Abstract
In an effort to define the biological functions of COMP, a functional genetic screen was performed. This led to the identification of extracellular matrix protein 1 (ECM1) as a novel COMP-associated partner. COMP directly binds to ECM1 both in vitro and in vivo. The EGF domain of COMP and the C-terminus of ECM1 mediate the interaction between them. COMP and ECM1 Colocalize in the Growth Plates in Vivo. ECM1 inhibits chondrocyte hypertrophy, matrix mineralization, and endochondral bone formation, and COMP overcomes the inhibition by ECM1. In addition, COMP-mediated neutralization of ECM1 inhibition depends on their interaction, since COMP largely fails to overcome the ECM1 inhibition in the presence of the EGF domain of COMP, which disturbs the association of COMP and ECM1. These findings provide the first evidence linking the association of COMP and ECM1 and the biological significance underlying the interaction between them in regulating endochondral bone growth.
Introduction
Cartilage oligomeric matrix protein (COMP), a prominent noncollagenous component of cartilage extra-cellular matrix, is a 524-kDa pentameric, disulfide-bonded, multidomain glycoprotein composed of approximately equal subunits (~110 kDa each) (Morgelin et al., 1992; Oldberg et al., 1992). It accounts for ~1% of the wet weight of tissue and has also been localized in tendon, bone (osteoblasts only), and synovium (Bosco et al., 2007; Di Cesare et al., 2000; Hedbom et al., 1992). Although COMP has been implicated in the regulation of chondrogenesis in a micromass culture of mesenchymal stem 10T1/2 cells and in limb development in vivo (Kipnes et al., 2003; Koelling et al., 2006; Xu et al., 2007), its function remains largely unknown. COMP has been shown to be upregulated after traumatic knee injury (Kuhne and Banks, 1998) and has been implicated in the pathogenesis of rheumatoid arthritis and osteoarthritis (Di Cesare et al., 1996; Neidhart et al., 1997; Saxne and Heinegard, 1992). Monitoring COMP levels in either joint fluid or serum can be used to assess the presence and progression of arthritis (Kraus et al., 2002; Lohmander et al., 1999; Mansson et al., 1995; Misumi et al., 2002; Neidhart, 1996; Petersson et al., 1998). Mutations in the human COMP gene have been linked to the development of pseudoachondroplasia and multiple epiphyseal dysplasia, autosomal-dominant forms of short limb dwarfism characterized by short stature, normal facies, epiphyseal abnormalities, and early onset osteoarthritis (Briggs et al., 1995; Briggs et al., 1998; Briggs et al., 1993; Hecht et al., 1993; Hecht et al., 1995; Susic et al., 1997).
COMP is expressed in the pericellular matrix of maturing articular chondrocytes (Susic et al., 1997) as well as growth plate (Xu et al., 2007) in vivo, which suggests that COMP may play an important role in chondrogenesis and endochondral bone growth. In addition, COMP associates with granulin-epithelin precursor (GEP), an autocrine growth factor, and potentiates GEP-stimulated chondrocyte proliferation (Xu et al., 2007). One of the aims of the present study was to isolate the novel proteins that associate with COMP in order to elucidate its biological functions in skeletogenesis. A yeast two-hybrid screen using the EGF domain of COMP as bait led to the isolation of extracellular matrix protein 1 (ECM1) as a novel COMP-binding protein.
ECM1 was first identified as a glycosylated 85-kDa protein secreted by a mouse osteogenic stromal cell line MN7 (Mathieu et al., 1994). Intriguingly, loss-of-function mutations in the ECM1 gene were discovered to be the cause of the rare autosomal recessive genodermatosis, lipoid proteinosis (Hamada et al., 2002; Hamada et al., 2003). Moreover, other studies have identified circulating autoantibodies against the ECM1 protein in most patients with lichen sclerosus, a common chronic inflammatory condition that shares some clinicopathological features with lipoid proteinosis (Chan, 2004; Chan et al., 2004; Oyama et al., 2003). Within the dermis, ECM1 binds to perlecan, the major heparan sulphate proteoglycan (Mongiat et al., 2003). Recent studies showed that ECM1 binds to other extracellular matrix proteins, including collagen type IV, fibronectin, laminin 332, fibulin-1C/1D and MMP-9 (Sercu et al., 2008a). Due to its promiscuous interaction with different matrix molecules it was hypothesized that ECM1 may act as a “biological glue” helping to regulate the basement membrane and interstitial collagen fibril macroassembly and growth factor binding in the skin. However, overexpression of ECM1a in vivo does not exert dramatic effects on epidermal structure (Sercu et al., 2007). A recent report provides genetic evidence linking ECM1 to ulcerative colitis and Crohn’s disease, debilitating inflammatory bowel diseases (Fisher et al., 2008).
ECM1 plays also a role in endochondral bone formation (Deckers et al., 2001), promotes proliferation of endothelial cells, and induces angiogenesis (Han et al., 2001; Mongiat et al., 2003). Here we provide evidence demonstrating a novel interaction between ECM1 and COMP and elucidate the biological significance of the association between these two critical extracellular matrix proteins in mediating endochondral bone growth.
Materials and Methods
Plasmid Constructs
Yeast expression vectors pDBleu and pPC86 (both from Invitrogen) are fusion vectors for the linkage of proteins to the Gal4 DNA binding domain and to the VP16 transactivation domain, respectively. The fragments encoding the four functional domains (i.e., the N-terminal domain (aa 20–83), EGF repeat domain (aa 84–261), type III repeat domain (aa 266–520), and C-terminal domain (aa 521–755; GenBank™ accession number AF257516) of mouse COMP were amplified by PCR and cloned in frame into the SalI/NotI sites of pDBleu (pDB-COMP-NT, pDB-COMP-epidermal growth factor, pDB-COMP-type III, and pDB-COMP-CT) to serve as bait in the screening assay.
The bacterial expression vector pGEX-3X (Invitrogen) was used to produce recombinant glutathione S-transferase (GST) fusion proteins in Escherichia coli. The cDNA fragments encoding the four functional domains (i.e., the N-terminal (aa 20–83), EGF repeat domain (aa 84–261), type III repeat domain (aa 266–520), and C-terminal domain (aa 521–755) of mouse COMP were inserted in frame into the BamHI/EcoRI sites of pGEX-3X to generate the plasmids pGEX-N-term, pGEX-EGF, pGEX-type III, and pGEX-C-term, respectively. The bacterial expression pBAD TOPO vector (Invitrogen) was used to produce His-tagged proteins in E. coli.
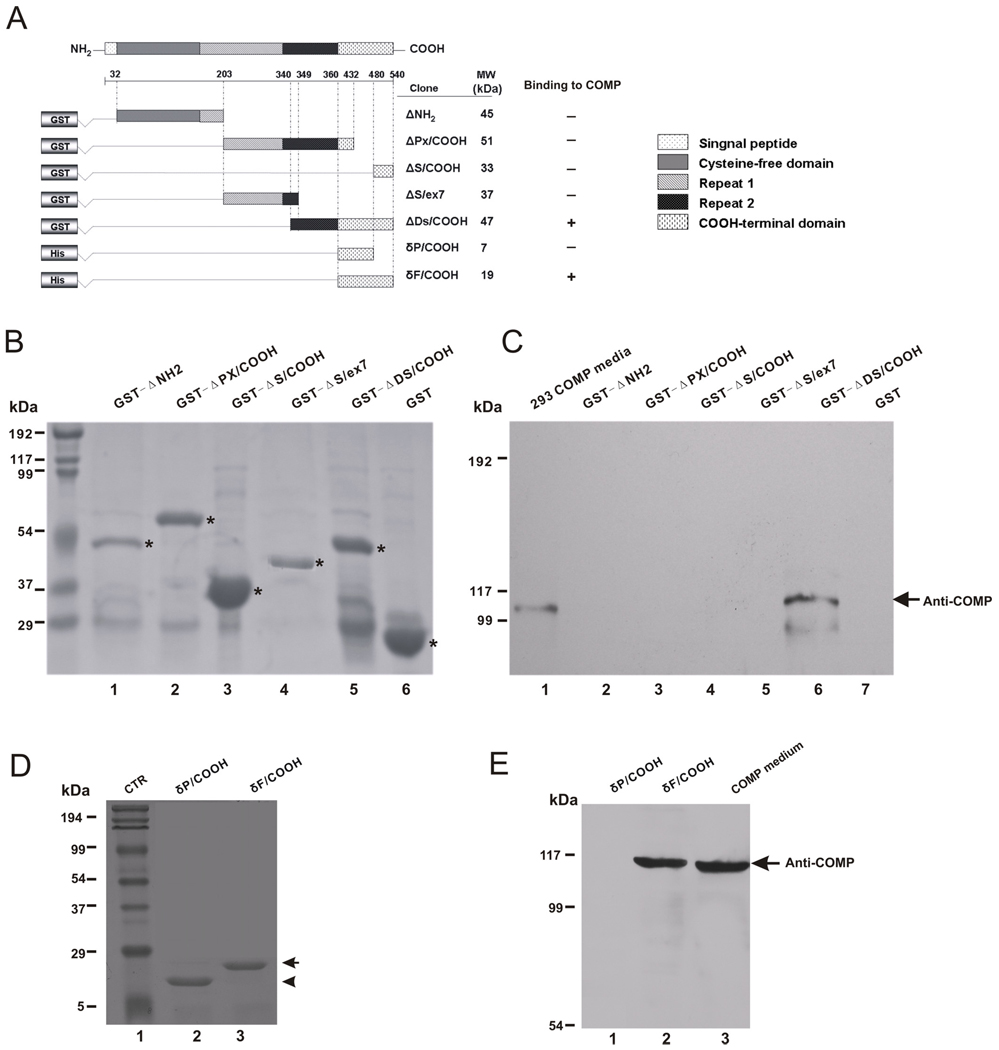
Constructs encoding different fragments of ECM1a fused to GST were described previously (Oyama et al., 2004). Fragment of intact δF/COOH (aa 360–540) and partial C δP/COOH (aa 360–480) terminal of ECM1 (Fig. 4A) were subcloned into the pBAD TOPO vector per the manufacturer’s instructions to generate the indicated plasmids.
Fig 4. C-terminus of ECM1 is required and sufficient for interaction with COMP.
