Abstract
Transposable elements are common in genomes and must be controlled. Many organisms use DNA methylation to silence such selfish DNA, but the mechanisms that restrict the methylation to appropriate regions are largely unknown. We identified a JmjC domain protein in Neurospora, DNA METHYLATION MODULATOR-1 (DMM-1), that prevents aberrant spreading of DNA and histone H3K9 methylation from inactivated transposons into nearby genes. Mutation of a conserved residue within the JmjC Fe(II)-binding site abolished dmm-1 function, as did mutations in conserved cysteine-rich domains. Mutants defective only in dmm-1 mutants grow poorly, but growth is restored by reduction or elimination of DNA methylation using the drug 5-azacytosine or by mutation of the DNA methyltransferase gene dim-2. DMM-1 relies on an associated protein, DMM-2, which bears a DNA-binding motif, for localization and proper function. HP1 is required to recruit the DMM complex to the edges of methylated regions.
Keywords: DNA methylation, JmjC domain, HP1, histone methylation, heterochromatin
A defining feature of epigenetic processes is that they propagate alternative regulatory states. Little is understood about how this is accomplished, however. The prototypical and perhaps best-characterized epigenetic mechanism, DNA methylation, is generally thought to reflect the operation of two distinct processes: (1) establishment of methylation de novo, and (2) maintenance of methylation patterns by a copying mechanism. Holliday and Pugh (1975) and Riggs (1975) pointed out that the symmetry of the sites of methylation in mammals—i.e., CpG dinucleotides—could serve as the basis of a copying system by a “maintenance methylase” specific for hemimethylated CpGs. Although hemimethylated CpGs are indeed preferentially methylated by certain methylases (Goll and Bestor 2005), and ample evidence for maintenance methylation has been reported, this model does not explain all aspects of observed maintenance methylation (Jones and Liang 2009). For example, it does not account for maintenance of incomplete methylation in clonal populations of cells. Moreover, it does not account for the observation that methylation that is not limited to symmetrical sites, such as in plants and in the fungus Neurospora crassa, can promote its own propagation (Singer et al. 1995; Irelan and Selker 1997; Selker et al. 2002; Chan et al. 2005).
One can imagine self-reinforcing mechanisms that should successfully propagate DNA methylation, histone modifications, or other epigenetic signals, but it is difficult to come up with schemes that are not expected to result in spreading into neighboring regions. It is therefore not surprising that spreading of silenced chromatin (e.g., heterochromatin) and its underlying marks, including DNA methylation, is occasionally observed and can also impact neighboring genes (Irelan and Selker 1997; Miao et al. 2000). It is perhaps more surprising that epigenetic marks do not always spread. Position Effect Variegation in Drosophila provides a classic example. Genes adjacent to heterochromatin in wild-type strains are normally safe from spreading, but chromosomal rearrangements that place heterochromatin next to euchromatin commonly result in spreading of the heterochromatin, which triggers silencing of nearby genes. It is generally assumed that “barriers” normally prevent spreading. Indeed, studies of silent domains of yeasts have identified sequences that can function as boundary elements (Rusche et al. 2003; Grewal and Jia 2007).
Neurospora has served as a useful model for elucidating the control of DNA methylation in eukaryotes. In contrast to the situation with plants and mammals, DNA methylation is dispensable in Neurospora (Kouzminova and Selker 2001), facilitating genetic studies of the process. In vegetative cells, one DNA methyltransferase (DNMT), DIM-2 (Defective In Methylation-2), is responsible for all DNA methylation at both symmetric and asymmetric sites. DIM-2 is directed by HP1, which recognizes trimethyl-Lys 9 on histone H3 (H3K9me3), deposited by the DIM-5 lysine methyltransferase (KMT1) (Kouzminova and Selker 2001; Tamaru and Selker 2001; Tamaru et al. 2003; Freitag et al. 2004; Honda and Selker 2008). DNA methylation is extensively distributed within centromeres, telomeres, transposon relics, and repetitive DNA, all of which are products of the genome defense system repeat-induced point mutation (RIP) (Selker et al. 2003; Lewis et al. 2008). RIP detects duplicate sequences during the sexual phase and generates numerous C:G-to-T:A transition mutations within both copies (Galagan and Selker 2004). The resulting products of RIP are A:T-rich and are potent signals for triggering H3K9 methylation and subsequent DNA methylation de novo (Tamaru and Selker 2003; Lewis et al. 2008).
Here we report finding a protein complex including a JmjC domain protein (DNA METHYLATION MODULATOR-1 [DMM-1]) and an associated protein (DMM-2) that is important for limiting the spread of DNA methylation in N. crassa. A number of JmjC domain proteins have been shown to catalyze demethylation of histone lysine and arginine residues through an oxidative reaction that requires Fe(II) and α-ketoglutarate (α-KG) (Klose et al. 2006). We show that the DMM complex is recruited to the edges of heterochromatin domains in an HP1-dependent manner. Consistent with the prior demonstration that DNA methylation interferes with transcription elongation in Neurospora (Rountree and Selker 1997), we show that abnormal spreading of methylation from inactivated transposons into nearby genes is detrimental.
Results
Loss of DMM-1 causes both hypo- and hypermethylation
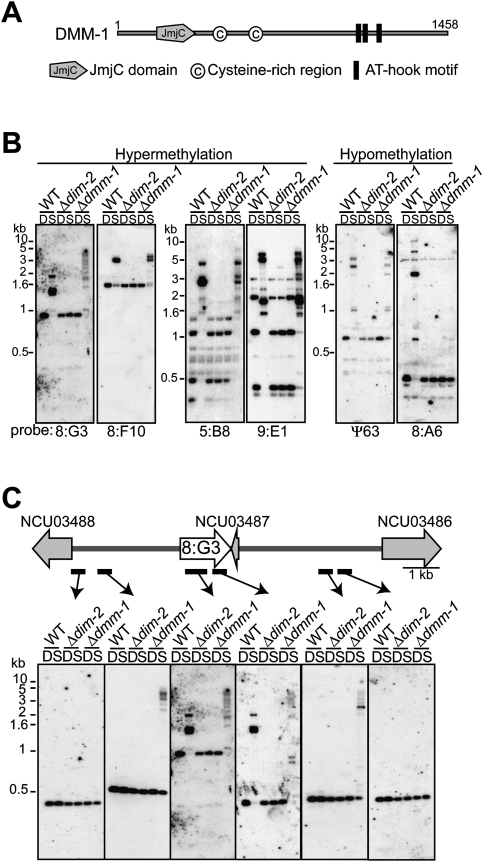
Knowledge that A:T-rich sequences typically trigger DNA methylation in Neurospora (Tamaru and Selker 2003) prompted us to explore the function of proteins bearing one or more AT hook motifs. The Neurospora genome includes at least 30 genes encoding proteins with at least one AT hook. We first focused on four predicted proteins, including two or more AT hook motifs: NCU01145, NCU01554, NCU01623, and NCU06791. We disrupted the corresponding genes and tested for effects on DNA methylation. None of the knockout mutants showed a dramatic reduction in DNA methylation, but disruption of one gene, NCU01554, resulted in novel phenotypes. This gene encodes a 1458-amino-acid protein with three AT hook motifs near its C terminus, a JmjC domain near its N terminus, and two cysteine-rich regions (CysR1, CysR2) (Fig. 1A). Genomic DNA was digested with the 5-methylcytosine (5mC)-sensitive restriction endonuclease Sau3AI or its 5mC-insensitive isoschizomer, DpnII, and used for Southern hybridization, probing for transposon relics that are normally methylated (Selker et al. 2003). The NCU01554 mutant primarily showed hypermethylation defects, but it also appeared to lose some or all DNA methylation at a small number of regions (e.g., 8:A6 and Ψ63) (Fig. 1B). Based on this phenotype, we named the gene dmm-1.
