Abstract
Activity of the sodium/iodide symporter (NIS) in lactating breast is essential for iodide (I–) accumulation in milk. Significant NIS upregulation was also reported in breast cancer, indicating a potential use of radioiodide treatment. All-trans-retinoic acid (tRA) is a potent ligand that enhances NIS expression in a subset of breast cancer cell lines and in experimental breast cancer models. Indirect tRA stimulation of NIS in breast cancer cells is very well documented; however, direct upregulation by tRA-activated nuclear receptors has not been identified yet. Aiming to uncover cis-acting elements directly regulating NIS expression, we screened evolutionary-conserved non-coding genomic sequences for responsiveness to tRA in MCF-7. Here, we report that a potent enhancer in the first intron of NIS mediates direct regulation by tRA-stimulated nuclear receptors. In vitro as well as in vivo DNA–protein interaction assays revealed direct association between retinoic acid receptor-α (RARα) and retinoid-X-receptor (RXR) with this enhancer. Moreover, using chromatin immunoprecipitation (ChIP) we uncovered early events of NIS transcription in response to tRA, which require the interaction of several novel intronic tRA responsive elements. These findings indicate a complex interplay between nuclear receptors, RNA Pol-II and multiple intronic RAREs in NIS gene, and they establish a novel mechanistic model for tRA-induced gene transcription.
INTRODUCTION
In alveolar cells of the lactating mammary gland, the activity of Na+/I– symporter (NIS) is required for the secretion of I– in mother’s milk (1,2). I– in milk is then used by the newborn in thyroid hormone biosynthesis, and therefore it has an essential role in postnatal development of the baby (3). The timing of NIS expression in mammary epithelia is precisely synchronized with the onset of gestation and with lactation, and it ends when newborns stop suckling (2). NIS is not expressed in non-lactating mammary gland tissue unless experimental animals were treated with combinations of steroid and lactogenic hormones (2,4,5). Yet, a functional overexpression of NIS was detected in transgenic mice bearing experimental mammary tumors induced by mouse mammary tumor virus (MMTV) promoter-driven activated Ras and Erb-B2/neu oncogenes, polyoma middle-T antigen (PyMT), or cyclooxygenase-2 gene (2,6,7).
Relevant with the human breast cancer, increased NIS expression was also detected in about 70–80% of ductal carcinoma in situ and invasive breast cancer samples, as compared to the absence of NIS expression in healthy breast samples obtained in reductive mammoplasty operations (2,8). On the one hand, these results have raised the possibility that radioiodide may be used for diagnosis and treatment of breast cancer, as it is routinely used against thyroid cancer and its metastasis (9). On the other hand, only a small fraction of these NIS-positive tumors has the capacity to accumulate radio-labeled substrates of the symporter (2,4,8), suggesting that further enhancement of NIS expression or post-translational activation of NIS are required for an effective use of radio-labeled substrates for imaging and/or treatment of malignant breast disease.
Related with this, a number of hormones or ligands, such as retinoids (natural or synthetic analogs of vitamin A), human chorionic gonadotropin (hCG), dexamethasone (Dex) and troglitazone (a PPAR-γ ligand), were identified as molecules having an upregulatory effect on NIS expression and I– transport in the ERα-positive (ERα+) breast cancer cell model, MCF-7 (6,10–12). Retinoids have the most robust effect on NIS transcription, and among this group of ligands, all-trans-retinoic acid (tRA) is the most potent as it leads to an increase up to 10-fold both at the transcriptional level, as well as at the level of I− transport (10–12). Moreover, in immunodeficient mice model with MCF-7 cell xenografts, systemic tRA treatment stimulated NIS expression and radioiodide uptake around 15-fold in the implemented graft (7).
We have previously shown that, tRA induces NIS expression only in ERα+ mammary cell line models, and neither basal nor inducible NIS transcription is observed in ERα-negative (ERα−) mammary cell lines (13). ERα directly interacts with a responsive element in NIS promoter, and independent from the presence of estradiols the presence of ERα is important for maximal tRA induced NIS transcription (13). tRA stimulates NIS transcription also in human follicular thyroid carcinoma cell lines FTC-133 and FTC-238, and in these cells the effect of tRA is mediated by a RA response element (RARE) that is located at position –1375 relative to human NIS start codon (14). Dentice et al. (15) have shown that the cardiac homeobox transcription factor Nkx2.5 is induced by tRA, and this factor is involved in the upregulation of NIS transcription by binding to two cis-acting elements in NIS promoter.
Conserved transcriptional expression patterns of orthologous and paralogous genes could reflect common molecular regulatory mechanisms, as well as the presence of conserved regulatory cis-acting elements. Results supporting this view have previously been published for the Drosophila homeotic (hox) gene complex (16), and similar observations were reported for NIS (17). For instance, when the entire 5′-flanking sequences starting from NIS coding region up to the upstream neighboring gene (RPL18A) were compared, it was found that the similarity between rat and human sequences is only 11.8%. Yet, the similarity ratios of previously established cis-acting elements that are located in the same sequence are remarkably high, for example, in the NIS upstream enhancer (NUE), which is a strong thyroid-stimulating hormone (TSH) responsive enhancer that is localized between −9470 and −9046 on the human NIS and −2611 and −2230 on the rat Nis is 70% similar (18,19). Again, as expected, the basal promoter of the human NIS (–475 to −393) has ∼72% similarity with the rat Nis promoter (17). In this study, we initially undertook a comparative genomics approach in order to identify evolutionary conserved sequences that could play a role in tRA-dependent NIS regulation. Subsequently, our experimental studies resulted in the identification of a potent intronic enhancer, which binds RARα and RXR, and upon stimulation by tRA activates NIS promoter both in vitro and in vivo. We also show that the initiation of transcription by tRA involves interactions between several identical RARE sequences found in multiple introns of the gene and RA stimulated nuclear receptors in vivo. We present a novel as well as a rather complex and dynamic interplay between putative RA responsive cis-acting elements, RNA Pol-II and RA-stimulated nuclear receptors during NIS transcription.
MATERIALS AND METHODS
Sequence information and databases
Genomic DNA sequences to be analyzed for DNA conservation in non-coding sequences were obtained from the Genome Browser database at the University of California Santa Cruz (20). The analysis was performed on 90-kb sequences from Homo sapiens (Release July 2003; Chr. 19: 17816282-17906905), Rattus norvegicus (release June 2003; Chr. 16: 19018698-19107000) and from Mus musculus (Release October 2003; Chr. 8: 71152446-71242000). Annotations for the human sequence were obtained from the Genome Vista database, and sequences were aligned with the mVista tool using the human sequence at the x-axis and at 50% conservation level and a window length of 75 bp.
Polymerase chain reaction amplification of conserved regions
Conserved regions clustered within about 1 kb were amplified by polymerase chain reaction (PCR) as one larger fragment and subsequently cloned into the reporter vector. PCR amplification was performed in 25-µl reaction volumes containing 0.8 × PCR buffer, 3 mM MgCl2, 200 µM dNTP, 10 pmol of each primer (see Supplementary Table S1 for primer sequences), 5% DMSO, 1 U of Taq DNA polymerase (Roche) and 100 ng of human genomic DNA. Thermal cycler conditions were an initial denaturation step at 94°C for 4 min; a loop cycle of 94°C, 30 s/62°C, 30 s/72°C, 40 s; and a final extension at 72°C for 7 min.
Reporter vector constructs
The reporter pGL3-E1b is a modified version of the original plasmid pGL3-Basic (Promega), which contains the E1b TATA element (5′-TCG AGT CTA GAG GGT ATA TAA TGG ATC-3′) between XhoI/BglII sites of pGL3-Basic; destroying the BglII site while keeping the XhoI site intact. PCR products containing conserved regions were cloned into MluI/XhoI sites of pGL3-E1b to produce the reporter plasmids named clusters (Cl) 1, 2, 3, 4, 5, 6 and 7.
Cluster 3 derivatives containing individual or combined conserved regions were prepared by restriction endonuclease digestion of internal sequences, or by PCR. The plasmid Cl3-1 was prepared by removing EspI/XhoI fragment from cluster 3, likewise plasmids Cl3-pm, Cl3 Δ3-4, Cl3-xp and Cl3-4 were prepared by removing the PvuII/MluI, the PstI/SmaI, the XhoI/PvuII and the SacI fragments, respectively. The plasmids Cl.3-2 and Cl3-3 were prepared by cloning PCR product containing each conserved region using the primers shown in Supplementary Table S2. Subsequently, PCR products were cloned into MluI/XhoI sites of pGL3-E1b.