A, Schematic representation of various ECM1a deletion mutants fused to either GST or His tag. The amino acid residue numbers are indicated. The predicted molecular weights (kDa) of each recombinant fusion fragment are shown on the right. B~E, Pull down assay. Purified ECM1 mutants fused to GST (B) and His-tagged ECM1 mutants (D) were separated on SDS-PAGE and visualized by Coomassie blue staining. These purified proteins were conjugated to either Glutathione-sepharose beads (C) or Pro-bond beads (E) and incubated with recombinant COMP. After washing, the bound proteins were immunoblotted using anti-COMP antibody. GST fused ECM1 mutants in panel B with expected sizes are indicated with “star”. Molecular weights are indicated on the left column.
All constructs were verified by nucleic acid sequencing; subsequent analysis was performed using BLAST software (available at ncbi.nlm.nih.gov/blast/).
Expression and Purification of Recombinant Proteins
For expression of GST fusion proteins, the appropriate plasmid pGEX-N-term, pGEX-EGF, pGEX-typeIII, pGEX-C-term, or different fragments of ECM1 subcloned into the pGEX-3X was transformed into E. coli DH5 (Invitrogen). Fusion proteins were affinity-purified on glutathione-agarose beads, as described previously (Liu et al., 1999). To cleave off and remove the GST moiety from the GST-fused catalytic domain, 50 µg of purified GST fusion protein was incubated with 1 µg of Xa factor (New England Biolabs, Beverly, MA) in 20 µl of 20mM Tris-HCl (pH 8.0), 100mM NaCl, and 2mM CaCl2 at 23°C for 8 h. The reaction was terminated by the addition of 2 µM dansyl-Glu-Gly-Arg-chloromethyl ketone (New England Biolabs) and incubated at room temperature for 1 min. The completion of the cleavage was established by SDS-PAGE, and the resultant GST moiety was removed using GSH-Sepharose 4B beads (Amersham, Piscatway, NJ).
His-δF/COOH and His-δP/COOH were purified by affinity chromatography using a HiTrap chelating column (Amersham Biosciences). Briefly, bacteria lysates supplemented with 20mM HEPES (pH 7.5) and 0.5M NaCl were applied to the HiTrap chelating column; the column was washed with HSB buffer (40mM HEPES, pH 7.5, 1M NaCl, and 0.05% Brij 35) containing 10mM imidazole; and the His-δF/COOH and His-δP/COOH were eluted with HSB buffer containing 300mM imidazole.
Recombinant intact COMP and ECM1 were expressed and purified from corresponding stable lines derived from HEK 293 cells (Liu et al., 2006a; Luan et al., 2008; Sercu et al., 2009). Flag-tagged COMP functional domains (i.e., the N-terminal (aa 20–83), EGF repeat domain (aa 84–261), type III repeat domain (aa 266–520), and C-terminal domain (aa 521–755) were expressed and purified in insect cells using The Bac-to-Bac Baculovirus Expression System (Invitrogen). Our previous procedure was followed (Liu et al., 2006a).
Generation and Characterization of Monoclonal Antibodies against Distinct COMP Functional Domains
Recombinant EGF and type III domain of COMP was used to inject mice for generating monoclonal antibodies. Briefly, Balb/c female mice were immunized with two intraperitoneal injections of 30 mg of recombinant EGF-COMP fragment solubilized in Titertec Gold adjuvant (Sigma), and spleens were fused with NS1 myeloma cells after detection of a sufficient level of circulating Igs (3–4 weeks after the initial immunization). Hybridoma clones were initially screened by ELISA and subsequently by immunoblotting. Positive clones were further subcloned by limiting dilution and ascertained to have maintained their reactivity by ELISA on the immunogen.
To examine the specificity of newly made monoclonal antibodies, Western blot assays using purified GST, GST-EGF, GST–type III, recombinant COMP, and COMP condition medium were performed. Protein samples were subjected to 10% SDS-PAGE and electrotransferred to nitrocellulose membranes for 2 h at 100 V using a standard transfer solution. Nonspecific bindings were blocked in TBS-T (25mM Tris, 140mM NaCl, 3mM KCl, 0.05% Tween-20, pH 8.0) containing 10% nonfat milk at 4°C. Membranes were incubated with primary anti-COMP monoclonal antibodies (2114B3-EGF, 2130E2 type III) at 1:1000 dilutions for 2 hr at room temperature, followed by antimouse IgG conjugated HRP at 1:1000 dilution. The proteins were visualized by chemiluminescence using an ECL kit (Amersham Pharmacia Biotech).
Yeast Two-hybrid (Y2H) Library Screen
Plasmid pDB-COMP-epidermal growth factor (see above) was used as bait to screen the Y2H rat brain cDNA library (Invitrogen) according to a modified manufacturer's protocol. Briefly, bait plasmid was introduced into a yeast MAV203 strain that contained three reporter genes, HIS+, URA+, and lacZ (Invitrogen), and transformants were selected on defined medium lacking leucine. The rat brain cDNA library in the vector pPC86 was then transformed into the resultant Leu+ yeast strain and plated on medium lacking tryptophan, leucine, histidine, and uracil but containing 25mM 3-amino-1,2,4-trizone that can specifically inhibit the activity of HIS3 gene product and block the basal concentration of HIS3 in yeast (SD-leu−/trp−/his−/ura−/3AT+). After incubation for 7–10 days at 30°C, colonies were screened for β-galactosidase by a filter lift assay (Hecht et al., 1995). Individual pPC86 recombinant plasmids that were identified in the initial screen were further verified for interaction with bait by repeating the Y2H assay.
Assay of Protein-Protein Interactions Using the Y2H System
Three independent colonies were analyzed for interaction in yeast of two proteins, one of which was fused to the Gal4 DNA binding domain and the other to the VP16 transactivation domain. The procedures of Vojtek et al. (Vojtek et al., 1993) and Hollenberg et al. (Hollenberg et al., 1995) were followed for (1) growing and transforming the yeast strain MAV203 with the selected plasmids; and (2) β-galactosidase activity and growth phenotypes on selective SD-leu−/trp−/his−/ura−/3AT+ plates.
In Vitro Binding Assay
For examination of the binding of COMP to ECM1 in vitro, glutathione-Sepharose beads (50 µl) preincubated with either purified GST (0.5 µg, serving as control) or the GST-fused EGF-like domain of COMP were incubated with a condition medium prepared from HEK293 cells stably transfected with an expression plasmid encoding full-length mouse ECM1. The beads were washed 5 times with washing buffer (0.3% Triton X-100 in phosphate-buffered saline). Bound proteins were resolved by 12% SDS-PAGE and detected by Western blotting with polyclonal rabbit anti-ECM1 antiserum (Rb469). To dissect the fragments of ECM1 required for interaction with COMP, glutathione-Sepharose beads (50 µl) preincubated with either purified GST (0.5 µg, serving as control) or the GST fused various fragments of ECM1 were incubated with COMP, respectively. Bound proteins were processed as described above.
In the case of the binding assay for determining whether whole C-terminal domain of ECM1a is required for its binding to COMP, ProBind resin (Invitrogen) bearing either His, His-tagged δF/COOH, or His-tagged δP/COOH was incubated with COMP and the bound proteins processed as described above.
Solid-phase Binding Assay
Microtiter plates (96-well EIA/RIA plates, Costar, Badhoevedorp, Netherlands) were coated with various amounts (0.001–5.000 µg) of purified ECM1 in 100 µl of TBS buffer (50mM Tris-HCl, 150mM NaCl, pH 7.4) overnight at 4°C. Wells were blocked with 1% bovine serum albumin in TBS buffer for 3 h at 37°C. After washing with TBS and 0.05% Tween, 100 µl of 50 µg/ml of COMP was added to each well, followed by the addition of 10mM CaCl2; samples were then allowed to bind overnight at 4°C. Bound protein from the liquid phase was detected by mouse monoclonal antibody against COMP, followed by a secondary antimouse antibody conjugated with horseradish peroxidase (HRP; Antigenix America, Huntington Station, NY) and 5-amino-2-hydroxybenzoic acid as a substrate, with absorbance measured at 490 nm in an ELISA reader.
Coimmunoprecipitation (co-IP) Assay
Approximately 1 ×107 human primary chondrocytes were harvested, washed twice with ice-cold PBS, suspended in 500 µl of a co-IP buffer (150mM NaCl, 10mM Tris-HCl [pH 7.4], 0.1% Nonidet P-40, 1mM phenylmethylsulfonyl fluoride, 50 mg leupeptin per ml), and briefly sonicated, and the cell debris was pelleted. Cell extracts were incubated with anti-ECM1 (Rb469) anti-COMP (2114B3) or control rabbit IgG (25 µg/ml) antibodies at room temperature for 2 h, followed by incubation with 30 µl of protein A-agarose (Invitrogen) at 4°C overnight. After washing 5 times with immunoprecipitation buffer, bound proteins were released by boiling in 20 µl of 2× SDS loading buffer for 3 min (Liu et al., 2002). Released proteins were examined by Western blotting with either anti-COMP (Di Cesare et al., 1999; Liu et al., 2006a) or anti-ECM1 antibodies (Rb469) and the signal detected using the ECL chemiluminescent system.
Immunohistochemistry
Five-micrometer-thick formalin-fixed paraffin sections of 18.5-day-old embryonic murine limbs were immunostained for ECM1. The sections were pretreated with 0.1% trypsin for 30 min at 37°C followed by 3% BSA, 20% goat serum block for 1 h at room temperature to reduce nonspecific staining. Polyclonal anti-ECM1 antibody (Rb471) or preimmune serum (serving as a negative control) was diluted at 1:100 and incubated overnight at 4°C. Binding of primary antibodies was detected using biotinylated antigoat secondary antibody (Vector ABC Elite kit) diluted at 1:200 and incubated for 1 h at 37°C followed by Vector ABC peroxidase (Vector) at 37°C for 1 h and developed with DAB (Sigma) for 2 min at room temperature. Sections were counterstained with methyl green (Dako). For the assay examining COMP expression, the same protocol was followed except that anti-COMP antibody was used in place of anti-ECM1 (Rb471).
Culture of 15.5-day-old Fetal Mouse Bone Explants and Histochemistry
Fetal mouse metatarsals were dissected from 15.5-day-old fetal BL/6 mice and cultured in αMEM (Invitrogen) containing 10% heat-inactivated fetal calf serum (Invitrogen) and 100 U penicillin/streptomycin (Gibco BRL) per ml (Deckers et al., 2001) in the presence of recombinant ECM1, COMP, EGF domain of COMP, and various combinations of these. After 6 days in culture, one group of explants were placed in 4% PFA in PBS for overnight fixation before being placed in staining solution (0.05% Alizarin red, 0.015% Alcian blue, 5% acetic acid in 70% ethanol) for 45–60 min; another group was fixed in 96% alcohol and processed for paraffin embedding for histological examination. Five-micrometer paraffin sections were cut and stained with Weigert’s Iron Hematoxylin 5 min followed by 0.02% Fast Green for 5 min and 0.1% Sa-franin O for 20 min.
