Abstract
Mutations of thymidine kinase 2 (TK2), an essential component of the mitochondrial nucleotide salvage pathway, can give rise to mitochondrial DNA (mtDNA) depletion syndromes (MDS). These clinically heterogeneous disorders are characterized by severe reduction in mtDNA copy number in affected tissues and are associated with progressive myopathy, hepatopathy and/or encephalopathy, depending in part on the underlying nuclear genetic defect. Mutations of TK2 have previously been associated with an isolated myopathic form of MDS (OMIM 609560). However, more recently, neurological phenotypes have been demonstrated in patients carrying TK2 mutations, thus suggesting that loss of TK2 results in neuronal dysfunction. Here, we directly address the role of TK2 in neuronal homeostasis using a knockout mouse model. We demonstrate that in vivo loss of TK2 activity leads to a severe ataxic phenotype, accompanied by reduced mtDNA copy number and decreased steady-state levels of electron transport chain proteins in the brain. In TK2-deficient cerebellar neurons, these abnormalities are associated with impaired mitochondrial bioenergetic function, aberrant mitochondrial ultrastructure and degeneration of selected neuronal types. Overall, our findings demonstrate that TK2 deficiency leads to neuronal dysfunction in vivo, and have important implications for understanding the mechanisms of neurological impairment in MDS.
INTRODUCTION
Mitochondrial DNA (mtDNA) depletion syndromes (MDS) are a group of severe autosomal recessive disorders of infancy or childhood, characterized by a quantitative reduction in mtDNA copy number in affected tissues (1). The clinical spectrum of MDS is heterogeneous and patients typically present with signs of progressive myopathy, hepatopathy and/or encephalopathy, depending in part on the underlying nuclear genetic defect (2). Currently there is no curative treatment, and the syndrome is often fatal by the age of 3 years, irrespective of the initial presentation (3).
The spectrum of MDS clinical presentations have been associated with mutations in several nuclear genes, encoding proteins which play key roles in deoxyribonucleotide triphosphate (dNTP) metabolism thymidine kinase 2 (TK2) (4), deoxyguanosine kinase (DGUOK) (5), ADP-forming beta and alpha-subunits of the succinate-co-enzyme-A-ligase (SUCLA2 and SUCLG1) (6,7) and mtDNA maintenance polymerase gamma (POLG) (8,9) and Twinkle (10). TK2 is an essential component of the mitochondrial nucleotide salvage pathway, the main source of nucleotides for mtDNA synthesis and maintenance in postmitotic cells, thus representing a barrier against the insurgence of mitochondria dysfunction. The first described TK2 mutations were associated with an isolated, progressive myopathic form of MDS (OMIM 609560) (4). More recently, neurological phenotypes have been identified in patients carrying TK2 mutations, suggesting that loss of TK2 results in neuronal dysfunction (11). Additionally, mice harboring an H126N TK2 mutation were shown to develop encephalomyelopathy with prominent vacuolar changes in the anterior horn of the spinal cord (12). While it is becoming increasingly clear that TK2 may play an important role in neuronal homeostasis, the relationship between alterations in TK2 and neuronal dysfunction has yet to be fully elucidated.
Here, we address the importance of TK2 in neuronal homeostasis. Using a TK2 knockout mouse model, we demonstrate that in vivo loss of TK2 activity leads to a severe ataxic phenotype, accompanied by reduced mtDNA copy number and decreased steady-state levels of electron transport chain (ETC) proteins in the brain. In TK2-deficient cerebellar neurons, these abnormalities are associated with impaired mitochondrial bioenergetic function, aberrant mitochondrial ultrastructure and degeneration of selected neuronal types. Overall, our findings demonstrate that TK2 deficiency leads to neuronal dysfunction in vivo, and have important implications for understanding the mechanisms of neurological impairment in MDS.
RESULTS
Neurological features have recently been described in patients carrying TK2 mutations (11). To address the role of TK2 in neuronal homeostasis we employed a TK2 knockout mouse line, which lack TK2 activity. These mice exhibit growth retardation, hypothermia and a high rate of mortality in the second week of life (13). Nonetheless, a neurological phenotype was not previously reported. We therefore embarked upon a detailed analysis of TK2-deficient mice for the insurgence of pathological features. Indeed, after 10–12 days of life the TK2−/− mice display an ataxic phenotype with severely impaired motor coordination and gait, and abnormal limb-clasping reflexes (Supplementary Material, Movies S1–S3), indicative of an underlying neuropathology.
