Abstract
Quantifying the variation in the human plasma proteome is an essential prerequisite for disease-specific biomarker detection. We report here on the longitudinal and individual variation in human plasma characterized by two-dimensional difference gel electrophoresis (2-D DIGE) using plasma samples from eleven healthy subjects collected three times over a two week period. Fixed-effects modeling was used to remove dye and gel variability. Mixed-effects modeling was then used to quantitate the sources of proteomic variation. The subject-to-subject variation represented the largest variance component, while the time-within-subject variation was comparable to the experimental variation found in a previous technical variability study where one human plasma sample was processed eight times in parallel and each was then analyzed by 2-D DIGE in triplicate. Here, 21 protein spots had larger than 50% CV, suggesting that these proteins may not be appropriate as biomarkers and should be carefully scrutinized in future studies. Seventy-eight protein spots showing differential protein levels between different individuals or individual collections were identified by mass spectrometry and further characterized using hierarchical clustering. The results present a first step toward understanding the complexity of longitudinal and individual variation in the human plasma proteome, and provide a baseline for improved biomarker discovery.
1. Introduction
Mapping the human proteome presents a significant scientific challenge, partly because of the complexity of the population and partly because of technological limitations [1]. However, potential rewards in the diagnosis and treatment of diseases make proteomic characterization of human plasma a very worthwhile endeavor. The Human Proteome Organization (HUPO) represents an international consortium of academic and industrial partners whose common goal is to foster collaboration and facilitate a better understanding of the human proteome. Recognizing the need for reproducibility, and following in the footsteps of the more mature field of gene expression analysis, proteomic standards are starting to emerge [2, 3]. The Human Plasma Proteome (HPP) project [4] of HUPO, which specifically targets plasma proteins, has made considerable progress while highlighting the complexity of plasma proteomics. For example, protein identification of the same specimen resulted in less than 50% agreement when repeated multiple times [5, 6], reflecting the challenges involved in biomarker discovery from human plasma [7, 8] and underlining the need for improvements in plasma proteomic characterization. Studies providing prefractionation and other sample preparation aspects are looking to improve this process [9–12].
A primary technological problem that needs to be addressed is the quantification of the experimental variation on a given proteomic platform. Next, the baseline variation within individuals over time and the variation between multiple individuals also need to be quantitated. Searching for disease-specific biomarkers makes sense only after these two steps are addressed. Our recent study, referred to as the Technical Variation Study (TVS) [13] throughout the manuscript, addressed the first question for two-dimensional difference gel electrophoresis (2-D DIGE) experiments by processing one human plasma sample eight times and analyzing each of the resulting eight technical replicates in triplicate on twelve gels [13–16]. The present study is a follow-up to the TVS, whereby plasma samples from eleven healthy volunteer subjects, taken at three time points separated by two weeks, were analyzed in triplicate on 50 gels.
The goal of this study was to assess longitudinal and individual variation in human plasma and to compare results to the experimental variation detected in the previously reported TVS [13]. While differences were detected in the plasma proteome within individuals over time, our analyses indicate that individual variation contributes the largest observed proteomic variability. Further, our results demonstrate that gender-related proteomic differences can be detected by 2-D DIGE and should also be considered in biomarker discovery. Overall, this work represents a first step in quantitating the variability in human plasma by addressing the individual and longitudinal proteomic variation in human plasma.
2. Materials and Methods
2.1. Sample Collection
Blood samples were collected from eleven healthy volunteers (five males, six females) at three time points separated by two weeks, with informed consent under Institutional Review Board approval from Lawrence Livermore National Laboratory. To minimize the effect of daily variations within an individual, the samples from a given subject were taken at approximately the same time, within a thirty-minute window, in the morning for each time point. Other variables were not controlled for in order to better mimic the variability in typical human plasma samples (age, fasting, illness, medication, etc.). To better examine the longitudinal variation and minimize the chance of an individual providing samples while experiencing an underlying condition such as a cold, two weeks between sample collection were chosen. Each individual was assigned an identification number to blind the samples and ease the experimental design.
2.2. Top-6 High-Abundance Protein Depletion and Sample Preparation
To increase the resolution of the 2-D DIGE technique, the six most abundant plasma proteins were depleted using affinity chromatography, as previously reported [12]. The sample cleanup and protein assay was performed as described previously [12, 13, 17, 18].
2.3. 2-D DIGE and Gel Imaging
The 33 top-6-depleted plasma samples from the 11 individuals were analyzed in triplicate in a 50-gel 2-D DIGE experiment [13–16] (see Table 1 in Supplementary Material available online at doi: 10.1155/2010/258494). Each gel contained three samples, one internal pooled standard and two experimental samples. The internal pooled standard consists of an equal amount of each of the 33 samples and was labeled with the Cy2 dye (GE Healthcare). Each experimental sample was dye-swapped and labeled with both the Cy3 and the Cy5 dyes (GE Healthcare) in the experimental design to mitigate the effect of potential dye-specific variations [19]. Samples from individuals obtained at different times were compared on some gels, while samples from two different individuals were compared on other gels. Gels were run randomly in batches of twelve in order to minimize batch-to-batch variability. Supplementary Table 1 shows the complete experimental design. Labeling first dimension (pI) separation, second dimension (mw) separation, and gel imaging was performed as described previously [13]. Mass spectrometry was carried out as previously described [20].
2.4. Data Analysis
The DeCyder Differential Analysis Software v5.01 (GE Healthcare) was used for quantitating differential abundance of proteins. The Differential In-gel Analysis (DIA) module was used to determine the optimal spot detection settings. Images were loaded into the Batch Processor module with the estimated number of spots set to 2,500. The master gel was assigned automatically to the gel with the most spots detected. Each sample was grouped for analysis in the Biological Variation Analysis (BVA) module. During batch processing, the Cy2 channel from each gel was used for normalization of the spot intensities and for automated matching between gels. For each spot on each gel, the software reported the standardized abundance (SA) as the ratio of the volume in the Cy3 (or Cy5) sample to the volume of the pooled standard sample labeled with Cy2, where the volumes were normalized across the gels. Standardized log abundance (SLA), defined as log 10 (SA), was used in quantifying differential expression. Fold change between groups was calculated as the ratio of the average SA in the two groups. If R denotes that ratio, the fold change F was defined as F = R if R ≧ 1 and F = −1/R otherwise. A k-fold expression increase/decrease corresponded to a +k/−k value of F.
Using DeCyder, all possible pairwise comparisons were made between the 33 groups defined by the eleven subjects at the three times. Within the BVA module each comparison was filtered to find the spots having (a) P-value ≤.05 and (b) greater than 1.5-fold change in expression between the groups. The analysis was converted into DeCyder 2D (v6.5), and the Extended Data Analysis (EDA) module (GE Healthcare) was used to perform expression pattern clustering [21–26]. Data from the TVS were integrated into the analysis, pooled standards were normalized and principal component analysis was conducted [27–30] on the spots that were successfully matched on >75% of the gels from the TVS and the present study [13].