Figure 1.
Mutants lacking the JmjC domain protein DMM-1 show aberrant DNA methylation. (A) Schematic representation of DMM-1 with predicted domains and amino acid coordinates indicated. (B) DNA from a Δdmm-1 strain is hypermethylated at 8:G3, 8:F10, 5:B8, and 9:E1, and hypomethylated at 8:A6 and Ψ63. Genomic DNA of a wild-type strain (WT), a DNA methyltransferase mutant (Δdim-2), and the dmm-1 mutant were digested with 5mC-sensitive Sau3AI (S) or its 5mC-insensitive isoschizomer, DpnII (D), and used for Southern hybridizations with the indicated probes for representative methylated regions (Selker et al. 2003). (C) Spreading of DNA methylation from the methylated 8:G3 region in the dmm-1 mutant. The genomic region is shown schematically above the results of a Southern hybridization probed for 8:G3 (white arrow) and the neighboring regions (probes indicated by bars), which include a RIP'd pseudogene (NCU03487) and two predicted genes (NCU03486 and NCU03488) (gray arrow).
To further characterize the DNA methylation defects in the dmm-1 mutant, we examined additional genomic regions that are normally methylated and their bordering regions. Probes for the 8:G3 and 8:F10 regions revealed spreading of methylation (Fig. 1C; Supplemental Figs. 1, 2A). For example, methylation extended from 3 kb upstream of to 2 kb downstream from the 8:G3 region, which includes a pseudogene that is a natural product of RIP (NCU03487). Two nearby predicted genes (NCU03488 and NCU03486) were not affected, however. Curiously, the dmm-1 mutant displayed lower-molecular-weight bands in the center of 8:G3 that were not observed in wild-type strains, indicating that the core of the region was hypomethylated (Fig. 1C). We verified the spreading of DNA methylation at 8:G3 by bisulfite sequencing, and found that all cytosines in both symmetric and asymmetric contexts were subject to methylation (Supplemental Fig. 2B).
The JmjC domain and cysteine-rich regions are required for DMM-1 function
Currently, 15 of 27 JmjC domain proteins in humans have been shown to have histone demethylase activity and can be placed into one of six groups based on their substrate specificity and amino acid sequence (Agger et al. 2008). To determine whether DMM-1 could be placed into one of these groups, we performed phylogenetic analyses using JmjC domains from various species, and found that the JmjC domain of DMM-1 does not have any close homologs in Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans, Schizosaccharomyces pombe, or Saccharomyces cerevisiae (Supplemental Fig. 3A). Nevertheless, dmm-1 is conserved in the filamentous fungi. Sequence alignments of Neurospora DMM-1 with its fungal homologs revealed that the Fe(II)- and αKG-binding sites include residues that are highly conserved in JmjC domains, and that are known to be required for demethylase activity (Supplemental Fig. 3B). The two cysteine-rich regions CysR1 and CysR2 are also conserved among the filamentous fungi. In contrast, the entire C-terminal region, which contains the AT hook motifs, is missing from about half of the predicted homologs (Supplemental Fig. 4A). The DMM-1 CysR2 domain is similar to a domain in DNMT3L that binds the N-terminal tail of histone H3 when it is unmodified at K4 (Supplemental Fig. 4B; Ooi et al. 2007).
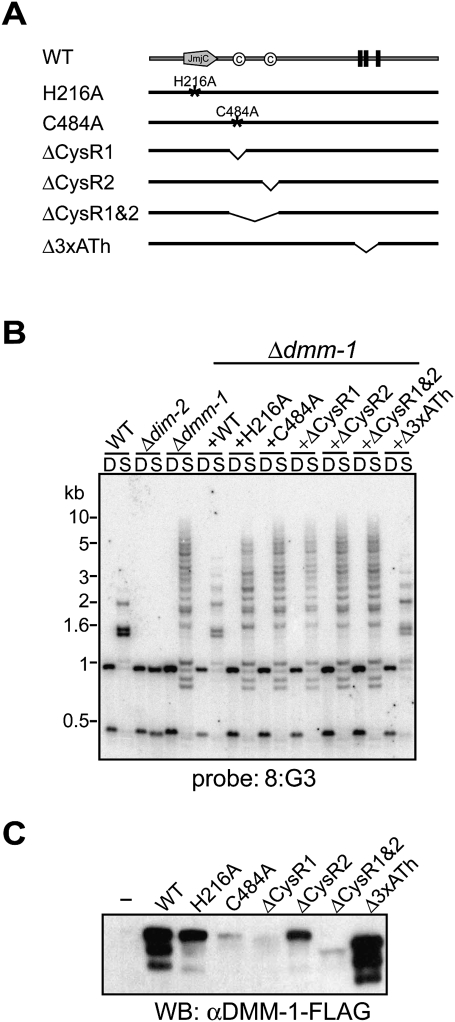
To evaluate the possible role of the identified domains/motifs in DMM-1, we generated a series of dmm-1 constructs with various deletions or point mutations and a 3xFlag epitope tag at the C terminus of the protein (dmm-1-Flag) (Fig. 2A; Supplemental Fig. 5A). All were driven by the native dmm-1 promoter and inserted into the his-3 locus of a dmm-1 mutant strain. Insertion of a wild-type dmm-1-Flag construct restored nearly normal patterns of DNA methylation, indicating that the DMM-1-Flag fusion is functional (Fig. 2B). Similarly, all constructs with mutations in the AT hook motifs complemented the methylation defect (Fig. 2B; Supplemental Fig. 5B), indicating that the DMM-1 AT hook motifs are not required for DMM-1 function. In contrast, a presumed catalytic dead mutant (DMM-1H216A) that contains a mutation in a conserved residue within the Fe(II)-binding site failed to complement the methylation defects even though it was well expressed (Fig. 2B,C). Similarly, mutation of either or both cysteine-rich regions (DMM-1C484A, DMM-1ΔCysR1, DMM-1ΔCysR2, and DMM-1ΔCysR1&2) failed to complement the methylation defects (Fig. 2B). The expression level of DMM-1ΔCysR2 was moderately reduced compared with that of wild-type DMM-1, whereas mutation of CysR1 (DMM-1C484A, DMM-1ΔCysR1, and DMM-1ΔCysR1&2) led to significantly reduced levels of expression (Fig. 2C), suggesting that CysR1 is important for DMM-1 stability.
Figure 2.
The DMM-1 JmjC domain and cysteine-rich (CysR) regions, but not AT hook motifs, are essential for normal DNA methylation. (A) Diagram of the DMM-1 point mutations and deletion mutations tested. His 216 is predicted to be critical for Fe(II) binding, and Cys 484 is conserved among fungal DMM-1 homologs (see Supplemental Figs. 3B, 4). (B) Effects of dmm-1 mutations on DNA methylation in 8:G3 region. DNA isolated from indicated strains was analyzed by Southern blot as described in Figure 1B. (C) Expression of dmm-1 mutants was assessed by Western blotting with antibodies against Flag.
To confirm that the methylation defect observed in DMM-1H216A was not due to its somewhat reduced expression, we overexpressed it using the Neurospora actin promoter. This construct also failed to complement the methylation defect (Supplemental Fig. 6), verifying the importance of the presumptive catalytic site for DMM-1 function. In attempts to determine whether DMM-1 is indeed a histone demethylase, we purified recombinant full-length DMM-1 from insect cells or, using tandem affinity epitope tags, from Neurospora extracts. We were unable to detect histone demethylase activity for any lysine or arginine residue on H3 or H4 peptides (Supplemental Table 1) or histones or nucleosomes isolated from Neurospora (data not shown). In addition, we examined the possibility that the DMM-1 cysteine-rich regions recognize methylated histones by testing binding of an N-terminal fragment of DMM-1 that contains the JmjC, CysR1, and CysR2 domains to various unmethylated and methylated histone peptides. We failed to detect any binding activity on any of the tested methylated or unmethylated lysines or arginines of histone H3 or H4 (Supplemental Table 2).