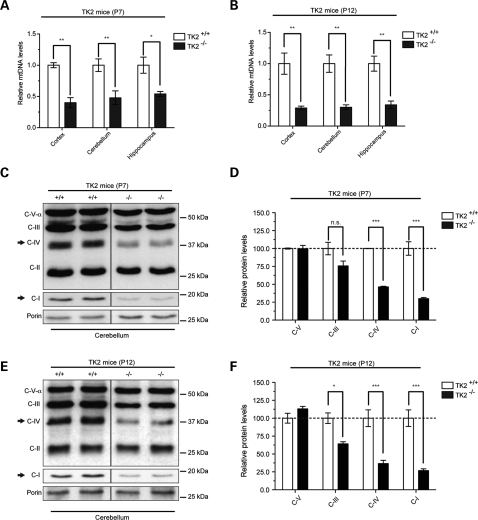
The loss of TK2 enzymatic activity in brain-protein extracts from newborn TK2−/− mice has previously been shown (13). To assess the effect of TK2 deficiency on mtDNA levels the brain, we quantified the relative amount of mtDNA in different brain regions of day 7 (P7, early symptomatic) and day 12 (P12, end-stage symptomatic) TK2−/− mice using quantitative PCR. Relative quantification of ND1 (mtDNA) gene and gapdh (nDNA) gene of DNA isolated from different areas of brains was performed. Using this approach, a ∼50% reduction in the mtDNA levels was observed in TK2−/− hippocampi, cerebella and cortices at P7 (Fig. 1A and B), falling to a more severe reduction of ∼70% by P12 in all brain regions examined (Fig. 1B).
Figure 1.
Reduction of mtDNA levels in brain due to TK2 deficiency leads to decreased steady-state levels of ETC proteins. (A) Relative quantification of mtDNA copy number in different brain regions of P7 and (B) P12 TK2+/+ and TK2−/− mice (n = 3). (*P < 0.05; **P < 0.01; Two-way ANOVA Bonferroni post-tests; error bars are SEM). Western blot and relative quantification of steady-state ETC proteins in the cerebellum of P7 (C and D) P12 (E and F) TK2+/+ and TK2−/− mice (n = 4 for each group; *P < 0.05; ***P < 0.001; Two-way ANOVA Bonferroni post-tests; error bars are SEM). Values are normalized to the TK2+/+ control from the same group. Arrows indicate mtDNA-encoded subunits.
We next analyzed the steady-state levels of mtDNA- and nDNA-encoded polypeptides of the electron transfer chain (ETC) in different brain regions of P7 and P12 TK2−/− mice. Western blot analysis revealed a reduction in mtDNA-encoded subunits of complex I (ND6) and complex IV [cytochrome oxidase (COX)] in the cerebellum (Fig. 1C–F) and in other brain regions (Supplementary Material, Fig. S1A–D and Supplementary Material, Fig. S2A–D) of both P7 and P12 TK2−/− mice. In contrast, levels of exclusively nuclear-DNA-encoded proteins (complex III: core 2 subunit and complex V: subunit Vα) were unaffected in TK2−/− mice when compared with TK2+/+ mice, for all brain regions at both time-points (Fig. 1C–F; Supplementary Material, Fig. S1A–D and Supplementary Material, Fig. S2A–D). Interestingly, the greatest reduction in the mtDNA-encoded ETC proteins was observed in brain tissue from the P12 TK2−/− mice (Fig. 1E and F and Supplementary Material, Fig. S2A–D), corresponding to the greatest reduction in mtDNA levels and the more severe phenotype.
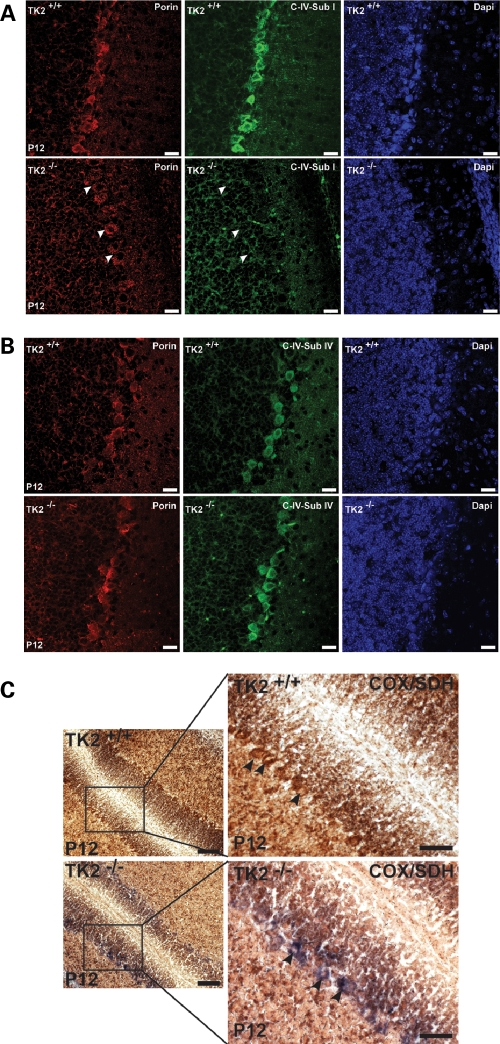
The cerebellum plays an important role in the control of motor coordination and balance. Given that TK2−/− mice appear ataxic and uncoordinated, and that mtDNA copy number and levels of mtDNA-encoded ETC proteins were reduced in the TK2−/− cerebellum, we next sought to establish whether changes in mtDNA-encoded ETC proteins were present at the cellular level. Immunofluorescence in coronal sections from symptomatic P12 TK2−/− cerebella revealed a large number of Purkinje cells (PCs) lacking the mtDNA-encoded COX I (Fig. 2A). In contrast, the steady-state levels of nDNA-encoded ETC proteins were unaffected in TK2−/− PCs, as indicated by the equal expression of the nDNA-encoded COX IV protein in both TK2+/+ and TK2−/− PCs (Fig. 2B). These results indicate that TK2 deficiency leads to a specific reduction of mtDNA-encoded ETC proteins in neurons by P12, while nDNA-encoded ETC proteins appear unaffected.