The plasmid DR2-1del was created by introducing a 490-bp deletion in Cl4Wt, deletion was created using restriction enzyme digestion with MluI and BalI. MluI digested overhangs were filled by incubating the digestion products with 5 U of Klenow fragment of DNA polymerase I (Fermentas) in the presence of 50 µM dNTP at 37°C for 1 h prior to ligation.
The plasmid Cl4-R was prepared by cloning the insert from Cl4Wt into BamHI and SalI sites downstream of the luciferase reporter. Cl3-pm was created by removing the sequence between PvuII and MluI from Cl3. The reporter Cl4-pm was created by replacing the MluI/PvuII fragment from Cl3 with KpnI/XhoI fragment containing Cl4 insert. Finally, Cl34-pm was prepared by cloning the insert from Cl4 into BamHI/SalI of Cl3-pm.
Site-directed mutagenesis
Site-directed mutagenesis was performed on constructs harboring the retinoic acid response elements in clusters 3 and 4. Summary of the oligonucleotides used in this procedure is presented in Supplementary Table S2. PCR-based mutagenesis was performed in 50-µl reaction volumes containing 40 ng of target plasmid DNA, 1× Pfu buffer, 200 µM dNTP mix, 100 ng of each primer and 2.5 U of Pfu-Turbo DNA polymerase (Stratagene). Reaction conditions were an initial denaturation step at 95°C for 30 s followed by 15 cycles of 95°C for 30 s/55°C for 1 min/68°C for 6 min and 20 s. Following the PCR reaction the tubes were cooled to 37°C on ice and then 1 µl of DpnI (10 U/µl; Fermentas) was added and incubated at 37°C for 1 h. Following the DpnI digestion of the parental (methylated) strand, 5 µl were used for transformation of super-competent E. coli (DH5α) cells. Plasmids were rescued from single colonies and checked for the presence of the mutation by automated DNA sequencing.
Luciferase reporter assay
MCF-7 cells were maintained in high glucose DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin (P/S) and 1% l-glutamine (Biochrom), at 37°C in a 5% CO2 incubator. Cells were transfected with plasmid DNAs using FuGENE-6 reagent (Roche). FuGENE:DNA ratios were determined experimentally to be 3:1. Cells were seeded in 24-well plates in DMEM; so that they would reach confluence at the time of the assay. Two days later, and 1 h prior to transfection, cells were washed twice with phosphate-buffered saline (PBS), and the medium was replaced with DMEM lacking antibiotics. Transfection was carried out with 200 ng of reporter vector plus 3 ng phRL-TK (Promega) to normalize for transfection efficiency. Two days post-transfection, medium was changed with fresh DMEM containing 1 µM tRA (Sigma) and continued incubation for 24 h, dimethyl sulfoxide (DMSO) was used as vehicle control. Then the cells were harvested and luciferase reporter assays were performed using the Dual-Glo Luciferase Assay system (Promega). Luciferase values for all samples were normalized by first subtracting the background of untransfected control, and then dividing firefly luciferase values over those of Renilla luciferase. Fold induction is relative to the value of the empty vector. Fold stimulation was calculated by dividing the values of the tRA-stimulated sample by the DMSO control sample and relative to the vector containing promoter only control. Experiments were performed at least three times in duplicates.
Electrophoretic mobility shift assay
Nuclear extracts were prepared using the NE-PER solution (Nuclear and Cytoplasmic Extraction Reagents – Pierce Biotechnology). Sub-confluent MCF-7 cells grown in 150-mm plates were treated with 10-nM E2 (for maximal RARα expression) for 6 h and then harvested by trypsinization and collected by centrifugation at 1200 r.p.m. Nuclear protein extraction was performed using 50 µl packed cell volume samples according to the manufacturer’s procedure. Oligonucleotide labeling was performed using the Biotin 3′ End Labeling Kit (Pierce Biotechnology). The labeling reactions contained 1× TdT reaction buffer, 100 nM unlabeled oligo, 0.5 µM Biotin-11-UTP and 10 U TdT in 50-µl reaction volumes. Electrophoretic mobility shift assay (EMSA) experiments were performed using the Lightshift Chemiluminescent EMSA kit (Pierce Biotechnology) according to the protocols provided. In brief, binding reactions included 1 × binding buffer, 2.5% glycerol, 5 mM MgCl2, 50 ng/µl Poly (dI•dC), 0.05% NP-40, 4 pmol of unlabeled oligo, 5 µl nuclear extract and 20 fmol of biotin labeled oligo. For the super shift reaction 1 µg of anti-RARα (C-20, Santa Cruz) or anti-RXR (F-1, Santa Cruz) antibodies were included to the reaction just before adding the Biotin labeled oligos (Supplementary Table S2). Binding reactions were incubated at room temperature for 40 min and then resolved on a 6% native polyacrylamide gel in 0.5× TBE in a minigel electrophoresis apparatus (dimensions 8 × 8 × 0.1 cm). After electrophoresis, complexes were electro-transferred to Hybond-N+ nylon membranes (Amersham), and then cross-linked in the GS Gene Linker (Bio-Rad) at an energy level of 150 mJ. Chemiluminescent detection of biotin-labeled DNA was performed according to the supplier’s protocol (Pierce Biotechnology).
Chromatin immunoprecipitation
MCF-7 cells (in 6-well plates) grown in steroid-free and phenol-red-free DMEM (Gibco) were treated with 1 µM tRA (Sigma) in the presence of 10 nM estradiol (Sigma). Formaldehyde cross-linking [1% (w/v)] was performed for 10 min at room temperature. Cross-linking was terminated by the addition of glycine to a final concentration of 125 mM. Cells were scraped in PBS, divided into two tubes and lysed in 400 µl lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris–HCl (pH 8.0) and 1 × protease inhibitor cocktail]. Cell lysates were then sonicated twice in the ultrasonic processor VCX-500 (Sonics and Materials, INC.), each for 15 min at 4°C using a pulse of 30 s and 80% amplitude. Cell lysates were cleared by centrifugation at 13 000 r.p.m. for 10 min, diluted to 6.1 ml in dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl (pH 8.0), 167 mM NaCl and 1× protease inhibitor cocktail]. At this point, 100 µl of chromatin solution was removed and labeled as ‘Input’. Chromatin solution was precleared by adding 50 µl of protein A-Sepharose CL-4B (GE Healthcare) as 50% slurry containing 0.5 mg/ml BSA (Sigma), 200 µg/ml sonicated salmon sperm DNA (Sigma) in TE (pH 8.0) for 60 min. at 4°C. Immunoprecipitation was performed at 4°C for 16 h using chromatin immunoprecipitation (ChIP)-grade antibodies against RARα (H1920, Abcam), RXR (F-1, Santa Cruz), Pol-II (N-20, Santa Cruz) and FGFR1 (C-15, Santa Cruz). Then, immunocomplexes were incubated with 50 µl protein A-Sepharose (50% slurry) for at least 16 h at 4°C. Afterwards, beads were collected, washed four times in wash buffer I [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl (pH 8.0) and 150 mM NaCl], and twice in wash buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl (pH 8.0) and 500 mM NaCl]. Precipitated DNA (as well as the inputs) was recovered using a solution of 10% Chelex (Bio-Rad) and Proteinase K, and was used for qPCR amplification. Primers used for qPCR amplification are presented in the Supplementary Table S3. Quantitative real-time PCR was performed using iQ™ SYBR® Green Supermix (BioRad) and amplification was performed in the Mx3005P® QPCR System (Stratagene) using the following cycling parameters: 95°C for 10 min, then 45 cycles of 95°C for 15 s followed by 64°C for 15 s and 72°C for 20 s, and a melt curve starting at 55°C with 0.5°C increments for 80 cycles, except for intron 8 element, annealing temperature was 58°C. Data was normalized to the background (No Ab) and calculated as fold enrichment over the control IgG (FGFR1) applying the formula [2−(normalized Ct sample - normalized Ct IgG)]. Parallel PCR reactions were performed for agarose gel electrophoresis analysis, PCR parameters were an initial denaturation at 95°C for 5 min and 19 cycles of 15 s at 95°C, 15 s at 64°C and 20 s at 72°C. PCR products were resolved on a 2% agarose gel stained with ethidium bromide. At least three qPCR reactions in triplicates were performed on two independent ChIP experiments.
mRNA expression analysis
A small fraction of the cells used for ChIP analysis was used for expression analysis. RNA was prepared using the Nucleospin RNA II kit (Macherey-Nagel) as recommended by the manufacturer. RNA quality was checked spectrophotometrically and quantified using NanoDrop 2000 (Thermo Scientific). Two micrograms of total RNA were used for cDNA synthesis using the Revert-Aid First Strand cDNA Synthesis Kit (Fermentas). NIS and GAPDH primers used for real-time PCR quantification are presented in Supplementary Table S3. Quantitative real-time PCR was performed as described above. Cycling parameters were similar with annealing temperature of 60°C. Quantification was performed using the ΔΔCt method. Data represent three qPCR experiments in triplicates from two independent samples.
