Abstract
Calpain 3 is a calcium-dependent cysteine protease that is primarily expressed in skeletal muscle and is implicated in limb girdle muscular dystrophy type 2A. To date, its best characterized function is located within the sarcomere, but this protease is found in other cellular compartments, which suggests that it exerts multiple roles. Here, we present evidence that calpain 3 is involved in the myogenic differentiation process. In the course of in vitro culture of myoblasts to fully differentiated myotubes, a population of quiescent undifferentiated “reserve cells” are maintained. These reserve cells are closely related to satellite cells responsible for adult muscle regeneration. In the present work, we observe that reserve cells express higher levels of endogenous Capn3 mRNA than proliferating myoblasts. We show that calpain 3 participates in the establishment of the pool of reserve cells by decreasing the transcriptional activity of the key myogenic regulator MyoD via proteolysis independently of the ubiquitin-proteasome degradation pathway. Our results identify calpain 3 as a potential new player in the muscular regeneration process by promoting renewal of the satellite cell compartment.
Keywords: Cell/Differentiation, Development Differentiation/Muscle, Diseases/Muscular Dystrophy, Protease/Calpain, Protein/Degradation, Stem Cells, Transcription/Helix-loop-helix, Transcription/Tissue-specific Factors
Introduction
Adult skeletal muscle is highly adaptable to stress, in part due to its remarkable regenerative capacity. This property is mostly attributed to the presence of satellite cells, which are undifferentiated mononucleated muscle precursor cells that are arrested in a G0-quiescent state and located beneath the basal lamina of myofibers (reviewed in Ref. 1). In response to injury, stretch, or exercise, satellite cells become activated, express MyoD, reenter the cell cycle, and proliferate (2–4). After several rounds of proliferation, the majority of cells differentiate and fuse to form new myofibers or to repair damaged ones. The remaining cells return to quiescence and participate in the replenishment of the initial population of satellite cells in which MyoD is down-regulated (5–8).
During in vitro differentiation of the C2C12 cell line, a large subpopulation of myoblasts undergoes terminal differentiation and forms myotubes. A subset of cells, called reserve cells (9, 10), remains undifferentiated and returns to quiescence. These reserve cells can reenter the cell cycle upon the addition of mitogenic serum and generate progeny that form myotubes and reserve cells. The characteristics of quiescence, self-renewal, and generation of myotubes are shared by both reserve cells and the resident satellite cells of muscle fibers (5, 10, 11).
The myogenic differentiation of satellite cells and their myoblast progeny is regulated by various transcription factors, including the paired box transcription factors Pax7 and Pax3 and the myogenic regulatory factors (MRFs) Myf5, MyoD, myogenin, and MRF4 (reviewed in Refs. 12–14). MyoD has been widely studied (reviewed in Ref. 15), and its tight regulation is crucial for the balance between proliferation and withdrawal from the cell cycle, leading to terminal differentiation or quiescence (16, 17). This regulation of MyoD involves various transcriptional and post-translational regulatory mechanisms that affect MyoD either directly or indirectly (18–23).
Calpain 3 is a member of the calpain family, a group of non-lysosomal calcium-dependent cysteine proteases (reviewed in Ref. 24). It is mainly expressed in skeletal muscle, but several differentially spliced variants are also present in skeletal muscle and non-muscle cells (25). Calpain 3 has four domains (designated I–IV) common to the ubiquitous calpain 1 and 2 (26, 27), and three additional specific sequence inserts (NS, IS1, and IS2) that confer characteristics such as autocatalysis and nuclear translocation (28–33).
Calpain 3 plays an important role in skeletal muscle homeostasis. Mutations in the human calpain 3 gene (CAPN3) were identified as being responsible for limb girdle muscular dystrophy type 2A, an autosomal recessive disorder that is mainly characterized by selective and progressive weakness and atrophy of proximal limb girdle and trunk muscles (34, 35). Numerous studies have attempted to identify potential substrates of calpain 3 (reviewed in Ref. 36); however, despite intense research, the physiological functions of calpain 3 and its pathophysiological implications remain elusive. Many studies have demonstrated the role of calpain 3 in the maintenance of sarcomere integrity via the regulation of sarcomeric protein turnover (37–40). Calpain 3 has also been observed in other cellular compartments besides the sarcomere, such as the subsarcolemmal membrane (41, 42), the triad-associated protein complex (43), or the nucleus (28, 31, 32, 44, 45), which suggests that calpain 3 has multiple functions within the muscle tissue. In this study, we present evidence that calpain 3 plays a role in myogenic differentiation, and more specifically in the maintenance of the pool of reserve cells.
Using the C2C12 cell line as a model to study muscle differentiation, we show that calpain 3 is implicated in the early phases of myogenic differentiation, where it participates in the generation of a pool of reserve cells. Calpain 3 is involved in the regulation of the myogenic regulatory factor MyoD by inducing its destabilization and leading myoblasts to quiescence. Altogether, our results strongly suggest that calpain 3 promotes the generation of reserve cells by down-regulating MyoD. In the context of muscular regeneration, these results highlight a potential role of calpain 3 in the maintenance of a pool of satellite cells.
EXPERIMENTAL PROCEDURES
Expression Constructs
The plasmids expressing MyoD, Myf5, MRF4, and myogenin were constructed in pEMSV as described previously (46). The plasmid pEMSV-CAPN3 was obtained by inserting into the EcoRI site of pEMSV vector the mouse Capn3 cDNA (generously provided by I. Richard (Généthon, CNRS-FRE 3018, France)). The plasmids encoding for the different forms of hemagglutinin-tagged MyoD proteins (pCDNA3.1-HA5-MyoD WT, pCDNA3.1-HA-MyoD K133R, pCDNA3.1-HA-MyoD S200A, and pCDNA3.1-HA-MyoD K133R/S200A) were obtained as described previously (18, 47). The plasmid pcDNA3.1-eGFP has been generously provided by M. Ventura (CNRS-Université Bordeaux 2, UMR-5234, France). The plasmid pMCK-Luc contains the muscle creatine kinase (MCK) gene regulatory sequences (from position −1266 to +1 bp relative to the start of transcription) driving luciferase expression (48). pTK-4E-CAT plasmid contains the thymidine kinase minimal promoter and four copies of the right E-boxes from the MCK enhancer driving the expression of the chloramphenicol acetyltransferase (49).
Cell Culture and Transfections
C2C12 myoblasts and C3H10T1/2 fibroblast cells were cultured in gelatin-coated dishes in growth medium (GM) supplemented with 10% fetal bovine serum (FBS), 1 mm l-glutamine, and antibiotics (penicillin/streptomycin, Invitrogen) in Dulbecco's modified Eagle's medium at 37 °C and 5% CO2 in a humidified chamber. Myogenic differentiation was induced on confluent cultured cells by changing the growth medium to differentiation medium, supplemented with 2% horse serum and antibiotics (penicillin/streptomycin, Invitrogen) in Dulbecco's modified Eagle's medium. Transient transfections of C2C12 and C3H10T1/2 cells with plasmids were performed using Lipofectamine 2000 reagent according to the manufacturer's instructions (Invitrogen).
Isolation of Reserve Cells and Myotubes
After 4 days of incubation in differentiation medium, C2C12 cultures were separated into undifferentiated reserve cells and myotubes by mild trypsinization (16). Briefly, myotubes were specifically detached by mild trypsinization (0.025% trypsin, 0.1 mm EDTA, 3 min, in Dulbecco's modified Eagle's medium), whereas undifferentiated reserve cells remained adherent to the dish. These cells were then all detached by trypsinization (0.25% trypsin/1 mm EDTA) and reseeded onto a new dish in growth medium. After 30 min, the fragments of resting floating myotubes were removed and reserve cells that had attached to the culture dish were collected by trypsinization (10).
Evaluation of Myoblast Fusion
At various times after inducing differentiation, C2C12 cells were rinsed and fixed at room temperature in 4% paraformaldehyde for 15 min, and nuclei were stained with Hansen's hemalun. The extent of fusion was determined as already reported (50). Fusion (%) = (number of nuclei in myotubes)/(total number of nuclei in myoblasts and myotubes) × 100.
Cell Cycle Analysis
12 h after transfection, the cells were trypsinized and redistributed at low density in the number of necessary plates for the experiment. For 5-bromo-2′-deoxyuridine (BrdUrd) labeling, cells were maintained in GM containing 5% FBS, and 10 μm BrdUrd was added in culture medium 6 h prior to analysis. In Ki-67 labeling experiments, cells were maintained in differentiation medium. In both cases, cells were harvested 36 h after replating, washed with PBS, fixed, and permeabilized with 3.7% paraformaldehyde, 0.05% Triton X-100 in PBS for 15 min at room temperature. The cells were washed with PBS plus 0.5% BSA and incubated for 2 h at 37 °C in PBS plus 0.5% BSA containing the corresponding primary antibodies as indicated in Tables 1 and 2: mouse anti-BrdUrd diluted 1:60 (Thermo Scientific) or goat anti-Ki-67 (M19) diluted 1:80 (Santa Cruz Biotechnology) and mouse anti-MyoD (5.8A) diluted 1:100 (Abcam). The cells were washed with PBS plus 0.5% BSA and then incubated for 30 min at room temperature with the corresponding secondary conjugated antibodies: Alexa Fluor 488-conjugated anti-mouse IgG antibody diluted 1:500 and Alexa Fluor 647-conjugated anti-rabbit IgG antibody diluted 1:500 (Molecular Probes). Finally, cells were washed with PBS plus 0.5% BSA, followed by flow cytometric analysis with a BD Biosciences FACSCanto II flow cytometer.