Spot characteristics calculated by DeCyder were exported for further statistical processing into the R statistical computing environment [19] (http://www.r-project.org/). Summary statistics for the spot matching across the gels were calculated. The high-quality spots, defined as the spots matched on at least 75% of the gels, were subjected to further analysis. Spotwise standard deviations (SDs) of the SLA values and coefficients of variations (CVs) of the SA values were calculated, first by using all the data at a given spot to obtain one SD and one CV for that spot, then by performing the same calculations separately for the eleven subjects, thus obtaining eleven SD and CV values for each spot. The former method estimated the protein expression variation among all subjects and time points, while the latter addressed the variation within individuals through time. The results were compared to the spotwise SD and CV values obtained from the TVS [13].
To quantitate the relative contribution of the components of variation, mixed-effects statistical modeling [31] was performed. Let yijk denote the SLA at spot i on gel j measured with dye k, with i = 1,…, I, where I represents the number of spots matched on 75% of the gels, j = 1,…, 50, and k = 1, 2. In addition, let l = 1,…, 11 and m = 1, 2, 3 indicate the subject and time indices, respectively. Let g = 1, 2 indicate male and female genders, respectively. The assumed model was of the form
| (1) |
where μ is the overall mean, αj denote the coefficients for the fixed gel effects, βk the coefficients for the fixed dye effects, (α β)jk the coefficients for the gel-dye interactions, γig the coefficient for the fixed gender effect at spot i, and ail, bil(m), and eijklm the random effects components for subject (individual), time (longitudinal), and error at spot i, independent and normally distributed [32] with mean zero and variance σsi2, σti2, and σei2, respectively. The gender, subject, and time subscripts in (1) are redundant, as for any given j and k, the identity of the sample, including the subject, gender and time, was known. However, we included them for the clarity of the model description. Since the gel and dye factors were balanced with respect to the spots (the first four terms in the model were common to all spots), (1) was fit in two stages, with results equivalent to, and computationally more efficient than, the full one-stage solution in (1) repeated at each spot. Similar methods have been established for microarrays [33]. In the first stage, the data from all I spots were used to estimate the global dye and gel effects; that is, only the first four terms in (1) plus error were included in the model. In the second stage, the last four terms in (1) were fit to the residuals from the first stage, one spot at a time. In essence, this first stage amounted to a normalization step, whereby the fixed dye and gel effects were estimated and removed by pooling the information across all the spots. In the second stage, a fixed gender effect and random variance components of subject, time, and error were estimated separately at each spot. Thus, at spot i, the total variance σyi2 was separated into its random components as σyi2 = σsi2 + σti2 + σei2.
The effect of additional statistical normalizations of the SLA on the variance component estimates and on the differentially expressed spots was investigated. The SLA values obtained from DeCyder were further normalized by statistical methods that corrected for potential dye biases within gels and range differences among the gels as previously described [34].
The spots that were determined to be of differential abundance (>1.5-fold difference with P-value <.05) were excised from the pick gel and identified by mass spectrometry as previously reported [12, 18, 20]. Identified spots were selected in DeCyder for additional expression pattern clustering.
3. Results and Discussion
3.1. Experimental Design
The experimental design is shown in Supplementary Table 1 and Supplementary Figure 1. Rather than randomly pairing the samples on the gels, the design was selected to minimize the experimental variation among the samples whose comparison was of most interest. By placing the samples from a subject across different time points on the same gel, gel-related variations for intrasubject comparisons were minimized. Essentially, our design was based on the requirements that (1) each of the 33 samples has three replicates and (2) comparisons of the samples from the same subject were of more interest than comparisons of different subjects across time points. This led to an experimental design that contained 22 gels used for comparing the same individual at different time points and 28 gels to compare two individuals at different time points. In addition, the use of dye swapping and triplicates also contributed to overall quality of the data. Our results suggest that gender variability may also be present. As individual and longitudinal variability was our main objective, we did not attempt to control for gender differences. We selected both males and females for our study to get a more appropriate human sample set. Future work looking at human proteome variability should account for gender-specific variability in the design of the experiment.
3.2. Spot Matching
Landmarks were placed manually on each gel to assist in the spot matching across the gels. Spots of interest identified through the analyses were verified to have the three-dimensional profile characteristics of a protein spot. Those spots with volumes close to background level and dust particles with very large slopes and small areas were eliminated. The total number of protein spots detected on the master gel was 2556. Three hundred and ninety seven (15%) spots were matched on at least 37 gels, and 1215 (46%) spots were matched on at least 25 gels. The following statistical analyses were restricted to the 397 high-quality spots matched on 75% of the gels. These high-quality spots were chosen to focus on spots that did not require warping or imputing of any missing data. Future data analysis may help determine if warping and data imputation can expand high-quality gel proteomic data. The latest version of DeCyder contains the ability to warp gels to potentially add missing data. These additions may provide additional spots that can be studied as high-quality, but this type of data manipulation may also create skewed expression values, as the results depend on the type of postrun imputation model that is utilized [35].
Our previous study with technical replicates of one human plasma sample [13] had 42% of the spots matched on 8 of 12 gels. The addition of biological samples from different subjects at varying time points added to the complexity of the current dataset and reduced the matching accuracy. While most of the decrease in the matching accuracy is expected to stem from the biological complexity of the experiment, part of it may be attributed to the larger number of gels, which inevitably increases the expected experimental variation. A study of five commercial software programs showed that an average of only 3% of the total analysis time was automated as opposed to manual, and as the number of gels increased, the percentage of automatically generated correct matches was dramatically reduced [36]. Taken together, these studies suggest that improved spot detection and matching software and algorithms are needed to increase the quality of spot matching. One such study was accomplished that created algorithms to improve spot matching with an integrated approach using hierarchical-based and optimization-based methods [37].
3.3. Differential Expression and Protein Identifications
The pairwise comparisons among the 33 samples identified over 1400 spots with P-value <.05 and fold-change > 1.5. Down selection using manual inspection eliminated most of these spots (due to three-dimensional profile characteristics not representative of protein or because of insufficient representation of the spot on enough gels) resulting in 427 spots of further interest. The majority of the spots that did not pass manual verification were similar to background levels, lacking visual characteristics of protein spots. The sensitive detection parameters used in this study, while allowing for the detection of low abundance proteins, results in increased detection of artifacts that require manual verification.