Statistical analysis
Statistical significance was determined by performing the Student’s t-test (Figures 4D, 6 and 7A) or paired Student’s t-test (Figures 2, 3, 4C and 7C) using a 95% confidence interval; P-values <0.05 were considered significant.
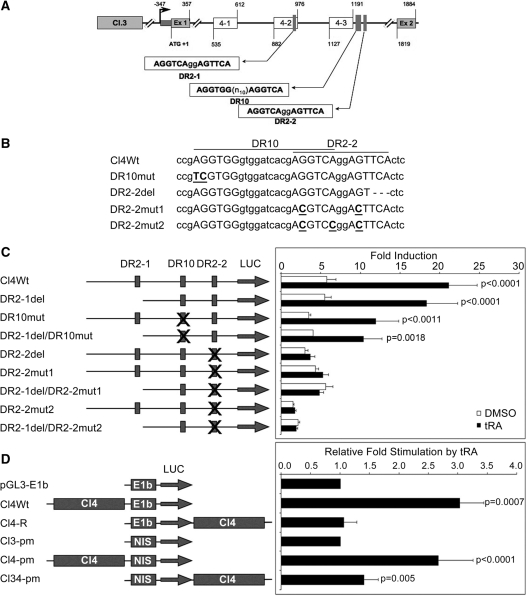
Figure 4.
A Functional RARE (DR2-2) is present in the first intron of NIS. (A) Representation of conserved regions and putative RARE sequences in cluster 4. The boxes labeled 4-1 to 4-3 represent the position of the three conserved regions; the positions of putative RARE sequences, transcription start site and the ATG are indicated. Ex 1 and Ex 2 indicate positions of exons 1 and 2, respectively. (B) Sequence representation of the Cl4 variants used, RARE half sites are capitalized and mutated nucleotides are underlined and bolded, deleted nucleotides are replaced with dashes. (C) MCF-7 cells were transiently transfected with Cl4Wt or its mutant derivatives, cells were treated with tRA (or DMSO) for 24 h and luciferase reporter assays were performed. Luciferase values were normalized to those of Renilla luciferase and the fold induction is represented relative to the empty vector. Reporter constructs are presented in the drawing on the left; putative RAREs are indicated by vertical boxes, an X on a box indicates the RARE being altered. P-values were calculated using paired Student’s t-test with 95% confidence interval, values <0.05 were considered significant. (D) MCF-7 cells were transfected with Cl4 controlling either the viral E1b promoter or the native NIS promoter (fragment NIS in Cl3-pm, see ‘Materials and Methods’ section). The sequence for Cl4 was located either 5′ of the promoter, or 3′ of the luciferase reporter. Luciferase assays were performed as mentioned above. Fold stimulation by tRA was calculated relative to the vector containing only the promoter element. P-values were calculated using Student’s t-test with 95% confidence interval, values <0.05 were considered significant. Drawings are not to scale.
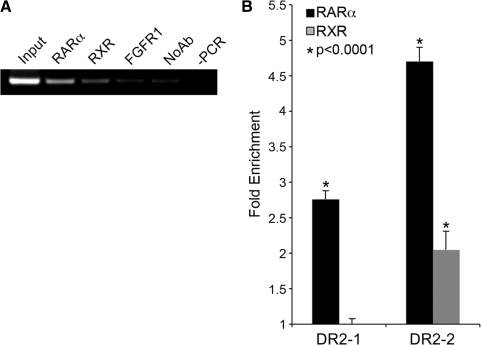
Figure 6.
RARα and RXR occupy the novel intronic element in vivo. MCF-7 cells grown in DMEM were treated with 1 µM tRA and used for ChIP analysis using RARα and RXR specific antibodies. (A) DNA isolated from immunocomplexes was used as a template for PCR amplification using primers specific for DR2-2. PCR products were resolved on a 2% agarose gel (containing Ethidium bromide) and visualized on a UV transilluminator, RARα; anti-RAR-α Ab precipitated DNA, RXR; anti-RXR Ab precipitated DNA, FGFR1; anti-FGFR1 Ab precipitated DNA, No Ab; bead-only control, and -PCR is a negative control with H2O as a template. (B) Quantitative PCR was performed using immunoprecipitated DNA using DR2-1 and DR2-2 specific primers. Ct values were normalized to background levels of bead-only controls (No Ab) using 2(δCt). Data are represented as fold enrichment compared to IgG control. *P-values were calculated using Student’s t-test (average of three experiments) with 95% confidence interval, values <0.05 were considered significant.
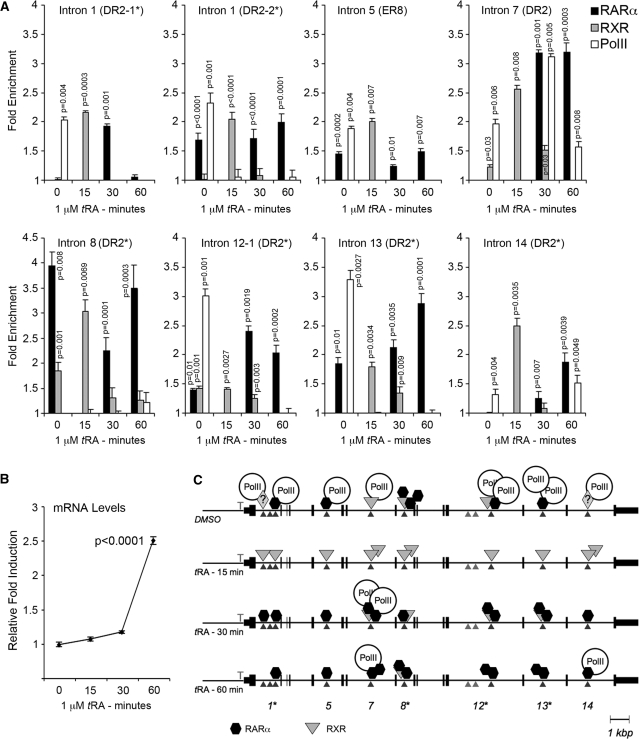
Figure 7.
Nuclear receptors and RNA Pol-II interact with NIS intronic elements in a dynamic manner during the initiation of transcription. MCF-7 cells grown in steroid-free and phenol-red-free DMEM were treated either with DMSO (time 0) or with 1 µM tRA for 15, 30 and 60 min and used for ChIP analysis using RARα and RXR and RNA Pol-II specific antibodies. (A) DNA isolated from immunocomplexes was used for quantitative PCR using primers described in Supplementary Table S3. Ct values were normalized to background levels of bead-only controls. Data are represented as fold enrichment compared to IgG control. (B) mRNA was isolated from a fraction of the cells used for ChIP analysis above. Two micrograms of total RNA was converted to cDNA and used for quantitative real-time PCR. Expression was normalized to the levels of GAPDH using the ΔΔCt method and presented as relative fold induction compared to DMSO-treated samples. (C) Schematic representation of ChIP data depicting the events of transcription initiation of NIS in response to tRA stimulation. Arrow heads indicate the position of the intronic element investigated, vertical small lines represent NIS exons, numbers below indicate introns studied, asterisks above numbers indicate identical sequences. Parallelograms with question mark represent unidentified interacting proteins. DR, direct repeat; ER, everted repeat. Statistical significance was determined by performing the Student’s t-test (Figure 7A) or paired Student’s t-test (Figure 7C) using a 95% confidence interval; P-values <0.05 were considered significant.