Statistical Test
Two-sample Student’s t-tests were used to determine significant differences (p < .05) in total and mineralized regions of cultured mouse fetal metatarsal bones.
RESULTS
Isolation of ECM1 as a COMP Binding Partner
To better understand the biological functions of COMP, we performed a Y2H screen. Briefly, we linked the four functional domains of COMP [the N-terminal pentamerizing domain (aa 20–83), the EGF-like domain (aa 84–261), type 3 repeats (aa 266–520), and the C-terminal domain (aa 521–755)] to the Gal4 DNA-binding domain in the plasmid pDBleu. We used the respective constructs as bait to screen a library of rat brain cDNA expressed as fusion proteins to the VP16 acidic activation domain in the vector pPC86.
A Y2H rat cDNA library was screened with the construct encoding the EGF-like domain of COMP .(Gomez-Barrena et al., 2006) We screened ~2.5 million clones and identified 21 that activated the three reporter genes. Further tests involved the retransformation of yeast with the purified target plasmids and bait. Only 12 of the original 21 yeast clones expressed hybrid proteins that still interacted with the EGF-like domain bait (not shown). Two of the positive clones encoded two N-terminal truncated mutants (aa 108–562 and aa 224–562) of the extracellular matrix protein 1 (ECM1).
Confirmation of Interaction between ECM1 and COMP in Yeast
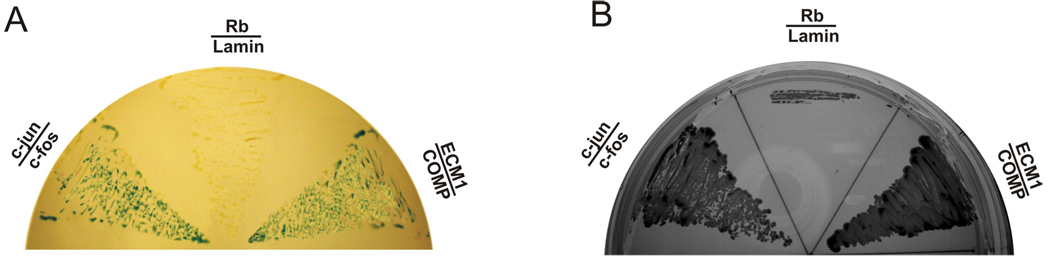
The Y2H assay was repeated to verify the interaction between the EGF-like domain of COMP and ECM1. The plasmid encoding the EGF-like domain of COMP linked to the Gal4 DNA-binding domain and the plasmid encoding the C-terminus of ECM1 fused to the VP16 acidic activation domain were used to cotransform the yeast. Like the c-Jun/c-Fos pair, which interacts and is used as a positive control, our assays indicated that COMP interacts with ECM1 in yeast, based on the activation of the lacZ reporter gene (Fig. 1A) and growth phenotypes on SD-leu−/trp−/his−/ura−/3AT+ plates (Fig. 1B).
Fig. 1. Binding of COMP to ECM1 in yeast.
Yeast two-hybrid assay to test the interaction of proteins fused to the VP16 AD and proteins fused to the Gal4 DBD. Each pair of plasmids, as indicated, encoding proteins fused to VP16 (below the line) in the vector pPC86 (i.e., pPC86-c-jun, pPC86-ECM1, and pPC86-Rb) and those encoding proteins fused to Gal4 (above the line) in the vector pDBleu (i.e., pDB-c-fos, pDB-COMP, and pDB-lamin) were cotransfected into yeast strain MAV203. Yeast transformants were selected on SD-leu−/trp− plates and tested for β-galactosidase activity (panel A), for growth on plates lacking histidine and uracil and containing 3AT (panel B, SD-leu−/trp−/his−/ura−/3AT+). The known interaction between c-Jun and c-Fos was used as a positive control, and the lack of interaction between Rb and lamin was used as a negative control.
COMP Directly Binds to ECM1
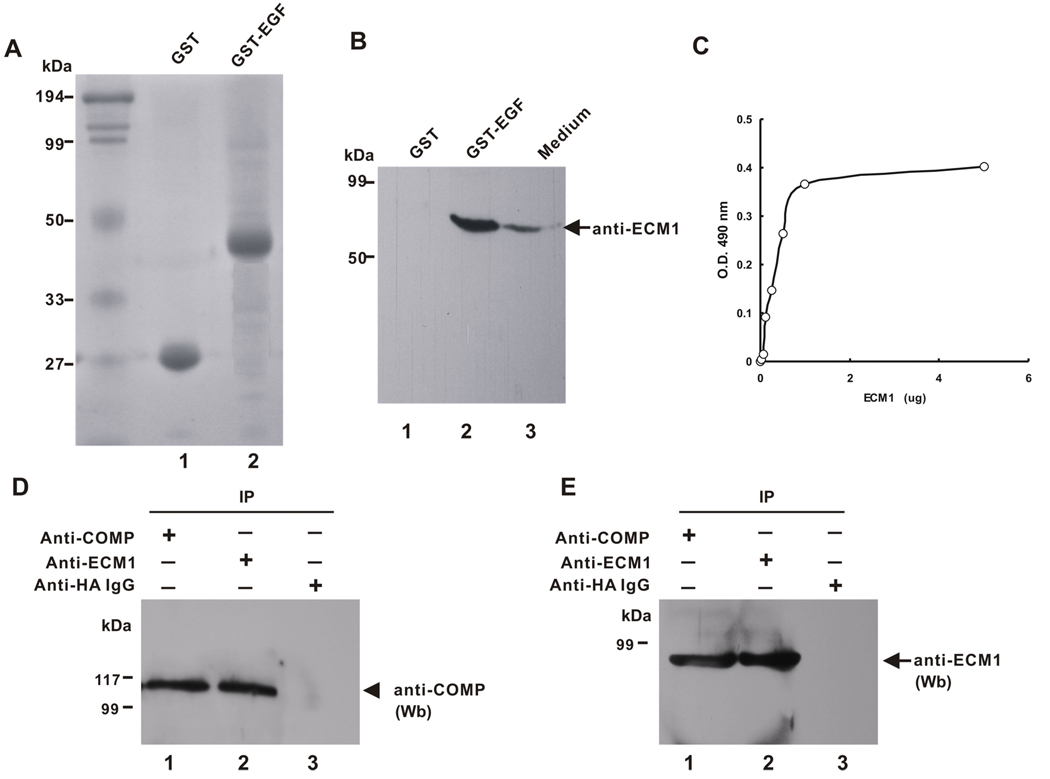
To verify the interaction between COMP and ECM1 that was first identified in yeast, a GST pull-down assay was performed to test whether the EGF-like domain of COMP binds to ECM1 in vitro. Purified GST or GST fused EGF domain of COMP (Fig. 2A) was conjugated to the glutathione-sepharose beads and incubated with a conditioned medium bearing ECM1. After washing, the bound proteins were detected with anti-ECM1 antibodies Rb469, which targeting to the carboxyl terminus of the mouse ECM1 protein (encoded by exon 11) and recognizing all ECM1 iso-forms (Bhalerao et al., 1995; Smits et al., 1999; Smits et al., 1997). As shown in Fig. 2B, the GST-fused EGF domain of COMP efficiently pulled down ECM1 (lane 2), whereas GST did not (lane 1). These results clearly indicate that the EGF domain of COMP binds to ECM1 in vitro.
Fig. 2. COMP binds to ECM1 in vitro and in vivo.
A and B, COMP associates with ECM1 in vitro. A, Expression of GST-fused EGF domain of COMP. Samples of affinity-purified GST and GST-EGF, as indicated, were examined by SDS-PAGE and Coomassie Blue staining. B, GST pulldown assay. Purified GST (lane 1) or GST-EGF fusion protein (lane 2) were conjugated to the glutathione-sepharose beads and incubated with ECM1-conditioned medium. Proteins trapped by the EGF domain of COMP fused to GST were examined by immunoblotting with anti-ECM1 antibodies. ECM1-conditioned medium (lane 3) was used as a positive control. C, Solid-phase assay. Various amounts of recombinant ECM1 protein, as indicated, were immobilized on 96-well microtiter plates. After blocking, COMP was added to each well, and bound protein from the liquid phase was detected using monoclonal antibodies against COMP. D and E, COMP interacts with ECM1 in vivo. Co-IP assay. Cell extracts prepared from human primary chondrocytes were incubated with anti-ECM1 (lane 1) anti-COMP (lane 2) or control IgG (lane 3) followed by protein A-agarose. The immunoprecipitated protein complexes were examined by immunoblotting with anti-COMP (D) and anti-ECM1 (E) antibodies.
The interaction between COMP and ECM1 was also characterized by an in vitro solid-liquid-phase titration experiment in which the dilution series of recombinant intact ECM1 and COMP showed dose-dependent binding and saturation to the liquid-phase COMP (Fig. 2C). The interaction between COMP and ECM1 was direct, since both COMP and the ECM1 were used as purified recombinant proteins.
Binding of COMP to ECM1 in Human Primary Chondrocytes
The in vivo interaction between COMP and ECM1 was verified using a Co-IP assay in order to determine whether these two proteins are bound in human primary chondrocytes. For the Co-IP assays, cell extracts prepared form human chondrocytes were incubated with either anti-ECM1 antiserum Rb469, anti-COMP antiserum 2114B3, or control IgG, and the immunoprecipitated complexes were subjected to a reducing SDS-PAGE and detected with anti-COMP antibody. A specific COMP band was present in the immunoprecipitated complexes brought down by anti-ECM1 (Fig. 2D, lane 2) but not control IgG (lane 3) antibodies. An opposite co-IP assay using anti-ECM1 antibody indicated that ECM1 also precipitated COMP (Fig. 2E, lane 1), while control IgG (lane 3) did not, clearly indicating that these two proteins associate in vivo.