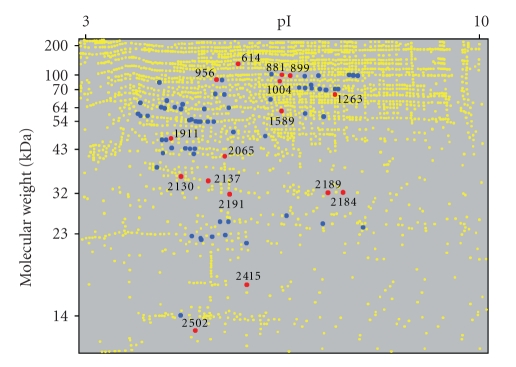
Of the 427 spots with differential abundance, those exhibiting the greatest differences in abundance levels were further characterized, and 78 proteins spots were identified by mass spectrometry. The identified proteins are listed in Table 2, along with the theoretical pI and molecular weight calculated from the full-length amino acid sequence of each protein. Figure 1 depicts the spatial distribution of the protein spots detected in human plasma by 2-D DIGE. Identified proteins are denoted by blue dots. Sixteen proteins (red dots) were found to have differences between the theoretical and observed molecular weights. The discrepancies are potentially due to posttranslational modifications or experimental processing; however, since all samples were treated identically, posttranslational modification is more likely. For example, spots 2189 and 2184, both identified as complement component C4A, were found to be statistically significant with at least a 1.5-fold difference between individuals. Complement component C4A has a theoretical molecular weight of 192.8 kDa; yet the protein spots identified indicate an approximate 32 kDa fragment. Since only C-terminal peptides were detected by mass spectrometry, the protein spot likely corresponds to the active Complement C4c fragment (mw = 33 kDa), which is a known cleavage product of Complement C4A [38, 39]. Variability in the amount of Complement C4c fragment between individuals could be a reflection of immune status, which may be a considerable variable when comparing human clinical subjects.
Table 2.
Variable proteins identified from human plasma.
| Protein Number | Protein Identity | Accession Number | mwa | pIa | Gene |
|---|---|---|---|---|---|
| 614a | clusterin | IPI00400826 | 57.8 | 6.25 | CLU |
| 614b | alpha-2-macroglobulin precursor | IPI00478003 | 163.3 | 6 | A2M |
| 835a | alpha-2-macroglobulin precursor | IPI00478003 | 163.3 | 6 | A2M |
| 835b | Complement C3 precursor | IPI00783987 | 187.1 | 6.02 | C3 |
| 849 | Plasminogen | IPI00019580 | 90.6 | 7.04 | PLG |
| 856 | Plasminogen | IPI00019580 | 90.6 | 7.04 | PLG |
| 881a | complement factor B preproprotein | IPI00019591 | 85.5 | 6.67 | CFB |
| 881b | complement protein C7 precursor | IPI00296608 | 93.5 | 6.09 | C7 |
| 881c | Complement C3 precursor | IPI00783987 | 187.1 | 6.02 | C3 |
| 884 | Plasminogen | IPI00019580 | 90.6 | 7.04 | PLG |
| 893 | complement factor B preproprotein | IPI00019591 | 85.5 | 6.67 | CFB |
| 899a | complement factor B preproprotein | IPI00019591 | 85.5 | 6.67 | CFB |
| 899b | complement protein C7 precursor | IPI00296608 | 93.5 | 6.09 | C7 |
| 899c | Complement C3 precursor | IPI00783987 | 187.1 | 6.02 | C3 |
| 910a | fibrinogen gamma | IPI00219713 | 49.5 | 5.7 | FGG |
| 910b | complement factor B preproprotein | IPI00019591 | 85.5 | 6.67 | CFB |
| 956a | complement component 1, r subcomponent | IPI00296165 | 80.2 | 5.89 | C1R |
| 956b | complement component C4A | IPI00032258 | 192.8 | 6.66 | C4A |
| 963 | complement component 1, r subcomponent | IPI00296165 | 80.2 | 5.89 | C1R |
| 1002 | complement component 1,s subcomponent | IPI00017696 | 76.7 | 4.86 | C1S |
| 1004a | gelsolin | IPI00646773 | 80.6 | 5.58 | GSN |
| 1004b | complement component 2 | IPI00303963 | 83.3 | 7.23 | C2 |
| 1004c | complement factor B preproprotein | IPI00019591 | 85.5 | 6.67 | CFB |
| 1004d | complement protein C7 precursor | IPI00296608 | 93.5 | 6.09 | C7 |
| 1004e | alpha-2-macroglobulin precursor | IPI00478003 | 163.3 | 6 | A2M |
| 1004f | Complement C3 precursor | IPI00783987 | 187.1 | 6.02 | C3 |
| 1004g | complement component C4A | IPI00032258 | 192.8 | 6.66 | C4A |
| 1027 | transferrin | IPI00022463 | 77 | 6.81 | TF |
| 1110 | IGHM protein | IPI00828205 | 65.3 | 8.1 | IGHM |
| 1113a | IGHM protein | IPI00828205 | 65.3 | 8.1 | IGHM |
| 1113b | transferrin | IPI00022463 | 77 | 6.81 | TF |
| 1128a | IGHM protein | IPI00828205 | 65.3 | 8.1 | IGHM |
| 1128b | transferrin | IPI00022463 | 77 | 6.81 | TF |
| 1129a | histidine-rich glycoprotein precursor | IPI00022371 | 59.6 | 7.09 | HRG |
| 1129b | coagulation factor XII precursor | IPI00019581 | 67.5 | 7.94 | F12 |
| 1129c | transferrin | IPI00022463 | 77 | 6.81 | TF |
| 1142a | histidine-rich glycoprotein precursor | IPI00022371 | 59.6 | 7.09 | HRG |
| 1142b | transferrin | IPI00022463 | 77 | 6.81 | TF |
| 1156 | transferrin | IPI00022463 | 77 | 6.81 | TF |
| 1185 | transferrin | IPI00022463 | 77 | 6.81 | TF |
| 1254 | hemopexin precursor | IPI00022488 | 51.5 | 6.57 | HPX |
| 1263a | histidine-rich glycoprotein precursor | IPI00022371 | 59.6 | 7.09 | HRG |
| 1263b | Complement C3 precursor | IPI00783987 | 187.1 | 6.02 | C3 |
| 1276a | hemopexin precursor | IPI00022488 | 51.5 | 6.57 | HPX |
| 1276b | Heparin cofactor II precursor | IPI00292950 | 57.1 | 6.41 | HCF2 |
| 1276c | peptidoglycan recognition protein L precursor | IPI00163207 | 62.2 | 7.25 | PGLYRP |
| 1382 | kininogen | IPI00215894 | 47.9 | 6.29 | KNG |
| 1394 | albumin | IPI00745872 | 69.1 | 5.85 | ALB |
| 1456 | alpha-1-antichymotrypsin precursor | IPI00550991 | 45.5 | 5.32 | AACT |
| 1471a | immunoglobulin alpha-1 heavy chain | IPI00166866 | 37.6 | 6.06 | IGHA1 |