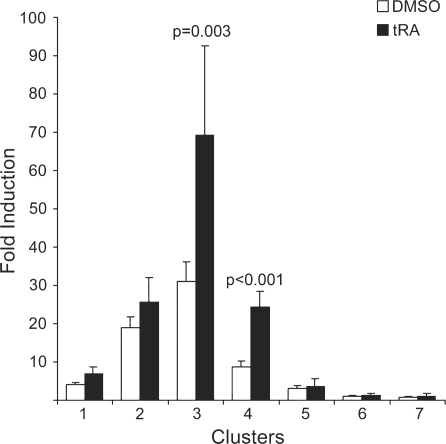
Figure 2.
Conserved clusters 3 and 4 respond to tRA in MCF-7. Conserved clusters were amplified by PCR and cloned into the luciferase reporter plasmid pGL3-E1b. MCF-7 cells were transfected with all conserved clusters and treated with tRA or DMSO for 24 h. Of the seven clusters analyzed, only clusters 3 and 4 were significantly responsive to tRA treatment. P-values for the tRA-induced samples were calculated using paired Student’s t-test with 95% confidence interval, values <0.05 were considered significant.
Figure 3.
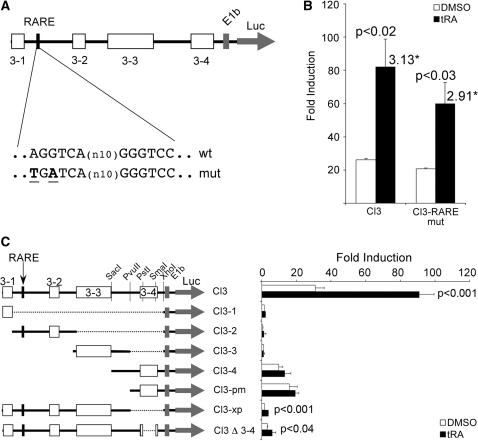
Stimulation of conserved cluster 3 is indirect and distinct from that in thyroid cells. (A) Map of Cl3, conserved regions are represented by boxes labeled 3-1 to 3-4, the previously characterized RARE is indicated by the black box, the wild type and the mutant sequences are shown below the map. (B) MCF-7 cells were transiently transfected with either the wild type or the mutant construct, cells were treated with tRA or DMSO and luciferase assays were performed. (C) Several deletion mutants were prepared to study the transcriptional potential in response to tRA stimulation of individual or combined conserved regions of Cl3. Plasmids were transfected to MCF-7 cells, followed by tRA (or the vehicle DMSO) treatment for 24 h, then luciferase assays were performed. The luciferase values were normalized to those of Renilla luciferase; fold induction was calculated relative to the empty vector. *Numbers above bars indicate the overall fold stimulation by tRA. P-values for the tRA-induced samples were calculated using paired Student’s t-test with 95% confidence interval, values <0.05 were considered significant.
RESULTS
Comparative genomics identifies potential cis-acting elements controlling NIS transcription
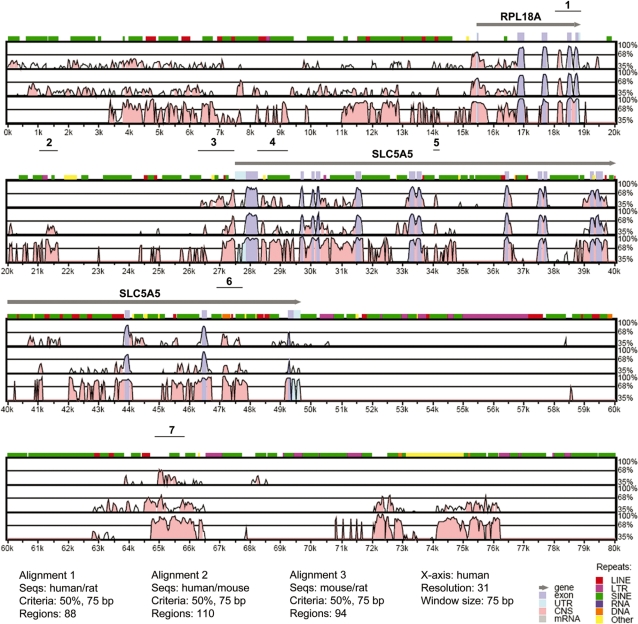
We analyzed the conservation of non-coding DNA sequences of NIS by comparing genomic sequences in three species, R. norvegicus, M. musculus and H. sapiens. Conserved sequences at non-coding regions (defined as windows of 75 bp with 50% nucleotide identity) were identified in pair-wise alignments using the VISTA program (21). Figure 1 shows the results of the VISTA alignment in which the short conserved regions that are localized in non-coding sequences and that meet the selected criteria were highlighted in pink. Conserved regions clustered within 1 kb were later studied as single blocks and they were named as cluster 1, cluster 2, cluster 3 and so forth (Supplementary Table S1). This grouping of conserved regions facilitated subsequent functional analyses.
Figure 1.
VISTA plot of sequence conservation in a 90-kb genomic DNA in human, mouse and rat. Percent nucleotide identities between human, mouse and rat DNA sequences are plotted as a function of position along the human sequence. Peaks of evolutionary conservation in overlapping exonic sequences are shaded blue. Aligned regions of >50% identity over 75 bases are shaded pink. Lines above each alignment indicate the position of the conserved cluster selected for PCR amplification. Only 80 kb of the alignment is shown.
Conserved clusters 3 and 4 respond to tRA in MCF-7 cells
Identified individual conserved regions that were located in clusters varied between 13 (as in cluster 6) and 244 bp (as in cluster 3) in length (Figure 1). We studied tRA responsive transcriptional activation capacities of these regions in MCF-7 cells in which, induction of NIS gene by tRA was previously reported (7,10,13). We amplified clusters of interest by PCR using human genomic DNA as a template, and we cloned these fragments into the luciferase reporter vector pGL3-E1b as described in ‘Materials and Methods’ section. Previously characterized NIS upstream enhancer, which contributes to the regulation of this gene in thyroid cells (18,19), was located in the coding sequence of the neighboring upstream gene, RPL18A. This region, named as ‘cluster 1’, was also included in our analysis in order to assess a possible function of this enhancer in a mammary cell system. All vector constructs were transiently transfected into MCF-7 cells, which were subsequently treated with either DMSO (as vehicle control) or 1 µM tRA for 24 h, and the regulatory potentials of these conserved clusters were assessed by luciferase reporter assays. As a result, we found that only clusters 3 and 4 significantly responded to the tRA stimulus (Figure 2).
Cluster 3 is a 1.1-kb sequence upstream of the human NIS, located from -1138 to -42 relative to the transcription start site (22,23). In a previous study, the tRA responsiveness of the human NIS promoter was attributed to a RARE which is located between bases 125 and 146 of this cluster [at −1375 relative to the ATG, (14)]. In order to assess the functionality of this RARE in MCF-7 cells, we created a mutant version of Cl3, in which we introduced the same mutation (Figure 3A) that completely abolished tRA response from this sequence in thyroid cell lines (14). Interestingly, this mutation had a minor effect on tRA stimulation of Cl3, reducing the tRA stimulation by only 7% (Figure 3B), indicating that this RARE is not the major RA responsive site in MCF-7. This result suggests the presence of an alternative site for tRA responsiveness in mammary cell systems. To this end, we created deletion mutants of Cl3 in reporter plasmids, which contained individual as well as combined conserved regions (Figure 3C and results not shown). These constructs were then tested for tRA responsiveness using luciferase reporter assays in MCF-7. Despite intensive analysis on a relatively large number of overlapping deletion mutants, a tRA responsive region other than the previously identified RARE could not be found in Cl3 (see Figure 3C for a summary of our results). On the other hand, removal of conserved region 3–4 (∼100 bp) has significantly reduced the magnitude of tRA response and the overall transcriptional potential of this construct (Figure 3C), indicating the presence of elements essential for maximum levels of transcription. These results suggest that tRA responsiveness of Cl3 requires a synergistic effect of multiple regulatory elements.