EGF-like and C-terminal domains of COMP bind to ECM1
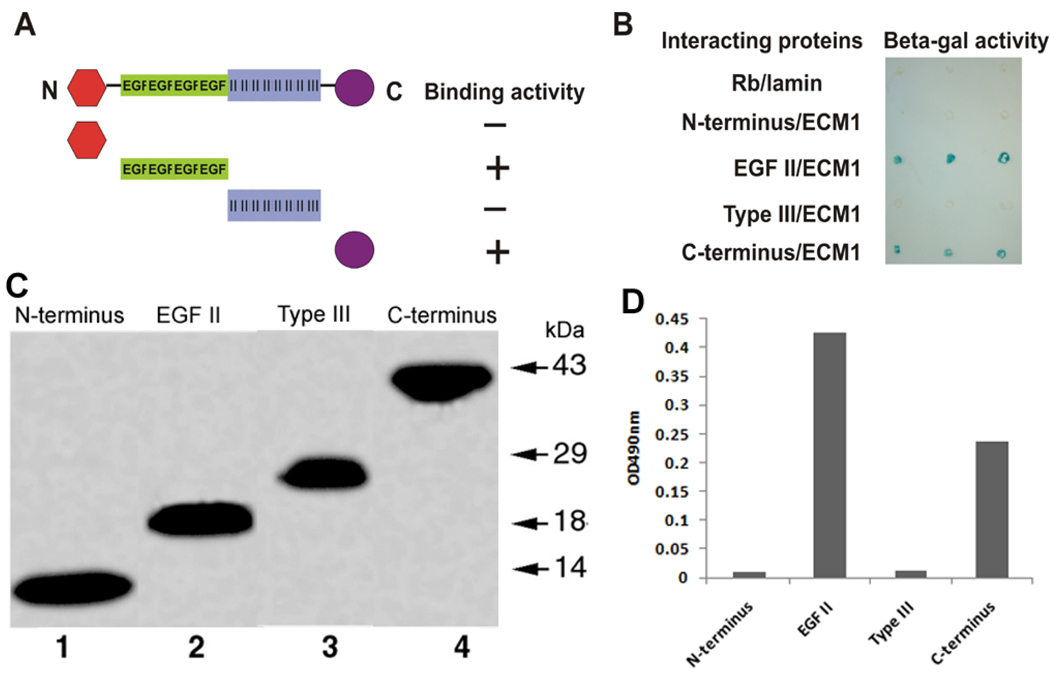
After ECM1 was identified as a COMP-binding protein using the Y2H screen, we sought to establish whether, in addition to the EGF domain, other domains of COMP associate with ECM1. A filter-based β-galactosidase assay was used to determine whether coexpression of the various domains of the COMP/Gal4 DNA-binding domain and ECM1/VP16 acidic activation domain fusion proteins activate the reporter lacZ gene. Each domain of COMP was cotransformed with ECM1 into yeast and an X-gal assay was performed (Figs. 3A, 3B). Similar to the EGF-like domain, the C-terminus of COMP also binds to ECM1 in yeast (Fig. 3B), indicating that there exist two ECM1-binding domains in COMP. In addition, recombinant COMP fragments corresponding to N-terminal, type II (aka EGF-like repeats), type III and C-terminal domains were produced in insect cells using baculovirus system (Fig. 3C), and their binding ability to ECM1 was examined using solid-phase binding assay (Fig. 3D). Note that EGF-like domain of COMP showed higher binding affinity to ECM1 than does C-terminal domain of COMP.
Fig. 3. ECM1 selectively binds to the EGF-like and C-terminal domain of COMP.
A, schematic structure of COMP constructs used to map those domains (N-terminal, EGF-like, type III, and C-terminal domain) that bind to ECM1. The presence or absence of binding between COMP domains and ECM1 is indicated as plus or minus sign, respectively. B, β-galactosidase activity was used to test the interaction between the ECM1 and one of four domains of COMP. Three independent yeast transformants for each pair of plasmids were transferred onto a nitrocellulose membrane, and β-galactosidase activity was determined. The known lack of interaction between Rb and lamin served as a negative control. C, Purified Flag-tagged COMP fragment, as indicated, were separated on SDS-PAGE and visualized by Coomassie blue staining. D, Solid-phase assay. Comparable amounts of purified COMP fragments, as indicated in panel C, were immobilized on 96-well microtiter plates. After blocking, recombinant ECM1 was added to each well, and bound protein from the liquid phase was detected using antibodies against ECM1.
C-terminus of ECM1 Is Required and Sufficient for Interaction with COMP
We next expressed and purified various deletion mutants of ECM1 as either GST-fused or His-tagged proteins (Fig. 4A), and performed an in vitro pull-down assay to dissect the COMP-binding region in ECM1. The purified deletion mutants of ECM1 fused to GST were verified by SDS-PAGE analysis (Fig. 4B). Results from in vitro GST pull-down assays of these mutants indicated that the fragment (aa340–540) containing the full-length C-terminus plus partial repeat 2 bound to COMP, while the other mutants did not (Fig. 4A,C). To further determine whether the C-terminus (aa 360–540) alone is sufficient for the binding to COMP, we performed a His pull-down assay using purified His-tagged mutants (Fig. 4D). As shown in Fig. 4E, the C-terminus (aa 360–540) of ECM1 clearly pulled down COMP while the partially C-terminal domain (aa 360–480) did not. Based on this set of assays, we concluded that the C-terminus (aa 360–540) of ECM1 is required and sufficient for its interaction with COMP.
COMP and ECM1 Colocalize in the Growth Plate in Vivo
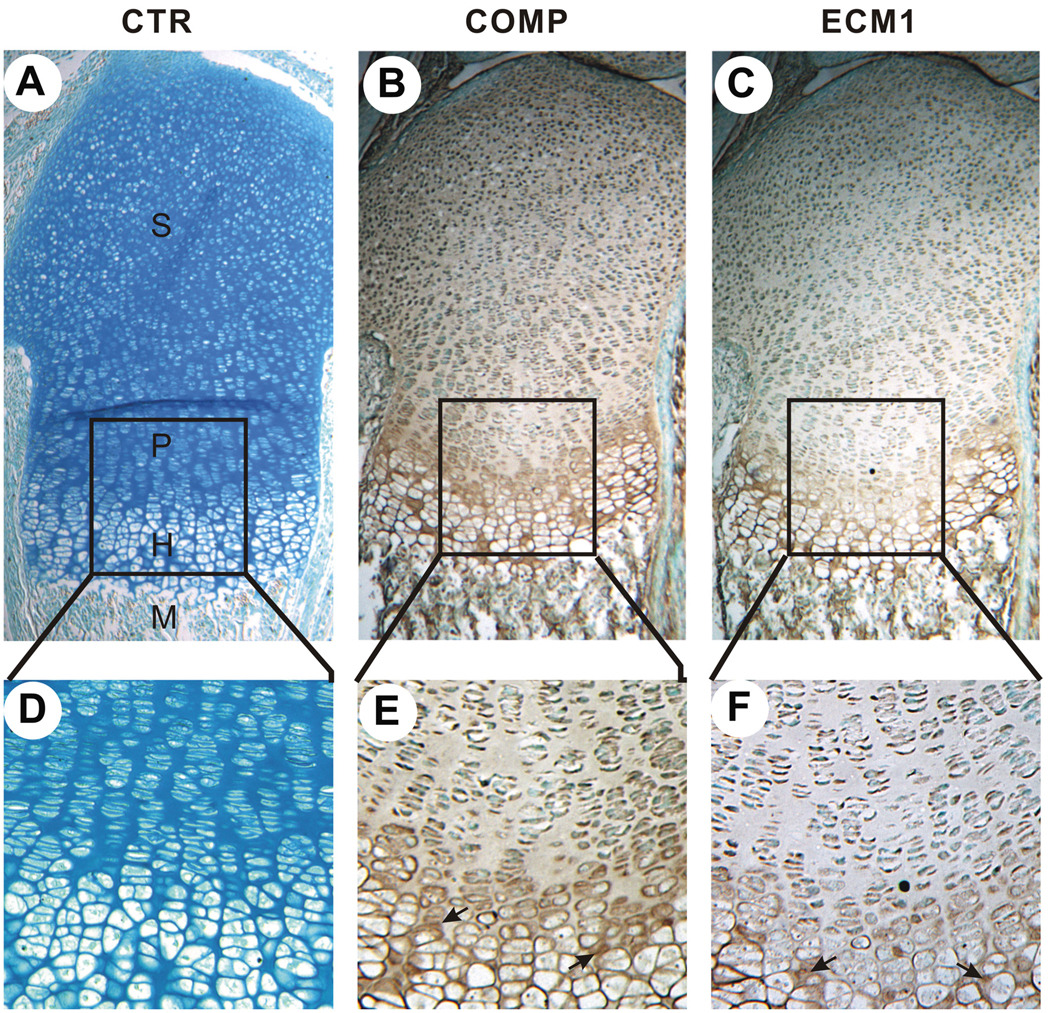
We next examined the in vivo expressions of COMP and ECM1 and sought to determine whether these two proteins show overlapping expression patterns in vivo using immunostaining assays on the 18.5-day-old embryonic murine growth plate. As shown in Fig. 5, both ECM1 and COMP are expressed throughout the growth plate chondrocytes. In addition, these two proteins overlapped in the pericellular matrix of hypertrophic chondrocytes.
Fig. 5. Immunohistochemistry of COMP and ECM1 in the section of long bone from a 18.5-day-old mouse embryo.
A, B, C, Low-power microphotograph of a section stained with preimmuno serum (A), anti-COMP (B) or anti-ECM1 (C) polyclonal antibody (brown) and counterstained with methyl green (blue). D, E, F, High-power microphotograph of sections in A, B, C. S, resting chondrocytes; P, proliferating chondrocytes; H, hypertrophic chondrocytes; M, bone metaphysis.arrow indicates the signal.