TABLE 1.
Effect of calpain 3 overexpression on the cell cycle progression and the induction of myogenic differentiation (n = 4)
| C2C12 transfected with | Percentage of total cells |
||
|---|---|---|---|
| Proliferation (5% FBS), BrdUrd+ | Differentiation (2% horse serum) |
||
| Ki-67−/MyoD+ | Ki-67−/MyoD− | ||
| % | % | % | |
| pEMSV | 61.2 ± 3.9 | 57.8 ± 2.8 | 15.1 ± 2.2 |
| pEMSV-CAPN3 | 30.6 ± 4.3a | 38.5 ± 2.5a | 45.7 ± 3.1a |
a Significantly different in the ANOVA test (p < 0.05).
TABLE 2.
Effect of the down-regulation of Capn3 expression level on the cell cycle progression and the induction of myogenic differentiation (n = 4)
| C2C12 transfected with | Percentage of total cells |
||
|---|---|---|---|
| Proliferation (5% FBS), BrdUrd+ | Differentiation (2% horse serum) |
||
| Ki-67−/MyoD+ | Ki-67−/MyoD− | ||
| % | % | % | |
| siRNA negative control | 59.4 ± 2.9 | 55.6 ± 3.4 | 15.4 ± 3.3 |
| siRNApool-CAPN3 | 61.7 ± 2.7 | 66.7 ± 3.1a | 6.3 ± 2.1a |
a Significantly different in the ANOVA test (p < 0.05).
Silencing of Capn3 by siRNA
To inhibit the expression of Capn3, short interfering RNA (siRNA) duplexes (Invitrogen) corresponding to three distinct regions of the DNA sequence of the mouse Capn3 gene (NM_007601) were used: ACCCUUCUCACCUUUGAACAGGGC, ACAGUUGGACUAAUAACCGUUGGC, and AUCCAUAUUUCUCACCAUCCGUGC. A nonspecific siRNA was used as negative control. (Stealth siRNA negative control duplex, medium GC, Invitrogen). Lamin A/C-targeting siRNA (siGLO lamina/C-mouse, Dharmacon) was used as a positive control and to measure transfection efficiency. C2C12 myoblasts at 80% confluence were transfected using DharmaFect 3 reagent (Dharmacon) with an equivalent mixed combination of Capn3 siRNA duplexes at 50 nm (siRNApool-CAPN3) or with siRNA negative control (50 nm) according to the manufacturer's instructions.
Western Blots
For immunoblot analysis, cells were solubilized in lysis buffer (20 mm Tris, pH 8, 150 mm NaCl, 0.25% deoxycholate, 1% Triton X-100, 10 mm orthovanadate, 2.5 mm EDTA, 2.5% EGTA, 30 mm sodium pyrophosphate, 2 mm phenylmethylsulfonyl fluoride, 1 μm dithiothreitol, 1× protease inhibitor mixture (Roche Applied Science) and then sonicated briefly and lysed for 30 min at 4 °C. Afterward, the cell lysate was centrifuged at 12,000 × g for 10 min at 4 °C to remove cellular debris. Protein concentration of each freshly prepared cell lysate was determined by the Bradford method (Coo Protein Assay Reagent, Interchim). After treatment with 5× SDS loading buffer, whole cell extracts were resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membrane (Immobilon-P, Millipore) by the semidry transfer method.
After blocking with Advance blocking agent (Amersham Biosciences) in Tris-buffered saline plus 0.1% Tween, membranes were probed overnight at 4 °C with the following primary antibodies: rabbit anti-MyoD (C20) diluted 1:1,000, mouse anti-myogenin (F5D) diluted 1:800, rabbit anti-MRF4 (C19) diluted 1:1,000, rabbit anti-Myf5 (C20) diluted 1:1,000, and rabbit anti-β-tubulin (H235) diluted 1:2,500 (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)); mouse anti-GAPDH (G8795) diluted 1:30,000, mouse anti-HA (H3663) diluted 1:5,000, and mouse anti-myosin heavy chain (M4276) diluted 1:700 (Sigma); mouse anti-calpain 3 (12A2) diluted 1:800 (Novocastra); and mouse anti-Pax7 diluted 1:500 (Developmental Studies Hybridoma Bank). After several washes in Tris-buffered saline, membranes were incubated with a secondary horseradish peroxidase-conjugated antibody: anti-mouse horseradish peroxidase diluted 1:100,000 and anti-rabbit horseradish peroxidase diluted 1:100,000 (Amersham Biosciences). After several washes with Tris-buffered saline, 0.1% Tween, proteins were detected by incubation with an enhanced chemiluminescence system (Advance ECL, Amersham Biosciences) according to the manufacturer's instructions and exposed to Hyperfilm ECL (Amersham Biosciences). Films were scanned (GelScan, Amersham Biosciences), and protein quantification was performed using TotalLab (Nonlinear Dynamics). The number of pixels for each protein of interest was normalized with respect to GAPDH or β-tubulin protein level.
RNA Extraction, Reverse Transcription, and Quantitative PCR
Total RNA was isolated from C2C12 cells using the SV total isolation system (Promega) according to the manufacturer's instructions. 100 ng of total RNA was used to perform reverse transcription with Improm-II reverse transcriptase, oligo(dT), and random hexamer primer according to the manufacturer's instructions (Promega). Real-time PCR was carried out using a SYBR protocol (Quantitect SYBR Green PCR kit and Quantitect Primer assay from Qiagen designed for amplifying mouse MyoD, myogenin, calpain 3, and GAPDH genes) according to the manufacturer's instructions, on a fluorescence temperature cycler (light cycler, Roche Applied Science). Each real-time reaction was carried out in triplicate and repeated several times. Results were calculated as relative differences in target threshold cycle (Ct) values normalized to GAPDH according to the method of Pfaffl (51).
Luciferase and Chloramphenicol Acetyltransferase (CAT) Assay
For the luciferase assay, C3H10T1/2 cells were collected 48 h after transfection, and luciferase activity was measured in triplicates using the Dual Luciferase reporter assay system (Promega) according to the manufacturer's instructions. Luciferase activity was estimated by measuring light emission resulting from the conversion of luciferin by luciferase with a luminometer (Lumat LB9507, Berthold). The values obtained for the luciferase assays were corrected for transfection efficiency by transfecting the plasmid pRLTK-Luc, included in the Dual Luciferase reporter assay system (Promega). For the CAT assay, C3H10T1/2 cells were collected, and the level of CAT protein was determined in triplicates using an enzymatic immunoassay (CAT ELISA, Roche Applied Science) according to the manufacturer's instructions. CAT expression was measured with a microplate reader (Molecular Devices).
Immunofluorescence Microscopy
Cells were cultured on coverslips or in the Lab-Tek™ Chamber Slide™ system in Permanox™ (Nunc) and fixed with 4% paraformaldehyde in PBS for 15 min and then permeabilized with 1% Triton X-100 in PBS for 15 min at room temperature. Cells were blocked in PBS plus 10% FBS for 2 h at room temperature. Cells were then incubated overnight 4 °C with primary antibodies mouse anti-MyoD (5.8A, Abcam) and rabbit anti-calpain 3 (RP1, Triple Point Technology) diluted 1:100 and 1:800, respectively, in PBS plus 2% FBS. After several washes in PBS, the cells were then incubated with secondary antibodies diluted in PBS plus 2% FBS for 1 h at room temperature: Alexa Fluor 488-conjugated donkey anti-rabbit IgG antibody diluted 1:2,500 and Alexa Fluor 594-conjugated donkey anti-mouse IgG antibody diluted 1:2,500 (Molecular Probes). After several washes in PBS, cells were mounted in low fluorescence mounting medium (Dako) and examined by laser-scanning confocal microscopy (Olympus BX512 Fluoview 500). Pictures were then analyzed with the MetaMorph software (Molecular Devices).
Cycloheximide Treatment
For half-life determination, 24 h after transfection, C2C12 cells were trypsinized and redistributed at low density in a 60-mm dish. Then C2C12 cells were cultured in GM for 24 h before being incubated with cycloheximide (Sigma) at 50 μg/ml for the indicated times and then harvested for Western blot detection of HA-MyoD and GAPDH as described above. The protein amount of HA-MyoD for each sample was normalized with respect to GAPDH protein level. Data were graphed using Statgraphics software (Statpoint Technologies).
Statistical Analysis
All statistical analyses were done using Statgraphics software (Statpoint Technologies). In most of the cases, the ANOVA test or Student's t test was used. In some cases, a Kruskal-Wallis test was used.
RESULTS
Role of Calpain 3 during Myogenic Differentiation of C2C12 Myoblastic Cells
The C2C12 muscle cell line was used as a model to investigate a potential role of calpain 3 in the myogenic differentiation process.