A functional RARE is present in the first intron of NIS
Cluster 4 is a 774-bp sequence located in the first intron of the human NIS. This cluster is composed of three conserved regions numbered from 4-1 to 4-3 (Figure 4A). A closer look at the sequence composition of this cluster revealed the presence of three putative RAREs. The first is a perfect DR2-type consensus RARE sequence (24) (DR2-1: AGGTCAggAGTTCA) located in the second conserved region (region 4-2) and starting at +927 relative to the ATG. The second is a DR10-type sequence [DR10: AGGTGG(n10)AGGTCA], located just after the third conserved region (region 4-3) and starting at +1206 relative to the ATG. The third is, again, a DR2-type sequence identical to DR2-1 (DR2-2: AGGTCAggAGTTCA) and is located at +1222 relative to the start codon, overlapping with the DR10-type sequence described above (Figure 4A). A series of pGL3-E1b-based luciferase reporter constructs containing different fragments of this intronic region, as well as in vitro mutagenized versions of putative RARE sequences were constructed. A summary of mutations created is presented in Figure 4B. Functional activity of these RARE sequences was assessed by transient transfections followed by luciferase reporter assays in MCF-7 cells treated either with DMSO (as vehicle control) or with 1 µM tRA for 24 h. In MCF-7 cells transfected with the wild-type construct, tRA treatment results in an increase of about 4-fold when compared to cells treated with DMSO. This result confirmed once again that the intronic region contains tRA responsive sequences (Figure 4C; Cl4Wt), which have the capacity to activate the transcription of the reporter gene. A very small deletion (3 bp) that changed the second half-site of DR2-2 but left the DR2-1 and DR10 sequences intact abolished entirely the tRA responsive activity (Figure 4C; DR2-2del), indicating that DR2-2 has a functional role in ligand responsive activation of reporter expression. Moreover, when single or double point mutations were introduced by in vitro mutagenesis in each half site of the DR2-2, the tRA response was completely lost (Figure 4C; DR2-2mut1 and DR2-2mut2). However, removal of the DR2-1 region from the construct by introducing a 490-bp deletion did not make any effect at all on tRA responsive activity of the reporter, showing that DR2-1 is fully dispensable for tRA response (Figure 4C; DR2-1del). Absence of the DR10 element both in an otherwise wild-type intronic fragment context, and in the context of a fragment missing the DR2-1 element (DR2-1del) did not abolish tRA responsiveness coming from this region (Figure 4C; DR10mut and DR2-1del/DR10mut), but they had a minor effect on the magnitude of the response (2- to 3-fold stimulation).
We also investigated the potential of this intronic RARE to activate transcription through the native NIS minimal promoter. For this purpose, we constructed reporter plasmids containing the minimal NIS promoter (218 bp of NIS upstream region between −260 bp and −42 bp relative to transcription start site, and capable of initiating transcription as assessed by functional reporter assays, Figure 3C) controlled by Cl4 sequence either upstream of the promoter element or downstream of the reporter sequence (mimicking its location in the intron of NIS). Similar constructs were also prepared using the viral TATA element, E1b. As anticipated, Cl4 significantly activated NIS promoter in response to tRA when positioned either at the 5′ (P < 0.0001, n = 8) or at 3′ of the reporter (P = 0.005, n = 8), in agreement with the function of an enhancer (Figure 4D). On the other hand, regarding E1b promoter activation, Cl4 responded to the tRA stimulus only when positioned at the 5′ of the promoter element. Altogether these findings clearly indicate that the DR2-2 sequence can act as a RA responsive element, and it is the major RARE that is present in the first intron of human NIS.
RARα and RXR physically interact with the intronic response element DR2-2 both in vitro and in vivo
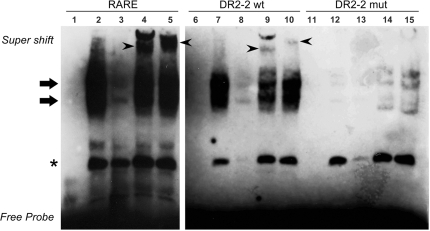
Retinoids exert their effects by binding to nuclear retinoic acid receptors (RARs) and retinoid-X-receptors (RXRs), which in turn interact with cis-acting regulatory elements, or RAREs, and stimulate transcription of target genes. Consistently, previous work of Kogai et al. (12) has indicated that tRA stimulation of human NIS is mediated by RAR and RXR. In the presence of tRA, RAR could form homo- or hetero-dimers with RXR, and the dimer mediates ligand-dependent gene activation by physically interacting with RAREs in RA responsive genes. In order to establish whether RAR and/or RXR have the potential to bind the DR2-2 element, we designed oligonucleotide probes corresponding to the wild-type DR2-2 sequence, or a variant of DR2-2 where single-point mutations were introduced in each half-site (Supplementary Table S2). We then studied in vitro binding capacities of the two nuclear receptors (RARα and RXR) to these probes by EMSA experiments (Figure 5). Initially, we have prepared nuclear extracts from 17-β-estradiol (E2)-treated MCF-7 cells, and we determined the abundance of both RARα and RXR in nuclear extracts by immunoblotting (data not shown). Subsequently, we incubated nuclear extracts with biotin-labeled probes containing either a consensus RARE that was known to interact with RARα and RXR, the wild-type DR2-2 sequences, or the mutant form of DR2-2. As expected, we have clearly observed retarded complexes when nuclear extracts were incubated with probes carrying the consensus RARE, or the novel element, DR2-2 (Figure 5, lanes 2 and 7). On the other hand, the introduction of point mutations into DR2-2 half-sites abolished this shift (Figure 5, compare lanes 7 and 12), unequivocally proving that these band shifts were observed as a result of direct interactions between the nuclear factors and the DR2-2 element (Figure 5, shown with bold arrows). A retarded band marked by an asterisk has indicated a DR2-2 independent interaction between nuclear proteins and the DNA probe. All bands were appropriately competed or shifted, confirming the specificity of the interactions (Figure 5, lanes 3, 8 and 13). In order to test whether the two nuclear transcription factors of interest, RARα and RXR, interact with DR2-2, we have added factor-specific monoclonal antibodies to our binding reactions. Both RARα and RXR antibodies caused a super shift in the consensus RARE and in DR2-2 probe (arrowheads), while they failed to do so when probes carrying point mutations on DR2-2 were used (Figure 5, for RARα compare lanes 4, 9 and 14; for RXR see lanes 5, 10 and 15). These results clearly indicate direct interactions between the nuclear transcription factors RARα/RXR and the novel intronic RARE, DR2-2.
Figure 5.
RARα and RXR interacts with the novel intronic RARE in gel retardation assays. Nuclear extracts from E2-treated MCF-7 cells were incubated with biotin labeled oligonucleotide probes representing a consensus RARE, the wild-type DR2-2 or the mutant variant DR2-2mut (DR2-2-Mut1-S in Supplementary Table S2). Samples were resolved on a 6% non-denaturing polyacrylamide gel in TBE, transferred to Hybond N+ membranes and then incubated with streptavidin, and biotin-labeled DNA probes were detected by chemiluminescence. The name of the probe used in each binding reaction is indicated on the top of each panel. All binding reactions were competed with 200-fold molar excess of the corresponding unlabeled probe. Arrows point out the nuclear receptor–DNA complexes, while arrowheads point out the super shift. The asterisk indicates a DR2-2 independent interaction with nuclear proteins. Labeled probes were incubated in the absence of nuclear extract (lanes 1, 6 and 11), in the presence of nuclear extract alone (lanes 2, 7 and 12), nuclear extract together with an excess of competing unlabeled probe (lanes 3, 8 and 13), in the presence of RARα antibodies (lanes 4, 9 and 14), or in the presence of RXR antibodies (lanes 5, 10 and 15). Experiments were repeated at least three times and a representative result is shown.
