervention studies employing various periodontal therapies produced inconsistent results among pregnant women (33, 38, 40, 42). One of the most prevalent forms of periodontal disease during gestation is pregnancy-associated gingivitis, affecting at least three-quarters of the expectant mothers (3). Gingivitis is characterized by inflamed gingiva and increased bacterial titers, including those of commensal species. These conditions may lead to frequent bacteremia, thus increasing the opportunity of hematogenous transmission. In the two cases of intrauterine infection with oral Bergeyella or F. nucleatum, both women showed no signs of periodontitis (a more advanced form of periodontal disease characterized by bone and attachment loss) during postpartum examinations but were suspected to have pregnancy-associated gingivitis (27, 28). Based on our findings, we postulate that periodontal therapies targeted at consistently reducing the total bacterial load in the mother's oral cavity may be effective in improving birth outcomes.
Supplementary Material
Acknowledgments
We thank Floyd Dewhirst for consultation on the use of the Human Oral Microbiome Database, Howard Kuramitsu for reading the manuscript, and Brian Schmotzer for assistance with revision.
This work was supported in part by NIH grants RO1 DE14924, KO2 DE16102, and R21 DE17165 to Y.W.H.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 1 February 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amsel, B. J., and A. C. Moulijn. 1996. Nonfebrile mitral valve endocarditis due to Neisseria subflava. Chest 109:280-282. [DOI] [PubMed] [Google Scholar]
- 3.Arafat, A. H. 1974. Periodontal status during pregnancy. J. Periodontol. 45:641-643. [DOI] [PubMed] [Google Scholar]
- 4.Bahrani-Mougeot, F. K., B. J. Paster, S. Coleman, J. Ashar, S. Barbuto, and P. B. Lockhart. 2008. Diverse and novel oral bacterial species in blood following dental procedures. J. Clin. Microbiol. 46:2129-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barak, S., O. Oettinger-Barak, E. E. Machtei, H. Sprecher, and G. Ohel. 2007. Evidence of periopathogenic microorganisms in placentas of women with preeclampsia. J. Periodontol. 78:670-676. [DOI] [PubMed] [Google Scholar]
- 6.Belanger, M., L. Reyes, K. von Deneen, M. K. Reinhard, A. Progulske-Fox, and M. B. Brown. 2008. Colonization of maternal and fetal tissues by Porphyromonas gingivalis is strain-dependent in a rodent animal model. Am. J. Obstet. Gynecol 199:86.e1-86.e7. [DOI] [PubMed] [Google Scholar]
- 7.Bhatti, M. A., and M. O. Frank. 2000. Veillonella parvula meningitis: case report and review of Veillonella infections. Clin. Infect. Dis. 31:839-840. [DOI] [PubMed] [Google Scholar]
- 8.Bhola, K., H. Al-Kindi, M. Fadia, A. L. Kent, P. Collignon, and J. E. Dahlstrom. 2008. Placental cultures in the era of peripartum antibiotic use. Aust. N. Z. J. Obstet. Gynaecol. 48:179-184. [DOI] [PubMed] [Google Scholar]
- 9.Boennelycke, M., J. J. Christensen, M. Arpi, and S. Krause. 2007. Leptotrichia amnionii found in septic abortion in Denmark. Scand. J. Infect. Dis. 39:382-383. [DOI] [PubMed] [Google Scholar]
- 10.Bongaerts, G. P., B. W. Schreurs, F. V. Lunel, J. A. Lemmens, M. Pruszczynski, and M. A. Merkx. 2004. Was isolation of Veillonella from spinal osteomyelitis possible due to poor tissue perfusion? Med. Hypotheses 63:659-661. [DOI] [PubMed] [Google Scholar]
- 11.Boo, T. W., B. Cryan, A. O'Donnell, and G. Fahy. 2005. Prosthetic valve endocarditis caused by Veillonella parvula. J. Infect. 50:81-83. [DOI] [PubMed] [Google Scholar]
- 12.Boutaga, K., P. H. Savelkoul, E. G. Winkel, and A. J. van Winkelhoff. 2007. Comparison of subgingival bacterial sampling with oral lavage for detection and quantification of periodontal pathogens by real-time polymerase chain reaction. J. Periodontol. 78:79-86. [DOI] [PubMed] [Google Scholar]
- 13.Brook, I. 1996. Veillonella infections in children. J. Clin. Microbiol. 34:1283-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cone, L. A., J. Hirschberg, C. Lopez, P. K. Kanna, E. J. Goldstein, A. Kazi, R. Gade-Andavolu, and B. Younes. 2008. Infective endocarditis associated with spondylodiscitis and frequent secondary epidural abscess. Surg. Neurol. 69:121-125. [DOI] [PubMed] [Google Scholar]
- 15.Darout, I. A., J. M. Albandar, and N. Skaug. 2002. Correlations between bacterial levels in autologous subgingival plaque and saliva of adult Sudanese. Clin. Oral Invest. 6:210-216. [DOI] [PubMed] [Google Scholar]