Figure 2.
ETC dysfunction in TK2−/− PCs. (A) Anti-complex IV subunit I (COX I; mtDNA-encoded), (B) anti-complex IV subunit IV (COX IV; nDNA-encoded) in PCs of P12 of TK2+/+ and TK2−/− mice. TK2−/− PCs have reduced expression of COX I whereas expression of COX IV is unaffected. Scale bars = 20 µm. (C) Sequential enzyme histochemistry for COX and SDH (complex II) activities in cerebella from P12 mice. TK2+/+ PCs have normal COX activity and appear brown (upper panel), whereas TK2−/− PCs lack COX activity and appear blue (low panel). Scale bar = 100 µm (low magnification) and 50 µm (high magnification), respectively.
To further characterize the ETC defect in TK2−/− neurons, we assayed for COX and succinate dehydrogenase (SDH; complex II) activities in fresh-frozen sections of cerebellum from P12 TK2+/+ and TK2−/− mice. Although SDH activity was unperturbed in TK2−/− PCs, COX activity was severely impaired in the majority of PCs, constituting a major disturbance in the ETC within these cells (Fig. 2C). These data indicate that the increasing reduction of mtDNA copy number and the subsequent loss of mtDNA-encoded ETC subunit expression observed in the cerebellum of P12 TK2−/− mice is sufficient to perturb mitochondrial bioenergetics within individual cerebellar neurons.
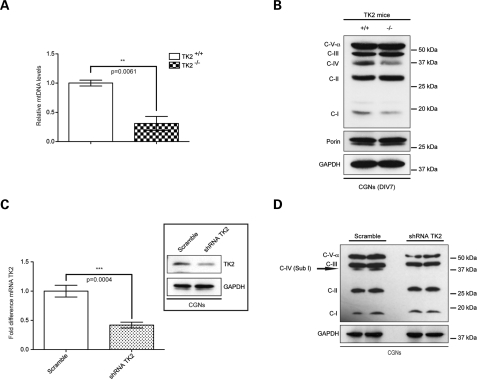
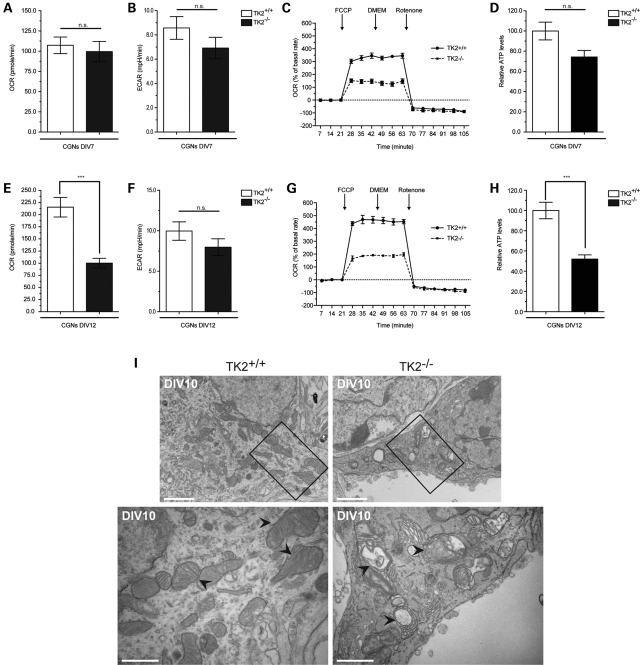
To further explore the disturbance in mitochondrial bioenergetics in the cerebellum, we prepared mixed cerebellar cultures (CGNs) from neonatal TK2+/+ and TK2−/− mice. At day in vitro 7 (DIV7), a reduction in both mtDNA levels and steady-state levels of mtDNA-encoded ETC subunits was observed in TK2−/− CGNs (Fig. 3A and B). Furthermore, shRNA-mediated downregulation of TK2 in wild-type cultured CGNs (Fig. 3C) led to a similar reduction in the level of mtDNA-encoded COX I (Fig. 3D). Changes in expression of ETC subunits may affect cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) (14,15). At DIV7, the basal OCR and ECAR were similar for both TK2+/+ and TK2−/− CGNs (Fig. 4A and B), suggesting that basal ETC activity was maintained in the DIV7 TK2−/− cells despite a reduction in mtDNA. In contrast, challenging DIV7 CGNs with carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP) revealed a 57% reduction in maximal respiration capacity in TK2−/− CGNs (Fig. 4C). Examination of mitochondrial bioenergetic function in DIV14 TK2−/− revealed a marked reduction also in basal OCR (Fig. 4E) without any compensatory rise in ECAR (Fig. 4F). A similar reduction in maximal respiration capacity was observed in the DIV14 TK2−/− CGNs, indicating that overall, DIV14 TK2−/− CGNs harbored a greater impairment of mitochondrial respiration capacity compared with DIV7 TK2−/− CGNs (Fig. 4G). To correlate these observed changes in OCR directly to ATP production, we measured cellular ATP concentration in DIV7 and DIV14 TK2−/− CGNs. The difference in the basal ATP levels in DIV7 TK2+/+ and TK2−/− CGNs were not significant (Fig. 4D), whereas significant reductions in ATP production were observed in DIV14 TK2−/− CGNs (Fig. 4H). These data are consistent with the increasing reduction of mtDNA copy number and subsequent loss of mtDNA-encoded ETC subunit expression observed in the cerebellum of P12, end-stage symptomatic, TK2−/− mice.