| 1471b | kininogen | IPI00215894 | 47.9 | 6.29 | KNG |
| 1471c | antithrombin III | IPI00032179 | 52.6 | 6.32 | AT3 |
| 1525 | Vitronectin precursor | IPI00298971 | 54.3 | 5.55 | VTN |
| 1526a | kininogen | IPI00215894 | 47.9 | 6.29 | KNG |
| 1526b | Angiotensinogen | IPI00032220 | 53.2 | 5.78 | AGT |
| 1555 | Vitronectin precursor | IPI00298971 | 54.3 | 5.55 | VTN |
| 1558 | immunoglobulin alpha-2 heavy chain | IPI00641229 | 36.4 | 5.71 | IGH |
| 1568a | kininogen | IPI00215894 | 47.9 | 6.29 | KNG |
| 1568b | antithrombin III | IPI00032179 | 52.6 | 6.32 | AT3 |
| 1568c | Angiotensinogen | IPI00032220 | 53.2 | 5.78 | AGT |
| 1577 | Alpha-2-HS-glycoprotein | IPI00022431 | 39.3 | 5.43 | AHSG |
| 1589a | immunoglobulin alpha-1 heavy chain | IPI00166866 | 37.6 | 6.06 | IGHA1 |
| 1589b | apolipoprotein H precursor | IPI00298828 | 38.3 | 8.34 | APOH |
| 1589c | prepro-plasma carboxypeptidase B | IPI00329775 | 48.4 | 7.61 | pCPB |
| 1589d | fibrinogen beta chain | IPI00298497 | 55.9 | 8.54 | FGB |
| 1589e | alpha-2-macroglobulin precursor | IPI00478003 | 163.3 | 6 | A2M |
| 1616a | apolipoprotein H precursor | IPI00298828 | 38.3 | 8.34 | APOH |
| 1616b | fibrinogen beta chain | IPI00298497 | 55.9 | 8.54 | FGB |
| 1626 | Alpha-2-HS-glycoprotein | IPI00022431 | 39.3 | 5.43 | AHSG |
| 1648 | fibrinogen beta chain | IPI00298497 | 55.9 | 8.54 | FGB |
| 1650a | apolipoprotein D | IPI00006662 | 28 | 5.14 | APOD |
| 1650b | Alpha-2-HS-glycoprotein | IPI00022431 | 39.3 | 5.43 | AHSG |
| 1650c | alpha-1-antichymotrypsin precursor | IPI00550991 | 45.5 | 5.32 | AACT |
| 1650d | fibrinogen gamma | IPI00219713 | 49.5 | 5.7 | FGG |
| 1652 | Alpha-2-HS-glycoprotein | IPI00022431 | 39.3 | 5.43 | AHSG |
| 1725 | vitamin D-binding protein precursor | IPI00742696 | 52.9 | 5.32 | GC |
| 1731 | vitamin D-binding protein precursor | IPI00742696 | 52.9 | 5.32 | GC |
| 1740 | vitamin D-binding protein precursor | IPI00742696 | 52.9 | 5.32 | GC |
| 1741 | vitamin D-binding protein precursor | IPI00742696 | 52.9 | 5.32 | GC |
| 1744 | vitamin D-binding protein precursor | IPI00742696 | 52.9 | 5.32 | GC |
| 1749 | vitamin D-binding protein precursor | IPI00742696 | 52.9 | 5.32 | GC |
| 1752 | vitamin D-binding protein precursor | IPI00742696 | 52.9 | 5.32 | GC |
| 1843a | pigment epithelial-differentiating factor | IPI00006114 | 46.3 | 5.84 | PEDF |
| 1843b | fibrinogen gamma | IPI00219713 | 49.5 | 5.7 | FGG |
| 1898 | vitamin D-binding protein precursor | IPI00742696 | 52.9 | 5.32 | GC |
| 1911a | serum paraoxonase | IPI00218732 | 37.8 | 4.96 | PON |
| 1911b | fibrinogen gamma | IPI00219713 | 49.5 | 5.7 | FGG |
| 1911c | Complement C3 precursor | IPI00783987 | 187.1 | 6.02 | C3 |
| 1918 | serum paraoxonase | IPI00218732 | 37.8 | 4.96 | PON |
| 1925a | serum paraoxonase | IPI00218732 | 37.8 | 4.96 | PON |
| 1925b | fibrinogen gamma | IPI00219713 | 49.5 | 5.7 | FGG |
| 1985a | serum paraoxonase | IPI00218732 | 37.8 | 4.96 | PON |
| 1985b | haptoglobin | IPI00641737 | 45.2 | 6.13 | HP |
| 1986a | apolipoprotein A-IV precursor | IPI00304273 | 43.4 | 5.22 | APOA4 |
| 1986b | haptoglobin | IPI00641737 | 45.2 | 6.13 | HP |
| 1986c | serum paraoxonase | IPI00218732 | 37.8 | 4.96 | PON |
| 1998 | apolipoprotein A-IV precursor | IPI00304273 | 43.4 | 5.22 | APOA4 |
| 2008 | apolipoprotein A-IV precursor | IPI00304273 | 43.4 | 5.22 | APOA4 |
| 2029 | alpha-2-glycoprotein 1 | IPI00166729 | 34.3 | 5.71 | AZGP1 |
| 2030a | apolipoprotein A-IV precursor | IPI00304273 | 43.4 | 5.22 | APOA4 |
| 2030b | haptoglobin | IPI00641737 | 45.2 | 6.13 | HP |
| 2065a | haptoglobin | IPI00641737 | 45.2 | 6.13 | HP |
| 2065b | Complement factor I | IPI00291867 | 65.8 | 7.72 | CFI |
| 2095a | alpha-1-antichymotrypsin precursor | IPI00550991 | 45.5 | 5.32 | AACT |
| 2095b | clusterin | IPI00400826 | 57.8 | 6.25 | CLU |
| 2130a | Proapolipoprotein | IPI00021841 | 29 | 5.45 | APOA1 |
| 2130b | Complement factor I | IPI00291867 | 65.8 | 7.72 | CFI |
| 2137 | clusterin | IPI00400826 | 57.8 | 6.25 | CLU |
| 2184 | complement component C4A | IPI00032258 | 192.8 | 6.66 | C4A |
| 2189 | complement component C4A | IPI00032258 | 192.8 | 6.66 | C4A |
| 2191 | transthyretin | IPI00022432 | 15.9 | 5.5 | TTR |
| 2236 | immunoglobulin kappa light chain | IPI00784070 | 26 | 8.16 | IGKC |
| 2259 | amyloid P component | IPI00022391 | 25.4 | 6.1 | APCS |
| 2260 | amyloid P component | IPI00022391 | 25.4 | 6.1 | APCS |
| 2272a | lambda-chain precursor | IPI00154742 | 24.7 | 7.54 | IGL |
| 2272b | immunoglobulin kappa light chain | IPI00784070 | 26 | 8.16 | IGKC |
| 2284 | immunoglobulin kappa light chain | IPI00784070 | 26 | 8.16 | IGKC |
| 2314 | Proapolipoprotein | IPI00021841 | 29 | 5.45 | APOA1 |
| 2325 | Proapolipoprotein | IPI00021841 | 29 | 5.45 | APOA1 |
| 2326 | Proapolipoprotein | IPI00021841 | 29 | 5.45 | APOA1 |
| 2338 | Proapolipoprotein | IPI00021841 | 29 | 5.45 | APOA1 |
| 2346 | plasma glutathione peroxidase | IPI00026199 | 16.7 | 8.93 | GPx-P |
| 2415 | haptoglobin | IPI00641737 | 45.2 | 6.13 | HP |
| 2468 | transthyretin | IPI00022432 | 15.9 | 5.5 | TTR |
| 2520 | haptoglobin | IPI00641737 | 45.2 | 6.13 | HP |
aTheoretical molecular weight (mw) in kDa and isoelectric point (pI) values.