DMM-1 interacts with DMM-2
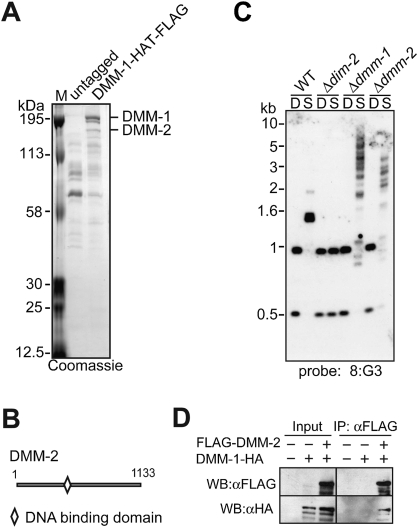
To gain insight into the function of DMM-1, we sought to identify DMM-1-interacting proteins. We created a DMM-1 fusion construct that contained a Flag-HAT (histidine affinity tag) tandem tag at its C terminus and was driven by the native dmm-1 promoter (dmm-1-HAT-Flag), and inserted it at the his-3 locus in a dmm-1 mutant strain. We confirmed that the DMM-1-HAT-Flag fusion protein was fully functional (Supplemental Fig. 6), and performed a two-step affinity purification (Honda and Selker 2009). The purified sample was gel-fractionated and visualized by Coomassie blue staining (Fig. 3A). Bands in the DMM-1-purified fraction that were not represented in the untagged control fraction were excised and digested with trypsin, and the resulting peptides were subjected to tandem mass spectrometry. The two brightly stained bands at the top of the gel corresponded to DMM-1 (Fig. 3A; Supplemental Fig. 7). The next band down was identified as a hypothetical protein encoded by NCU08289 and was named DMM-2. The dmm-2 gene encodes a 1133-amino-acid protein containing a fungal-specific Zn(II)2Cys6 DNA-binding domain (Fig. 3B; MacPherson et al. 2006). DMM-2 is not conserved in all filamentous fungi, but is conserved in Podospora anserina, Chaetomium globosum, Magnaporthe grisea, and Gibberella zeae, all of which have a homolog of the DNMT DIM-2.
Figure 3.
DMM-1 is associated with DMM-2. (A) Purification and mass spectrometric analyses of proteins associated with DMM-1. Extracts from strains containing untagged or HAT-Flag-tagged DMM-1 were subjected to a two-step purification, separated by SDS-PAGE, and visualized by Coomassie blue staining, and proteins from individual bands were digested with trypsin and analyzed by tandem mass spectrometry. (B) Schematic representation of the predicted 1133-amino-acid DMM-2 protein showing the position of a Zn(II)2Cys6 putative DNA-binding domain. (C) A mutant lacking DMM-2 shows hypermethylation at 8:G3 region. DNA isolated from the indicated strains was analyzed by Southern blot as described in Figure 1B. (D) DMM-2 associates with DMM-1 in vivo. Extracts from strains with (+) or without (−) the Flag-tagged dmm-2 gene and/or the HA-tagged dmm-1 gene were immunoprecipitated with anti-Flag antibodies. Input and immunoprecipitation samples were fractionated and analyzed by Western blotting with the indicated antibodies.
The possibility that DMM-2 interacts with DMM-1 prompted us to examine DNA methylation in mutants lacking DMM-2. The dmm-2 mutants showed hypermethylation defects at 8:G3 and 8:F10, but these defects were less severe than those observed in dmm-1 mutants (Fig. 3C; Supplemental Fig. 8). Interestingly, dmm-2 mutants also showed subtle hypermethylation defects at 8:A6 and Ψ63, two regions that display DNA hypomethylation in dmm-1 mutants (Supplemental Fig. 8). To verify that DMM-2 interacts with DMM-1, we created an N-terminal 3xFlag-tagged copy of dmm-2 driven by its native promoter (Flag-dmm-2) and inserted this construct into the his-3 locus of a dmm-2 mutant strain that also contained a C-terminal 3xHA-tagged copy of dmm-1 (dmm-1-HA) at its native locus. We confirmed that both tagged proteins were functional (Supplemental Fig. 9), and carried out a coimmunoprecipitation (co-IP) assay to test for an interaction. Indeed, DMM-1 coimmunoprecipitated with DMM-2 (Fig. 3D), verifying that DMM-2 and DMM-1 interact in vivo.
DNA methylation is responsible for defective growth of dmm-1 mutant strains
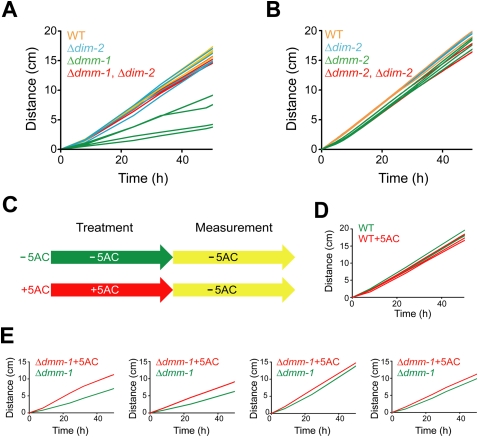
Although dmm-1 and dmm-2 mutants both exhibit DNA hypermethylation, as noted above, there are slight differences in the phenotypes of these two mutant strains. For example, the degree of hypermethylation at 8:G3 is higher for dmm-1 (Fig. 3C). Interestingly, whereas dmm-1 strains show severe growth defects, dmm-2 strains grow nearly normally. As DNA methylation can prevent gene expression in Neurospora (Rountree and Selker 1997), we hypothesized that massive spreading of DNA methylation beyond transposon relics in dmm-1 strains might down-regulate important and perhaps essential genes, resulting in the observed growth defects. To examine this possibility, we tested whether elimination of DNA methylation, by introduction of a null mutation in the DNMT gene, dim-2, would relieve the growth defect. We isolated and analyzed four classes of progeny from a cross of dmm-1 and dim-2 mutants: wild-type strains, dim-2 strains, dmm-1 strains, and dmm-1, dim-2 double-mutant strains. Tellingly, the dmm-1, dim-2 double-mutant strains displayed growth rates that were equivalent to those wild-type and dim-2 strains. In contrast, dmm-1 single-mutant strains grew poorly (Fig. 4A). As expected, dmm-1, dim-2 double mutants showed complete loss of DNA methylation (Supplemental Fig. 10). The growth defects of dmm-1 mutants were not suppressed by mutations in dim-5 or hpo (Supplemental Fig. 11), which display severe growth defects themselves, perhaps due to defects in centromere function (Tamaru and Selker 2001; Freitag et al. 2004). We also examined single dmm-2 mutants and dmm-2, dim-2 double mutants and found that they showed nearly the same growth rate (Fig. 4B), consistent with our observation that dmm-2 strains do not show spreading comparable with that observed in dmm-1 strains (Fig. 3C; see below).
Figure 4.