Recombinant EGF domain of COMP is able to disturb the interaction between COMP and ECM1
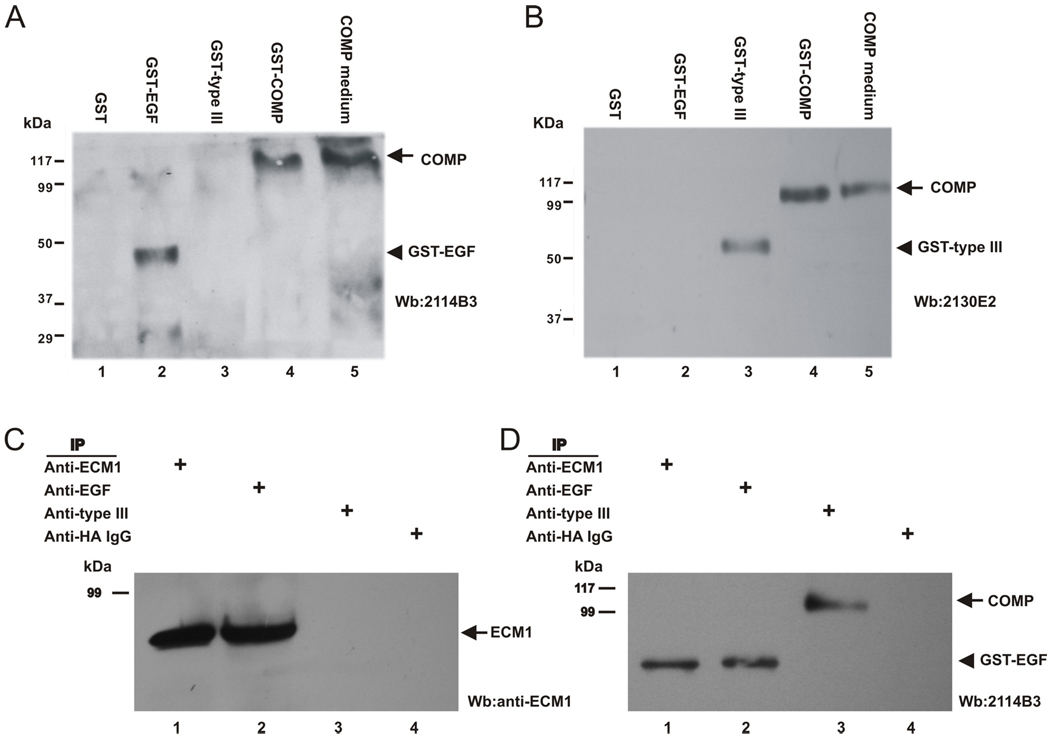
Since we have shown that the EGF-like domain of COMP demonstrates higher binding affinity to ECM1 (Fig. 3), next we sought to determine whether the recombinant EGF domain of COMP affects the binding of ECM1 to full-length COMP. For this purpose, we first generated a panel of monoclonal antibodies using purified COMP functional domains as antigens. Briefly, both the EGF-like and type III domains of COMP (see Fig. 3 for structure) were purified as GST fusion proteins and the GST moiety was further removed by an Xa factor. Two monoclonal antibodies, 2114B3-EGF and 2130E2–type III, generated using the recombinant EGF and type III domains of COMP as antigens, respectively, were employed in this experiment. As revealed in Fig. 6A, 2114B3-EGF specifically recognized the full-length and EGF domains of COMP, but not the type III domain, whereas 2130E2–type III reacted with the full-length and type III domains of COMP, but not the EGF domain of COMP (Fig. 6B). We next performed Co-IP using these monoclonal antibodies together with anti-ECM1, which efficiently precipitates COMP (Fig. 2B). Briefly, the conditioned medium collected from HEK293 cells cotransfected with ECM1 and COMP expression plasmid was supplemented with an excess amount of purified recombinant GST-EGF domain of COMP and immunoprecipitated with various antibodies, as indicated in Fig. 6 (panel C and D). Although anti-ECM1 efficiently precipitated COMP (Fig. 2), the same anti-ECM1 antibody failed to do so in the presence of the recombinant EGF domain of COMP (lane 1 in Fig. 6C), indicating that the recombinant EGF domain of COMP is able to disturb the association between ECM1 and COMP. This finding was further verified with the 2130E2–type III monoclonal antibody that reacts with the type III domains of COMP, since it efficiently precipitated COMP but not ECM1 in the presence of excess of recombinant EGF domain. In addition, 2114B3-EGF monoclonal antibody that recognizes the EGF domains of COMP precipitated ECM1 but not COMP, demonstrated that ECM1 was occupied by excess recombinant EGF domain.
Fig. 6. EGF domain of COMP disturbs the interaction between ECM1 and COMP.
A, B, Characterization of anti-COMP monoclonal antibodies using Western blotting assay. Bacteria extracts bearing GST, GST-EGF or GST-type III, as well as the recombinant COMP or COMP conditioned medium, were subjected to 10% SDS-PAGE and detected with either 2114B3-EGF (A) or 2130E2–type III (B) monoclonal antibody. C, D, Co-IP assay. Cell extracts prepared from 293 cells cotransfected with ECM1 and COMP expression plasmids supplemented with recombinant EGF domain of COMP were incubated with anti-ECM1 (lane 1), 2114B3-EGF that recognized EGF domain of COMP (lane 2), 2130E2–type III that bound to the type III domain of COMP (lane 3), or control HA probe (serving as a control IgG (lane 4), followed by protein A-agarose. The immunoprecipitated protein complexes were examined by immunoblotting with anti-ECM1 (Rb469) (C) and anti-COMP (D) antibodies.
Effect of Recombinant ECM1 and COMP on Endochondral Bone Formation
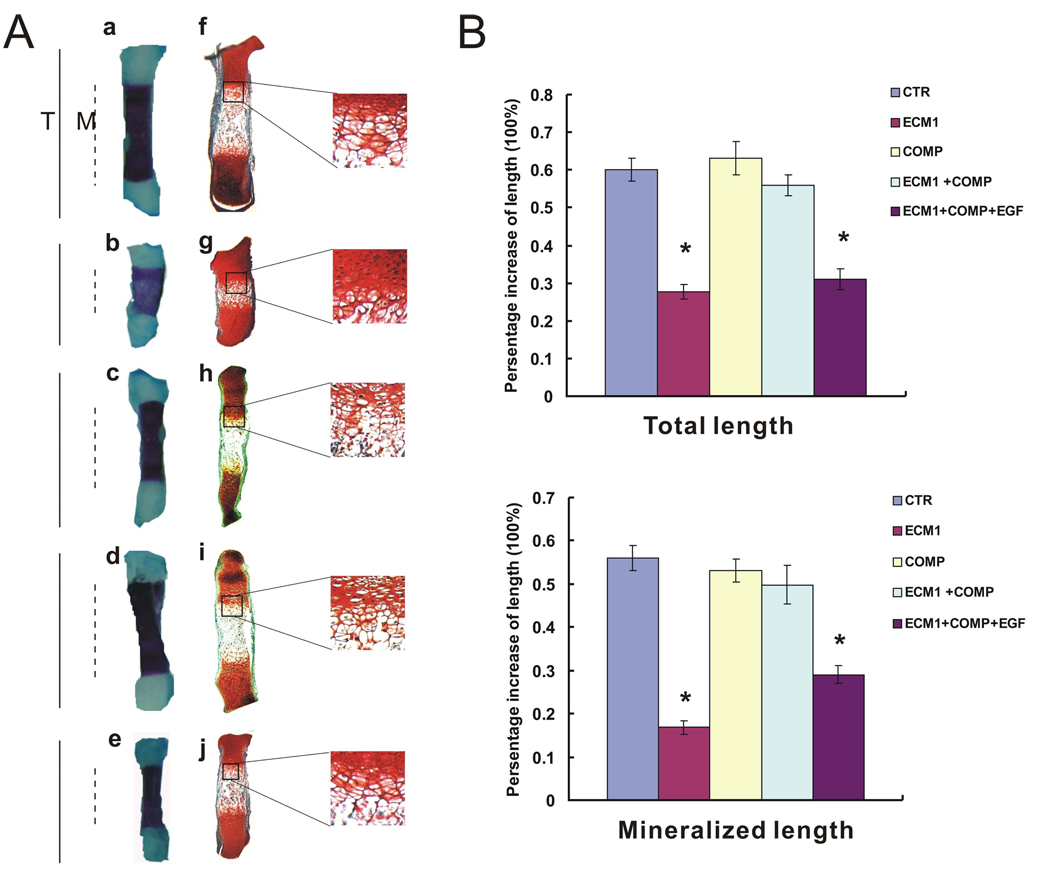
The effect of recombinant ECM1 and COMP on endochondral bone formation was studied in an in vitro model using cultures of fetal mouse metatarsals. At time of explantation, explants consisted of undifferentiated cartilage. Over a 6-day culture period, these explants proceeded through all the stages of endochondral bone formation. The hypertrophic condition of growth plate chondrocytes was confirmed by Alizarin red/Alcian blue staining, which identifies calcified chondrocyte matrix. Consistent with a previous report (Deckers et al., 2001), culturing explants in the presence of ECM1 strongly decreased mineralization in and length of the hypertrophic chondrocyte zone of cultured metatarsals (Fig. 7A:b) when compared to controls (Fig. 7A:a). Interestingly, addition of COMP in the medium produced negligible effects (Fig. 7A: c); however, addition of COMP corrected ECM1-mediated inhibition of chondrocyte hypertrophy, matrix mineralization, and endochondral bone growth (Fig. 7A: d). In contrast, COMP largely failed to overcome ECM1 inhibition of endochondral bone formation in the presence of the recombinant EGF domain of COMP that was found to disturb the interaction between COMP and ECM1 (Fig. 6 and Fig. 7A: e). These findings suggest that the association of ECM1 and COMP is important for appropriate regulation of endochondral bone formation. The regulation of chondrocyte hypertrophy by ECM1/COMP interaction was confirmed by staining of histological sections with Safranin O (Fig. 7A: f–j). Note that each group of explants contained 6 different metatarsals from different mouse feet and that mean total length and mean mineralized cartilage were measured at the start and end time points during the experiment (Fig. 7B).
Fig. 7. COMP overcomes ECM1-mediated inhibition of endochondral bone growth.
Metatarsals were explanted from fetal mice and cultured in the presence of stimuli in 6-tuple. After 6 days, the explants were fixed and processed for either Alizarin red/Alcian blue staining or histochemical analysis for Safranin O staining as described in “Experimental Procedures.” A, Representative photograph (a–e) and Safraninin O staining (f–j) of an explanted metatarsal after 6 days of culture in the presence of vehicle (a, f), 250 ng/mL ECM1a (b, g), 250 ng/mL COMP (c, h), 250 ng/mL ECM1a plus 250 ng/mL COMP in the absence (d, i), or presence of 250 ng/mL purified EGF domain of COMP (e, j). Straight line represents the total length (T) and the broken line indicates the zone of mineralized cartilage (M). B, Percentage increase in T (top) and M (bottom) length of metatarsal bones after 6 cultures for 6 days. (Percentage of increase = [length at day 6 − length at day 0] / length at day 0). *Significantly different from control (p < .05).
DISCUSSION
Yeast two-hybrid screening has proven to be an effective tool in identifying protein interaction (Liu et al., 2001; Liu et al., 2003; Liu et al., 2006a). To identify protein interaction partners of COMP, an extra-cellular matrix protein that has been implicated in the regulation of chondrogenesis and endochondral bone formation, we screened the yeast expression cDNA library using the EGF repeat domain of COMP as bait and identified ECM1 as a direct binding protein of COMP.