Subsequently, in order to assess whether RARα and/or RXR occupies the DR2-2 element in response to tRA in vivo, we carried out a series of ChIP experiments in MCF-7 cells. In the presence of RARα or RXR antibodies, a DNA fragment containing the DR2-2 element was precipitated from formaldehyde cross-linked total cell lysates (Figure 6). In contrast, the unrelated control IgG against fibroblast growth factor receptor 1 (FGFR1) did not precipitate the DR2-2 element above background levels (No Ab) of protein-A Sepharose beads (Figure 6A). We also investigated the presence of the two nuclear receptors on DR2-1 using the same immunoprecipitated samples. Despite being non-responsive to tRA stimulation in reporter assays (Figure 4C), we found that DR2-1 is actually occupied by both nuclear receptors in MCF-7 cells (Figure 6B), but with lower abundance when compared to DR2-2 (∼50% less). These findings demonstrate that, endogenous RARα and RXR bind to both DR2-1 and DR2-2 that are present in the first intron of NIS in vivo, thereby strongly suggesting that at least part of the regulatory effects of the two nuclear receptors, RARα and RXR, on NIS expression in MCF-7 cells were due to a direct interaction between the receptors and the intronic RAREs (Figure 6).
Initiation of NIS transcription in response to tRA involves dynamic interactions between multiple intronic elements, nuclear receptors and the RNA Pol-II
Besides the intronic enhancer identified above (DR2-2 in intron 1), there exists several other putative intronic RARE sequences within NIS gene (25). As we found a functional RARE in intron 1, we were intrigued to study a possible involvement of other intronic putative RARE sequences in NIS gene transcription. As nuclear receptors interact with cognate response elements in a dynamic manner (26), we decided to study the dynamics of interactions taking place between cis- and trans-acting factors. To this end, we used ChIP assays to screen for the involvement of intronic sequences in NIS transcription in response to tRA treatment in MCF-7 cells (Figure 7).
MCF-7 cells were cultured in charcoal-treated steroid-free medium lacking phenol-red, and they were either treated with DMSO, or with tRA for 15, 30 or 60 min, and interactions of RARα, RXR and/or RNA Pol-II with sequences located in introns 1, 5, 7, 8, 12, 13 and 14 (Figure 7) were investigated. Prior to tRA stimulation (time point 0) all intronic sequences, except the one in intron 8, were engaged in an interaction with basal transcriptional machinery (based on precipitation of these sequences with RNA Pol-II, Figure 7A). By 15 min of stimulation, interactions were limited to RXR but then they were replaced by RARα (30 min), with strong appearance together with RNA Pol-II on the element from intron 7 (Figure 7A). This presence of RARα persists at 60 min post-stimulation in the absence of RXR, accompanied by an acute increase in mRNA levels (Figure 7B). It appears that interactions with transcription machinery are sustained by sequences in intron 7 (30 and 60 min.) and in intron 14 (60 min.). Figure 7C recapitulates the dynamic events taking place during initiation of transcription in response to tRA stimulation.
DISCUSSION
The identification of elements that control NIS regulation in mammary gland cells is of particular importance in the context that mammary-tumor-specific stimulation of NIS activity followed by radioiodide substrate (131I, 123I or 99mTcO4) administration could have an important potential as a novel alternative in the diagnosis and treatment of breast tumors (2,7,8,17,27). Yet, in a recent publication (28), it was reported that NIS-related intracellular staining in immunohistochemistry experiments on thyroid cancers was due to non-specific binding of the antibodies. These authors also conclude that the intracellular signal in breast tumors might not always be specific. Therefore, in our opinion, establishing clinical methods of increasing expression levels of NIS would be essential if the activity of this symporter is to be used for a radio-labeled substrate-based diagnosis and therapy of a large set of breast tumors (29,30). In a series of publications, Kogai et al. (7,17,25,31) has demonstrated that tRA stimulates NIS expression and the selective cytotoxic effect of radioiodide both in MCF-7 cells as well as in MCF-7-derived xenografts in nude mice. Yet, the molecular mechanisms of tRA effect on NIS in mammary gland cells are not completely revealed. Although tRA upregulates both rodent and human NIS expression at the transcriptional level in a variety of thyroid and mammary gland cell lines, molecular mechanisms and elements that were reported to mediate tRA responsive NIS regulation varied depending on the organism and the cell origin. A better understanding of molecular determinants, mechanisms and ligands that have a role in NIS regulation would be essential for successful implementation of possible NIS-activity-based novel methods for the management of malignant breast diseases. In this respect, our description regarding the molecular interplay between retinoid receptors, intronic cis-acting elements and RNA Pol-II brings important clarifications about precise molecular mechanisms and interactions controlling tRA-dependent NIS gene upregulation (see below).
In an initial effort to identify mammary-gland-specific enhancers of NIS, we adopted a comparative genomics approach followed by functional assays of conserved DNA sequences in different cell types (data not shown). As widely appreciated, interspecies genome comparisons were in general very useful for a rather accurate prediction of protein encoding genes in different organisms (32). On the other hand, prediction of gene regulatory regions by genome comparisons tends to be relatively more difficult as they tolerate much more sequence divergence than coding sequences while retaining their original function. Yet, several reports have indicated that interspecies genome comparisons of close relatives could lead to correct predictions of regulatory sequences (33–35). A similar comparative analysis of a 90-kb region, including and flanking NIS loci in mice, rat and human genomes, followed by functional tests in MCF-7 cells, has led us to identify a novel tRA responsive region in the first intron of the human NIS.
A conserved 5′ upstream region of the human NIS (referred to as cluster 3 in this work) contains a tRA responsive element that was previously identified as a RARE in human follicular thyroid carcinoma cell lines, FTC-133 and FTC-238 (14). As expected, we have observed that this region has a remarkable tRA responsive regulatory capacity (Figure 2) in MCF-7 cells. Surprisingly, point mutations that were shown to abolish tRA responsiveness of this element in thyroid cells (14) had no significant suppressive effect on tRA stimulation in MCF-7 cells (Figure 3B). Our intensive search both by bioinformatics and by experimental methods, as well as studies carried out by other groups (25), did not reveal another RARE in this region. This could imply that first, the mechanism of RA stimulation of human NIS expression in mammary cell models is distinct from that of thyroid cells; and, second, the stimulatory effect of RA on this sequence is actually an indirect one. Moreover, analysis of overlapping deletion mutations in Cl3 did not reveal any RA responsive sequence (Figure 3C), suggesting that RA stimulation of the human NIS gene requires the synergistic effect of multiple factors.
Indirect stimulation of NIS by tRA was previously reported; tRA was shown to upregulate the rat Nis promoter in MCF-7 cells by activating the homeobox-containing transcription factor, Nkx2.5 (15). In this cell line, tRA treatment stimulates the expression of this NK2-type transcription factor, which in turn, activates Nis promoter by binding to two cis-acting elements located at −446 and −154 relative to ATG (15). Despite extensive search for similar binding sites in the human NIS locus, Nkx2.5 responsive cis-acting sequences were not found (data not shown). In a more recent study, Kogai et al. (25) described an indirect effect of RA on NIS expression that is mediated by IGF-1/PI3K signaling pathways in MCF-7 but not in the rat normal thyroid cell line FRTL-5. In MCF-7 cells, RA exerts its effect by activating PI3K signaling pathways, which result in the activation of an unknown transcription factor, leading to an enhanced NIS expression (36). It would be interesting to find out whether the binding site of this yet-uncharacterized factor maps in Cl3 or not.
Intronic sequences could contain motifs and cis-acting elements with varying functions, such as exon/intron recognition, regulation of alternative splicing events, activation of transcription, stimulation of polyadenylation and, surprisingly enough, in certain cases elements regulating the stability of encoded proteins (37–42). In this report, we describe a novel enhancer that is located in intron 1, and that mediates retinoic acid responsive NIS gene expression. We have initially shown evidences for a direct interaction between RARα/RXR and a novel RARE within the first intron of the human NIS in MCF-7 cells (Figures 5–7). This DR-2 type RARE (DR2-2) was functional in reporter assays and had the capacity to activate both the native NIS promoter (regardless of position or orientation) as well as the viral E1b TATA element (Figure 4D). Recently, Kogai et al. (25) reported putative RARE sequences present within several introns of the human NIS, including elements found in the first intron, but they could not attribute a functional role to these elements (25). Thus, it seems that other than its native promoter, NIS intronic enhancer could activate only a subset of heterologous promoters. Interestingly, another DR-2-type element (DR2-1) of identical sequence in the same intron was occupied by tRA stimulated nuclear receptors in vivo despite being non-responsive to tRA in reporter assays. The difference in the regulatory potential of these sequence-wise identical elements in intron 1 could be related to initial (time 0) and persistent (60 min) interactions with RARα that we observe in DR2-2 but not DR2-1 (Figure 7A). This temporal regulation of RARα interaction with its cognate response elements could well be governed by epigenetic factors modifying local chromatin structure of this region. Related to this, we have clearly observed tri-methylation of H3K4 residues both on DR2-1, DR2-2 as well as on other intronic RAREs (see below, and also Supplementary Figure S1), a characteristic histone H3 modification found at enhancer regions of transcriptionally active genes (43). We have previously shown that tRA-upregulated NIS expression is restricted to ERα-positive mammary gland cell lines. In the absence of ERα, neither NIS promoter, nor the luciferase reporter gene under control of cluster 3 were stimulated in response to tRA stimulation, even though tRA signaling was operative in these cells (13, and Supplementary Figure S2). Re-introduction of ERα in previously ERα (−) cells was required for the recovery of NIS expression (13). It is therefore plausible to anticipate that intronic elements that were described in this work could only stimulate initiation of NIS gene transcription only in the presence of active ERα.