The growth defect in Δdmm-1 strains is relieved by loss of DNA methylation. (A) Growth rates at 32°C on solidified Vogel's medium in “race tubes” are shown for four wild-type strains (WT; orange), four Δdim-2 mutants (blue), four Δdmm-1 mutants (green), and four Δdmm-1, Δdim-2 double mutants (red). (B) Δdmm-2 strains do not show a growth defect under the same conditions. Four Δdmm-2 mutants (green) and four Δdmm-2, Δdim-2 double mutants (red) along with control strains as in A. (C) Schematic diagram of 5AC treatments. Strains were grown on solidified Vogel's medium for 3 d, followed by liquid culture for 5 h with (+) or without (−) 5AC, and linear growth rates were determined in race tubes containing solidified Vogel's medium without 5AC. (D,E) Growth rates are shown for four wild-type strains (WT) and four Δdmm-1 strains pretreated with (red) or without (green) 5AC.
We also tested the effect of treatment of dmm-1 and control strains with the DNA methylation inhibitor 5-azacytidine (5AC), as diagrammed in Figure 4C. While wild-type strains, dim-2 mutants, and dmm-1, dim-2 double mutants showed slight growth inhibition due to known toxic effects of 5AC (Fig. 4D; Supplemental Fig. 12; Selker and Stevens 1985), the growth rate of the dmm-1 mutants was moderately or slightly restored by 5AC treatment in four replicate experiments (Fig. 4E). We confirmed that 5AC treatment caused a nearly complete loss of DNA methylation in the dmm-1 strain, and that de novo DNA methylation occurred in dmm-1 mutants after further growth in the absence of 5AC (Supplemental Fig. 13). Taken together, these data support our hypothesis that inactivation of important or essential genes by ectopic DNA methylation impairs growth in dmm-1 strains.
DNA methylation profiling reveals hypermethylated genes in dmm-1 but not dmm-2 strains
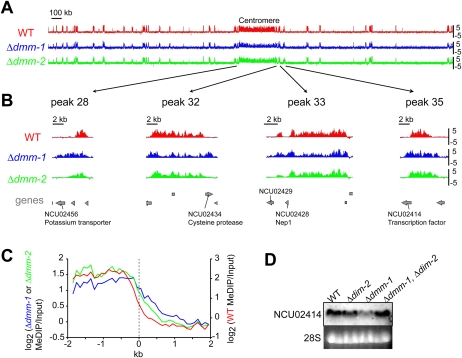
To further investigate the possibility that spreading of methylation from inactivated transposons may affect nearby genes in dmm-1 strains, we performed methylated DNA immunoprecipitation (MeDIP) on DNA from various strains and analyzed the MeDIPs with a high-density microarray covering Neurospora Linkage Group VII (LGVII). For wild-type, dmm-1, and dmm-2 strains, input and immunoprecipitation fractions were differentially labeled and hybridized to the array, which includes oligonucleotides representing essentially all LGVII sequences (Lewis et al. 2008). LGVII has 45 methylated regions including the centromere and the telomeres, all defined by the presence of RIP'd transposon relics and repeats (Lewis et al. 2008). We found that almost all peaks of 5mC in dmm-1 and dmm-2 mutants colocalized with those of wild-type strains (Fig. 5A; Supplemental Fig. 14A). As expected, however, close inspection of the data revealed that many regions were hypermethylated in dmm-1 and dmm-2 strains. Specifically, 17 of the 45 methylated regions showed spreading from the normally methylated, A:T-rich DNA into adjacent G:C-rich, genic DNA in dmm-1 strains; 10 of the 45 methylated regions were hypermethylated in mdm-2 strains. Conversely, four of the 45 regions showed hypomethylation in dmm-1 strains and three regions showed hypomethylation in dmm-2 strains. Data from four representative regions are shown in Figure 5B, and data for all methylated regions are presented in Supplemental Figure 14B. We performed Southern hybridization experiments to validate these data, and found the results fully consistent (Supplemental Fig. 15). Notably, we found that, in dmm-1 but not dmm-2 mutants, DNA methylation spread into 10 genes on LGVII. We aligned all noncentromeric methylated regions from LGVII at their edges and determined the average MeDIP enrichment for wild-type, dmm-1, and dmm-2 strains for 2 kb on either side of the edge (see the Materials and Methods; Fig. 5C; Zilberman et al. 2007). The average spreading of DNA methylation was ∼1 kb for mdm-1 and ∼0.5 kb for mdm-2, consistent with a more severe hypermethylation defect in mdm-1. Extrapolating from our analysis of LGVII, we expect that scores of genes are aberrantly methylated in the absence of functional DMM-1, which might result in down-regulation of a critical gene. Nevertheless, we were curious whether a Northern analysis of a few methylated genes might reveal any down-regulation. Indeed, examination of a few aberrantly methylated genes on LGVII revealed reduced expression of NCU02414 in dmm-1 strains (Fig. 5D); normal transcription was restored in dmm-1, dim-2 double-mutant strains.
Figure 5.
DNA methylation spreads into neighboring genes in Δdmm-1 but not Δdmm-2 strains. (A) DNA methylation profile of wild-type (WT), Δdmm-1, and Δdmm-2 strains across entire chromosome VII. Methylated DNA were immunoprecipitated (MeDIP) from DNA of wild-type (red), Δdmm-1 (blue), and Δdmm-2 (green) strains and used to probe ∼40,000 oligonucleotide sequences on an Agilent slide (Lewis et al. 2008). Results are shown as log2[IP/input] values (Y-axis). A scale bar is shown, and the centromere region is identified. (B) Data for four representative methylated regions of chromosome VII (positions indicated by arrows) are shown magnified ∼50-fold. (Bottom) The positions of predicted genes are shown in gray, and those that are methylated in Δdmm-1 but not Δdmm-2 are indicated. (C) Averaged levels of DNA methylation across the ∼80 edges of methylated regions on chromosome VII in wild-type (WT, red), Δdmm-1 (blue), and Δdmm-2 (green) strains. (D) Down-regulation of NCU02414 gene in a Δdmm-1 strain is relieved by loss of DNA methylation. Total RNA of the indicated strains were isolated and used for Northern hybridization with an NCU02414 probe. 28S rRNA was visualized by ethidium bromide (EtBr) staining.
H3K9me3 spreads in dmm-1 but not in dmm-1, dim-2 double mutants
In Neurospora, H3K9me3 is colocalized with 5mC and is apparently required for all DNA methylation (Tamaru et al. 2003; Honda and Selker 2008; Lewis et al. 2008). In contrast, previous studies suggested that most H3K9 methylation is independent of 5mC in this fungus. To elucidate the relationship between H3K9me3 and DNA methylation in dmm-1 mutants, we isolated histones from various mutants and performed Western blotting with antibodies against H3K9me3. As expected, global levels of H3K9me3 appeared abolished in dim-5 mutants but normal in wild-type, hpo, and dim-2 strains. Similarly, global H3K9me3 levels were normal in dmm-1, dmm-2, and dmm-1, dim-2 double mutants. Global levels of H3K4me2 were unchanged and are shown as a loading control (Supplemental Fig. 16).