Figure 3.
TK2 deficiency leads to reduction of mtDNA-encoded ETC subunits in cerebellar granule neurons (CGNs) in vitro. Dissociated CGNs were cultured from TK2+/+ and TK2−/− mice. (A) qPCR was used to confirm the decrease in mtDNA levels in DIV7 CGNs from TK2−/− mice (student's t-test; n = 3 **P < 0.01; error bars are SEM). (B) Western blot analysis of ETC subunits in TK2−/− CGNs, demonstrating a reduction of mtDNA-encoded subunits of complex IV and complex I. (C) CGNs from wild-type mice infected with a Turbo-GFP lentivector containing either a sequence targeted to a unique site of the mouse TK2 gene (shRNA-TK2) or non-silencing sequence (Scramble). qPCR was used to confirm the decrease in TK2 mRNA expression and the decrease of the protein level was confirmed by Western blot (student's t-test; n = 3 ***P < 0.005; error bars are SEM). (D) Western blot analysis of subunits of respiratory chain complexes of CGNs shown a lack of COX I subunit in shRNA-TK2 CGNs.
Figure 4.
Decreased mitochondrial bioenergetic capacity and aberrant mitochondrial structure in TK2−/− neurons. (A and E) OCR and (B and F) ECAR in DIV7 and DIV14 CGNs, cultured from TK2+/+ and TK2−/− mice (student's t-test n = 3; ***P < 0.005; error bars are SEM). Both rates are normalized against cell number and expressed as rate per minutes. (C and G) Real-time analysis of OCR in CGNs. Mitochondrial uncoupler FCCP and mitochondrial complex I inhibitor rotenone were injected sequentially at the indicated time points into each well after baseline rate measurement. (D and H) Relative ATP levels in DIV7 and DIV14 CGNs, cultured from TK2+/+ and TK2−/− mice. Measurements were made in triplicate (student's t-test n = 3; ***P < 0.005; error bars are SEM) with two animals per group. (I) EM images showing mitochondria in the cell body of DIV10 CGNs from TK2+/+ and TK2−/− mice. TK2−/− mitochondria have dramatic abnormalities in morphology and cristae organization. Scale bars = 2 and 1 µm, respectively.
To determine whether changes in mitochondria bioenergetics are the direct consequence of TK2 loss, we knocked down TK2 expression in wild-type culture CGNs using a lentiviral vector expressing a specific shRNA. Here, a similar reduction in basal OCR was observed without any compensatory rise in ECAR (Supplementary Material, Fig. S3). Additionally, the loss in maximal respiration capacity was similar to that observed in the DIV14 TK2−/− CGNs (Supplementary Material, Fig. S3B). Taken together, these data provide strong evidence that loss of TK2 activity leads to a disturbance in mitochondrial bioenergetic function within neurons of the cerebellum.
We then investigated whether impaired ETC activity results in alterations of mitochondria morphology and cristae organization, as suggested previously (16). DIV7 TK2−/− CGNs showed normal mitochondrial structure and cristae formation (Supplementary Material, Fig. S4). In contrast, by DIV10, TK2−/− CGNs contained large, swollen mitochondria with abnormal cristae structure (Fig. 4I). Furthermore, the mitochondrial matrix of the TK2−/− CGNs appeared to be empty with distorted cristae lying in concentric double membrane rings. These mitochondrial structural abnormalities most likely reflect impaired ETC function, consistent with aberrant mitochondrial structure previously observed in MDS and neurodegenerative disorders (5,17,18).
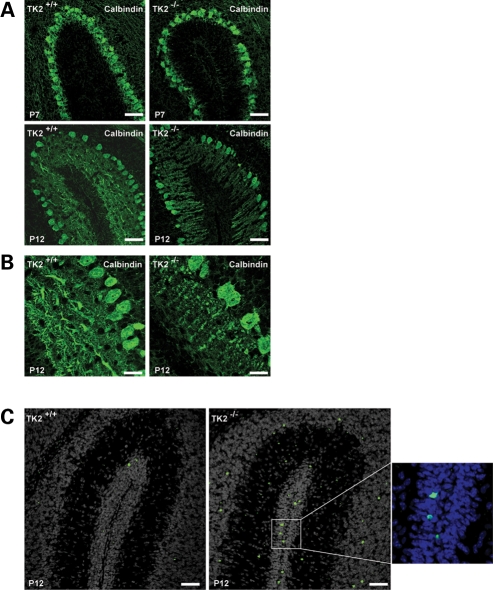
We next analyzed the architecture of cerebella of TK2−/− mice at P7 and P12 searching for any evidence of aberrations of architecture. Calbindin immunofluorescence staining of PCs from the early-symptomatic P7 TK2−/− mice revealed a reduction in the number of dendrites and a decreased dendritic arborization (Fig. 5A). These alterations became more severe in P12 TK2−/− mice (Fig. 5B). Given that the PCs are crucial to cerebellar function, constituting the sole efferents from the cerebellum to the rest of the central nervous system, it is likely these alterations underlie the ataxic phenotype observed in the TK2−/− mice. Furthermore, increased apoptosis was apparent throughout the TK2−/− cerebella at P12. While no TUNEL-positive PCs were seen in P12 TK2−/− cerebella, many granule cells were clearly apoptotic, likely secondary to insufficient trophic support from COX deficient PCs (Fig. 5C). These findings suggest that TK2-associated reductions in neuronal ETC activity impede the development of correctly arborized neuronal networks in vivo, thus leading to neuron demise.