Our ChIP assays have revealed that at the onset of NIS gene transcription, sequences located in introns 1, 5, 7, 8, 12, 13 and 14 (Figure 7) interact temporally with nuclear receptors and with RNA Pol-II. Interestingly, all intronic elements (except the one in intron 8) were found to interact intensively with RNA Pol-II in the absence of ligand, tRA (Figure 7A and C), which might keep the polymerase ready for the initiation. With the tRA-dependent onset of transcription, interactions between cis-acting sequences and RNA Pol-II become undetectable in our assay system, and in the first 15 min of tRA induction RXR solely occupies all intronic positions (Figure 7C), indicating the early role of this trans-acting factor in tRA dependent transcription. At later time points (30 and 60 min) while mRNA levels increase in a steady manner (Figure 7B), interactions between sequences in intron 7 and RNA Pol-II (and later in intron 14 and RNA Pol-II) becomes detectable once again, suggesting frequent interactions between general transcription apparatus and elements in intron 7 and 14 during maximal levels of transcription (Figure 7A–C). At the same time points, RARα replaces RXR and dominates interactions in intronic sites, which implies the principal role of RARα at relatively late stages of tRA-dependent transcription initiation. Positions of intronic sequences found to interact with the RNA Pol-II and tRA-stimulated nuclear receptors matches exactly with locations where H3K4 tri-methylations were observed (Supplementary Figure S1). In fact, in most cases, persistent RARα interactions at later stages (30–60 min) with intronic elements coincide with increased H3K4 tri-methylation (Figures 7 and S1). Nuclear receptors, including RARα and RXR, are well known to recruit histone-modifying co-regulators facilitating conversion of chromatin regions to transcriptionally permissive states (44). For instance, MLL5, which is a histone lysine methyltransferase, was recently described to be one of the H3K4 modifying enzymes enhancing RARα-dependent gene transcription (45); one of the epigenetic mechanisms that could also operate during NIS expression.
Earlier work and results presented in this manuscript clearly indicate that molecular mechanisms presiding NIS expression involve a number of classical and intronic elements that regulate temporal and spatial expression, and they are likely to be more complex than previously anticipated. Future experiments will show whether the activities of intronic enhancers lead also to a tissue- or cell-type-specific regulation of NIS. A better understanding of this mechanism of regulation, as well as the ability to selectively enhance NIS expression in tumor cells will, eventually, be of great importance for potential future applications in breast cancer diagnosis and treatment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The EU 7th Framework Program Project UNAM-REGPOT Grant no: 203953 and by an EMBO Short Term Fellowship (to H.A.); Associazione Italiana per la Ricerca sul Cancro (to R.D.L. and D.S.); Turkish Scientific and Technical Research Council (104-T-231 to U.H.T); the Turkish Academy of Sciences-Young Scientist Award (GEBIP/2001-2-18 to U.H.T); Bilkent University Research Funds; DPT-KANILTEK Project Funds; Feyzi Akkaya Research Fund for Scientific Activities (FABED; to U.H.T.). Funding for open access charge: partially waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Mehmet Öztürk and Dr. Khemais Benhaj for valuable discussions.
REFERENCES
- 1.Spitzweg C, Joba W, Eisenmenger W, Heufelder AE. Analysis of human sodium iodide symporter gene expression in extrathyroidal tissues and cloning of its complementary deoxyribonucleic acids from salivary gland, mammary gland, and gastric mucosa. J. Clin. Endocrinol. Metab. 1998;83:1746–1751. doi: 10.1210/jcem.83.5.4839. [DOI] [PubMed] [Google Scholar]
- 2.Tazebay UH, Wapnir IL, Levy O, Dohan O, Zuckier LS, Zhao QH, Deng HF, Amenta PS, Fineberg S, Pestell RG, et al. The mammary gland iodide transporter is expressed during lactation and in breast cancer. Nat. Med. 2000;6:871–878. doi: 10.1038/78630. [DOI] [PubMed] [Google Scholar]
- 3.Stubbe P, Schulte FJ, Heidemann P. Iodine deficiency and brain development. Bibl. Nutr. Dieta. 1986:206–208. doi: 10.1159/000412616. [DOI] [PubMed] [Google Scholar]
- 4.Cho JY, Leveille R, Kao R, Rousset B, Parlow AF, Burak W.E., Jr, Mazzaferri EL, Jhiang SM. Hormonal regulation of radioiodide uptake activity and Na+/I- symporter expression in mammary glands. J. Clin. Endocrinol. Metab. 2000;85:2936–2943. doi: 10.1210/jcem.85.8.6727. [DOI] [PubMed] [Google Scholar]
- 5.Perron B, Rodriguez AM, Leblanc G, Pourcher T. Cloning of the mouse sodium iodide symporter and its expression in the mammary gland and other tissues. J. Endocrinol. 2001;170:185–196. doi: 10.1677/joe.0.1700185. [DOI] [PubMed] [Google Scholar]
- 6.Knostman KA, Cho JY, Ryu KY, Lin X, McCubrey JA, Hla T, Liu CH, Di Carlo E, Keri R, Zhang M, et al. Signaling through 3′,5′-cyclic adenosine monophosphate and phosphoinositide-3 kinase induces sodium/iodide symporter expression in breast cancer. J. Clin. Endocrinol. Metab. 2004;89:5196–5203. doi: 10.1210/jc.2004-0907. [DOI] [PubMed] [Google Scholar]
- 7.Kogai T, Kanamoto Y, Che LH, Taki K, Moatamed F, Schultz JJ, Brent GA. Systemic retinoic acid treatment induces sodium/iodide symporter expression and radioiodide uptake in mouse breast cancer models. Cancer Res. 2004;64:415–422. doi: 10.1158/0008-5472.can-03-2285. [DOI] [PubMed] [Google Scholar]
- 8.Wapnir IL, van de Rijn M, Nowels K, Amenta PS, Walton K, Montgomery K, Greco RS, Dohan O, Carrasco N. Immunohistochemical profile of the sodium/iodide symporter in thyroid, breast, and other carcinomas using high density tissue microarrays and conventional sections. J. Clin. Endocrinol. Metab. 2003;88:1880–1888. doi: 10.1210/jc.2002-021544. [DOI] [PubMed] [Google Scholar]
- 9.Mazzaferri EL. An overview of the management of papillary and follicular thyroid carcinoma. Thyroid. 1999;9:421–427. doi: 10.1089/thy.1999.9.421. [DOI] [PubMed] [Google Scholar]
- 10.Kogai T, Curcio F, Hyman S, Cornford EM, Brent GA, Hershman JM. Induction of follicle formation in long-term cultured normal human thyroid cells treated with thyrotropin stimulates iodide uptake but not sodium/iodide symporter messenger RNA and protein expression. J. Endocrinol. 2000;167:125–135. doi: 10.1677/joe.0.1670125. [DOI] [PubMed] [Google Scholar]
- 11.Tanosaki S, Ikezoe T, Heaney A, Said JW, Dan K, Akashi M, Koeffler HP. Effect of ligands of nuclear hormone receptors on sodium/iodide symporter expression and activity in breast cancer cells. Breast Cancer Res. Treat. 2003;79:335–345. doi: 10.1023/a:1024064424855. [DOI] [PubMed] [Google Scholar]
- 12.Kogai T, Kanamoto Y, Li AI, Che LH, Ohashi E, Taki K, Chandraratna RA, Saito T, Brent GA. Differential regulation of sodium/iodide symporter gene expression by nuclear receptor ligands in MCF-7 breast cancer cells. Endocrinology. 2005;146:3059–3069. doi: 10.1210/en.2004-1334. [DOI] [PubMed] [Google Scholar]