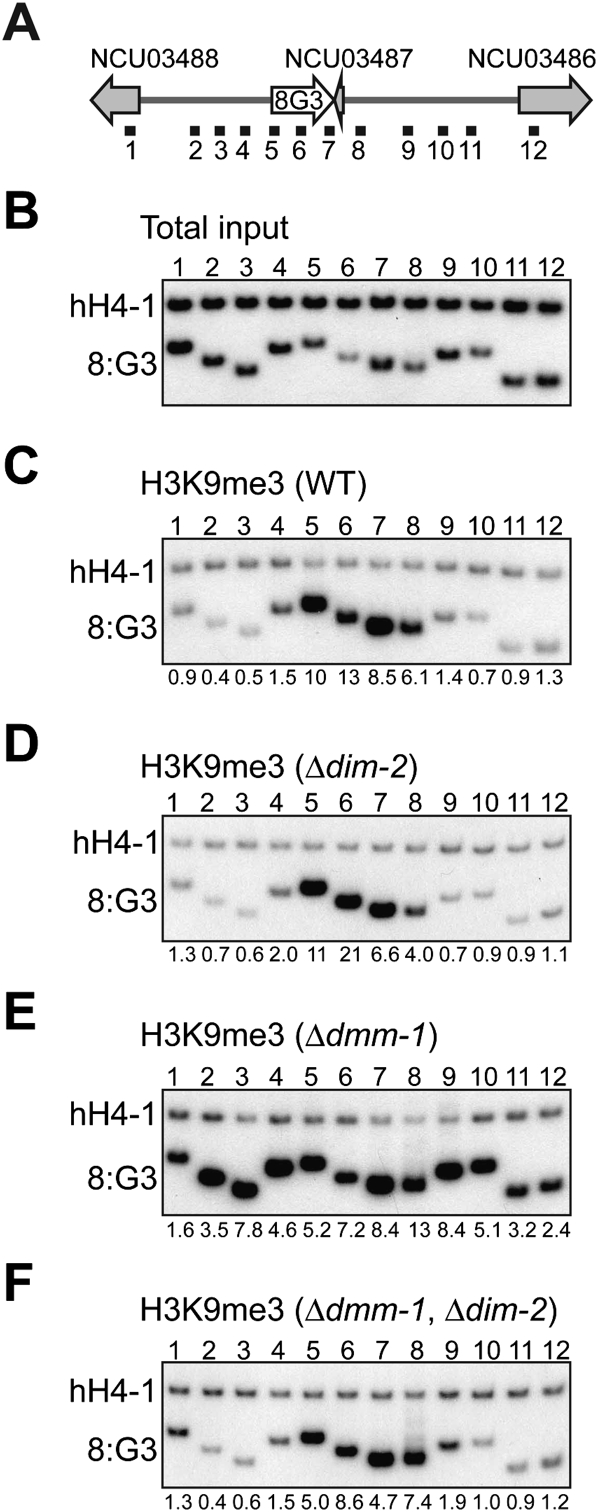
We next performed conventional chromatin immunoprecipitation (ChIP) with antibodies against H3K9me3 to examine the local distribution of H3K9me3 associated with the 8:G3 region and its flanking sequences. These experiments revealed a striking change in the distribution of H3K9me3 in dmm-1 mutant strains. While H3K9me3 was strictly localized to 8:G3 in wild-type and dim-2 strains, it spread into neighboring regions in dmm-1 mutants (Fig. 6C–E). Indeed, a ChIP–chip analysis with anti-H3K9me3 antibodies revealed that the distribution of H3K9me3 in dmm-1 mutants was virtually identical to that of 5mC (Supplemental Fig. 17). Interestingly, the spreading of H3K9me3 around 8:G3 in dmm-1 mutants was suppressed in dmm-1, dim-2 double mutants (Fig. 6F). This indicates that this ectopic H3K9me3 depends on DNA methylation, whereas normal H3K9me3 does not. The inhibition of H3K9me3 spreading may contribute to the nearly normal growth rate of dmm-1, dim-2 mutant strains. As expected, H3K4me2 was similarly distributed within neighboring genes but not within the 8:G3 transposon relic in wild-type and dmm-1 strains (Supplemental Fig. 18).
Figure 6.
H3K9me3 spreads in Δdmm-1 mutants but not in Δdmm-1, Δdim-2 double mutants. (A) Schematic representation of the 8:G3 region (see Fig. 1B). The numbered horizontal black bars indicate the regions tested on total input DNA (example for wild type shown in B) or by ChIP using antibodies against H3K9me3 (C–F) using chromatin from the indicated strains. A euchromatic gene lacking DNA methylation (hH4-1) was used as an internal control. The ratios of intensities measured for hH4-1 and the indicated probe were normalized to the ratios obtained without immunoprecipitation (total input) and are presented below the lanes. ChIP experiments were repeated at least twice with equivalent results (data not shown).
DMM-1 is preferentially localized to edges of methylated regions in an HP1-dependent manner
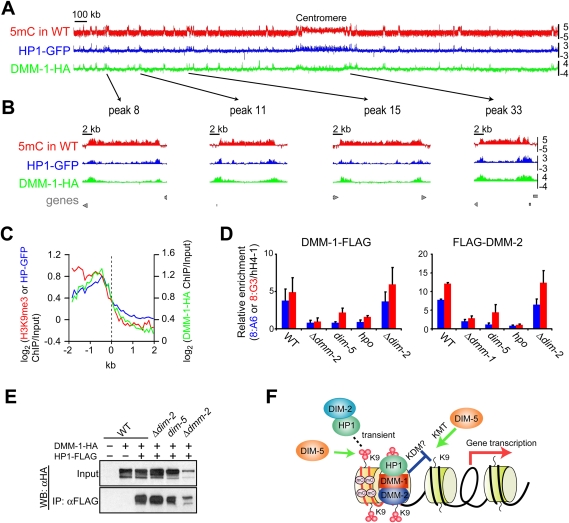
To elucidate how DMM-1 prevents the spreading of H3K9me3 and DNA methylation, we examined the localization of DMM-1 by ChIP–chip analysis. To do this, we built strains expressing DMM-1-HA under control of its native promoter and performed ChIP using antibodies specific for the HA epitope. This provided evidence that DMM-1 associates with regions of DNA methylation (Supplemental Figure 19). Interestingly, we observed lower levels of enrichment at the centromere relative to regions along the chromosome arms (Fig. 7A; Supplemental Fig. 20A), and a close inspection of the data revealed that DMM-1 is preferentially localized to the edges of methylated regions. This was most obvious in regions >5 kb (Fig. 7B; Supplemental Fig. 20B). We note that localization of DMM-1 to the edges of methylated domains resembles the localization pattern of HP1 observed previously (Lewis et al. 2008). To test whether the edge-specific localization of DMM-1-HA is a general feature of Neurospora heterochromatin domains, we aligned noncentromeric hetrochromatin domains at their edges and determined the average enrichment for H3K9me3, HP1-GFP, and DMM-1-HA for 2 kb in either direction of the heterochromatin boundary. Plotting the average distribution revealed peaks of DMM-1 and HP1, but not H3K9me3, at the edges of heterochromatin domains (Fig. 7C). To further confirm the localization pattern observed in the ChIP–chip experiments, conventional ChIP experiments were performed using a strain harboring copies of DMM-1, DMM-2, and HP1, each tagged with a different epitope. These data were fully consistent with the microarray results; all three proteins were found at the edges of each methylated region, while H3K9me3 was localized uniformly throughout each region (Supplemental Fig. 21).
Figure 7.
DMM-1 is localized preferentially at edges of methylated regions via associations with HP1 and DMM-2. (A) MeDIP (red) and ChIP–chip analyses with antibodies against HP1-GFP (blue) and DMM-1-HA (green) in otherwise wild-type (WT) strains are shown as log2[IP/input] values (Y-axis). (B) Data for four representative methylated regions of chromosome VII (positions indicated by arrows) are shown magnified ∼50-fold. (Bottom) The positions of predicted genes are shown in gray. (C) Average enrichment of H3K9me3 (red), HP1-GFP (blue), and DMM-1-HA (green) on the 80 edges of methylated regions of chromosome VII. (D) Conventional ChIP analyses with antibodies against DMM-1-Flag or Flag-DMM-2 in indicated strains at two methylated regions: 8:A6 and the center of 8:G3 (segment 6 in Fig. 6A). Data from duplicate ChIP experiments (Supplemental Fig. 22B) were averaged and are presented graphically with error bars to indicate variation. (E) DMM-1 association with HP1 is independent of DIM-2, DIM-5, and DMM-2. Extracts from strains with (+) or without (−) the HA-tagged dmm-1 gene and/or the Flag-tagged hpo gene in the indicated strains were immunoprecipitated with anti-Flag antibodies. Input and immunoprecipitation samples were fractionated and analyzed by Western blotting with anti-HA antibodies. (F) Model for the involvement of HP1/DMM-1/2 complex in normal distribution of DNA methylation within transposon relics. (Red lines) RIP'd/A:T-rich DNA; (black lines) unRIP'd DNA; (small red circles) methyl groups; (mC) methyl-cytosine; (KDM) lysine demethylase.