- 16.de Vries, J. J., N. L. Arents, and W. L. Manson. 2008. Campylobacter species isolated from extra-oro-intestinal abscesses: a report of four cases and literature review. Eur. J. Clin. Microbiol. Infect. Dis. 27:1119-1123. [DOI] [PubMed] [Google Scholar]
- 17.DiGiulio, D. B., R. Romero, H. P. Amogan, J. P. Kusanovic, E. M. Bik, F. Gotsch, C. J. Kim, O. Erez, S. Edwin, and D. A. Relman. 2008. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One 3:e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douvier, S., C. Neuwirth, L. Filipuzzi, and J. P. Kisterman. 1999. Chorioamnionitis with intact membranes caused by Capnocytophaga sputigena. Eur. J. Obstet. Gynecol. Reprod. Biol. 83:109-112. [DOI] [PubMed] [Google Scholar]
- 19.Ebinger, M., T. Nichterlein, U. K. Schumacher, B. Manncke, D. Schmidt, and I. Bohn. 2000. Isolation of Capnocytophaga granulosa from an abscess in an immunocompetent adolescent. Clin. Infect. Dis. 30:606-607. [DOI] [PubMed] [Google Scholar]
- 20.Forbes, J. C., and D. W. Scheifele. 1987. Early onset Campylobacter sepsis in a neonate. Pediatr. Infect. Dis. J. 6:494. [PubMed] [Google Scholar]
- 21.Fujihara, N., S. Takakura, T. Saito, Y. Iinuma, and S. Ichiyama. 2006. A case of perinatal sepsis by Campylobacter fetus subsp. fetus infection successfully treated with carbapenem-case report and literature review. J. Infect. 53:e199-e202. [DOI] [PubMed] [Google Scholar]
- 22.Garnier, F., G. Masson, A. Bedu, P. Masson, E. Decroisette, V. Guigonis, V. Fermeaux, M. C. De Barbentane, F. Denis, and M. C. Ploy. 2009. Maternofetal infections due to Eikenella corrodens. J. Med. Microbiol. 58:273-275. [DOI] [PubMed] [Google Scholar]
- 23.Gibbs, R. S., S. Schrag, and A. Schuchat. 2004. Perinatal infections due to group B streptococci. Obstet. Gynecol. 104:1062-1076. [DOI] [PubMed] [Google Scholar]
- 24.Goldenberg, R. L., J. C. Hauth, and W. W. Andrews. 2000. Intrauterine infection and preterm delivery. N. Engl. J. Med. 342:1500-1507. [DOI] [PubMed] [Google Scholar]
- 25.Goldenberg, R. L., and C. Thompson. 2003. The infectious origins of stillbirth. Am. J. Obstet. Gynecol. 189:861-873. [DOI] [PubMed] [Google Scholar]
- 26.Gribble, M. J., I. E. Salit, J. Isaac-Renton, and A. W. Chow. 1981. Campylobacter infections in pregnancy. Case report and literature review. Am. J. Obstet. Gynecol. 140:423-426. [DOI] [PubMed] [Google Scholar]
- 27.Han, Y. W., Y. Fardini, C. Chen, K. G. Iacampo, V. A. Peraino, J. M. Shamonki, and R. W. Redline. 2010. Term stillbirth caused by oral Fusobacterium nucleatum. Obstet. Gynecol. 115:442-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han, Y. W., A. Ikegami, N. F. Bissada, M. Herbst, R. W. Redline, and G. G. Ashmead. 2006. Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth. J. Clin. Microbiol. 44:1475-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han, Y. W., R. W. Redline, M. Li, L. Yin, G. B. Hill, and T. S. McCormick. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect. Immun. 72:2272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han, Y. W., T. Shen, P. Chung, I. A. Buhimschi, and C. S. Buhimschi. 2009. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J. Clin. Microbiol. 47:38-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill, G. B. 1998. Preterm birth: associations with genital and possibly oral microflora. Ann. Periodontol. 3:222-232. [DOI] [PubMed] [Google Scholar]
- 32.Ikegami, A., P. Chung, and Y. W. Han. 2009. Complementation of the fadA mutation in Fusobacterium nucleatum demonstrates that the surface-exposed adhesin promotes cellular invasion and placental colonization. Infect. Immun. 77:3075-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeffcoat, M. K., N. C. Geurs, M. S. Reddy, S. P. Cliver, R. L. Goldenberg, and J. C. Hauth. 2001. Periodontal infection and preterm birth: results of a prospective study. J. Am. Dent. Assoc. 132:875-880. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338-344. [DOI] [PubMed] [Google Scholar]
- 35.Kutlu, S. S., S. Sacar, N. Cevahir, and H. Turgut. 2008. Community-acquired Streptococcus mitis meningitis: a case report. Int. J. Infect. Dis. 12:e107-e109. [DOI] [PubMed] [Google Scholar]