Figure 5.
PC degeneration in TK2−/− cerebellar sections. (A) Anti-calbindin immunocytochemistry to highlight PCs. TK2−/− PC show reduced dentritic arbors at P7 and degeneration at P12. Scale bars = 50 µm. (B) Higher magnification of P12 PCs. TK2+/+ PCs dendrites spread into molecular layer, whereas mutant PCs shown reduced dendritic arbors and cell loss. Scale bars = 20 µm. (C) TUNEL staining (green for apoptotic cells). Increased TUNEL positive cells are present throughout the cerebellar layers in P12 TK2−/− mice. DAPI counterstaining (grey or blue) was used to demonstrate the cellular architecture. Scale bars = 50 µm.
DISCUSSION
The regulation of mtDNA copy number is key to preserving mitochondrial function. In order to sustain mtDNA copy number, the DNA polymerase gamma requires a balanced supply of dNTP substrates, maintained either through direct transport from the cytosol where they are synthesized de novo or through salvaging deoxyribonucleosides in the mitochondrial matrix by the salvage pathway (19). The relative importance of the cyotsolic de novo dNTP synthesis versus the mitochondrial salvage pathway varies between proliferating and non-proliferating cells. In this respect, non-proliferating cells such as neurons are particularly dependent on the mitochondrial salvage pathway due to reduced expression of enzymes involved in cytosolic dNTP salvage and de novo nucleotide synthesis (20). TK2, a mitochondrial deoxyribonucleoside kinase, is an essential enzyme of the mitochondrial salvage pathway and is responsible for the phosphorylation of deoxythymidine, deoxycytidine and deoxyuridine. Previous work has shown that loss of TK2 expression causes a deficiency of pyrimidine dNTPs within the mitochondria, leading to impaired mtDNA replication due to a lack of dNTP substrates. In turn, this results in a progressive depletion of structurally intact mtDNA (4). Accordingly, mutations in TK2 are recognized an important cause of mtDNA depletion syndromes. TK2 mutations were first identified in patients with the myopathic form of mtDNA depletion syndrome, and subsequently over 20 different pathogenic TK2 mutations have now been identified (21). While patients with TK2 mutations typically present with proximal weakness and hypotonia during early infancy and childhood, additional clinical features may include hepatopathy and/or encephalopathy (21,22). Recently, patients with homozygous and compound heterozygous mutations in the TK2 gene were shown to harbor TK2-associated mtDNA depletion in brain and other non-muscle tissue, confirming the clinical suspicion that TK2 mutations can lead to multi-tissue pathology (11). In particular, the observation of neurological features in these patients, including terminal-phase seizures, epilepsia partialis continua and cortical laminar necrosis, strengthens the suggestion that TK2 activity may be critical for neuronal functioning.
In the present study, we have addressed the role of TK2 in neuronal homeostasis using a TK2 knock-out mouse model. Despite appearing normal at birth, the TK2−/− mice rapidly display an ataxic phenotype after 10–12 days of life, with severely impaired motor coordination and gait, and abnormal limb-clasping reflexes indicative of an underlying neuropathology. Severe reductions in mtDNA levels (∼50%) were observed throughout the brain of P7 TK2−/− mice falling to more severe reductions (∼70%) by P12 in all brain regions examined. We have shown that this loss of mtDNA copy is associated with reduced expression of mtDNA-encoded ETC subunits in the cerebellum, hippocampus and cortex by P7, corresponding to the onset of neurological impairment. Thus, in vivo the threshold level at which the phenotype is expressed in brain appears to be ∼50% below normal levels, consistent with the finding that below 60% of normal levels, mtDNA depletion causes a dramatic decrease of the respiratory rate (23). Importantly, a more severe reduction in expression was observed in the brain regions of P12 mice, correlating with the increased loss of mtDNA during later stages of disease progression. Additionally, we have shown that TK2-associated mtDNA depletion results in a severe reduction of mtDNA-encoded ETC subunit expression within individual neurons, as demonstrated by the large number of PCs in the P12 TK2−/− mice lacking expression of the mtDNA-encoded complex IV (COX) subunit I (COX I). While mtDNA-encoded subunits constitute only 3 of the 13 subunits of COX, the TK2-associated reduction in mtDNA-encoded proteins was sufficient to perturb mitochondrial bioenergetics in these neurons, as indicated by deficient COX activity in vivo, and reduced respiration capacity and cellular OCR in vitro. Our findings that P12 TK2−/− PCs suffer a reduction in the number of dendrites and decreased dendritic arborization suggest that lack of TK2 activity impedes the development of correctly arborized neuronal networks in vivo, thus leading to neuron demise.