The ECM1 gene is located adjacent to the epidermal differentiation complex region on chromosome 1q21 (Johnson et al., 1997; Smits et al., 1997). The ECM1 protein contains a signal peptide of 19 amino acids followed by 4 functional domains: a cysteine-free N terminus, two tandem repeats, and a C terminus (Mongiat et al., 2003; Smits et al., 1997). The latter three cysteine-containing domains all have the typical CC-(X7–10)C arrangement that is capable of forming protein double loops involved in protein–protein interactions (Bhalerao et al., 1995; Smits et al., 1997). The CC-(X7−10)C motif is also present in the serum albumin family of proteins and shows structural similarities to the Endo 16 calcium-binding protein of sea urchin (Bhalerao et al., 1995; Godin et al., 1996). These could enable ECM1 to serve as a transporter protein or to be involved in binding growth or differentiation factors (Fujimoto et al., 2005; Mongiat et al., 2003). Indeed, several ECM1-binding partners have been reported; for instance, the perlecan-ECM1 interaction was suggested to modulate endochondral bone formation, angiogenesis, and tumorgenesis (Mirancea et al., 2006; Mongiat et al., 2003; Sercu et al., 2008a). ECM1 associated with and inhibited MMP9 proteolytic activity, which may have relevance to pathogenesis of lipoid proteinosis and tumor progression (Fujimoto et al., 2006; Sercu et al., 2008b). Recent studies have demonstrated that ECM1 utilizes different regions to bind to a variety of extracellular matrix components, such as laminin 332, collagen type IV, fibronectin, hyaluronan, heparin, and chondroitin sulfate A (Sercu et al., 2008a). Our global screen led to the isolation of ECM1 as a novel binding partner of COMP, and the interaction between these two molecules appears to be important for endochondral bone growth (Fig. 1, Fig. 2, Fig. 7). In addition, the EGF-like and C-terminal domains of COMP and the C-terminal region of ECM1 are involved in the association between COMP and ECM1 (Fig. 3 and Fig.4). Interestingly, EGF-like domain of COMP that exhibits higher binding affinity to ECM1 (Fig. 3) efficiently disturbs the binding of COMP to ECM1 (Fig. 6), although both EGF-like and C-terminal domains are able to interact to ECM1, this is probably due to the fact the binding of EGF-like domain to ECM1 affects the confirmation of ECM1 and in turn inhibits the binding of full length COMP to ECM1.
COMP has been reported to interact with multiple protein partners, and these interactions are important for its physiologic functions and cytoplasmic processing and transport. COMP appears to mediate chondrocyte attachment via an integrin receptor (Di Cesare et al., 2000; Di Cesare et al., 1994), and several reports suggest that COMP may function to stabilize the cartilage extracellular matrix by specific cation-dependent interactions with matrix components, including collagen types II and IX, fibronectin, aggrecan, and matrilin-1, -3, and -4 (Chen et al., 2005; Di Cesare et al., 2002; Mann et al., 2004; Mansson et al., 1995; Rosenberg et al., 1998). COMP has also been shown to associate with several chaperone proteins, including BiP, calreticulin, protein disulfide, ERp72, Grp94, HSP47, and calnexin. It has been proposed that these associations facilitate the processing and transport of wildtype COMP in normal chondrocytes and in the retention of mutant COMP in pseudoachondroplasia chondrocytes (Duke et al., 2003; Hecht et al., 2001; Vranka et al., 2001). In addition to the interactions between COMP and its protein partners, the five-stranded N-terminal domain of COMP forms a complex with vitamin D3, indicating that COMP has a storage function for hydrophobic compounds, including prominent cell-signaling molecules (Ozbek et al., 2002). We previously reported that ADAMTS-7 (Liu et al., 2006a) and ADAMTS-12 (Liu et al., 2006b), two members of a family of metalloproteinases with similar domain structure and organization, bind to the same EGF domain of COMP to which ECM1 binds and degrade COMP in vitro. In addition, cleavage of COMP by ADAMTS-7 and ADAMTS-12 are important factors in COMP degradation in osteoarthritis (Luan et al., 2008) and COMP homeostasis in chondrogenesis (Bai et al., 2009a; Bai et al., 2009b). It remains to determine whether ECM1 affects COMP degradation and homeostasis by ADAMTS-7 and ADAMTS-12.
COMP and ECM1 colocalize in growth plate in the day-18.5 mouse embryo (Fig. 5). These results suggest an in vivo association between COMP and ECM1. ECM1 has been shown to stimulate epithelial cell proliferation (Mirancea et al., 2007; Wang et al., 2003). ECM1-mediated cell growth appears to be cell-type specific, since ECM1 was found to potently inhibit chondrocyte proliferation in vitro; further-more, COMP neutralizes ECM1-mediated inhibition of chondrocyte proliferation (data not shown). ECM1 is involved in regulating endochondral bone formation (Chan, 2004; Deckers et al., 2001; Mongiat et al., 2003). In this study we showed that ECM1 and COMP play opposite roles in regulating chondrocyte hypertrophy, matrix calcification, and endochondral bone growth and that balanced regulation depends on their interaction (Fig. 7). Although the molecular mechanism by which ECM1 and COMP regulate endochondral bone formation in an opposite manner remains unknown, their association with and modulation of granulin-epithelin precursor (GEP), an autocrine growth factor known to be a stimulator of chondrocyte proliferation and differentiation (Xu et al., 2007), may represent one of the molecular events in regulating endochondral bone growth by ECM1 and COMP. We have reported that COMP associates with GEP and potentiates GEP-stimulated chondrocyte proliferation (Xu et al., 2007). Interestingly, ECM1 was also found to interact with GEP. COMP and ECM1, however, exert an opposite effect on GEP cell surface localization: COMP enhances whereas ECM1 inhibits cell surface appearance of GEP (data not shown), suggesting that COMP might present GEP growth factor to its receptor(s) whereas ECM1 might sequester GEP from its receptor(s).
In conclusion, we first identified ECM1 in cartilage as a COMP-binding protein and subsequently characterized this novel association and conducted functional assays showing that endochondral bone formation is mediated by the interaction between ECM1 and COMP. These findings advance our understanding of matrix/matrix interactions in skeletal biology and may also provide a potential target for developing and optimizing a therapeutic application for cartilage repair and treatment of arthritic disorders.
Acknowledgments
We thank Dr. Mary B. Goldring for providing human C28I2 chondrocytes, Drs. Marc Fajardo and Kirill Ilalov for assisting in collecting mouse fetal metatarsal bones. This work was aided by NIH research grants AR050620, AR053210, and AG029388 and a grant from the Arthritis National Research Foundation to C. J. Liu. This work was partially supported by a grant from the Fund for Scientific Research–Flandres (FWO-G.0133.05) to J. Merregaert
Abbreviations
- COMP
cartilage oligomeric matrix protein
- ECM1
extracellular matrix protein 1
- EGF
epidermal growth factor
- ADAMTS
a disintegrin and metalloproteinase with thromospondin motifs
- GEP
granulin-epithelin precursor
- GST
glutathione S-transferase
- PCR
polymerase chain reaction
- Co-IP
coimmunoprecipitation
- HRP
horseradish peroxidase
- Y2H
yeast 2-hybrid
- PBS
phosphate-buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bai XH, Wang DW, Kong L, Zhang Y, Luan Y, Kobayashi T, Kronenberg HM, Yu XP, Liu CJ. ADAMTS-7, a direct target of PTHrP, adversely regulates endochondral bone growth by associating with and inactivating GEP growth factor. Mol Cell Biol. 2009a;29:4201–4219. doi: 10.1128/MCB.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XH, Wang DW, Luan Y, Yu XP, Liu CJ. Regulation of chondrocyte differentiation by ADAMTS-12 metalloproteinase depends on its enzymatic activity. Cell Mol Life Sci. 2009b;66:667–680. doi: 10.1007/s00018-008-8633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao J, Tylzanowski P, Filie JD, Kozak CA, Merregaert J. Molecular cloning, characterization, and genetic mapping of the cDNA coding for a novel secretory protein of mouse. Demonstration of alternative splicing in skin and cartilage. J Biol Chem. 1995;270:16385–16394. doi: 10.1074/jbc.270.27.16385. [DOI] [PubMed] [Google Scholar]
- Bosco EE, Wang Y, Xu H, Zilfou JT, Knudsen KE, Aronow BJ, Lowe SW, Knudsen ES. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest. 2007;117:218–228. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MD, Hoffman SM, King LM, Olsen AS, Mohrenweiser H, Leroy JG, Mortier GR, Rimoin DL, Lachman RS, Gaines ES. Pseudoachondroplasia and multiple epiphyseal dysplasia due to mutations in the cartilage oligomeric matrix protein gene. Nature. 1995;10:330–336. doi: 10.1038/ng0795-330. [DOI] [PubMed] [Google Scholar]