- 36.Lau, S. K., P. C. Woo, M. Y. Mok, J. L. Teng, V. K. Tam, K. K. Chan, and K. Y. Yuen. 2004. Characterization of Haemophilus segnis, an important cause of bacteremia, by 16S rRNA gene sequencing. J. Clin. Microbiol. 42:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, H., R. W. Redline, and Y. W. Han. 2007. Fusobacterium nucleatum induces fetal death in mice via stimulation of TLR4-mediated placental inflammatory response. J. Immunol. 179:2501-2508. [DOI] [PubMed] [Google Scholar]
- 38.López, N. J., I. Da Silva, J. Ipinza, and J. Gutierrez. 2005. Periodontal therapy reduces the rate of preterm low birth weight in women with pregnancy-associated gingivitis. J. Periodontol. 76:2144-2153. [DOI] [PubMed] [Google Scholar]
- 39.Madianos, P. N., S. Lieff, A. P. Murtha, K. A. Boggess, R. L. Auten, Jr., J. D. Beck, and S. Offenbacher. 2001. Maternal periodontitis and prematurity. Part II: maternal infection and fetal exposure. Ann. Periodontol. 6:175-182. [DOI] [PubMed] [Google Scholar]
- 40.Michalowicz, B. S., J. S. Hodges, A. J. DiAngelis, V. R. Lupo, M. J. Novak, J. E. Ferguson, W. Buchanan, J. Bofill, P. N. Papapanou, D. A. Mitchell, S. Matseoane, and P. A. Tschida. 2006. Treatment of periodontal disease and the risk of preterm birth. N. Engl. J. Med. 355:1885-1894. [DOI] [PubMed] [Google Scholar]
- 41.Moyo, S. R., I. Hagerstrand, L. Nystrom, S. A. Tswana, J. Blomberg, S. Bergstrom, and A. Ljungh. 1996. Stillbirths and intrauterine infection, histologic chorioamnionitis and microbiological findings. Int. J. Gynaecol Obstet. 54:115-123. [DOI] [PubMed] [Google Scholar]
- 42.Offenbacher, S., J. D. Beck, H. L. Jared, S. M. Mauriello, L. C. Mendoza, D. J. Couper, D. D. Stewart, A. P. Murtha, D. L. Cochran, D. J. Dudley, M. S. Reddy, N. C. Geurs, and J. C. Hauth. 2009. Effects of periodontal therapy on rate of preterm delivery: a randomized controlled trial. Obstet. Gynecol. 114:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez-Chaparro, P. J., P. Gracieux, G. I. Lafaurie, P. Y. Donnio, and M. Bonnaure-Mallet. 2008. Genotypic characterization of Porphyromonas gingivalis isolated from subgingival plaque and blood sample in positive bacteremia subjects with periodontitis. J. Clin. Periodontol. 35:748-753. [DOI] [PubMed] [Google Scholar]
- 45.Rabe, L. K., K. K. Winterscheid, and S. L. Hillier. 1988. Association of viridans group streptococci from pregnant women with bacterial vaginosis and upper genital tract infection. J. Clin. Microbiol. 26:1156-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richard, P., G. Amador Del Valle, P. Moreau, N. Milpied, M. P. Felice, T. Daeschler, J. L. Harousseau, and H. Richet. 1995. Viridans streptococcal bacteraemia in patients with neutropenia. Lancet 345:1607-1609. [DOI] [PubMed] [Google Scholar]
- 47.Romero, R., A. L. Scioscia, S. C. Edberg, and J. C. Hobbins. 1986. Use of parenteral antibiotic therapy to eradicate bacterial colonization of amniotic fluid in premature rupture of membranes. Obstet. Gynecol. 67:15S-17S. [DOI] [PubMed] [Google Scholar]
- 48.Shukla, S. K., P. R. Meier, P. D. Mitchell, D. N. Frank, and K. D. Reed. 2002. Leptotrichia amnionii sp. nov., a novel bacterium isolated from the amniotic fluid of a woman after intrauterine fetal demise. J. Clin. Microbiol. 40:3346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simor, A. E., M. A. Karmali, T. Jadavji, and M. Roscoe. 1986. Abortion and perinatal sepsis associated with campylobacter infection. Rev. Infect. Dis. 8:397-402. [DOI] [PubMed] [Google Scholar]
- 50.Sinave, C. P., and K. R. Ratzan. 1987. Infective endocarditis caused by Neisseria flavescens. Am. J. Med. 82:163-164. [DOI] [PubMed] [Google Scholar]
- 51.Singh, N., and V. L. Yu. 1992. Osteomyelitis due to Veillonella parvula: case report and review. Clin. Infect. Dis. 14:361-363. [DOI] [PubMed] [Google Scholar]
- 52.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 53.Somers, C. J., B. C. Millar, J. Xu, D. P. Moore, A. M. Moran, C. Maloney, B. Keogh, P. G. Murphy, and J. E. Moore. 2003. Haemophilus segnis: a rare cause of endocarditis. Clin. Microbiol. Infect. 9:1048-1050. [DOI] [PubMed] [Google Scholar]
- 54.Strach, M., M. Siedlar, D. Kowalczyk, M. Zembala, and T. Grodzicki. 2006. Sepsis caused by Veillonella parvula infection in a 17-year-old patient with X-linked agammaglobulinemia (Bruton's disease). J. Clin. Microbiol. 44:2655-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaura, E., B. J. Keijser, S. M. Huse, and W. Crielaard. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.