Interestingly, neurological features appear to be more severe in the TK2−/− mice compared with patients with TK2 deficiency. This may, in part, reflect a difference in tissues specific dNTP pool regulatory pathways between species. Indeed, previous work has identified that in wild-type mice, TK2 accounts for more than 60% of total brain TK activity, suggesting that the mouse brain is more dependent on the mitochondrial nucleotide salvage pathway than other tissues (12). Additionally, TK2−/− mice have a total lack of TK2 enzyme activity. In contrast, TK2 deficiency in patients is associated with missense mutations in the TK2 gene, resulting in defective TK2 enzymes with some degree of residual enzymatic activity (4,21,24). At present, no patients with TK2 non-sense mutations or total lack of TK2 protein have been identified, suggesting that total loss of TK2 expression in humans may also cause a more severe phenotype that may not be compatible with life (13). Nonetheless, the presence of neurodegeneration and ataxic phenotypes in mice lacking TK2 activity indicates the importance of this enzyme in neuronal homeostasis.
In summary, our findings provide direct evidence that lack of TK2 activity causes a decline in mtDNA levels in neurons in vivo and in vitro. In turn, this leads to a reduction of normal steady-state levels of ETC proteins, impaired mitochondrial bioenergetic function and degeneration of selected neuronal types. Taken together, our findings have important implications for understanding the mechanisms of neurological dysfunction observed in MDS, and indicate the importance of considering neurological phenotypes in the differential diagnosis of MDS due to TK2 deficiency.
MATERIALS AND METHODS
Animals
TK2−/− mice, in which the TK2 gene has been disrupted by homologous recombination, were developed by Zhou et al. (13) in A. Karlsson's laboratory (Karolinska Institute, Stockholm).
Cell culture
Mixed cerebellar cultures were established using a modified version of the protocol described previously by Hatten (25). Briefly, cerebella were dissected from P7 pups and individually digested with trypsin and DNase. Trituration in DNase was followed by a mechanical trituration of the tissue with a 10 ml syringe with an 18 gauge needle before filter the cells suspension through 100 µm plastic mesh to remove clumps. The cell suspension was plated on poly-d-lysine-coated plate at a density of 0.8 × 106 cells/ml.
shRNA-TK2 mouse lentiviral vector
To knockdown TK2 expression in the mouse cerebella granule neurons (CGNs), we used a commercially available pGIPZ-lentiviral shRNAmir vector containing a hairpin sequence targeting TK2 (Open Biosystems, Huntsville, AL, USA). The TK2 hairpin sequence was as follows with underlined sequencing corresponding to region of the mouse TK2 gene targeted: TGCTGTTGACAGTGAGCGCGCTGCTCTTGACATTCTAGAATAGTGA AGCCACAGATGTATTCTAGAATGTCAAGAGCAGCA TGCCTACTGCCTCGGA. The shRNA-containing lentiviral vector was cotransfected with lentiviral packaging into HEK-293T cells (Open Biosystems) to produce shRNA-carrying lentivirus particles. Culture supernatants were collected at 48 h after transfection and lentivirus particles were concentrated using PEG (System Biosciences). CGNs cells were transduced by the resulting concentrated viral particles (MOI = 1) at day in vitro 1 (DIV1).
Antibodies
Primary antibodies used include the following: rabbit anti-calbindin (Chemicon), anti-gapdh (Santa Cruz), anti complex IV subunit IV (Santa Cruz), anti-TK2 (Primm), anti-complex IV subunit I, anti-OXPHOS complexes and anti-porin (Mitoscience).
Immunohistochemistry and immunocytochemistry
P7 and P12 brains were fixed in 4% paraformaldehyde (wt/vol) and embedded in paraffin. The sections were de-waxed, re-hydrated in graded alcohols, rinsed in distilled water and antigen retrieval-treated for 20 min (1 mm EDTA pH 8.0) in a pressure cooker at 700 W. The sections were washed three times in 1× phosphate-buffered saline (PBS) for 10 min each, blocked in 5% goat serum in 0.1% PBS-Tween (vol/vol), incubated overnight with primary antibody, washed three times for 10 min each with 1× PBS, incubated for 1 h with secondary antibody, washed three times with 1× PBS and counterstained with DAPI and mounted for confocal microscopy. Terminal deoxynucleotidyl transferase-mdiated biotinylated UTP nick labeling (TUNEL) (Roche) was performed according to the instructions of the manufacturer. For immunocytochemistry experiments cell were fixed with 4% paraformaldehyde in PBS, pH 7.4, for 15 min at RT. Excess buffer was drained and the cells were washed three times in PBS. Cells were then post-fixed in ice-cold methanol 5 min or permeabilized using 0.5% Triton X-100 in PBS with 3% bovine serum albumin. Fixed cells were incubated with the appropriate primary antibody in PBS with 10% goat serum and 0.1% Triton X-100 overnight at 4°C. Cells were washed and incubated with anti-mouse or anti-rabbit Alexa conjugated antibodies (Invitrogen) 1–2 h at room temperature. Coverslips were washed and mounted. All imaging was performed on a Zeiss LSM510 confocal microscope using a 25× or a 63× objective.