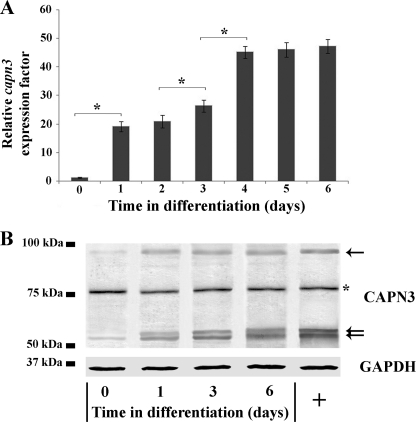
First, the endogenous mRNA level of Capn3 was measured by quantitative RT-PCR (qRT-PCR) in the course of C2C12 differentiation. As shown in Fig. 1A, Capn3 expression was biphasic. During myoblast proliferation, Capn3 mRNA was faintly detectable; however, expression increased by over 19-fold on day 1 at the onset of differentiation. Between days 3 and 4, Capn3 mRNA expression further increased by about 1.8-fold and reached a plateau at terminal differentiation (day 6). Elsewhere, our Western blot experiments showed that calpain 3 protein increased overall during the first 24 h after the induction of differentiation (Fig. 1B). These observations show that calpain 3 is subjected to a transcriptional activation, leading to an accumulation of the calpain 3 protein from early C2C12 myoblast differentiation.
FIGURE 1.
Endogenous expression of Capn3 during myogenic differentiation of C2C12 cells. A, proliferating (day 0) and differentiating C2C12 cells maintained in differentiation for various lengths of time (days 1–6) were harvested, and total RNA was extracted. The expression level of Capn3 was determined by qRT-PCR, as described under “Experimental Procedures.” Relative Capn3 expression is expressed so that 1 represents the Capn3 expression level at day 0 (proliferation). Data are presented as mean ± S.D. (n = 3). Asterisks indicate data that are significantly different according to the ANOVA test for p < 0.05. B, proliferating (day 0) and differentiating C2C12 cells maintained in differentiation for various times (days 1–6) were harvested, and total cell extracts were prepared. 40 μg of cell extract was analyzed by SDS-PAGE and immunoblotting with antibodies specific for calpain 3 and GAPDH. In our experiments, calpain 3 protein is visualized in Western blot as three major specific bands (black arrows): one band at 94 kDa corresponding to the full-length protein and two autoproteolytic products between 60 and 55 kDa corresponding to the autolyzed and active forms of the protease (fragments IS1-Cter). The asterisk indicates a nonspecific band around 75 kDa, which could correspond to the ubiquitous calpains 1 and 2 (71). Specificity of the calpain 3 antibody used has been evaluated by the comparison of Western blot profiles that we obtained with calpain 3-overexpressing cells (right lane, +) and control cells.
To study the impact of calpain 3 in the early phases of differentiation, we transiently overexpressed calpain 3 in C2C12 myoblasts using a plasmid coding for mouse calpain 3 (supplemental Fig. S1) and then induced myogenic differentiation. After 6 days of differentiation, C2C12 cells transfected with a calpain 3 expression vector showed a reduced final number of myotubes, whereas the number of resting cells increased (Fig. 2A). These findings were further detailed with the analysis of the fusion index. In control experiments (Fig. 2B, gray curve), the C2C12 myoblasts began to fuse after 2 days; 50% of myotubes were formed (C50) by day 4, and they were fully differentiated by day 7, where 60% of the nuclei were in multinucleated myotubes. In contrast, C2C12 myoblasts overexpressing calpain 3 started to fuse on day 3, reached the C50 value by day 5, and had a lower maximal level of fusion (46%) on day 8 (Fig. 2B, black curve).
FIGURE 2.
Effect of calpain 3 on myogenic differentiation of C2C12 cells. A–F, C2C12 cells were transfected with plasmid encoding calpain 3 (pEMSV-CAPN3) or a pool of three Capn3 siRNAs (siRNApool-CAPN3) and the respective negative controls that are the empty corresponding vector pEMSV and nonspecific siRNA controls (siRNA negative control). A and E, phase-contrast microscopy images were taken and compared after 6 days of differentiation. Two independent experiments were performed, and about 10 images were compared in each experiment and for each condition. Scale bars, 50 μm. B and F, the effects of calpain 3 overexpression (B) and Capn3 knockdown (F) were evaluated by analyzing the fusion curves obtained as described under “Experimental Procedures.” The slopes of fusion curves were determined by the tangential at the C50 point, when 50% of the myotubes were formed. Fusion curves were obtained from three independent experiments. C, differentiation was induced 24 h after transfection, and cells were harvested at different times, as indicated in the figures. J1, J3, and J4, and total cell extract were prepared and analyzed by SDS-PAGE and immunoblotting with antibodies specific for myosin heavy chain (MHC) and myogenin, which serve as differentiation markers of the myogenic lineage, and β-tubulin, which serves as a control. D, relative amounts of the studied proteins were expressed such that 100% is the protein amount in control cells at day 3. All values were normalized with regard to the β-tubulin protein level (n = 3). Asterisks indicate data that are significantly different from control cells by Student's t test, with p values of <0.05 determining statistical significance.
This defect in myogenic differentiation was confirmed by the analysis of several muscle markers during the course of differentiation. Results present in Fig. 2, C and D, showed that calpain 3 overexpression leads to a delay in the expression of both early (myogenin) and late differentiation markers (myosin heavy chain). Hence, calpain 3 overexpression negatively affects the differentiation process from the earliest steps.
To further determine the endogenous role of calpain 3 in myogenic differentiation, we down-regulated its expression level in myoblasts using siRNA (supplemental Fig. S2). Down-regulation of Capn3 led to a greater number of myotubes observed after 6 days of differentiation (Fig. 2E). We then observed the impact on myogenic differentiation by measuring the fusion index in differentiating C2C12 cells treated or not with calpain 3-specific siRNA (Fig. 2F). When calpain 3 was down-regulated, C2C12 started to fuse earlier (from day 1) with a C50 reached more rapidly (day 3), and the maximum level of fusion reached was increased to 79%. This increase in the myogenic differentiation was also confirmed by a Western blot analysis of myogenin and myosin heavy chain as myogenic differentiation markers, showing that they were both expressed earlier, when Capn3 was down-regulated (data not shown). Altogether, these results show an involvement of calpain 3 in the early steps of myogenic differentiation.
Calpain 3 Promotes the Generation of Reserve Cells during C2C12 Myogenic Differentiation
The initiation of differentiation is closely related to the regulation of myoblast cell cycle, and cells in the G1 phase have distinct cell fate choices between proliferation, commitment to differentiation, and entrance into quiescence. C2C12 myoblasts that had been transfected with a plasmid encoding calpain 3 and maintained in a proliferation state were incubated with BrdUrd, a marker of S phase entry, and analyzed by flow cytometry. Calpain 3 overexpression induced a significant decrease of 50% in the proportion of BrdUrd-labeled cells (from 61.2 to 30.6%; Table 1), suggesting that calpain 3 could be implicated in G0/G1 arrest. Cell cycle arrested cells may have entered either into differentiation or into quiescence. To distinguish between these two possibilities, cells were analyzed by flow cytometry for MyoD level. Indeed, MyoD is expressed at high levels in differentiating cells (16) but drops to undetectable levels in quiescent undifferentiating cells (10). As a control, we analyzed the expression of Ki-67, which is a marker of cell proliferation expressed during G1, S, G2, and M phases but absent in cell cycle-arrested cells. Flow cytometry analysis of differentiating C2C12 cells revealed that calpain 3 overexpression decreased by 1.5-fold the proportion of differentiating cells (Ki-67−/MyoD+ cells, from 57.8 to 38.5%; Table 1), whereas the proportion of non-differentiating cells that have exited the cell cycle was increased of about 3-fold (Ki-67−/MyoD−, from 15.1 to 45.7%; Table 1). These data suggest that overexpression of calpain 3 in myoblasts inhibits early differentiation by inducing cell cycle arrest without commitment to differentiation.
Moreover, to study the impact of Capn3 down-regulation on myogenic differentiation commitment, similar flow cytometric analyses were performed. Myoblasts transfected with or without Capn3 siRNA were induced to differentiate, and cells were analyzed for expression of Ki-67 and MyoD. We found that down-regulation of Capn3 reduced by nearly 2.5-fold the proportion of non-differentiating cells that have exited the cell cycle, probably corresponding to quiescent cells (Ki-67−/MyoD−, from 15.4 to 6.3%; Table 2), whereas the proportion of cells that have initiated differentiation was increased of about 1.2-fold (Ki-67−/MyoD+, from 55.6 to 66.7%; Table 2). Elsewhere, down-regulation of Capn3 did not significantly affect the myoblast proliferation as measured by BrdUrd labeling (Table 2).
Altogether, these results show that calpain 3 plays a role in the balance between myoblast differentiation and the maintenance of undifferentiated quiescent cells in vitro.
To investigate further this role of calpain 3, we focused on three different subpopulations of proliferating and differentiating C2C12 cells, myoblasts, myotubes, and reserve cells (10) that are undifferentiated quiescent cells (Fig. 3A).
FIGURE 3.