- Briggs MD, Mortier GR, Cole WG, King LM, Golik SS, Bonaventure J, Nuytinck L, De Paepe A, Leroy JG, Biesecker L, Lipson M, Wilcox WR, Lachman RS, Rimoin DL, Knowlton RG, Cohn DH. Diverse mutations in the gene for cartilage oligomeric matrix protein in the pseudoachondroplasia-multiple epiphyseal dysplasia disease spectrum. Am J Hum Genet. 1998;62:311–319. doi: 10.1086/301713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MD, Rasmussen IM, Weber JL, Yuen J, Reinker K, Garber AP, Rimoin DL, Cohn DH. Genetic linkage of mild pseudoachondroplasia (PSACH) to markers in the pericentromeric region of chromosome 19. Genomics. 1993;18:656–660. doi: 10.1016/s0888-7543(05)80369-6. [DOI] [PubMed] [Google Scholar]
- Chan I. The role of extracellular matrix protein 1 in human skin. Clin Exp Dermatol. 2004;29:52–56. doi: 10.1111/j.1365-2230.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- Chan I, Oyama N, Neill SM, Wojnarowska F, Black MM, McGrath JA. Characterization of IgG autoantibodies to extracellular matrix protein 1 in lichen sclerosus. Clin Exp Dermatol. 2004;29:499–504. doi: 10.1111/j.1365-2230.2004.01573.x. [DOI] [PubMed] [Google Scholar]
- Chen FH, Thomas AO, Hecht JT, Goldring MB, Lawler J. Cartilage Oligomeric Matrix Protein/Thrombospondin 5 Supports Chondrocyte Attachment through Interaction with Integrins. J Biol Chem. 2005;280:32655–32661. doi: 10.1074/jbc.M504778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckers MM, Smits P, Karperien M, Ni J, Tylzanowski P, Feng P, Parmelee D, Zhang J, Bouffard E, Gentz R, Lowik CW, Merregaert J. Recombinant human extracellular matrix protein 1 inhibits alkaline phosphatase activity and mineralization of mouse embryonic metatarsals in vitro. Bone. 2001;28:14–20. doi: 10.1016/s8756-3282(00)00428-2. [DOI] [PubMed] [Google Scholar]
- Di Cesare E, Costanzi A, Fedele F, Di Renzi P, D'Eusanio G, Lupattelli L, Passariello R. MRI postoperative monitoring in patients surgically treated for aortic dissection. Magn Reson Imaging. 1996;14:1149–1156. doi: 10.1016/s0730-725x(96)00221-4. [DOI] [PubMed] [Google Scholar]
- Di Cesare PE, Chen FS, Moergelin M, Carlson CS, Leslie MP, Perris R, Fang C. Matrix-matrix interaction of cartilage oligomeric matrix protein and fibronectin. Matrix Biol. 2002;21:461–470. doi: 10.1016/s0945-053x(02)00015-x. [DOI] [PubMed] [Google Scholar]
- Di Cesare PE, Fang C, Leslie MP, Della Valle CJ, Gold JM, Tulli H, Perris R, Carlson CS. Localization and expression of cartilage oligomeric matrix protein by human rheumatoid and osteoarthritic synovium and cartilage. J Orthop Res. 1999;17:437–445. doi: 10.1002/jor.1100170321. [DOI] [PubMed] [Google Scholar]
- Di Cesare PE, Fang C, Leslie MP, Tulli H, Perris R, Carlson CS. Expression of cartilage oligomeric matrix protein (COMP) by embryonic and adult osteoblasts. J Orthop Res. 2000;18:713–720. doi: 10.1002/jor.1100180506. [DOI] [PubMed] [Google Scholar]
- Di Cesare PE, Nimni ME, Yazdi M, Cheung DT. Effects of lathyritic drugs and lathyritic demineralized bone matrix on induced and sustained osteogenesis. J Orthop Res. 1994;12:395–402. doi: 10.1002/jor.1100120312. [DOI] [PubMed] [Google Scholar]
- Duke J, Montufar-Solis D, Underwood S, Lalani Z, Hecht JT. Apoptosis staining in cultured pseudoachondroplasia chondrocytes. Apoptosis. 2003;8:191–197. doi: 10.1023/a:1022926811397. [DOI] [PubMed] [Google Scholar]
- Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, Nimmo ER, Massey D, Berzuini C, Johnson C, Barrett JC, Cummings FR, Drummond H, Lees CW, Onnie CM, Hanson CE, Blaszczyk K, Inouye M, Ewels P, Ravindrarajah R, Keniry A, Hunt S, Carter M, Watkins N, Ouwehand W, Lewis CM, Cardon L, Lobo A, Forbes A, Sanderson J, Jewell DP, Mansfield JC, Deloukas P, Mathew CG, Parkes M, Satsangi J. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto N, Terlizzi J, Aho S, Brittingham R, Fertala A, Oyama N, McGrath JA, Uitto J. Extracellular matrix protein 1 inhibits the activity of matrix metalloproteinase 9 through high-affinity protein/protein interactions. Exp Dermatol. 2006;15:300–307. doi: 10.1111/j.0906-6705.2006.00409.x. [DOI] [PubMed] [Google Scholar]
- Fujimoto N, Terlizzi J, Brittingham R, Fertala A, McGrath JA, Uitto J. Extracellular matrix protein 1 interacts with the domain III of fibulin-1C and 1D variants through its central tandem repeat 2. Biochem Biophys Res Commun. 2005;333:1327–1333. doi: 10.1016/j.bbrc.2005.06.046. [DOI] [PubMed] [Google Scholar]
- Godin RE, Urry LA, Ernst SG. Alternative splicing of the Endo16 transcript produces differentially expressed mRNAs during sea urchin gastrulation. Dev Biol. 1996;179:148–159. doi: 10.1006/dbio.1996.0247. [DOI] [PubMed] [Google Scholar]
- Gomez-Barrena E, Lindroos L, Ceponis A, Lopez-Franco M, Sanchez-Pernaute O, Monkkonen J, Salo J, Herrero-Beaumont G, Konttinen Y. Cartilage oligomeric matrix protein (COMP) is modified by intra-articular liposomal clodronate in an experimental model of arthritis. Clin Exp Rheumatol. 2006;24:622–628. [PubMed] [Google Scholar]
- Hamada T, McLean WH, Ramsay M, Ashton GH, Nanda A, Jenkins T, Edelstein I, South AP, Bleck O, Wessagowit V, Mallipeddi R, Orchard GE, Wan H, Dopping-Hepenstal PJ, Mellerio JE, Whittock NV, Munro CS, van Steensel MA, Steijlen PM, Ni J, Zhang L, Hashimoto T, Eady RA, McGrath JA. Lipoid proteinosis maps to 1q21 and is caused by mutations in the extracellular matrix protein 1 gene (ECM1) Hum Mol Genet. 2002;11:833–840. doi: 10.1093/hmg/11.7.833. [DOI] [PubMed] [Google Scholar]
- Hamada T, Wessagowit V, South AP, Ashton GH, Chan I, Oyama N, Siriwattana A, Jewhasuchin P, Charuwichitratana S, Thappa DM, Jeevankumar B, Lenane P, Krafchik B, Kulthanan K, Shimizu H, Kaya TI, Erdal ME, Paradisi M, Paller AS, Seishima M, Hashimoto T, McGrath JA. Extracellular matrix protein 1 gene (ECM1) mutations in lipoid proteinosis and genotype-phenotype correlation. J Invest Dermatol. 2003;120:345–350. doi: 10.1046/j.1523-1747.2003.12073.x. [DOI] [PubMed] [Google Scholar]
- Han Z, Ni J, Smits P, Underhill CB, Xie B, Chen Y, Liu N, Tylzanowski P, Parmelee D, Feng P, Ding I, Gao F, Gentz R, Huylebroeck D, Merregaert J, Zhang L. Extracellular matrix protein 1 (ECM1) has angiogenic properties and is expressed by breast tumor cells. Faseb J. 2001;15:988–994. doi: 10.1096/fj.99-0934com. [DOI] [PubMed] [Google Scholar]
- Hecht JT, Francomano CA, Briggs MD, Deere M, Conner B, Horton WA, Warman M, Cohn DH, Blanton SH. Linkage of typical pseudoachondroplasia to chromosome 19. Genomics. 1993;18:661–666. doi: 10.1016/s0888-7543(05)80370-2. [DOI] [PubMed] [Google Scholar]
- Hecht JT, Hayes E, Snuggs M, Decker G, Montufar-Solis D, Doege K, Mwalle F, Poole R, Stevens J, Duke PJ. Calreticulin, PDI, Grp94 and BiP chaperone proteins are associated with retained COMP in pseudoachondroplasia chondrocytes. Matrix Biol. 2001;20:251–262. doi: 10.1016/s0945-053x(01)00136-6. [DOI] [PubMed] [Google Scholar]
- Hecht JT, Nelson LD, Crowder E, Wang Y, Elder FF, Harrison WR, Francomano CA, Prange CK, Lennon GG, Deere M. Mutations in exon 17B of cartilage oligomeric matrix protein (COMP) cause pseudoachondroplasia. Nat Genet. 1995;10:325–329. doi: 10.1038/ng0795-325. [DOI] [PubMed] [Google Scholar]
- Hedbom E, Antonsson P, Hjerpe A, Aeschlimann D, Paulsson M, Rosa-Pimentel E, Sommarin Y, Wendel M, Oldberg A, Heinegard D. Cartilage matrix proteins. An acidic oligomeric protein (COMP) detected only in cartilage. J Biol Chem. 1992;267:6132–6136. [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MR, Wilkin DJ, Vos HL, Ortiz de Luna RI, Dehejia AM, Polymeropoulos MH, Francomano CA. Characterization of the human extracellular matrix protein 1 gene on chromosome 1q21. Matrix Biol. 1997;16:289–292. doi: 10.1016/s0945-053x(97)90017-2. [DOI] [PubMed] [Google Scholar]
- Kipnes J, Carlberg AL, Loredo GA, Lawler J, Tuan RS, Hall DJ. Effect of cartilage oligomeric matrix protein on mesenchymal chondrogenesis in vitro. Osteoarthritis Cartilage. 2003;11:442–454. doi: 10.1016/s1063-4584(03)00055-4. [DOI] [PubMed] [Google Scholar]
- Koelling S, Clauditz TS, Kaste M, Miosge N. Cartilage oligomeric matrix protein is involved in human limb development and in the pathogenesis of osteoarthritis. Arthritis Res Ther. 2006;8:R56. doi: 10.1186/ar1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus VB, Huebner JL, Fink C, King JB, Brown S, Vail TP, Guilak F. Urea as a passive transport marker for arthritis biomarker studies. Arthritis Rheum. 2002;46:420–427. doi: 10.1002/art.10124. [DOI] [PubMed] [Google Scholar]
- Kuhne C, Banks L. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J Biol Chem. 1998;273:34302–34309. doi: 10.1074/jbc.273.51.34302. [DOI] [PubMed] [Google Scholar]