- 13.Alotaibi H, Yaman EC, Demirpençe E, Tazebay UH. Unliganded estrogen receptor-alpha activates transcription of the mammary gland Na+/I- symporter gene. Biochem. Biophys. Res. Commun. 2006;345:1487–1496. doi: 10.1016/j.bbrc.2006.05.049. [DOI] [PubMed] [Google Scholar]
- 14.Schmutzler C, Schmitt TL, Glaser F, Loos U, Köhrle J. The promoter of the human sodium/iodide-symporter gene responds to retinoic acid. Mol. Cell. Endocrinol. 2002;189:145–155. doi: 10.1016/s0303-7207(01)00735-3. [DOI] [PubMed] [Google Scholar]
- 15.Dentice M, Luongo C, Elefante A, Romino R, Ambrosio R, Vitale M, Rossi G, Fenzi G, Salvatore D. Transcription factor Nkx-2.5 induces sodium/iodide symporter gene expression and participates in retinoic acid- and lactation-induced transcription in mammary cells. Mol. Cell. Biol. 2004;24:7863–7877. doi: 10.1128/MCB.24.18.7863-7877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negre B, Casillas S, Suzanne M, Sánchez-Herrero E, Akam M, Nefedov M, Barbadilla A, de Jong P, Ruiz A. Conservation of regulatory sequences and gene expression patterns in the disintegrating Drosophila Hox gene complex. Genome Res. 2005;15:692–700. doi: 10.1101/gr.3468605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kogai T, Taki K, Brent GA. Enhancement of sodium/iodide symporter expression in thyroid and breast cancer. Endocr. Relat. Cancer. 2006;13:797–826. doi: 10.1677/erc.1.01143. [DOI] [PubMed] [Google Scholar]
- 18.Ohno M, Zannini M, Levy O, Carrasco N, di Lauro R. The paired-domain transcription factor Pax8 binds to the upstream enhancer of the rat sodium/iodide symporter gene and participates in both thyroid-specific and cyclic-AMP-dependent transcription. Mol. Cell. Biol. 1999;19:2051–2060. doi: 10.1128/mcb.19.3.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taki K, Kogai T, Kanamoto Y, Hershman JM, Brent GA. A thyroid-specific far-upstream enhancer in the human sodium/iodide symporter gene requires Pax-8 binding and cyclic adenosine 3′,5′-monophosphate response element-like sequence binding proteins for full activity and is differentially regulated in normal and thyroid cancer cells. Mol. Endocrinol. 2002;16:2266–2282. doi: 10.1210/me.2002-0109. [DOI] [PubMed] [Google Scholar]
- 20.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 22.Behr M, Schmitt TL, Espinoza CR, Loos U. Cloning of a functional promoter of the human sodium/iodide-symporter gene. Biochem. J. 1998;331(Pt 2):359–63. doi: 10.1042/bj3310359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endo T, Kaneshige M, Nakazato M, Ohmori M, Harii N, Onaya T. Thyroid transcription factor-1 activates the promoter activity of rat thyroid Na+/I- symporter gene. Mol. Endocrinol. 1997;11:1747–1755. doi: 10.1210/mend.11.11.0012. [DOI] [PubMed] [Google Scholar]
- 24.Giguère V. Retinoic acid receptors and cellular retinoid binding proteins: complex interplay in retinoid signaling. Endocr. Rev. 1994;15:61–79. doi: 10.1210/edrv-15-1-61. [DOI] [PubMed] [Google Scholar]
- 25.Kogai T, Ohashi E, Jacobs MS, Sajid-Crockett S, Fisher ML, Kanamoto Y, Brent GA. Retinoic acid stimulation of the sodium/iodide symporter in MCF-7 breast cancer cells is mediated by the insulin growth factor-I/phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase signaling pathways. J. Clin. Endocrinol. Metab. 2008;93:1884–1892. doi: 10.1210/jc.2007-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 27.Wapnir IL, Goris M, Yudd A, Dohan O, Adelman D, Nowels K, Carrasco N. The Na+/I- symporter mediates iodide uptake in breast cancer metastases and can be selectively down-regulated in the thyroid. Clin. Cancer Res. 2004;10:4294–4302. doi: 10.1158/1078-0432.CCR-04-0074. [DOI] [PubMed] [Google Scholar]
- 28.Peyrottes I, Navarro V, Ondo-Mendez A, Marcellin D, Bellanger L, Marsault R, Lindenthal S, Ettore F, Darcourt J, Pourcher T. Immunoanalysis indicates that the sodium iodide symporter is not overexpressed in intracellular compartments in thyroid and breast cancers. Eur. J. Endocrinol. 2009;160:215–225. doi: 10.1530/EJE-08-0505. [DOI] [PubMed] [Google Scholar]
- 29.Daniels GH, Haber DA. Will radioiodine be useful in treatment of breast cancer? Nat. Med. 2000;6:859–860. doi: 10.1038/78604. [DOI] [PubMed] [Google Scholar]
- 30.Welcsh PL, Mankoff DA. Taking up iodide in breast tissue. Nature. 2000;406:688–689. doi: 10.1038/35021173. [DOI] [PubMed] [Google Scholar]
- 31.Kogai T, Schultz JJ, Johnson LS, Huang M, Brent GA. Retinoic acid induces sodium/iodide symporter gene expression and radioiodide uptake in the MCF-7 breast cancer cell line. Proc. Natl Acad. Sci. USA. 2000;97:8519–8524. doi: 10.1073/pnas.140217197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dolinski K, Botstein D. Orthology and functional conservation in eukaryotes. Annu. Rev. Genet. 2007;41:465–507. doi: 10.1146/annurev.genet.40.110405.090439. [DOI] [PubMed] [Google Scholar]
- 33.Dubchak I, Brudno M, Loots GG, Pachter L, Mayor C, Rubin EM, Frazer KA. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 35.Haubold B, Wiehe T. Comparative genomics: methods and applications. Naturwissenschaften. 2004;91:405–421. doi: 10.1007/s00114-004-0542-8. [DOI] [PubMed] [Google Scholar]
- 36.Ohashi E, Kogai T, Kagechika H, Brent GA. Activation of the PI3 kinase pathway by retinoic acid mediates sodium/iodide symporter induction and iodide transport in MCF-7 breast cancer cells. Cancer Res. 2009;69:3443–3450. doi: 10.1158/0008-5472.CAN-08-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lou H, Gagel RF, Berget SM. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- 38.Müller F, Chang B, Albert S, Fischer N, Tora L, Strähle U. Intronic enhancers control expression of zebrafish sonic hedgehog in floor plate and notochord. Development. 1999;126:2103–2116. doi: 10.1242/dev.126.10.2103. [DOI] [PubMed] [Google Scholar]
- 39.Noé V, MacKenzie S, Ciudad CJ. An intron is required for dihydrofolate reductase protein stability. J. Biol. Chem. 2003;278:38292–38300. doi: 10.1074/jbc.M212746200. [DOI] [PubMed] [Google Scholar]
- 40.Khandekar M, Brandt W, Zhou Y, Dagenais S, Glover TW, Suzuki N, Shimizu R, Yamamoto M, Lim KC, Engel JD. A Gata2 intronic enhancer confers its pan-endothelia-specific regulation. Development. 2007;134:1703–1712. doi: 10.1242/dev.001297. [DOI] [PubMed] [Google Scholar]
- 41.Feng W, Huang J, Zhang J, Williams T. Identification and analysis of a conserved Tcfap2a intronic enhancer element required for expression in facial and limb bud mesenchyme. Mol. Cell. Biol. 2008;28:315–325. doi: 10.1128/MCB.01168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hertel KJ. Combinatorial control of exon recognition. J. Biol. Chem. 2008;283:1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- 43.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 44.McEwan IJ. Gene regulation through chromatin remodelling by members of the nuclear receptor superfamily. Biochem. Soc. Trans. 2000;28:369–373. [PubMed] [Google Scholar]
- 45.Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.