Characterization of reserve cells and endogenous expression of Capn3. A, schematic representation of protocol used to study the different subpopulations of C2C12 cells. C2C12 myoblasts, transfected or not, were maintained in proliferation for 24 h. Differentiation was then induced over 4 days, and the populations of myotubes and reserve cells were separated, as described under “Experimental Procedures.” B, 40 μg of total cell extract of each subpopulation (myoblasts, reserve cells, and myotubes), just as reserve cells that had been put back in proliferative medium for 12 h (reserve cells + 12 h), were analyzed by SDS-PAGE and immunoblotting with antibodies specific for MyoD, Pax7, myogenin, and β-tubulin. C and D, the relative endogenous Capn3 and myogenin expression levels were quantified in the three populations by qRT-PCR, as described under “Experimental Procedures.” The relative expression level of Capn3 or myogenin is presented such that 1 indicates the expression level in myoblasts. The data are presented as mean ± S.D. (n = 3). The single asterisks indicate data that are significantly different according to an ANOVA test for p < 0.05.
These subpopulations were first characterized. Myotubes and reserve cells can be easily separated by brief and mild trypsinization as described previously (16). Western blot analyses of these three different subpopulations revealed that the reserve cell fraction contained significantly smaller amounts of MyoD and a higher amount of Pax7 than the myoblast fraction. Moreover, unlike myotubes, reserve cells did not express myogenin (Fig. 3B). We thus confirmed the major hallmarks of the expression pattern of these already described reserve cells (6, 10, 11). Moreover, as previously reported (10), when these reserve cells were put back in proliferative medium and allowed to grow for 12 h, they changed their phenotype and gave rise to proliferative myoblasts characterized by an up-regulation of MyoD expression (Fig. 3B). At last, these new myoblasts were able to differentiate, showing that reserve cells maintained the capacity for self-renewal. Thus, these reserve cells can be considered as a peculiar subpopulation of C2C12 cells and do not correspond to cells presenting a defect in the differentiation program.
To gain insight into the role of endogenous calpain 3, we performed qRT-PCR analysis on the different subpopulations (i.e. myoblasts, myotubes, and reserve cells). In agreement with our previous results (Fig. 1A), the Capn3 expression level was found to be 40-fold higher in myotubes than in proliferative myoblasts (Fig. 3C). Interestingly, the Capn3 relative expression level was about 7-fold higher in reserve cells than in proliferating myoblasts (Fig. 3C). Analysis of myogenin expression level showed that both reserve cells and myoblasts did not express a significant level of it, contrary to myotubes, demonstrating that the myotube and the reserve cells fractions had been properly separated (Fig. 3D). Besides its strong expression level in myotubes, these results could suggest a peculiar role for calpain 3 in reserve cells.
To define the role of calpain 3, we up- or down-regulated the cellular content of calpain 3 in myoblasts (Fig. 3A) and analyzed the effects on the subpopulations of myoblasts, reserve cells, and myotubes. After isolation, these subpopulations were characterized as described above before further analyses.
As we have previously observed (Fig. 2, A and E, and Tables 1 and 2), besides its effect on myoblast differentiation, the ectopic modification of Capn3 expression level affected the number of undifferentiated resting cells, which probably corresponded to reserve cells. Thus, we first evaluated the effect of such forced expression or repression on the generation of reserve cells by counting them after 4 days of differentiation. We found that calpain 3 overexpression induced a 23% increase in the number of reserve cells (Fig. 4A), whereas down-regulation led to a 30% decrease (Fig. 4B). Thus, calpain 3 appears to be positively implicated in the establishment of this peculiar subpopulation.
FIGURE 4.
Effect of calpain 3 on the apparition of reserve cells during C2C12 differentiation. A and B, C2C12 cells were transfected with plasmid encoding calpain 3 (pEMSV-CAPN3) or a pool of three Capn3 siRNAs (siRNApool-CAPN3) and the respective negative controls. After 4 days of differentiation, reserve cells were isolated and counted with a hemocytometer. Data were analyzed using a box plot graph (n = 3). The double asterisks indicate data that are significantly different according to the Kruskal-Wallis test for p < 0.05.
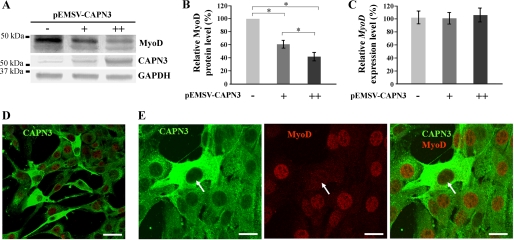
Then, after ectopic up- and down-regulation of Capn3 in myoblasts, we monitored the Capn3 expression levels in the three different subpopulations obtained as described in Fig. 3A. Interestingly, by comparing these subpopulations, drastic differences appeared in the induction -fold of the endogenous Capn3 expression level, resulting from the ectopic overexpression. Indeed, Capn3 was mainly overexpressed in reserve cells (2- and 5-fold greater than in myoblasts and myotubes, respectively) (Fig. 5A). Given that in transient transfection experiments, not all of the cells are effectively transfected (supplemental Fig. S1A), our results indicate that myoblasts, which overexpress calpain 3, will preferentially form reserve cells. In order to support this finding, myoblasts were co-transfected with calpain 3-expressing vector and an expression vector encoding for eGFP. In co-transfected cells, eGFP expression was exclusively found among the population of reserve cells and never in myotubes (Fig. 5C), whereas fluorescence was randomly distributed in control cells transfected with the eGFP plasmid alone (Fig. 5D). These results illustrate the impact of ectopic overexpression of calpain 3 on myoblast fate upon myogenic differentiation and confirm the role of calpain 3 in the subpopulation of reserve cells.
FIGURE 5.
Transcriptional analysis of Capn3 expression level after ectopic up- or down-regulation. A and B, for each subpopulation (i.e. myoblasts, myotubes, and reserve cells) obtained as described in Fig. 3A, the relative mRNA level of Capn3, as quantified by qRT-PCR, was compared between cells in which Capn3 expression had been up-regulated (A) or down-regulated (B) and the respective control cells. For each subpopulation, the -fold induction or repression was expressed such that endogenous Capn3 expression level measured in respective controls cells was 1. In overexpression experiments (A), the -fold induction in myotubes, myoblasts, and reserve cells was 13.2 ± 0.62, 27 ± 2.1, and 68.2 ± 2.2, respectively. In down-regulation experiments (B), the -fold repression in myotubes, myoblasts, and reserve cells was 12.4 ± 0.63, 4.2 ± 0.21, and 1.2 ± 0.25, respectively. The data are presented as mean ± S.D. (n = 3). The asterisks indicate data that are significantly different according to an ANOVA test for p < 0.05. C, C2C12 cells were co-transfected with pEMSV-CAPN3 and a plasmid encoding a reporter gene (eGFP). At 24 h after transfection, C2C12 cells were induced to differentiate over 4 days and observed by fluorescent microscopy in order to visualize the eGFP protein. D, the same experiment was repeated using cells that had been transfected with only pcDNA3.1-eGFP. The images shown in C have been intentionally selected in order to show the same number of myotubes with respect to the control experiment (D). Four independent experiments were performed, and about 20 images were compared in each experiment and for each condition (C and D). Scale bars, 50 μm.
In addition, the -fold reduction of the endogenous Capn3 expression levels induced by ectopic down-regulation (siRNA) was studied in these subpopulations. Fig. 5B indicates that Capn3 was essentially down-regulated in myotubes (3.5- and 10-fold greater than in myoblasts and reserve cells, respectively). As for overexpression experiments, these results mean that myoblasts, in which Capn3 is down-regulated, will mainly give rise to myotubes. Altogether, these results implicate a peculiar role of calpain 3 among the population of reserve cells. We have shown that endogenous calpain 3 expression is induced to a greater extent in reserve cells when compared with proliferating myoblasts. This induction appears to be necessary for the establishment of the pool of quiescent reserve cells. Thus, calpain 3 affects the balance between myoblast differentiation and self-renewal by promoting the establishment of reserve cells.
Calpain 3 Specifically Inhibits the Transactivation Function of MyoD
Because down-regulation of MyoD is a causal event in the generation of reserve cells (10), and because calpain 3 is involved in the apparition of this cell subpopulation, we wondered whether calpain 3 could play a role in the inhibition of MyoD function in myoblasts.
First, we tested if calpain 3 was able to affect the protein level of MyoD. MyoD was expressed in fibroblastic cells (C3H10T1/2) alone or with calpain 3. The three other MRFs (Myf5, MRF4, and myogenin) were also individually assayed. Western blot analyses showed that co-expression of calpain 3 resulted in a significant decrease of the MyoD protein level, whereas the other MRFs were not affected (Fig. 6A).
FIGURE 6.
Effect of calpain 3 on the MRFs. A, C3H10T1/2 cells were transfected with a plasmid encoding one of the four MRFs (pEMSV-Myf5, pEMSV-MRF4, pEMSV-myogenin, or pEMSV-MyoD) by itself or along with pEMSV-CAPN3. Cells were maintained in proliferation medium for 24 h before being harvested. Total cell extract was prepared, and 40 μg was analyzed by SDS-PAGE and immunoblotting with antibodies specific for MyoD, Myf5, MRF4, myogenin, and β-tubulin. B–E, the transcriptional activity of each MRF was measured by analysis of MCK promoter-driven expression of luciferase. At 48 h after transfection, the C3H10T1/2 cells were harvested, and transactivation of the reporter gene was determined by a luciferase assay, as described under “Experimental Procedures.” Luciferase activity is expressed such that 100% indicates the maximum activity measured for each MRF tested without calpain 3. F and G, the transcriptional activity of MyoD was measured by use of a skeletal muscle reporter gene (CAT) under the control of a minimal promoter consisting of four repeats of the E-box element. At 48 h after transfection, the C3H10T1/2 cells were harvested, and transactivation of the reporter gene was determined by a CAT-ELISA assay, as described under “Experimental Procedures.” The amount of CAT is expressed such that 100% indicates the maximum value measured for MyoD without calpain 3. The MRFs and calpain 3 expression plasmids that were transiently transfected into the fibroblast cells (C3H10T1/2) are indicated below the graphs, and +, ++, and +++ indicate the relative amount of plasmid DNA used. The data represent mean ± S.D. (n = 3). The asterisks indicate data that are significantly different according to an ANOVA test for p < 0.05.