Figure 1.
The spatial distribution of proteins spots (yellow dots) detected in human plasma by 2-D DIGE. Identified proteins (blue dots) and those showing differences between the theoretical and observed molecular weights (numbered red dots) are highlighted.
3.4. Spotwise Variation
The distribution of the SLA was consistent across the gels. The spot-wise SD values of the SLA for the 397 high-quality spots, when considering all samples, ranged from 0.04 to 0.53, with a median of 0.10. When broken down separately by subject, the range was 0.0002 to 0.50, with 0.06 as the median, reflecting the lower variation of time-within-subjects than variation between the different subjects. Both sets of values represented an increase from the spot-wise SDs observed among technical replicates of the same human plasma sample [13], where the maximum was 0.20 and the median 0.04. In the previous TVS work [13], the CV values of the SA had a median of 10% and a maximum of 42%. Here, the range of the spot-wise CVs was 10% to 93%, with a median CV of 23%. The higher CVs of the present study reflect the additional complexity due to the heterogeneity of the samples from multiple human subjects. These results are comparable to the recently reported 6% (min), 108% (max), and 19% (median) CVs found in a 2-D DIGE study of normal liver samples from ten human subjects [40]. Here, about 90% of the spots had less than 40% CV, and only 21 spots (5% of the 397 high-quality spots) had higher than 50% CV. The spots are likely not good biomarker candidates due to their high individual or longitudinal variability. These spots showing relatively high variation may correspond to a single isoform of individual proteins and do not represent all isoforms of any given protein. Notably, several of the proteins identified (Albumin, Transferrin, Haptoglobin, IgG, and IgA) are proteins removed by the Top-6 depletion process [12], which was subsequently found to result in variability when processing multiple samples in series. In future studies, column equilibration steps are recommended between samples to reduce this variability and ensure more complete depletion of high-abundant proteins.
In summary, the majority of the spots had small enough CV to indicate that the corresponding protein expressions were relatively constant across individuals, and thus could be potentially used as biomarkers. The minimum, maximum, and median CVs, when calculated separately for the subjects were 0.05%, 131%, and 14%, respectively. Over 95% of the CVs were below 35%, indicating that for most subjects, the variation over the three timepoints was comparable to the experimental variation in the previously published TVS data [13].
3.5. Statistical Normalization, Gel and Dye Effect Removal, and Variance Decomposition
The SLA values were further normalized as explained in the methods. The effect of the normalization on the results is addressed as appropriate in the following sections. The F-tests for the analysis of variance calculations corresponding to (1) indicated significant gel (P-value < 2.2e−16), dye (P-value < 3.2e−16), and gel-dye interaction (P-value < 2.2e−16) effects. The residual diagnostic plots did not reveal major departures from the assumptions, thus indicating the validity of the model. Similar analyses using the normalized SLA resulted in slightly higher P-values (gel effect P-value < 2.2e−16, dye effect P-value.003, gel-dye interaction P-value < 3.0e−09) but were consistent with the conclusions based on the calculations using the SLA.
The standard deviations corresponding to the random variance component estimates from the mixed-effects model (Figure 2) show the relative contribution of the three components at each of the spots matched on 75% of the gels. Overall, the time-within-subject component was found to have the smallest contribution to the total variance, while subject-related variation had the highest. The corresponding frequency distributions of the three variance components (Table 1) confirm that for most spots (89%) the contribution of the time component was less than 30% of the total variance. Only 5% of the spots had 70% to 80% of their variation explained by the time component, and no spot had the time component greater than 80% of the total variance. For 21% of the spots, the contribution of the subject component comprised over 70% of the total variance. For about 44% of the spots, the contribution of the subject component represented over 50% of the total variation.
Figure 2.
The subject (σs), time-within-subject (σt), and random error (σe) variance component estimates (on SD scale) for the 397 protein spots matched on at least 75% of the gels, ordered by the magnitude of the subject component.
Table 1.
Frequency distribution of the variance component estimates.
| % contribution to total variance | Subject | Time in subject | Error | |||
|---|---|---|---|---|---|---|
| (a) | (b) | (a) | (b) | (a) | (b) | |
| 0–10 | 6.31 | 6.31 | 63.89 | 63.89 | 6.57 | 6.57 |
| 10–20 | 9.34 | 15.66 | 17.68 | 81.57 | 17.17 | 23.74 |
| 20–30 | 12.89 | 28.50 | 7.83 | 89.39 | 16.67 | 40.40 |
| 30–40 | 14.14 | 42.68 | 4.29 | 93.69 | 9.34 | 49.75 |
| 40–50 | 13.63 | 56.31 | 2.27 | 95.96 | 10.86 | 60.61 |
| 50–60 | 11.36 | 67.68 | 3.03 | 98.99 | 11.87 | 72.47 |
| 60–70 | 11.36 | 79.04 | 0.51 | 99.49 | 10.37 | 82.83 |
| 70–80 | 11.61 | 90.66 | 0.50 | 100.00 | 8.08 | 90.91 |
| 80–90 | 7.83 | 98.48 | 4.80 | 95.71 | ||
| 90–100 | 1.51 | 100.00 | 4.29 | 100.00 | ||
The components of subject (σs), time-within-subject (σt), and random error (σe) are shown separately as (a) the percentage of spots and (b) the cumulative percentage of spots with contribution to the total variance indicated in the first column.
To further elucidate the contributing factors involved in spot variance, we performed a meta-analysis on these data. Essentially, all the variances for the 397 high-quality spots were summed and the total variance that could be explained by the sum of the spot-wise subject, time within-subject, and error components was determined, respectively. A pooled estimate of the variance components was obtained by taking the average of the corresponding variance components over the spots. When aggregating the total variance over all the spots matched on at least 75% of the gels, the sum of the subject components explained 59% of the total variation and the sum of the time-within-subject components explained 12% of the total variation. The average subject variance component across the spots was 0.0097 (corresponding to σs = 0.098 on the Sd scale), and the average time-within-subject variance component was 0.0019 (σt = 0.044).