To elucidate possible interdependencies among DMM-1, DMM-2, and other key components of the DNA methylation machinery, we performed ChIP assays with strains expressing DMM-1-Flag or Flag-DMM-2 in various mutant backgrounds. Localization was examined at two methylated regions: 8:A6 and the center of 8:G3. Expression of the fusion proteins was confirmed (Supplemental Fig. 22A), and duplicate ChIP experiments (Supplemental Fig. 22B) were averaged and are presented graphically (Fig. 7D). Enrichment of DMM-1 and DMM-2 was dramatically reduced at both regions in dim-5 and hpo mutants, but not dim-2 mutants (Fig. 7D), indicating that binding of the DMM-1/DMM-2 complex depends on H3K9 methylation and HP1, but not DNA methylation. In addition, we found that localization of DMM-1 and DMM-2 are interdependent (Fig. 7D). Using primer pairs interrogating the entire 8:G3 region and adjacent sequences, we found that DMM-1 displayed significantly reduced levels of enrichment at the center of the 8:G3 region in the dmm-2 mutant, but spread into the adjacent sequences (Supplemental Fig. 23A). In contrast, we detected no enrichment of DMM-2 in dmm-1 strains (Supplemental Fig. 23B), suggesting that association of DMM-2 with chromatin depends on DMM-1 but not vice versa. The spreading of DMM-1 was also observed in the catalytic site mutant DMM-1H216A (Supplemental Fig. 19). We next examined localization of HP1 in dmm-1 and dmm-2 mutants. Although microscopic examination of HP1-GFP revealed typical heterochromatic foci in dmm-1 and dmm-2 mutants (Supplemental Fig. 24A; Freitag et al. 2004), the ChIP analysis revealed that HP1 spread from 8:G3 into adjacent sequences in both dmm-1 and dmm-2 mutants (Supplemental Fig. 24B).
The observations that localization of DMM-1 resembles that of HP1 and that DMM-1 localization requires HP1 suggests that DMM-1 might interact with HP1. Our biochemical purification with DMM-1-HAT-Flag failed to detect HP1 peptides, however. Because of the possibility of a weak interaction between DMM-1 and HP1, we tested for interaction of HP1 and DMM-1 in total cell extracts by co-IP experiments. We observed an interaction between HP1 and DMM-1 that was dependent on the salt concentration in the extraction buffer (Fig. 7E); interaction was detected in buffer containing 100 mM but not 150 mM NaCl (data not shown). Co-IP assays in various mutant backgrounds revealed that the HP1/DMM-1 interaction occurs in mutants lacking DIM-2, DIM-5, and DMM-2 (Fig. 7E), indicating that the interaction does not depend on DNA methylation, H3K9me3, or DMM-2.
Discussion
Our studies of the dmm-1 gene provide the first example of a Neurospora mutant displaying DNA hypermethylation. Mutants lacking DMM-1 or its associated protein, DMM-2, cause ectopic spreading of DNA methylation, H3K9me3, and HP1 from numerous dispersed transposon relics, indicating that Neurospora has a mechanism to segregate methylated and unmethylated regions. The importance of this function can be inferred from the poor growth of dmm-1 mutants. We showed that normal growth of dmm-1 strains can be largely or completely restored by elimination of DNA methylation, and presented evidence that the poor growth of single dmm-1 mutants is due to inappropriate silencing of genes that reside near transposon relics. Interestingly, normal distribution of H3K9me3 was observed for some regions in dmm-1, dim-2 double mutants, indicating that spreading of H3K9me3 can depend on DNA methylation. Understanding how DNA methylation domains can be contained is not only an interesting academic problem, but it is also medically relevant, as some disease states, such as cancer, are associated with aberrant DNA methylation. Moreover, control of DNA methylation appears to be related to the control of heterochromatin domains, generally. Mutations in dmm-1 or dmm-2 predominantly resulted in DNA hypermethylation, but some hypomethylation was also detected, typically in short (∼1-kb) methylated regions. Hypomethylation may be an indirect effect of the more widespread hypermethylation; e.g., because of limited availability of methylation machinery such as the DNA methyltransferase DIM-2. Alternatively, these contrasting phenotypes could reflect different activities of the DMM-1/DMM-2 complex.
Although no proteins structurally or functionally equivalent to DMM-1 or DMM-2 have been described, parallels can be drawn to two partially characterized JmjC domain proteins, Epe1 in fission yeast (Ayoub et al. 2003; Zofall and Grewal 2006; Isaac et al. 2007; Trewick et al. 2007) and IBM1 in Arabidopsis (Saze et al. 2008; Miura et al. 2009). While JmjC domains are frequently involved in demethylation of histones, demethylase activity has not been detected for either of these proteins, as with DMM-1. Epe1 has been implicated in the control of heterochromatin in S. pombe, an organism with no DNA methylation. Mutation of S. pombe epe1 leads primarily to spreading of heterochromatin, but has also been reported to destabilize heterochromatin, similar to how mutation of dmm-1 leads primarily to spreading of DNA methylation, but also causes hypomethylation in some regions.
Unlike the situation in Neurospora, in S. pombe, heterochromatin is flanked by boundary elements with particular DNA sequences, and loss of the boundary elements cause spreading of heterochromatin into neighboring euchromatin (Grewal and Jia 2007). Aside from sharing a JmjC domain, Epe1 and DMM-1 share little similarity, but both rely on chromo/chromoshadow domain proteins (Swi6 or HP1). It is not known whether Epe1 associates with other proteins—e.g., comparable with Neurospora DMM-2—but likely homologs of dmm-2 have not been found in yeasts. Indeed, it appears that dmm-2 is found specifically in fungi that have homologs of the DIM-2 DNMT; DMM-1 appears conserved in all filamentous fungi. Epe1 lacks cysteine-rich domains comparable with those in DMM-1, which we showed are essential for the normal distribution of DNA methylation. A somewhat similar cysteine-rich region of mammalian DNMT3L has been shown to bind unmethylated H3K4 (Ooi et al. 2007), but we were unable to detect an interaction between the DMM-1 cysteine-rich regions and various methylated and unmethylated histone H3 and H4 tails (Supplemental Table S2). Of course, it is possible that such an association depends on one or more unidentified factors.
As a first step to investigate how DMM-1 controls spreading of DNA methylation, we investigated its cellular localization. We found that the DMM complex is stably bound to chromatin in genomic regions that include DNA methylation and that almost exclusively consist of relics of transposable elements (Selker et al. 2003; Lewis et al. 2008). Like HP1, the localization of DMM-1 and DMM-2 depend on methylation of K9 of histone H3 by the DIM-5 KMTase, but their localization does not depend on DNA methylation. Interestingly, DMM-1, DMM-2, and HP1 all appear most concentrated at the boundaries of heterochromatic regions. Association of DMM-2 to chromatin depends on DMM-1, consistent with our observation that dmm-1, dmm-2 double mutants phenocopy dmm-1 single mutants (data not shown). DMM-2 contains a fungal-specific Zn(II)2Cys6 binuclear cluster DNA-binding domain. In other proteins, this domain is responsible for binding to simple DNA sequences such as hyphenated trinucleotides (e.g., CGG and CCG) (MacPherson et al. 2006). Our finding that DMM-1 spreads in dmm-2 mutants suggests that DMM-2 might bind to specific DNA sequences near the borders of methylated regions, leading to the observed localization of the DMM complex. Thus, it is possible that HP1/DIM-2 transiently binds at the core of methylated regions, spreads to the edges to introduce 5mC throughout transposon relics, and then recruits DMM-1 along with DMM-2. The specific localization of the DMM complex would be controlled by binding of DMM-2 to specific sequences enriched at the edge and/or outside of methylated regions, and perhaps by recognition of undefined chromatin marks by the cysteine-rich domains of DMM-1 (model presented in Fig. 7F).