Sequential cytochrome c oxidase and sodium succinate dehydrogenase histochemistry
Sequential histochemical assay for cytochrome c oxidase (COX) (three catalytic subunits encoded by mitochondrial genome) and SDH (all subunits nuclear encoded) was performed as previously described (26).
Isolation of total DNA
Dry pellets of 1 × 106 cells were resuspended in 200 µl of PBS and processed using a QIAamp DNA Mini Kit (Qiagen®, Germany) according to the manufacturer's protocol. DNA was eluted in distilled water keeping the final concentration in the range 50–200 ng/μl.
RNA extraction and quantitative real-time PCR
Total RNA was derived from cells using the Qiagen RNAeasy kit (Qiagen) and tested for purity (A260/280 ratios) and integrity (denaturing gel electrophoresis). Quantitative real-time PCR (qPCR) amplification was performed using specific primers and probes for mouse TK2, ND1, GAPDH (TaqMan® Gene Expression Assays, Applied Biosystems). Thermal cycling was performed using the ABI PRISM® 7000 Sequence Detector System (AB, Applied Biosystems).
Immunoblotting
Whole-cell lysates were prepared by lysing cells and tissue in RIPA lysis buffer. The insoluble material was excluded by centrifugation. The resulting supernatant was mixed with SDS–PAGE loading buffer 5X and was subjected to SDS-12%-PAGE gel. After electrophoresis, the proteins were transferred to nitrocellulose membrane, and the membrane was incubated with the appropriate primary antibody. Protein-antibody interactions were detected with a peroxidase-conjugated secondary antibody, subjected to Pierce® ECL Chemiluminescent reagent (Thermo Fisher). Quantification of the proteins was carried out using NIH Image J 1.37V software.
XF24 oxygen consumption analysis and ATP determination
OCR and ECAR measurements were made using the XF24 Analyzer (Seahorse Bioscience) as described by Wu et al. (15) following the manufacture's instructions. Briefly, the mitochondrial uncoupler FCCP and the respiration inhibitor rotenone were loaded in drug delivery system and, after measuring basal OCR and ECAR, compounds were added sequentially into the wells during the assay. The effects on OCR were measured after each compound addition. XF assay medium was low buffered bicarbonate free DMEM (pH 7.4). ATP concentration were determined using the CellTiter-Glo® Luminescent assay (Promega), and value were normalized to microgram of proteins.
Statistical analysis
All experiments were performed in triplicates for each condition, and from at least three different cell culture preparations. Results are expressed as mean ± SEM. Statistical analysis was performed by one-way ANOVA with Bonferroni post-test to compare all conditions or by paired or unpaired Students’ t-test using Prism 5.0 software (Graph Pad Software, San Diego, CA, USA). P < 0.05 were regarded as significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Medical Research Council (MRC). P.S. is also supported by Samantha Dickson Brain Tumour Trust and by the Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
We particularly thank J. Edwards (Medical Research Council) for assistance with immunohistochemistry, J. McWilliams and T. Smith (Medical Research Council) for preparation of samples for EM, and R. Snowden, S. Galavotti and M. Guerra-Martin (Medical Research Council) for technical support. We also thank D. Read and members of the P.N. laboratory for critical discussion and support for image analysis.
Conflicts of Interest statement. None declared.
REFERENCES
- 1.Moraes C.T., Shanske S., Tritschler H.J., Aprille J.R., Andreetta F., Bonilla E., Schon E.A., DiMauro S. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am. J. Hum. Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]
- 2.Alberio S., Mineri R., Tiranti V., Zeviani M. Depletion of mtDNA: syndromes and genes. Mitochondrion. 2007;7:6–12. doi: 10.1016/j.mito.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 3.McFarland R., Taylor R.W., Turnbull D.M. The neurology of mitochondrial DNA disease. Lancet Neurol. 2002;1:343–351. doi: 10.1016/s1474-4422(02)00159-x. [DOI] [PubMed] [Google Scholar]
- 4.Saada A., Shaag A., Mandel H., Nevo Y., Eriksson S., Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet. 2001;29:342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- 5.Mandel H., Szargel R., Labay V., Elpeleg O., Saada A., Shalata A., Anbinder Y., Berkowitz D., Hartman C., Barak M., et al. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat. Genet. 2001;29:337–341. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- 6.Elpeleg O., Miller C., Hershkovitz E., Bitner-Glindzicz M., Bondi-Rubinstein G., Rahman S., Pagnamenta A., Eshhar S., Saada A. Deficiency of the ADP-forming succinyl-CoA synthase activity is associated with encephalomyopathy and mitochondrial DNA depletion. Am. J. Hum. Genet. 2005;76:1081–1086. doi: 10.1086/430843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostergaard E., Hansen F.J., Sorensen N., Duno M., Vissing J., Larsen P.L., Faeroe O., Thorgrimsson S., Wibrand F., Christensen E., et al. Mitochondrial encephalomyopathy with elevated methylmalonic acid is caused by SUCLA2 mutations. Brain. 2007;130:853–861. doi: 10.1093/brain/awl383. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari G., Lamantea E., Donati A., Filosto M., Briem E., Carrara F., Parini R., Simonati A., Santer R., Zeviani M. Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-gammaA. Brain. 2005;128:723–731. doi: 10.1093/brain/awh410. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen K.V., Ostergaard E., Ravn S.H., Balslev T., Danielsen E.R., Vardag A., McKiernan P.J., Gray G., Naviaux R.K. POLG mutations in Alpers syndrome. Neurology. 2005;65:1493–1495. doi: 10.1212/01.wnl.0000182814.55361.70. [DOI] [PubMed] [Google Scholar]