3.6. Multivariate Analysis of Expression Patterns
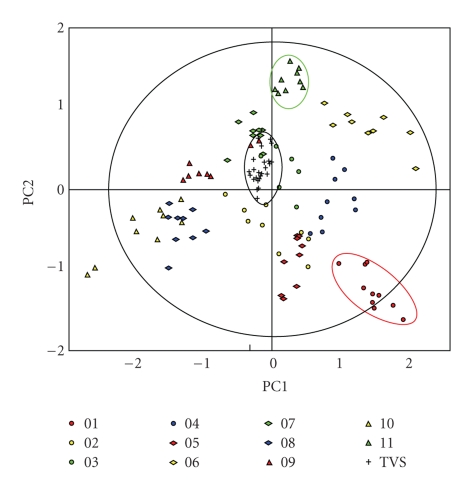
The EDA module of DeCyder 2D was used to visually display the results of the current study and the previous TVS study [13] (Figure 3). The multivariate expression profiles of the samples across 328 spots that were matched on >75% of the spot maps from both studies were transformed into the principal component basis and the projection of the samples onto the first two principal components displayed (Figure 3). The tight scatter of the samples from the TVS (encircled in black) indicates the small magnitude of the experimental variability when analyzing technical replicates of the same human sample. The magnitude of the longitudinal variation exceeded the technical variation, as evidenced by the larger scatter of the sample points of a given subject at the different time points. For example, the two red and green ellipses (Figure 3) highlight the longitudinal variation for subjects 1 and 11, respectively. The differential scattering of the samples, from the subjects into varying regions of the principal components plot, indicates that the subject-to-subject variation exceeded the longitudinal variation within subjects.
Figure 3.
Principal component analysis of the 33 samples from the present study and the 8 replicates from the previous Technical Variation Study (TVS) [9], color-coded according to the legend, projected onto the first two principal components. Ellipses highlighting subjects 1 (red), 11 (green), and the TVS (black) are added for illustrative purposes only.
Hierarchical clustering was used to group the 33 samples based on the similarity of their protein expression profiles along the 397 high-quality spots that were matched on >75% of the gels (Figure 4). Clustering was performed in on the proteins and experimental samples, using Euclidean distance and average linkage to define similarity. For all subjects, the first clustering step placed the three samples of the given subject into one cluster. Samples of the same subject collected at the three time points were most similar to each other, as evidenced by the succession of self-similar bands of three rows (highlighted by the yellow lines in Figure 4). The clustering also shows a general trend of clustering the samples based on gender appeared (highlighted by the blue and red bars for males and females, resp., in Figure 4).
Figure 4.
Hierarchical clustering of the 33 samples (y-axis) based on the abundance of the 397 high-quality protein spots on the x-axis, using Euclidean distance and average linkage. The samples are in SubjectNumberTime format, where SubjectNumber ranges from 01 to 11, and the Time values {x, y, and z } correspond to {T1, T2, and T3}. The intensities range from −1.5-fold change (bright green) to 1.5-fold change (bright red). The dendrogram on the right indicates the order of the sample grouping, with more similar samples being grouped together first. The color band on the left shows the genders of the samples, with red for females, and blue for males.
3.7. Gender Effects
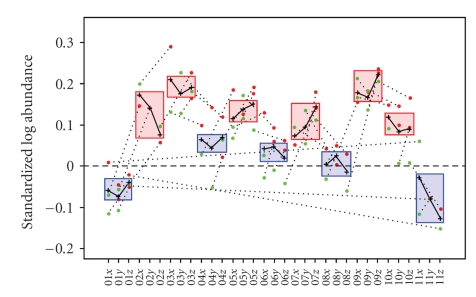
In addition to the results seen in the hierarchical clustering (Figure 4), after fitting the mixed-effects model to the residuals from the SLA at the spots matched on 75% of the gels, 17 spots showed gender-effect P-value <.01. None of these spots were found to be significant (P-value <.01) after the False Discovery Rate (FDR) method [41] for multiple comparisons was applied suggesting that larger numbers of samples are needed to validate gender differences in the human plasma proteome. Despite the lack of statistically significant data on gender differences, trends in this dataset suggest that future, larger datasets might enable the differentiation of protein expression levels due to gender. One spot, 1659, had FDR-adjusted gender-effect P-value equal to.055 with a 1.49-fold-change between the male and female groups (Figure 5). Five additional spots (466, 1626 alpha-2-HS-glycoprotein, 1650 alpha-2-HS-glycoprotein, 1652 alpha-2-HS-glycoprotein, 1678) had adjusted P-values of.11. The results were similar when fitting the same model to the residuals from the statistically normalized SLA, albeit with P-values that slightly exceeded their corresponding values based on the SLA. Three of the spots exhibiting gender effects were identified as alpha-2-HS-glycoprotein, which has been shown to vary between males and females. The concentration of alpha-2-HS-glycoprotein has been found to undergo a progressive age-related decrease in women, while men show no noticeable change [36].
Figure 5.
Expression data for alpha-2-HS-glycoprotein, with an average increase of 1.49-fold between the female and male groups, and FDR-adjusted gender-effect P-value =.055. The samples are in SubjectNumberTime format, where SubjectNumber ranges from 01 to 11, and the Time values {x, y, and z } correspond to {T1, T2, and T3}. The annotations indicate the gels (numbers) and the dyes (red for Cy5, green for Cy3) corresponding to the samples. Dotted lines connect samples multiplexed on the same gel. Crosses indicate sample averages over the technical replicates. The solid line connects all sample averages. Boxes around the three Time values for each SubjectNumber highlight male and female genders (blue and red respectively) added for illustrative purposes only.
The removal of the dye and gel effects using the model in (1) proved to be a beneficial preprocessing step. Without this step, when the mixed-effect model was fit to the original SLA, the smallest FDR-adjusted gender-effect P-value was.19 (spot 1659). When the same model was fit to the statistically normalized SLA, the smallest FDR-adjusted P-value was also.19 (spot 1659). The statistical normalization improved the quality of the data slightly, but it did not reduce dramatically the observed P-values. On the other hand, pooling the information across the gels to remove the common dye and gel effects strengthened the signal and reduced markedly the FDR-adjusted P-values. As explained in the previous paragraph, for spot 1659, the new P-value was close to.05.
3.8. Multivariate Analysis of Identified Proteins
Hierarchical clustering of the identified proteins (Figure 6) was conducted using the Euclidean distance metric and average linkage methods. For all subjects, other than subject 10, the first clustering step placed the three samples of that subject into one cluster. Subject 10 had two time points grouped together (x and z) with the third point (y) separated by Subject 3. Because the 78 identified proteins were the most differential between time and subjects, it is not unexpected to see clustering results that may not perfectly align all subjects or time points. For example, all protein spots identified as transferrin (TF) clustered together due to their similar expression patterns, while the vitamin D-binding protein (GC) spots were found in multiple clusters due to differences in expression patterns between the individuals. Multiple proteins may cluster together due to coregulation and similar functions, and in the case of APOA4 and APOA1 (Figure 6), coregulation has been reported [37, 42].