The molecular details of how the DMM complex blocks the spread of H3K9 methylation and DNA methylation remains unknown. In contrast to the situation in S. pombe, where heterochromatin is flanked by boundary elements with particular DNA sequences, in Drosophila, heterochromatin/euchromatin boundaries are thought to result from competition between opposing histone modifications, such as H3K4me and H3K9me, without relying on specific DNA sequences (Gaszner and Felsenfeld 2006). Neurospora may provide a third paradigm, as sequence-specific boundary elements have not been identified and H3K4me2/3 is not found consistently at the edges of methylated transposon relics (Lewis et al. 2008). It is attractive to suppose that the JmjC domain of DMM-1 imparts a demethylase activity, which might function by removing excessive methylation by DIM-5. Interestingly, the Arabidopsis JmjC domain protein IBM1 has been implicated recently in the normal localization of DNA methylation (Saze et al. 2008; Miura et al. 2009), but, as noted above, no activity has been reported for this protein. It is noteworthy that the JmjC domain of Epe1 lacks a conserved residue within the catalytic domain. (Klose et al. 2006; Zofall and Grewal 2006). Thus, Epe1 function may not function as a histone demethylase. The DMM-1 JmjC domain has all of the conserved residues within the Fe(II)- and αKG-binding sites reportedly required for oxidative lysine demethylation (Klose et al. 2006), and we determined that one of these residues, His 216, is essential for DMM-1 function, consistent with the possibility that DMM-1 is a demethylase. However, we could not detect demethylase activity of the DMM-1 JmjC domain on any of the numerous tested methylated lysine or arginine residues in histone H3 and H4 in vitro (Supplemental Table 1; data not shown). Recent work revealed that activity of the Drosophila KDM4A JmjC histone demethylase is enhanced by HP1 (Lin et al. 2008). It is possible that demethylase activity of DMM-1 depends on loosely associated proteins, such as HP1. Thus, it is possible that DMM-1 is stimulated to demethylate H3K9me3, recognized by HP1, when it is in the vicinity of DNA sequences recognized by DMM-2. In this manner, the DMM-1/DMM-2 complex would serve to protect genes that reside near transposon relics from inappropriate gene silencing resulting from spreading of DNA methylation/heterochromatin (Fig. 7F).
In summary, our results uncovered a molecular mechanism that serves to protect endogenous genes from aberrant silencing by the DNA methylation machinery. It is noteworthy that HP1 mediates two opposing reactions via recruitment of DIM-2 and DMM-1/2 to introduce DNA methylation and to antagonize excessive DNA methylation, respectively. Arabidopsis may use a similar mechanism (Saze et al. 2008; Miura et al. 2009). It will be interesting to learn whether mammals do as well. The fact that transposable elements can be toxic to their hosts is of general importance. Not only can transposons inactivate genes by insertion and lead to chromosome rearrangements as a result of recombination between nonallelic copies, but they can also impact neighboring genes (Martienssen et al. 1990; Cambareri et al. 1996; Slotkin and Martienssen 2007). Ironically, the negative effects on neighboring genes can be regarded as side-effects of host defense mechanisms that serve to silence these “selfish” elements. In eukaryotes that sport DNA methylation, including mammals, plants, and some fungi such as Neurospora, methylation is used to control transposable elements. Indeed, this may be the primary function of DNA methylation (Selker et al. 2003; Goll and Bestor 2005). Interestingly, methylated transposable elements tend to be depleted near genes (Hollister and Gaut 2009). Nevertheless, as we showed, spreading of methylation from transposable elements can still be a serious problem. Thus, organisms have developed mechanisms to prevent inappropriate spreading of DNA methylation and heterochromatin generally.
Materials and methods
Neurospora strains and isolation of genomic DNA and total RNA
Neurospora strains and primers used in this study are listed in Supplemental Tables S3 and S4, respectively. Strains were grown, maintained, and crossed according to standard published procedures (Davis 2000). For isolation of genomic DNA, Neurospora strains were grown with shaking in Vogel's minimal medium with required supplements for 2 d at 32°C; genomic DNA was isolated and used for PCR and Southern hybridizations as described (Freitag et al. 2004). For isolation of total RNA, Neurospora strains were grown with shaking in Bird medium (Metzenberg 2004) with required supplements overnight at 32°C; total RNA was isolated as described (Rountree and Selker 1997). Northern hybridization was performed in ULTRAhyb (Ambion) following manufacturer's instructions.
Gene disruption of dmm-1 and dmm-2
The dmm-1 gene was knocked out as described previously for dap-1 (Honda and Selker 2008). The hph gene and the 5′ and 3′ flanking fragments of the dmm-1 ORF (NCU01554.3) were amplified by PCR and assembled with linearized pRS416 in yeast. The plasmid containing the linear knockout cassette was recovered in Escherichia coli and transformed into a Δmus-52 strain (N2930) of Neurospora by electroporation, and resulting strains were crossed with a mus-52+ strain to recover progeny with the wild-type allele of mus-52. A dmm-2-null (NCU08289.3) mutant was generated through the Neurospora gene knockout project (Colot et al. 2006) and was obtained from the Fungal Genetic Stock Center.
Generation of strains with modified dmm-1 and dmm-2 alleles
A fragment of the dmm-1 coding region with its promoter was amplified by PCR and assembled with linearized pRS416-10xGly in yeast as described (Honda and Selker 2009). A fragment containing the dmm-1 gene with a 10xGly tail was transferred by digestion with NotI and PacI from the assembled plasmid to pCCG-C-3xFlag, pMF270, and pCCG-C-HAT-Flag, yielding pDMM-1-3xFlag, pDMM-1-3xHA, and pDMM-1-HAT-Flag, respectively. The plasmids were linearized with NheI and inserted at the his-3 locus of N3064 by gene replacement (Margolin et al. 1997). Replacements and deletions in DMM-1 were made with a QuickChange site-directed mutagenesis kit (Stratagene). To generate strains expressing DMM-1-HA at its native locus, a 1-kb fragment including the 3′ end of the dmm-1 coding region, without the stop codon, and a 500-base-pair (bp) fragment of the 3′ dmm-1 flanking region were amplified by PCR. The two PCR products were assembled in yeast with linearized pRS416 and a 3xFlag knock-in module. The assembled plasmids were transformed into a Δmus-52 strain (N2930) by electroporation, and a resulting transformant was characterized and then crossed with mus-52+ strains to recover progeny with the wild-type mus-52 allele. Fragments of the dmm-2 coding region and its promoter were amplified separately by PCR and inserted into pCCG-N-3xFlag, yielding p3xFlag-DMM-2. p3xFlag-DMM-2 was linearized and inserted at the his-3 locus of N3388.