- Liu C, Dib-Hajj SD, Waxman SG. Fibroblast growth factor homologous factor 1B binds to the C terminus of the tetrodotoxin-resistant sodium channel rNav1.9a (NaN) J Biol Chem. 2001;276:18925–18933. doi: 10.1074/jbc.M101606200. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Dib-Hajj SD, Renganathan M, Cummins TR, Waxman SG. Modulation of the Cardiac Sodium Channel Nav1.5 by Fibroblast Growth Factor Homologous Factor 1B. J Biol Chem. 2003;278:1029–1036. doi: 10.1074/jbc.M207074200. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Ding B, Wang H, Lengyel P. The MyoD-inducible p204 protein overcomes the inhibition of myoblast differentiation by Id proteins. Mol Cell Biol. 2002;22:2893–2905. doi: 10.1128/MCB.22.9.2893-2905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Kong W, Ilalov K, Yu S, Xu K, Prazak L, Fajardo M, Sehgal B, Di Cesare PE. ADAMTS-7: a metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. 2006a;20:988–990. doi: 10.1096/fj.05-3877fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Kong W, Xu K, Luan Y, Ilalov K, Sehgal B, Yu S, Howell RD, Di Cesare PE. ADAMTS-12 associates with and degrades cartilage oligomeric matrix protein. J Biol Chem. 2006b;281:15800–15808. doi: 10.1074/jbc.M513433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Wang H, Lengyel P. The interferon-inducible nucleolar p204 protein binds the ribosomal RNA- specific UBF1 transcription factor and inhibits ribosomal RNA transcription. Embo J. 1999;18:2845–2854. doi: 10.1093/emboj/18.10.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmander LS, Ionescu M, Jugessur H, Poole AR. Changes in joint cartilage aggrecan after knee injury and in osteoarthritis. Arthritis Rheum. 1999;42:534–544. doi: 10.1002/1529-0131(199904)42:3<534::AID-ANR19>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Luan Y, Kong L, Howell DR, Ilalov K, Fajardo M, Bai XH, Di Cesare PE, Goldring MB, Abramson SB, Liu CJ. Inhibition of ADAMTS-7 and ADAMTS-12 degradation of cartilage oligomeric matrix protein by alpha-2-macroglobulin. Osteoarthritis Cartilage. 2008;16:1413–1420. doi: 10.1016/j.joca.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HH, Ozbek S, Engel J, Paulsson M, Wagener R. Interactions between the cartilage oligomeric matrix protein and matrilins. Implications for matrix assembly and the pathogenesis of chondrodysplasias. J Biol Chem. 2004;279:25294–25298. doi: 10.1074/jbc.M403778200. [DOI] [PubMed] [Google Scholar]
- Mansson B, Carey D, Alini M, Ionescu M, Rosenberg LC, Poole AR, Heinegard D, Saxne T. Cartilage and bone metabolism in rheumatoid arthritis. Differences between rapid and slow progression of disease identified by serum markers of cartilage metabolism. J Clin Invest. 1995;95:1071–1077. doi: 10.1172/JCI117753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu E, Meheus L, Raymackers J, Merregaert J. Characterization of the osteogenic stromal cell line MN7: identification of secreted MN7 proteins using two-dimensional polyacrylamide gel electrophoresis, western blotting, and microsequencing. J Bone Miner Res. 1994;9:903–913. doi: 10.1002/jbmr.5650090616. [DOI] [PubMed] [Google Scholar]
- Mirancea N, Hausser I, Beck R, Metze D, Fusenig NE, Breitkreutz D. Vascular anomalies in lipoid proteinosis (hyalinosis cutis et mucosae): basement membrane components and ultrastructure. J Dermatol Sci. 2006;42:231–239. doi: 10.1016/j.jdermsci.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Mirancea N, Hausser I, Metze D, Stark HJ, Boukamp P, Breitkreutz D. Junctional basement membrane anomalies of skin and mucosa in lipoid proteinosis (hyalinosis cutis et mucosae) J Dermatol Sci. 2007;45:175–185. doi: 10.1016/j.jdermsci.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Misumi K, Vilim V, Hatazoe T, Murata T, Fujiki M, Oka T, Sakamoto H, Carter SD. Serum level of cartilage oligomeric matrix protein (COMP) in equine osteoarthritis. Equine Vet J. 2002;34:602–608. doi: 10.2746/042516402776180205. [DOI] [PubMed] [Google Scholar]
- Mongiat M, Fu J, Oldershaw R, Greenhalgh R, Gown AM, Iozzo RV. Perlecan protein core interacts with extracellular matrix protein 1 (ECM1), a glycoprotein involved in bone formation and angiogenesis. J Biol Chem. 2003;278:17491–17499. doi: 10.1074/jbc.M210529200. [DOI] [PubMed] [Google Scholar]
- Morgelin M, Engel J, Heinegard D, Paulsson M. Proteoglycans from the swarm rat chondrosarcoma. Structure of the aggregates extracted with associative and dissociative solvents as revealed by electron microscopy. J Biol Chem. 1992;267:14275–14284. [PubMed] [Google Scholar]
- Neidhart M. Elevated serum prolactin or elevated prolactin/cortisol ratio are associated with autoimmune processes in systemic lupus erythematosus and other connective tissue diseases. J Rheumatol. 1996;23:476–481. [PubMed] [Google Scholar]
- Neidhart M, Hauser N, Paulsson M, DiCesare PE, Michel BA, Hauselmann HJ. Small fragments of cartilage oligomeric matrix protein in synovial fluid and serum as markers for cartilage degradation. Br J Rheumatol. 1997;36:1151–1160. doi: 10.1093/rheumatology/36.11.1151. [DOI] [PubMed] [Google Scholar]
- Oldberg A, Antonsson P, Lindblom K, Heinegard D. COMP (cartilage oligomeric matrix protein) is structurally related to the thrombospondins. J Biol Chem. 1992;267:22346–22350. [PubMed] [Google Scholar]
- Oyama N, Chan I, Neill SM, Hamada T, South AP, Wessagowit V, Wojnarowska F, D'Cruz D, Hughes GJ, Black MM, McGrath JA. Autoantibodies to extracellular matrix protein 1 in lichen sclerosus. Lancet. 2003;362:118–123. doi: 10.1016/S0140-6736(03)13863-9. [DOI] [PubMed] [Google Scholar]
- Oyama N, Chan I, Neill SM, South AP, Wojnarowska F, Kawakami Y, D'Cruz D, Mepani K, Hughes GJ, Bhogal BS, Kaneko F, Black MM, McGrath JA. Development of antigen-specific ELISA for circulating autoantibodies to extracellular matrix protein 1 in lichen sclerosus. J Clin Invest. 2004;113:1550–1559. doi: 10.1172/JCI20373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbek S, Engel J, Stetefeld J. Storage function of cartilage oligomeric matrix protein: the crystal structure of the coiled-coil domain in complex with vitamin D(3) Embo J. 2002;21:5960–5968. doi: 10.1093/emboj/cdf628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson IF, Boegard T, Svensson B, Heinegard D, Saxne T. Changes in cartilage and bone metabolism identified by serum markers in early osteoarthritis of the knee joint. Br J Rheumatol. 1998;37:46–50. doi: 10.1093/rheumatology/37.1.46. [DOI] [PubMed] [Google Scholar]
- Rosenberg K, Olsson H, Morgelin M, Heinegard D. Cartilage oligomeric matrix protein shows high affinity zinc-dependent interaction with triple helical collagen. J Biol Chem. 1998;273:20397–20403. doi: 10.1074/jbc.273.32.20397. [DOI] [PubMed] [Google Scholar]
- Saxne T, Heinegard D. Cartilage oligomeric matrix protein: a novel marker of cartilage turnover detectable in synovial fluid and blood. Br J Rheumatol. 1992;31:583–591. doi: 10.1093/rheumatology/31.9.583. [DOI] [PubMed] [Google Scholar]
- Sercu S, Lambeir AM, Steenackers E, El Ghalbzouri A, Geentjens K, Sasaki T, Oyama N, Merregaert J. ECM1 interacts with fibulin-3 and the beta 3 chain of laminin 332 through its serum albumin subdomain-like 2 domain. Matrix Biol. 2009;28:160–169. doi: 10.1016/j.matbio.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Sercu S, Poumay Y, Herphelin F, Liekens J, Beek L, Zwijsen A, Wessagowit V, Huylebroeck D, McGrath JA, Merregaert J. Functional redundancy of extracellular matrix protein 1 in epidermal differentiation. Br J Dermatol. 2007;157:771–775. doi: 10.1111/j.1365-2133.2007.08114.x. [DOI] [PubMed] [Google Scholar]
- Sercu S, Zhang L, Merregaert J. The extracellular matrix protein 1: its molecular interaction and implication in tumor progression. Cancer Invest. 2008a;26:375–384. doi: 10.1080/07357900701788148. [DOI] [PubMed] [Google Scholar]
- Sercu S, Zhang M, Oyama N, Hansen U, Ghalbzouri AE, Jun G, Geentjens K, Zhang L, Merregaert JH. Interaction of extracellular matrix protein 1 with extracellular matrix components: ECM1 is a basement membrane protein of the skin. J Invest Dermatol. 2008b;128:1397–1408. doi: 10.1038/sj.jid.5701231. [DOI] [PubMed] [Google Scholar]
- Smits P, Bhalerao J, Merregaert J. Molecular cloning and characterization of the mouse Ecm1 gene and its 5' regulatory sequences. Gene. 1999;226:253–261. doi: 10.1016/s0378-1119(98)00558-7. [DOI] [PubMed] [Google Scholar]
- Smits P, Ni J, Feng P, Wauters J, Van Hul W, Boutaibi ME, Dillon PJ, Merregaert J. The human extracellular matrix gene 1 (ECM1): genomic structure, cDNA cloning, expression pattern, and chromosomal localization. Genomics. 1997;45:487–495. doi: 10.1006/geno.1997.4918. [DOI] [PubMed] [Google Scholar]
- Susic S, McGrory J, Ahier J, Cole WG. Multiple epiphyseal dysplasia and pseudoachondroplasia due to novel mutations in the calmodulin-like repeats of cartilage oligomeric matrix protein. Clin Genet. 1997;51:219–224. doi: 10.1111/j.1399-0004.1997.tb02458.x. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Vranka J, Mokashi A, Keene DR, Tufa S, Corson G, Sussman M, Horton WA, Maddox K, Sakai L, Bachinger HP. Selective intracellular retention of extracellular matrix proteins and chaperones associated with pseudoachondroplasia. Matrix Biol. 2001;20:439–450. doi: 10.1016/s0945-053x(01)00148-2. [DOI] [PubMed] [Google Scholar]
- Wang L, Yu J, Ni J, Xu XM, Wang J, Ning H, Pei XF, Chen J, Yang S, Underhill CB, Liu L, Liekens J, Merregaert J, Zhang L. Extracellular matrix protein 1 (ECM1) is overexpressed in malignant epithelial tumors. Cancer Lett. 2003;200:57–67. doi: 10.1016/s0304-3835(03)00350-1. [DOI] [PubMed] [Google Scholar]
- Xu K, Zhang Y, Ilalov K, Carlson CS, Feng JQ, Di Cesare PE, Liu CJ. Cartilage oligomeric matrix protein associates with granulin-epithelin precursor (GEP) and potentiates GEP-stimulated chondrocyte proliferation. J Biol Chem. 2007;282:11347–11355. doi: 10.1074/jbc.M608744200. [DOI] [PubMed] [Google Scholar]