These observations prompted us to study more in depth the relationship between calpain 3 and MyoD. We evaluated whether calpain 3 affects MyoD function by measuring its ability to transactivate muscle-specific gene expression; the effect of calpain 3 on Myf5, MRF4, and myogenin was also assayed. We used a luciferase reporter gene containing the mouse muscle creatine kinase promoter (MCK-Luc), which is known to be activated by all MRFs (52–54). In order to avoid possible interference by endogenous proteins, these experiments were performed in C3H10T1/2 fibroblasts, which contain either no or very low levels of MRFs and calpain 3. As expected, all four MRFs enhance the activity of the MCK-Luc reporter gene in C3H10T1/2 cells (Fig. 6, B–E). Calpain 3 overexpression had no effect on the transcriptional stimulation of the luciferase reporter gene by Myf5, myogenin, or MRF4 (Fig. 6, B–D). Interestingly, the transactivation activity of MyoD showed a 60% down-regulation in the presence of calpain 3, suggesting that MyoD might be a primary target of calpain 3 (Fig. 6E).
To confirm this result, we tested the effect of calpain 3 on a reporter construct that contains the chloramphenicol acetyltransferase gene under the control of a minimal promoter consisting of only four repeated E-box elements (49). Overexpression of calpain 3 did not affect the transactivation activity of Myf5, myogenin, and MRF4 (data not shown). However, overexpression of calpain 3 dramatically reduced the activity of MyoD in a dose-dependent manner (Fig. 6F). This calpain-dependent decrease of MyoD transactivation potential was blocked by co-transfection of an increased amount of MyoD expression vector (Fig. 6G).
The ability of MyoD to trigger the myogenic conversion of C3H10T1/2 cells (55) was then evaluated in the presence or absence of overexpressed calpain 3. Myogenic conversion was first assessed by Western blot analysis of the expression of MyoD and myogenin as an early differentiation marker. At day 5 of differentiation, calpain 3-transfected cells showed a 60 and 74% decrease in MyoD and myogenin protein levels, respectively (Fig. 7, A and B). The efficiency of myogenic conversion was also estimated by phase-contrast microscopy analysis. After 24 h of culture in differentiation medium, cells that overexpressed MyoD alone showed the characteristic alignment of differentiating myoblasts (Fig. 7C), whereas cells overexpressing MyoD and calpain 3 showed a stochastic repartition with no marked evidence of aligned cells (Fig. 7D).
FIGURE 7.
Effect of calpain 3 on myogenic conversion of fibroblastic cells induced by ectopic expression of MyoD. A–D, C3H10T1/2 cells were transfected with a plasmid encoding MyoD (pEMSV-MyoD) by itself (control cells) or along with pEMSV-CAPN3. Cells were maintained in proliferation medium for 24 h before inducing differentiation. A and B, after 5 days of differentiation, the cells were harvested, and 40 μg of total extract was analyzed by SDS-PAGE and immunoblotting with antibodies specific for MyoD, myogenin, and GAPDH. All values were normalized to GAPDH protein expression level. Relative protein levels are expressed such that 100% indicates the protein amount in control cells. The data are presented as mean ± S.D. (n = 3). The asterisks indicate data that are significantly different according to Student's test for p < 0.05. C and D, at 24 h after transfection, cells were observed by phase-contrast microscopy. This experiment was repeated three times, and representative images are shown of 20 images captured per experiment. Scale bars, 50 μm.
Altogether, our data demonstrate that calpain 3 can affect the regulation of MyoD, but not Myf5, MRF4, and myogenin, by specific inhibition of the transactivation ability of MyoD. Our data strongly suggest that calpain 3 acts through the direct regulation of MyoD rather than through its numerous cofactors and that calpain 3 affects the protein level of MyoD. Moreover, we have demonstrated that calpain 3 affects the biological function of MyoD in its ability to initiate the myogenic program.
Calpain 3 Down-regulates MyoD Protein Levels
Next, we employed Western blot analysis to evaluate the effect of calpain 3 overexpression on endogenous MyoD levels in C2C12 myoblasts. Quantification of Western blot experiments showed that calpain 3 overexpression induced an overall decrease in the level of MyoD protein (Fig. 8A). This decrease is dose-dependent and ranges from 39 ± 5.9 to 57 ± 6.5% (Fig. 8B). In these experiments, there was no significant effect on the expression of MyoD mRNA, as shown by qRT-PCR analysis (Fig. 8C).
FIGURE 8.
Calpain 3 specifically induces a decrease in the protein level of endogenous MyoD in C2C12 cells. A–E, C2C12 proliferating myoblasts were transfected with either plasmid encoding calpain 3 (pEMSV-CAPN3) or the corresponding empty vector (pEMSV) as control cells. Cells were maintained in proliferation for 24 h and then induced to differentiate before being harvested 24 h later. A and B, total cell extract was prepared, and 40 μg was analyzed by SDS-PAGE and immunoblotting with antibodies specific for MyoD, calpain 3, and GAPDH. All values are normalized to the GAPDH protein level. The relative amount of MyoD protein is expressed such that 100% indicates the protein amount of MyoD in control cells. The data are presented as the mean ± S.D. (n = 6), and asterisks indicate data that are significantly different according to Student's test for p < 0.05. C, the relative MyoD mRNA level was quantified by qRT-PCR, as described under “Experimental Procedures.” The relative expression level of MyoD is expressed such that 100% represents the expression level in control cells. The data are presented as mean ± S.D. (n = 6). D, calpain 3 is visualized by confocal microscopy following immunostaining, as described under “Experimental Procedures.” Cells that effectively overexpress calpain 3 are easily distinguishable (highly green-staining cells) among the total population of transiently transfected C2C12 cells. Scale bar, 50 μm. E, the cellular amount and localization of calpain 3 and endogenous MyoD were studied by immunocytochemical staining and confocal microscopy, as described under “Experimental Procedures.” The white arrows indicate calpain 3-overexpressing cells. Scale bars, 25 μm.
In an attempt to draw a link between MyoD disappearance and calpain 3 overexpression, confocal microscopy analyses were performed. As shown in Fig. 8E, MyoD remained exclusively nuclear, in accordance with its already described pattern (47). As seen in Fig. 8D, cells that overexpressed calpain 3 were easily distinguishable, appearing as highly green stained cells. In these calpain 3-overexpressing cells, the total amount of MyoD decreased, and statistical analysis showed that 94 ± 3.8% of the cells overexpressing calpain 3 presented a dramatic concomitant decrease in MyoD signal intensity (Fig. 8E, white arrows). These results support our previous findings and suggest that calpain 3 may modulate MyoD protein stability.
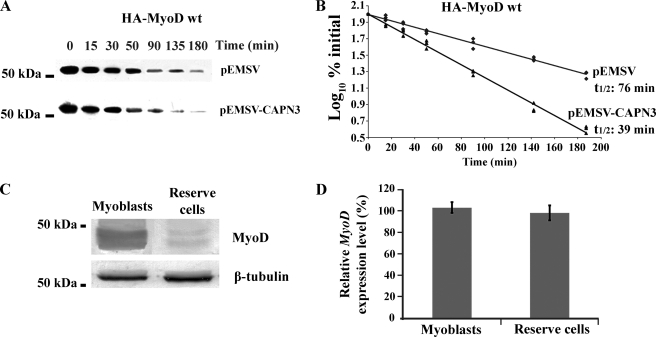
To confirm the increased destabilization of MyoD by calpain 3, we analyzed the effect of calpain 3 overexpression on the half-life of MyoD protein. An expression vector encoding HA-MyoD was transfected into C2C12 cells alone or together with a calpain 3 expression plasmid. Transfected cells were then treated with cycloheximide in order to inhibit further protein synthesis, and the amount of HA-MyoD was determined at various times post-treatment (Fig. 9A). The addition of three HA tags to MyoD (HA-MyoD) did not significantly modify its degradation rate, as was previously shown (18). Co-expression of HA-MyoD with calpain 3 significantly decreased the half-life of HA-MyoD from 76 to 39 min when compared with cells transfected with plasmid vector alone (Fig. 9B). Taken together, these results demonstrate that calpain 3 can increase the rate of MyoD degradation.
FIGURE 9.