Figure 6.
Hierarchical clustering of the 33 samples (x-axis) based on the abundance of the 78-identified protein spots on the y-axis, using Euclidean distance metrics and average linkage methods. The samples are in SubjectNumberTime format, where SubjectNumber ranges from 01 to 11, and the Time values (x, y, and z) correspond to (T1, T2, and T3). The intensities range from −1.5 (bright green) to 1.5 (bright red). The dendrogram on the top indicates the order of the sample grouping, with samples corresponding to the lower leaves being grouped together first. Similarly, the dendrogram on the left indicates the ordering of the protein spots. All transferrin (TF) and vitamin D-binding protein (GC) identifications are highlighted in blue and red, respectively.
4. Conclusion
Statistical analysis of a 2-D DIGE experiment involving triplicate plasma samples from eleven human subjects taken at three time points separated by several weeks demonstrated that the subject-to-subject variation exceeded the time-within-subject variation. The variation in the human plasma proteome reported here was greater than a previous technical variation study wherein one plasma sample was processed multiple times [13].
Here, for 70% of the high-quality protein spots, the coefficient of variation of the SLA was less than 30% across all subjects and time points, thus indicating that the baseline expression levels of those proteins are relatively stable in the population represented by the subjects in this study. Only 21 spots had larger than 50% CV, suggesting that these protein isoforms should be avoided as biomarker candidates. Many of these protein spots represent medium to high abundance plasma proteins. Since they are higher abundance, they might bias LC/MS datasets, but since the total number of these spots relative to the total plasma proteome is small, their total influence on a sample is likely also small. In addition, protein spots with gender-related differences should be considered separately for males and females. However, more thorough studies, including the use of a larger population set with additional time points over longer periods of time, are recommended to more fully address individual, longitudinal, and gender variability as related to biomarker discovery.
We noted that preprocessing the data by first removing the fixed effects of the gels and dyes was important in data analysis and improved the quality of the data. This step resulted in six protein spots showing a statistically significant gender effect at an FDR-adjusted 11% significance level. Without the preprocessing step, the smallest gender effect P-value was.19. While removing the gel and dye effects lead to stronger conclusions, the additional statistical normalization of the SLA had only marginal effects and did not alter the conclusions.
Spot matching confounds gel- and software-related protein differences with real biological effects. In the present study, we only considered spots that were matched on at least 75% of the gels. Spots with lower matching quality can be investigated separately, as they may correspond to proteins that are absent or have very low expression in certain individuals, but which may have biological significance. We envision that such studies will become more relevant as the field of personalized medicine matures, and as detection and matching algorithms continue to improve.
This study represents a first step toward quantitating the longitudinal and individual variation in the human plasma proteome, as measured on the 2-D DIGE platform. Interestingly, gender-related variations were also detected suggesting that gender variability should also be considered in biomarker discovery. Future, larger-scale experiments that include more subjects representative of various population segments, encompassing differences in ethnicity, age, gender, disease status, and other relevant factors, have the potential to define baseline proteomic similarities and differences in the human population, which will in turn facilitate improved biomarker discovery.
Supplementary Material
The supplementary material contains one table and one figure. Both of these are used to better illustrate the experimental design used in this project. In Supplementary Table 1, each individual sample is listed as it appears on one of the 33 gels run for this experiment. Each individual was randomly assigned a number (1, 2, 3,…, 11) and each time point was assigned a letter (x, y, or z). Each gel contained three samples, one internal pooled standard and two experimental samples. Each experimental sample was dye-swapped and labeled with both the Cy3 and the Cy5 dyes, while the pooled standard consisting of an equal amount of each of the 33 samples was labeled with Cy2 and included on every gel. Supplementary Figure 1 shows a graphical representation of how the experimental design was conducted and how any number of samples could be incorporated into this experimental design to obtain similar statistically valid results. Rather than randomly pairing the samples on the gels, the design was selected to minimize the experimental variation among the samples whose comparison was of most interest. Samples represented as circles are joined by arrows to represent the two samples that are directly compared on a given gel.
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Security, LLC under Contract DE-AC52-07NA27344, with support from the Department of Homeland Security and LLNL Laboratory Directed Research and Development funding UCRL-JRNL-229654. Todd H. Corzett and Imola K. Fodor contributed equally to this work.
References
- 1.Nedelkov D. Population proteomics: addressing protein diversity in humans. Expert Review of Proteomics. 2005;2(3):315–324. doi: 10.1586/14789450.2.3.315. [DOI] [PubMed] [Google Scholar]
- 2.Hermjakob H. The HUPO proteomics standards initiative—overcoming the fragmentation of proteomics data. Proteomics. 2006;1(S2):34–38. doi: 10.1002/pmic.200600537. [DOI] [PubMed] [Google Scholar]
- 3.Taylor CF. Minimum reporting requirements for proteomics: a MIAPE primer. Proteomics. 2006;1(S2):39–44. doi: 10.1002/pmic.200600549. [DOI] [PubMed] [Google Scholar]
- 4.Omenn GS. Exploring the human plasma proteome: editorial. Proteomics. 2005;5(13):3223–3225. doi: 10.1002/pmic.200590056. [DOI] [PubMed] [Google Scholar]
- 5.Omenn GS, Paik Y-K, Speicher D. The HUPO plasma proteome project: a report from the Munich congress. Proteomics. 2006;6(1):9–11. doi: 10.1002/pmic.200500802. [DOI] [PubMed] [Google Scholar]
- 6.Omenn GS, States DJ, Adamski M, et al. Overview of the HUPO plasma proteome project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5(13):3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 7.Cottingham K. HUPO plasma proteome project: challenges and future directions. Journal of Proteome Research. 2006;5(6):p. 1298. doi: 10.1021/pr062733+. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs JM, Adkins JN, Qian W-J, et al. Utilizing human blood plasma for proteomic biomarker discovery. Journal of Proteome Research. 2005;4(4):1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 9.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 10.Han MJ, Speicher DW. Microscale isoelectric focusing in solution: a method for comprehensive and quantitative proteome analysis using 1-D and 2-D DIGE combined with MicroSol IEF prefractionation. Methods in Molecular Biology. 2008;424:241–256. doi: 10.1007/978-1-60327-064-9_20. [DOI] [PubMed] [Google Scholar]