- 10.Hakonen A.H., Isohanni P., Paetau A., Herva R., Suomalainen A., Lonnqvist T. Recessive Twinkle mutations in early onset encephalopathy with mtDNA depletion. Brain. 2007;130:3032–3040. doi: 10.1093/brain/awm242. [DOI] [PubMed] [Google Scholar]
- 11.Gotz A., Isohanni P., Pihko H., Paetau A., Herva R., Saarenpaa-Heikkila O., Valanne L., Marjavaara S., Suomalainen A. Thymidine kinase 2 defects can cause multi-tissue mtDNA depletion syndrome. Brain. 2008;131:2841–2850. doi: 10.1093/brain/awn236. [DOI] [PubMed] [Google Scholar]
- 12.Akman H.O., Dorado B., Lopez L.C., Garcia-Cazorla A., Vila M.R., Tanabe L.M., Dauer W.T., Bonilla E., Tanji K., Hirano M. Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum. Mol. Genet. 2008;17:2433–2440. doi: 10.1093/hmg/ddn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X., Solaroli N., Bjerke M., Stewart J.B., Rozell B., Johansson M., Karlsson A. Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum. Mol. Genet. 2008;17:2329–2335. doi: 10.1093/hmg/ddn133. [DOI] [PubMed] [Google Scholar]
- 14.Jekabsons M.B., Nicholls D.G. In situ respiration and bioenergetic status of mitochondria in primary cerebellar granule neuronal cultures exposed continuously to glutamate. J. Biol. Chem. 2004;279:32989–33000. doi: 10.1074/jbc.M401540200. [DOI] [PubMed] [Google Scholar]
- 15.Wu M., Neilson A., Swift A.L., Moran R., Tamagnine J., Parslow D., Armistead S., Lemire K., Orrell J., Teich J., et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol. Cell. Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 16.Kukat A., Kukat C., Brocher J., Schafer I., Krohne G., Trounce I.A., Villani G., Seibel P. Generation of rho0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses. Nucleic Acids Res. 2008;36:e44. doi: 10.1093/nar/gkn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H., McCaffery J.M., Chan D.C. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Mandel H., Hartman C., Berkowitz D., Elpeleg O.N., Manov I., Iancu T.C. The hepatic mitochondrial DNA depletion syndrome: ultrastructural changes in liver biopsies. Hepatology. 2001;34:776–784. doi: 10.1053/jhep.2001.27664. [DOI] [PubMed] [Google Scholar]
- 19.Mathews C.K. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 20.Rylova S.N., Mirzaee S., Albertioni F., Eriksson S. Expression of deoxynucleoside kinases and 5′-nucleotidases in mouse tissues: implications for mitochondrial toxicity. Biochem. Pharmacol. 2007;74:169–175. doi: 10.1016/j.bcp.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 21.Oskoui M., Davidzon G., Pascual J., Erazo R., Gurgel-Giannetti J., Krishna S., Bonilla E., De Vivo D.C., Shanske S., DiMauro S. Clinical spectrum of mitochondrial DNA depletion due to mutations in the thymidine kinase 2 gene. Arch. Neurol. 2006;63:1122–1126. doi: 10.1001/archneur.63.8.1122. [DOI] [PubMed] [Google Scholar]
- 22.Mancuso M., Salviati L., Sacconi S., Otaegui D., Camano P., Marina A., Bacman S., Moraes C.T., Carlo J.R., Garcia M., et al. Mitochondrial DNA depletion: mutations in thymidine kinase gene with myopathy and SMA. Neurology. 2002;59:1197–1202. doi: 10.1212/01.wnl.0000028689.93049.9a. [DOI] [PubMed] [Google Scholar]
- 23.Rocher C., Taanman J.W., Pierron D., Faustin B., Benard G., Rossignol R., Malgat M., Pedespan L., Letellier T. Influence of mitochondrial DNA level on cellular energy metabolism: implications for mitochondrial diseases. J. Bioenerg. Biomembr. 2008;40:59–67. doi: 10.1007/s10863-008-9130-5. [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Limongelli A., Vila M.R., Carrara F., Zeviani M., Eriksson S. Molecular insight into mitochondrial DNA depletion syndrome in two patients with novel mutations in the deoxyguanosine kinase and thymidine kinase 2 genes. Mol. Genet. Metab. 2005;84:75–82. doi: 10.1016/j.ymgme.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Hatten M.E. Neuronal regulation of astroglial morphology and proliferation in vitro. J. Cell. Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betts J., Jaros E., Perry R.H., Schaefer A.M., Taylor R.W., Abdel-All Z., Lightowlers R.N., Turnbull D.M. Molecular neuropathology of MELAS: level of heteroplasmy in individual neurones and evidence of extensive vascular involvement. Neuropathol. Appl. Neurobiol. 2006;32:359–373. doi: 10.1111/j.1365-2990.2006.00731.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.