Study of MyoD degradation by calpain 3. A and B, C2C12 cells were transfected with pcDNA3.1-HA-MyoD WT, encoding wild type HA-MyoD protein, alone or together with pEMSV-CAPN3, and the half-life of HA-MyoD WT was determined following cycloheximide treatment, as described under “Experimental Procedures.” A, total cell extract was prepared at the indicated times, and 40 μg of extract was analyzed by SDS-PAGE and immunoblotting with antibodies specific for HA and GAPDH. B, HA-MyoD WT protein levels, corrected with respect to GAPDH expression, are expressed such that 100% indicates the protein amount before cycloheximide treatment (t0). For each time point, the log10 of the percentage of pixels (y) is plotted versus the time in minutes (x). The half-life (t½) is determined from the log10 of 50% from the linear regression of three independent experiments (regression equations in the absence or in the presence of calpain 3 overexpression are as follows y = −0.00390x + 1.99712 with R2 = 0.98, and y = −0.00772x + 2.00301 with R2 = 0.99, respectively). C and D, the protein and mRNA expression levels of endogenous MyoD were compared in myoblasts and in reserve cells. C, total cell extract was prepared, and 40 μg was analyzed by SDS-PAGE and immunoblotting with antibodies specific for MyoD and β-tubulin (n = 3). D, the relative mRNA level of MyoD was quantified by qRT-PCR, as described under “Experimental Procedures.” The relative expression level of MyoD is expressed such that 100% represents the expression level in myoblasts. The data are presented as mean ± S.D. (n = 3). The asterisks indicate data that are significantly different according to an ANOVA test for p < 0.05.
Elsewhere, we confirmed that MyoD expression, at the protein level but not at the mRNA level, was subjected to a drastic decrease in reserve cells as compared with myoblasts (Fig. 9, C and D), indicating that down-regulation of MyoD during the establishment of reserve cells is due to translational or post-translational regulation of MyoD.
Calpain 3 Affects the Protein Level of MyoD Independently of Its Post-translational Modifications and of Degradation by the Ubiquitin-proteasome Pathway
Similar to many short lived transcriptional regulators, MyoD is subjected to ubiquitin-proteasome-dependent degradation (22, 56, 57). We asked whether this pathway is involved in MyoD proteolysis upon calpain 3 overexpression. To answer this question, we used MyoD mutants in which specific residues implicated in MyoD degradation via the ubiquitin-proteasome pathway have been mutated (18, 47). C2C12 cells were co-transfected with plasmids expressing calpain 3 and the MyoD mutants, and the protein level of each HA-MyoD mutant was evaluated by Western blot analysis (Fig. 10A). Calpain 3 induced a 65% decrease in HA-MyoD WT level, but, surprisingly, it also affected all mutated forms of MyoD (i.e. HA-MyoD K133R, HA-MyoD K133R/S200A, and HA-MyoD S200A), with a 51, 48, and 41% decrease, respectively (Fig. 10B). Thus, the ubiquitin-proteasome pathway and phosphorylation of MyoD on serine 200 do not appear to modulate its decrease in protein level induced by calpain 3.
FIGURE 10.
Study of post-translational modifications and degradation of MyoD by the ubiquitin-proteasome pathway. A–D, C2C12 cells were transfected with plasmid encoding different forms of HA-MyoD protein harboring point mutations (18, 47), with or without pEMSV-CAPN3. A, at 48 h after transfection, cells were harvested, and total cell extract was prepared. 40 μg of cell extract was analyzed by SDS-PAGE and immunoblotting with antibodies specific for HA and GAPDH. B, the relative amount of each HA-MyoD protein, corrected with respect to GAPDH expression, is expressed such that 100% indicates the protein amount of each form of HA-MyoD expressed alone (control cells). The data are presented as mean ± S.D. (n = 3), and asterisks indicate data that are significantly different from control cells according to Student's test for p < 0.05. C and D, the half-life of HA-MyoD K133R was determined following cycloheximide treatment, as described under “Experimental Procedures.” C, total cell extract was prepared at the indicated times, and 40 μg of extract was analyzed by SDS-PAGE and immunoblotting with antibodies specific for HA and GAPDH. D, HA-MyoD K133R protein levels, corrected with respect to GAPDH expression, are expressed such that 100% indicates the protein amount before cycloheximide treatment (t0). For each time point, the log10 of the percentage of pixels (y) is plotted versus the time in minutes (x). The half-life (t½) is determined from the log10 of 50% from the linear regression of three independent experiments (regression equations in the absence or in presence of calpain 3 overexpression are as follows, y = −0.00137x + 1.94542 with R2 = 0.84, and y = −0.00300x + 1.94914 with R2 = 0.94, respectively).
We next determined the half-life of HA-MyoD K133R in the presence or absence of calpain 3. As shown in Fig. 10, C and D, the half-life of HA-MyoD K133R was increased from 76 to 180 min when compared with that of HA-MyoD WT (see Fig. 9B), consistent with its inability to be degraded by the ubiquitin-proteasome pathway. Upon overexpression of calpain 3, the half-life of HA-MyoD K133R protein was clearly reduced from 180 to 83 min, in a manner similar to that found for HA-MyoD WT (Fig. 10D). Therefore, we conclude that calpain 3-induced MyoD degradation occurs independently of the ubiquitin-proteasome pathway. Together, our results suggest the existence of an additional degradation pathway for MyoD that is specifically driven by calpain 3.
DISCUSSION
In this study, we have described a role for calpain 3 in the process of myogenic differentiation as a new regulator of MyoD activity. Calpain 3 induces the proteolysis of MyoD and promotes the generation of a pool of reserve cells.
Experiments performed in C3H10T1/2 cells allowed us to show that overexpression of calpain 3 drastically decreased the transactivation function of MyoD and hampered its ability to induce myogenic conversion. Furthermore, our data show that calpain 3 restricts MyoD activity through proteolysis, thus revealing MyoD as a new target for the protease.
MyoD plays a crucial role during the early steps of myogenic differentiation, and its importance is evident during the regeneration process (17, 58–61).
First, a tight control of MyoD protein amount during the progression of myoblasts through the cell cycle appears to be required to direct muscle cell fate toward differentiation or proliferation (22, 23, 47). In the presence of proliferative signals during the G1 phase, MyoD activity is suppressed, allowing the progression through a new division cycle (16, 22). This inhibition of MyoD activity has been attributed to its degradation by the ubiquitin-proteasome system (18, 62). A recent study suggests collaboration between calpain 3 and this proteolytic system because calpain 3-deficient muscles show down-regulation of genes involved in the ubiquitin-proteasome pathway (63). In the present study, we demonstrated that MyoD degradation induced by calpain 3 is independent of the ubiquitin-proteasome pathway. Moreover, we suggest that the post-translational modifications of MyoD that modulate its degradation during the G1 phase (9, 18, 22, 57) do not modify calpain 3-dependent degradation of MyoD, as shown with phosphorylation on serine 200 (Fig. 10). Elsewhere, a role for calpain 3 in the maintenance of proliferation through destabilization of MyoD is unlikely because calpain 3 mRNA is faintly detectable in proliferating myoblasts (Fig. 1) (64). Moreover, calpain 3 overexpression provokes a G0/G1 arrest rather than cell cycle progression (Table 1), and its knockdown does not modify the proliferation rate of myoblasts (Table 2). Altogether, our results demonstrate a role of calpain 3 among differentiating myoblasts.
In the presence of differentiating signals during the G1 phase, the level of MyoD increases in most cells and induces an irreversible arrest of the cell cycle and terminal differentiation (22). Concurrently, a peculiar subpopulation of cells, termed reserve cells, is also formed. In these cells, the level of MyoD is down-regulated, and cells reversibly exit the cell cycle to enter a quiescent state (10).
An early role for endogenous calpain 3 in myogenic differentiation was first suggested by its strong induction of expression on day 1 at the onset of differentiation. Then gain-of-function and loss-of-function experiments demonstrated in several ways that calpain 3 affects the balance between myoblast differentiation and the maintenance of undifferentiated cells. Further studies have demonstrated that this population of undifferentiated cells corresponded to the peculiar subpopulation of reserve cells. We have shown that the number of reserve cells was affected when calpain 3 was down-regulated or overexpressed. Hence, overexpression of calpain 3 induces differentiating myoblasts to form reserve cells rather than pursue differentiation, whereas down-regulation of Capn3 has the opposite effect. We have also noted that endogenous calpain 3 is specifically up-regulated in the subpopulation of reserve cells compared with proliferating myoblasts.
Altogether, our results and more especially those from our knockdown experiments have clearly demonstrated the physiological implication of calpain 3 within the establishment of reserve cells. The apparition of these reserve cells represents an active mechanism and does not result from a differentiation defect. Hence, results obtained in our first experiments showing that calpain 3 affected the myogenic differentiation process by modifying the proportion of fully differentiated cells (Fig. 2) certainly represent an indirect consequence. Indeed, the role of calpain 3 within reserve cells will indirectly affect the differentiation process by modifying the number of cells available to pursue myogenic differentiation, as we have seen in our flow cytometry experiments (Tables 1 and 2, proportion of Ki-67−/MyoD+ cells).