- 11.Cho SY, Lee E-Y, Kim H-Y, et al. Protein profiling of human plasma samples by two-dimensional electrophoresis. Methods in Molecular Biology. 2007;428:57–75. doi: 10.1007/978-1-59745-117-8_4. [DOI] [PubMed] [Google Scholar]
- 12.Chromy BA, Gonzales AD, Perkins J, et al. Proteomic analysis of human serum by two-dimensional differential gel electrophoresis after depletion of high-abundant proteins. Journal of Proteome Research. 2004;3(6):1120–1127. doi: 10.1021/pr049921p. [DOI] [PubMed] [Google Scholar]
- 13.Corzett TH, Fodor IK, Choi MW, et al. Statistical analysis of the experimental variation in the proteomic characterization of human plasma by two-dimensional difference gel electrophoresis. Journal of Proteome Research. 2006;5(10):2611–2619. doi: 10.1021/pr060100p. [DOI] [PubMed] [Google Scholar]
- 14.Alban A, David SO, Bjorkesten L, et al. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3(1):36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 15.Lilley KS, Friedman DB. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Review of Proteomics. 2004;1(4):401–409. doi: 10.1586/14789450.1.4.401. [DOI] [PubMed] [Google Scholar]
- 16.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Analytical and Bioanalytical Chemistry. 2005;382(3):669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 17.Echan LA, Tang H-Y, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5(13):3292–3303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 18.Mahnke RC, Corzett TH, McCutchen-Maloney SL, Chromy BA. An integrated proteomic workflow for two-dimensional differential gel electrophoresis and robotic spot picking. Journal of Proteome Research. 2006;5(9):2093–2097. doi: 10.1021/pr050465u. [DOI] [PubMed] [Google Scholar]
- 19.Venables WN, Smith DM. An Introduction to R: Notes on R, A Programming Environment for Data Analysis and Graphics, v.2.0.1. Bristol, UK: Network Theory; 2004. [Google Scholar]
- 20.Chromy BA, Perkins J, Heidbrink JL, et al. Proteomic characterization of host response to Yersinia pestis and near neighbors. Biochemical and Biophysical Research Communications. 2004;320(2):474–479. doi: 10.1016/j.bbrc.2004.05.199. [DOI] [PubMed] [Google Scholar]
- 21.Bolshakova N, Azuaje F. Cluster validation techniques for genome expression data. Signal Processing. 2003;83(4):825–833. [Google Scholar]
- 22.Dunn JC. Well separated clusters and optimal fuzzy partitions. The Journal on Systemics, Cybernetics and Informatics. 1974;4:95–104. [Google Scholar]
- 23.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. New York, NY, USA: John Wiley & Sons; 1990. [Google Scholar]
- 25.Sokal RR, Michener CD. A statistical method for evaluating systematic relationships. University of Kansas Science Bulletin. 1958;38(6):1409–1438. [Google Scholar]
- 26.Tibshirani R, Walther G, Hastie T. Estimating the Number of Clusters in a Dataset via the Gap Statistic. Stanford, Calif, USA: Department of Biostatistics, Stanford University; 2000. [Google Scholar]
- 27.Eriksson L, Kettaneh-Wold N, Trygg J, Wold S. Multi- and Megavariate Data Analysis. Vol. 533. Umeå, Sweden: Umetrics Academy; 2001. [Google Scholar]
- 28.Jolliffe IT. Principal Component Analysis. Berlin, Germany: Springer; 2002. [Google Scholar]
- 29.Umetrics SIMCA,10.5. Sweden.
- 30.Wold H. Estimation of principal components and related models by iterative least squares. In: Krishnaiah P, editor. Multivariate Analysis. New York, NY, USA: Academic Press; 1966. pp. 391–420. [Google Scholar]
- 31.Pinheiro JC, Bates DM. Mixed-Effects Models in S and S-PLUS. New York, NY, USA: Springer; 2000. (Statistics and Computing). [Google Scholar]
- 32.Kuehl RO. Design of Experiments: Statistical Principles of Research Design and Analysis. 2nd edition. Belmont, Calif, USA: Duxbury Press; 2000. [Google Scholar]
- 33.Cui X, Churchill GA. Statistical tests for differential expression in cDNA microarray experiments. Genome Biology. 2003;4(4, article 210) doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fodor IK, Nelson DO, Alegria-Hartman M, et al. Statistical challenges in the analysis of two-dimensional difference gel electrophoresis experiments using DeCyderTM. Bioinformatics. 2005;21(19):3733–3740. doi: 10.1093/bioinformatics/bti612. [DOI] [PubMed] [Google Scholar]
- 35.Pedreschi R, Hertog MLATM, Carpentier SC, et al. Treatment of missing values for multivariate statistical analysis of gel-based proteomics data. Proteomics. 2008;8(7):1371–1383. doi: 10.1002/pmic.200700975. [DOI] [PubMed] [Google Scholar]
- 36.Dickson IR, Bagga M, Paterson CR. Variations in the serum concentration and urine excretion of α2HS-glycoprotein, a bone-related protein, in normal individuals and in patients with osteogenesis imperfecta. Calcified Tissue International. 1983;35(1):16–20. doi: 10.1007/BF02405000. [DOI] [PubMed] [Google Scholar]
- 37.Schamaun O, Olaisen B, Mevag B, et al. The two apolipoprotein loci apoA-I and apoA-IV are closely linked in man. Human Genetics. 1984;68(2):181–184. doi: 10.1007/BF00279311. [DOI] [PubMed] [Google Scholar]
- 38.Gigli I, von Zabern I, Porter RR. The isolation and structure of C4, the fourth component of human complement. Biochemical Journal. 1977;165(3):439–446. doi: 10.1042/bj1650439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Press EM, Gagnon J. Human complement component C4. Structural studies on the fragments derived from C4b by cleavage with C3b inactivator. Biochemical Journal. 1981;199(2):351–357. doi: 10.1042/bj1990351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Guo Y, Song Y, et al. Proteomic analysis of individual variation in normal livers of human beings using difference gel electrophoresis. Proteomics. 2006;6(19):5260–5268. doi: 10.1002/pmic.200600006. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57(1):289–300. [Google Scholar]
- 42.Vergnes L, Taniguchi T, Omori K, Zakin MM, Ochoa A. The apolipoprotein A-I/C-III/A-IV gene cluster: ApoC-III and ApoA-IV expression is regulated by two common enhancers. Biochimica et Biophysica Acta. 1997;1348(3):299–310. doi: 10.1016/s0005-2760(97)00071-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary material contains one table and one figure. Both of these are used to better illustrate the experimental design used in this project. In Supplementary Table 1, each individual sample is listed as it appears on one of the 33 gels run for this experiment. Each individual was randomly assigned a number (1, 2, 3,…, 11) and each time point was assigned a letter (x, y, or z). Each gel contained three samples, one internal pooled standard and two experimental samples. Each experimental sample was dye-swapped and labeled with both the Cy3 and the Cy5 dyes, while the pooled standard consisting of an equal amount of each of the 33 samples was labeled with Cy2 and included on every gel. Supplementary Figure 1 shows a graphical representation of how the experimental design was conducted and how any number of samples could be incorporated into this experimental design to obtain similar statistically valid results. Rather than randomly pairing the samples on the gels, the design was selected to minimize the experimental variation among the samples whose comparison was of most interest. Samples represented as circles are joined by arrows to represent the two samples that are directly compared on a given gel.