Purification of DMM-1 complex
Conidia from four 250-mL flasks of a DMM-1-HAT-Flag strain (N3269) were inoculated into four 2.8-L flasks, each containing 1000 mL of medium and grown for 20 h at 32°C with shaking. The harvested tissue was frozen in liquid nitrogen, ground to a fine powder, suspended in 500 mL of ice-cold lysis buffer (50 mM Hepes at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF), and centrifuged at 10,000g for 20 min at 4°C. The pellets were washed twice in ice-cold PBS (10 mM phosphate at pH 7.4, 137 mM NaCl, 2.7 mM KCl) and suspended in 500 mL of ice-cold nuclear extraction buffer without EDTA (15 mM Hepes at pH 7.5, 300 mM NaCl, 5 mM MgCl2, 20 mM ZnCl2, 5% glycerol, 1 mM PMSF [Sigma], 1 μg/mL leupeptin [Roche], 1 μg/mL pepstatin [Roche], 1 μg/mL E-64 [Roche]). Extracts were sonicated (Branson Sonifier-450) five times at 10-min intervals for 2 min with a duty cycle 80 and output set to 3 and centrifuged at 10,000g for 20 min at 4°C. The subsequent steps were performed as described previously to purify HP1 (Honda and Selker 2009).
Mass spectrometric analysis
Gel bands were excised from Coomassie-stained gels, washed twice for 30 min in 50 mM ammonium bicarbonate and 50% (v/v) acetonitrile, reduced in 10 mM DTT/100 mM ammonium bicarbonate, and alkylated in 55 mM iodoacetamide and 100 mM ammonium bicarbonate, and dehydrated gels were reswelled in digestion buffer containing 50 mM ammonium bicarbonate, 5 mM CaCl2, and 12.5 ng/mL sequencing grade modified trypsin (Promega). Following an overnight incubation at 37°C, extracted peptides were identified by collection of MS/MS data using a LTQ linear ion trap mass spectrometer (ThermoFisher). Peptides were separated using an Agilent 1100 series capillary LC system (Agilent Technologies) and 0.5 × 250 mm Zorbax SB-C18 column (Agilent Technologies). Proteins were identified by collection of three data-dependent MS/MS scans following each survey MS scan, matching results to a N. crassa protein database containing 9845 entries, specifying a static +57 mass for cysteines and no enzyme specificity using the program Sequest (version 27, revision 12, ThermoFisher). Protein identification was then validated using the program Scaffold (version 2_04_00, Proteome Software), which uses a probabilistic model to estimate the peptide and protein false discovery rate (Keller et al. 2002; Nesvizhskii et al. 2003). Minimum peptide and protein probabilities were set at 80% and 95%, respectively, with a minimum of two unique peptides matches per protein.
Co-IP, ChIP, and histone isolation
Co-IP, ChIP, and histone extraction were performed as described previously (Honda and Selker 2008). For detection of the DMM-1/HP1 interaction, we used low-salt lysis buffer (20 mM Hepes at pH 7.5, 100 mM NaCl, 0.1% NP40, 1 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL pepstatin) for extraction and washes.
Microarray analysis
Microarray design, MeDIP, ChIP, sample labeling, microarray hybridization, and data analysis were carried out as described previously (Lewis et al. 2008). Nonspecific (noise) peaks were removed by subtraction of those detected in dim-5 mutants. To determine the average enrichment at the edges of methylated regions, we used the sequence coordinates of all wild-type methylated domains defined by Lewis et al. (2008), excluding those spanning the centromere. Each edge was given position 0, and averages were calculated in 100-bp increments moving toward the center of the methylated domains or moving away from the methylated domains. Arbitrarily, negative values indicate the average within methylated domains and positive values indicate the average of data for sequences adjacent to methylated domains.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM025690-22 to E.U.S., NCI118487 to Y.S., and EY10572 and CA069533 to L.L.D). S.H. is supported by a post-doctoral fellowship from the Uehara Foundation. Z.A.L. is supported by a Leukemia and Lymphoma Society Special Fellowship (GM3295-09).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1893210.
Supplemental material is available at http://www.genesdev.org.
References
- Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:159–168. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Ayoub N, Noma K, Isaac S, Kahan T, Grewal SI, Cohen A. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol Cell Biol. 2003;23:4356–4370. doi: 10.1128/MCB.23.12.4356-4370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri EB, Foss HM, Rountree MR, Selker EU, Kinsey JA. Epigenetic control of a transposon-inactivated gene in Neurospora is dependent on DNA methylation. Genetics. 1996;143:137–146. doi: 10.1093/genetics/143.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH. Neurospora: Contributions of a model organism. Oxford University Press; Oxford, UK: 2000. [Google Scholar]
- Freitag M, Hickey PC, Khlafallah TK, Read ND, Selker EU. HP1 is essential for DNA methylation in Neurospora. Mol Cell. 2004;13:427–434. doi: 10.1016/s1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Selker EU. RIP: The evolutionary cost of genome defense. Trends Genet. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Selker EU. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol Cell Biol. 2008;28:6044–6055. doi: 10.1128/MCB.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Selker EU. Tools for fungal proteomics: Multifunctional Neurospora vectors for gene replacement, protein expression and protein purification. Genetics. 2009;182:11–23. doi: 10.1534/genetics.108.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irelan JT, Selker EU. Cytosine methylation associated with repeat-induced point mutation causes epigenetic gene silencing in Neurospora crassa. Genetics. 1997;146:509–523. doi: 10.1093/genetics/146.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac S, Walfridsson J, Zohar T, Lazar D, Kahan T, Ekwall K, Cohen A. Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics. 2007;175:1549–1560. doi: 10.1534/genetics.106.068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Kouzminova E, Selker EU. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001;20:4309–4323. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZA, Honda S, Khlafallah TK, Jeffress JK, Freitag M, Mohn F, Schubeler D, Selker EU. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2008;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Li B, Swanson S, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell. 2008;32:696–706. doi: 10.1016/j.molcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol Mol Biol Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin BS, Freitag M, Selker EU. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet Newsl. 1997;44:34–36. [Google Scholar]
- Martienssen R, Barkan A, Taylor WC, Freeling M. Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize. Genes & Dev. 1990;4:331–343. doi: 10.1101/gad.4.3.331. [DOI] [PubMed] [Google Scholar]
- Metzenberg RL. Bird Medium: An alternative to Vogel medium. Fungal Genet Newsl. 2004;51:19–20. [Google Scholar]
- Miao VP, Freitag M, Selker EU. Short TpA-rich segments of the z-h region induce DNA methylation in Neurospora crassa. J Mol Biol. 2000;300:249–273. doi: 10.1006/jmbi.2000.3864. [DOI] [PubMed] [Google Scholar]
- Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 2009;28:1078–1086. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Rountree MR, Selker EU. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes & Dev. 1997;11:2383–2395. doi: 10.1101/gad.11.18.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- Selker EU, Stevens JN. DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc Natl Acad Sci. 1985;82:8114–8118. doi: 10.1073/pnas.82.23.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, Freitag M, Kothe GO, Margolin BS, Rountree MR, Allis CD, Tamaru H. Induction and maintenance of nonsymmetrical DNA methylation in Neurospora. Proc Natl Acad Sci. 2002;99:16485–16490. doi: 10.1073/pnas.182427299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, Tountas NA, Cross SH, Margolin BS, Murphy JG, Bird AP, Freitag M. The methylated component of the Neurospora crassa genome. Nature. 2003;422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
- Singer MJ, Marcotte BA, Selker EU. DNA methylation associated with repeat-induced point mutation in Neurospora crassa. Mol Cell Biol. 1995;15:5586–5597. doi: 10.1128/mcb.15.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. Synthesis of signals for de novo DNA methylation in Neurospora crassa. Mol Cell Biol. 2003;23:2379–2394. doi: 10.1128/MCB.23.7.2379-2394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 2007;26:4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]