Nevertheless, this role of calpain 3 in the establishment of reserve cells does not exclude a later role of this protease among differentiating myotubes. In addition to the increased expression of calpain 3 in reserve cells compared with proliferative myoblasts, we observed a strong induction of its expression within differentiating myotubes (Figs. 1 and 3C), as was previously described (64). This result is consistent with several previous studies that demonstrate a role of calpain 3 during terminal stages of muscle differentiation and show that a down-regulation of Capn3 causes a perturbation in the myofibrillar integrity and that calpain 3 is implicated in sarcomere assembly (39, 40, 65). Our series of experiments allowed us to describe an early role for calpain 3 in the differentiation process. However, our transient transfection experiments done in myoblasts were not designed to fully study the role of calpain 3 during terminal stages of muscle differentiation. At least it seems improbable that calpain 3 affects the cell fusion process. As we have shown, although the kinetics and the maximum level of formed myotubes were clearly affected, the full process of differentiation progressed similarly, as suggested by the identical slopes of the fusion curves obtained in our gain-of-function and loss-of-function experiments compared with control cells (Fig. 2, B and F). Moreover, although the strong expression level of Capn3 within myotubes suggests an important role of the protease within these cells, it is unlikely that calpain 3 affects MyoD function within myotubes via its proteolysis, given that high levels of MyoD are also present. We propose that a regulation of calpain 3 activity takes place within these cells and could occur via the sequestration of calpain 3 within the sarcomeres during myofibrillogenesis (29, 64). Calpain 3 activity regulation may also take place through post-translational regulation of MyoD, which would prevent its calpain 3-induced degradation (57, 66).
Elsewhere, our study has revealed that calpain 3 induces degradation of MyoD, independently of the ubiquitin-proteasome pathway, suggesting that MyoD could be a direct substrate of calpain 3. Bioinformatic analysis of the MyoD sequence is not informative because no consensus cleavage site exists for this protease family (67, 68). Moreover, due to the high instability of calpain 3 once isolated from the cellular context (32), experiments to test the in vitro digestion of MyoD by calpain 3 were not feasible. In mature muscle fibers, the main pool of calpain 3 has been observed in the skeletal muscle sarcomere (69); however, calpain 3 has also been observed in other cellular compartments, such as the subsarcolemmal membrane (41, 42), the triad-associated protein complex (43), and the nucleus (26, 28, 31, 32, 44, 45). Moreover, during the myogenic process, calpain 3 localization is dynamic. Indeed, during early myofibrillogenesis, localization of calpain 3 is not sarcomeric but cytosolic (64), in accordance with our experiments (Fig. 8, D and E), which moreover showed that calpain 3 immunostaining was inclined to be more important at the perinuclear level. It is thus tempting to speculate that calpain 3 may degrade MyoD either at the perinuclear level before its entry inside the nucleus or in the nucleus, thanks to its nuclear localization signal (31), thereby preventing its physiological function as a transcription factor. Studies are in progress to draw a link between the respective localization of these two proteins. These experiments and others should allow us to reveal a possible direct proteolytic action of calpain 3 on MyoD.
Altogether, our results demonstrate that calpain 3 plays an important role in myogenic cell fate determination by controlling the level of MyoD protein within the myoblast population induced to differentiate, leading to the establishment of a pool of reserve cells necessary for self-renewal. Reserve cells share many characteristics with the satellite cells present within adult skeletal myofibers (11). Moreover, the generation of these cells during C2C12 cell differentiation is closely related to satellite cell self-renewal, which occurs during muscle regeneration. In both cases, down-regulation of MyoD is observed and appears to be a causal event (5, 6, 8, 10). Thus, these results could reveal a role of calpain 3 in the regeneration process, through the replenishment of the initial pool of quiescent satellite cells via the early degradation of MyoD in activated satellite cells.
Several studies have shown that in the absence of MyoD, satellite cells show an increased propensity for self-renewal rather than differentiation, which results in a deficit in muscle regeneration (17, 59, 60). Conversely, a calpain 3 deficiency would lead to an overall perturbed regenerative response via an accelerated myofiber neoformation, followed by a gradual exhaustion of the pool of satellite cells after repeated periods of muscle regeneration. Consequently, it follows that deficiencies in calpain 3 would lead to a progressive muscular atrophy that could be involved in the etiology of limb girdle muscular dystrophy type 2A, in accordance with the late onset of apparition and the progressive nature of the disease (70). This study provides support for a new potential pathophysiological mechanism underlying the apparition of muscular dystrophy limb girdle muscular dystrophy type 2A, which is caused by a defective calpain 3 function.
Supplementary Material
Acknowledgments
We thank Drs. Pierre Thiebaud and Paul Keire for critical reading of the manuscript. We thank Amélie Pires-Alves and Pierre Lochet for technical assistance and Vincent Pitard for help with cell cycle analysis. We are also grateful to Juliette Bitard for helpful discussions.
This work was supported by grants from the Institut National de la Recherche Agronomique, the Region Aquitaine, and the “Association Française contre les Myopathies.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- HA
- hemagglutinin
- WT
- wild type
- MCK
- muscle creatine kinase
- GM
- growth medium
- FBS
- fetal bovine serum
- BrdUrd
- bromodeoxyuridine
- BSA
- bovine serum albumin
- siRNA
- small interfering RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- CAT
- chloramphenicol acetyltransferase
- PBS
- phosphate-buffered saline
- ANOVA
- analysis of variance
- MRF
- myogenic regulatory factor
- eGFP
- enhanced green fluorescent protein.
REFERENCES
- 1.Hawke T. J., Garry D. J. (2001) J. Appl. Physiol. 91, 534–551 [DOI] [PubMed] [Google Scholar]
- 2.Cooper R. N., Tajbakhsh S., Mouly V., Cossu G., Buckingham M., Butler-Browne G. S. (1999) J. Cell Sci. 112, 2895–2901 [DOI] [PubMed] [Google Scholar]
- 3.Cornelison D. D., Wold B. J. (1997) Dev. Biol. 191, 270–283 [DOI] [PubMed] [Google Scholar]
- 4.Smith C. K., 2nd, Janney M. J., Allen R. E. (1994) J. Cell. Physiol. 159, 379–385 [DOI] [PubMed] [Google Scholar]
- 5.Baroffio A., Bochaton-Piallat M. L., Gabbiani G., Bader C. R. (1995) Differentiation 59, 259–268 [DOI] [PubMed] [Google Scholar]
- 6.Olguin H. C., Olwin B. B. (2004) Dev. Biol. 275, 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olguin H. C., Yang Z., Tapscott S. J., Olwin B. B. (2007) J. Cell Biol. 177, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zammit P. S., Golding J. P., Nagata Y., Hudon V., Partridge T. A., Beauchamp J. R. (2004) J. Cell Biol. 166, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitzmann M., Vandromme M., Schaeffer V., Carnac G., Labbé J. C., Lamb N., Fernandez A. (1999) Mol. Cell. Biol. 19, 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida N., Yoshida S., Koishi K., Masuda K., Nabeshima Y. (1998) J. Cell Sci. 111, 769–779 [DOI] [PubMed] [Google Scholar]
- 11.Benchaouir R., Rameau P., Decraene C., Dreyfus P., Israeli D., Piétu G., Danos O., Garcia L. (2004) Exp. Cell Res. 294, 254–268 [DOI] [PubMed] [Google Scholar]
- 12.Buckingham M. (2007) C. R. Biol. 330, 530–533 [DOI] [PubMed] [Google Scholar]
- 13.Chargé S. B., Rudnicki M. A. (2004) Physiol. Rev. 84, 209–238 [DOI] [PubMed] [Google Scholar]
- 14.Yablonka-Reuveni Z., Day K., Vine A., Shefer G. (2008) J. Anim. Sci. 86, E207–E216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tapscott S. J. (2005) Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 16.Kitzmann M., Carnac G., Vandromme M., Primig M., Lamb N. J., Fernandez A. (1998) J. Cell Biol. 142, 1447–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yablonka-Reuveni Z., Rudnicki M. A., Rivera A. J., Primig M., Anderson J. E., Natanson P. (1999) Dev. Biol. 210, 440–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batonnet S., Leibovitch M. P., Tintignac L., Leibovitch S. A. (2004) J. Biol. Chem. 279, 5413–5420 [DOI] [PubMed] [Google Scholar]
- 19.Molkentin J. D., Black B. L., Martin J. F., Olson E. N. (1995) Cell 83, 1125–1136 [DOI] [PubMed] [Google Scholar]
- 20.Puri P. L., Avantaggiati M. L., Balsano C., Sang N., Graessmann A., Giordano A., Levrero M. (1997) EMBO J. 16, 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sartorelli V., Puri P. L., Hamamori Y., Ogryzko V., Chung G., Nakatani Y., Wang J. Y., Kedes L. (1999) Mol. Cell 4, 725–734 [DOI] [PubMed] [Google Scholar]
- 22.Tintignac L. A., Leibovitch M. P., Kitzmann M., Fernandez A., Ducommun B., Meijer L., Leibovitch S. A. (2000) Exp. Cell Res. 259, 300–307 [DOI] [PubMed] [Google Scholar]
- 23.Tintignac L. A., Sirri V., Leibovitch M. P., Lécluse Y., Castedo M., Metivier D., Kroemer G., Leibovitch S. A. (2004) Mol. Cell. Biol. 24, 1809–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goll D. E., Thompson V. F., Li H., Wei W., Cong J. (2003) Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 25.Herasse M., Ono Y., Fougerousse F., Kimura E., Stockholm D., Beley C., Montarras D., Pinset C., Sorimachi H., Suzuki K., Beckmann J. S., Richard I. (1999) Mol. Cell. Biol. 19, 4047–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorimachi H., Imajoh-Ohmi S., Emori Y., Kawasaki H., Ohno S., Minami Y., Suzuki K. (1989) J. Biol. Chem. 264, 20106–20111 [PubMed] [Google Scholar]
- 27.Sorimachi H., Suzuki K. (1992) Biochim. Biophys. Acta 1160, 55–62 [DOI] [PubMed] [Google Scholar]
- 28.Benayoun B., Baghdiguian S., Lajmanovich A., Bartoli M., Daniele N., Gicquel E., Bourg N., Raynaud F., Pasquier M. A., Suel L., Lochmuller H., Lefranc G., Richard I. (2008) FASEB J. 22, 1521–1529 [DOI] [PubMed] [Google Scholar]
- 29.Hayashi C., Ono Y., Doi N., Kitamura F., Tagami M., Mineki R., Arai T., Taguchi H., Yanagida M., Hirner S., Labeit D., Labeit S., Sorimachi H. (2008) J. Biol. Chem. 283, 14801–14814 [DOI] [PubMed] [Google Scholar]
- 30.Ono Y., Hayashi C., Doi N., Tagami M., Sorimachi H. (2008) FEBS Lett. 582, 691–698 [DOI] [PubMed] [Google Scholar]
- 31.Sorimachi H., Ohmi S., Emori Y., Kawasaki H., Saido T. C., Ohno S., Minami Y., Suzuki K. (1990) Biol. Chem. Hoppe Seyler 371, 171–176 [PubMed] [Google Scholar]
- 32.Sorimachi H., Toyama-Sorimachi N., Saido T. C., Kawasaki H., Sugita H., Miyasaka M., Arahata K., Ishiura S., Suzuki K. (1993) J. Biol. Chem. 268, 10593–10605 [PubMed] [Google Scholar]
- 33.Taveau M., Bourg N., Sillon G., Roudaut C., Bartoli M., Richard I. (2003) Mol. Cell. Biol. 23, 9127–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono Y., Shimada H., Sorimachi H., Richard I., Saido T. C., Beckmann J. S., Ishiura S., Suzuki K. (1998) J. Biol. Chem. 273, 17073–17078 [DOI] [PubMed] [Google Scholar]
- 35.Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., Hillaire D., Passo-Bueno M., Zatz M., Tischfield J. A., Ferdeau M., Jackson C. E., Cohen D., Beckmann J. S. (1995) Cell 81, 27–40 [DOI] [PubMed] [Google Scholar]
- 36.Beckmann J. S., Spencer M. (2008) Neuromuscul. Disord. 18, 913–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramerova I., Kudryashova E., Tidball J. G., Spencer M. J. (2004) Hum. Mol. Genet. 13, 1373–1388 [DOI] [PubMed] [Google Scholar]
- 38.Kramerova I., Kudryashova E., Venkatraman G., Spencer M. J. (2005) Hum. Mol. Genet. 14, 2125–2134 [DOI] [PubMed] [Google Scholar]
- 39.Poussard S., Duvert M., Balcerzak D., Ramassamy S., Brustis J. J., Cottin P., Ducastaing A. (1996) Cell Growth Differ. 7, 1461–1469 [PubMed] [Google Scholar]
- 40.Spencer M. J., Guyon J. R., Sorimachi H., Potts A., Richard I., Herasse M., Chamberlain J., Dalkilic I., Kunkel L. M., Beckmann J. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8874–8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y., de Morrée A., van Remoortere A., Bushby K., Frants R. R., Dunnen J. T., van der Maarel S. M. (2008) Hum. Mol. Genet. 17, 1855–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y., Verheesen P., Roussis A., Frankhuizen W., Ginjaar I., Haldane F., Laval S., Anderson L. V., Verrips T., Frants R. R., de Haard H., Bushby K., den Dunnen J., van der Maarel S. M. (2005) Eur. J. Hum. Genet. 13, 721–730 [DOI] [PubMed] [Google Scholar]
- 43.Kramerova I., Kudryashova E., Wu B., Ottenheijm C., Granzier H., Spencer M. J. (2008) Hum. Mol. Genet. 17, 3271–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baghdiguian S., Martin M., Richard I., Pons F., Astier C., Bourg N., Hay R. T., Chemaly R., Halaby G., Loiselet J., Anderson L. V., Lopez de Munain A., Fardeau M., Mangeat P., Beckmann J. S., Lefranc G. (1999) Nat. Med. 5, 503–511 [DOI] [PubMed] [Google Scholar]
- 45.Richard I., Roudaut C., Marchand S., Baghdiguian S., Herasse M., Stockholm D., Ono Y., Suel L., Bourg N., Sorimachi H., Lefranc G., Fardeau M., Sébille A., Beckmann J. S. (2000) J. Cell Biol. 151, 1583–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis R. L., Weintraub H., Lassar A. B. (1987) Cell 51, 987–1000 [DOI] [PubMed] [Google Scholar]
- 47.Batonnet-Pichon S., Tintignac L. J., Castro A., Sirri V., Leibovitch M. P., Lorca T., Leibovitch S. A. (2006) Exp. Cell Res. 312, 3999–4010 [DOI] [PubMed] [Google Scholar]
- 48.Reynaud E. G., Pelpel K., Guillier M., Leibovitch M. P., Leibovitch S. A. (1999) Mol. Cell. Biol. 19, 7621–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weintraub H., Davis R., Lockshon D., Lassar A. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5623–5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brustis J. J., Elamrani N., Balcerzak D., Safwate A., Soriano M., Poussard S., Cottin P., Ducastaing A. (1994) Eur. J. Cell Biol. 64, 320–327 [PubMed] [Google Scholar]
- 51.Pfaffl M. W. (2001) Nucleic Acids Res. 29, 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buskin J. N., Hauschka S. D. (1989) Mol. Cell. Biol. 9, 2627–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. (1990) Cell 60, 733–746 [DOI] [PubMed] [Google Scholar]
- 54.Shield M. A., Haugen H. S., Clegg C. H., Hauschka S. D. (1996) Mol. Cell. Biol. 16, 5058–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Auradé F., Pinset C., Chafey P., Gros F., Montarras D. (1994) Differentiation 55, 185–192 [DOI] [PubMed] [Google Scholar]
- 56.Abu Hatoum O., Gross-Mesilaty S., Breitschopf K., Hoffman A., Gonen H., Ciechanover A., Bengal E. (1998) Mol. Cell. Biol. 18, 5670–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song A., Wang Q., Goebl M. G., Harrington M. A. (1998) Mol. Cell. Biol. 18, 4994–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cornelison D. D., Olwin B. B., Rudnicki M. A., Wold B. J. (2000) Dev. Biol. 224, 122–137 [DOI] [PubMed] [Google Scholar]
- 59.Megeney L. A., Kablar B., Garrett K., Anderson J. E., Rudnicki M. A. (1996) Genes Dev. 10, 1173–1183 [DOI] [PubMed] [Google Scholar]
- 60.Sabourin L. A., Girgis-Gabardo A., Seale P., Asakura A., Rudnicki M. A. (1999) J. Cell Biol. 144, 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seale P., Ishibashi J., Holterman C., Rudnicki M. A. (2004) Dev. Biol. 275, 287–300 [DOI] [PubMed] [Google Scholar]
- 62.Ciechanover A., Breitschopf K., Hatoum O. A., Bengal E. (1999) Mol. Biol. Rep. 26, 59–64 [DOI] [PubMed] [Google Scholar]
- 63.Combaret L., Béchet D., Claustre A., Taillandier D., Richard I., Attaix D. (2003) Int. J. Biochem. Cell Biol. 35, 676–684 [DOI] [PubMed] [Google Scholar]
- 64.Ojima K., Ono Y., Doi N., Yoshioka K., Kawabata Y., Labeit S., Sorimachi H. (2007) J. Biol. Chem. 282, 14493–14504 [DOI] [PubMed] [Google Scholar]
- 65.Chae J., Minami N., Jin Y., Nakagawa M., Murayama K., Igarashi F., Nonaka I. (2001) Neuromuscul. Disord. 11, 547–555 [DOI] [PubMed] [Google Scholar]
- 66.Duquet A., Polesskaya A., Cuvellier S., Ait-Si-Ali S., Héry P., Pritchard L. L., Gerard M., Harel-Bellan A. (2006) EMBO Rep. 7, 1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rey M. A., Davies P. L. (2002) FEBS Lett. 532, 401–406 [DOI] [PubMed] [Google Scholar]
- 68.Tompa P., Buzder-Lantos P., Tantos A., Farkas A., Szilágyi A., Bánóczi Z., Hudecz F., Friedrich P. (2004) J. Biol. Chem. 279, 20775–20785 [DOI] [PubMed] [Google Scholar]
- 69.Sorimachi H., Kinbara K., Kimura S., Takahashi M., Ishiura S., Sasagawa N., Sorimachi N., Shimada H., Tagawa K., Maruyama K., Suzuki K. (1995) J. Biol. Chem. 270, 31158–31162 [DOI] [PubMed] [Google Scholar]
- 70.Kramerova I., Beckmann J. S., Spencer M. J. (2007) Biochim. Biophys. Acta 1772, 128–144 [DOI] [PubMed] [Google Scholar]
- 71.Huebsch K. A., Kudryashova E., Wooley C. M., Sher R. B., Seburn K. L., Spencer M. J., Cox G. A. (2005) Hum. Mol. Genet. 14, 2801–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.