Abstract
In this issue of Chemistry & Biology, Astle et al. report the rapid identification of the most active hits from a large one-bead-one-compound peptoid library by magnetic sorting and without the need for labor-intensive resynthesis of the hits.
Combinatorial chemistry has become a powerful tool for the pharmaceutical industry to speed up the process of drug discovery and optimization. Among the various library forms, the one-bead-one-compound (OBOC) library, where each bead carries many copies of a single compound, holds the greatest potential for the rapid identification of novel hits against a drug target. However, this potential has not yet been fully realized due to a number of technical obstacles. One of the difficulties is that, while a library of millions of compounds can be readily synthesized by the split-and-pool method (Lam et al. 1991), screening millions of beads by the conventional method, which involves viewing the beads under a microscope and manually picking out the “hits” (e.g., fluorescently labeled beads), is impractical for industrial applications.
Another problem with OBOC libraries is that during on-bead screening, the signal strengths (e.g., fluorescence intensities) do not always correlate with the potency of the ligands on these beads. Several factors may contribute to this problem. First, immobilization of a ligand onto a surface may change its binding properties. Second, the commercial resins typically used for library synthesis have high ligand loading (e.g., 90-μm TentaGel resin with a loading capacity of 0.3 mmol/g has a ligand density of ~100 mM), which is necessary to provide a sufficient amount of material for subsequent hit identification. However, high ligand density makes it possible for a target molecule to bind to beads that contain moderate- or even low-affinity ligands. High ligand density may also result in unintended multi-dentate interactions (i.e., a single target molecule interacts with more than one ligand), leading to false positives and screening biases. Therefore, in order to identify the most active hit(s), a common practice is to resynthesize and test part or all of the initial hits individually. Since screening of a large OBOC library can easily produce hundreds of hits or more, one is often faced with this dilemma: testing just a fraction of the hits runs the risk of missing out on the most active compound, whereas testing all of them is expensive and time-consuming.
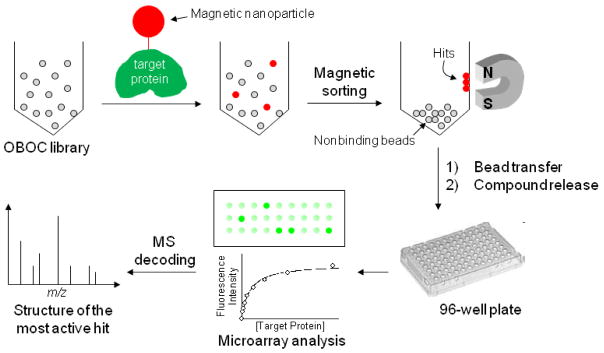
Fortunately, over the past year or so, several research groups including the authors of the featured article, have made big strides in improving the OBOC technology. On page xxx of this issue (Astle et al. 2010), Kodadek and co-workers greatly simplified the process of hit isolation through magnetic bead sorting. They labeled their target protein with a magnetic nanoparticle. Binding of the target protein to a positive bead makes the bead magnetic, allowing it to be separated from the rest of the library beads by simply placing a magnet on the side of the tube (Figure 1). To identify the most active hit(s) without resynthesis of the initial hits, the authors interfaced on-bead screening with a microarray technique. Thus, positive beads isolated by magnetic sorting were separated into individual wells of a microtiter plate and the compounds were released by treatment with CNBr (which cleaves after methionine). The resulting samples (about 50%) were spotted onto a glass slide to generate a covalently attached small-molecule microarray, which was then incubated with varying concentrations of the fluorescently labeled target protein. A plot of the fluorescence intensity against the protein concentration gave the dissociation constant for each protein-ligand pair. The authors tested their technique on a 64-million mixed peptide/peptoid library for binding to an anti-FLAG antibody. Initial on-bead screening produced 63 hits. Microarray analysis of the 63 hits revealed 27 low-nanomolar ligands against the antibody. The identity of the most active hits was then revealed by MALDI mass spectrometry using the samples remaining in the microtiter plate.
Figure 1. OBOC library screening by magnetic bead sorting coupled with microarray analysis.
Library beads incubated with the target protein are rendered as a suspension in a tube. A magnet is placed next to the tube and the beads are allowed to settle. Negative beads settle to the bottom while the hits remain on the wall of the tube.
Among other notable recent developments in this area, the Lam (Wang et al., 2005) and Pei groups (Chen et al., 2009) synthesized OBOC libraries on spatially segregated TentaGel beads that featured a reduced ligand density on the bead surface but a normal loading in the bead interior. They found that the lower surface ligand density greatly reduced the amount of nonspecific binding and binding by weak ligands. The normal ligand loading inside the bead (which is inaccessible to macromolecular targets such as proteins) still provided enough material for hit identification. To achieve the goal of identifying the most active hit without hit resynthesis, Hintersteiner et al. (2009) developed another miniaturized system to determine the binding constant of each hit compound by using the materials directly released from the positive beads. Briefly, the hits were labeled with a fluorescent dye through click chemistry (while still bound to bead), released into solution, and analyzed for binding to the target protein by fluorescence anisotropy in the 384-well plate format. By using the material derived from a 90-μm TentaGel bead (which carries ~100 pmol of compounds), they were able to reliably determine the KD value of each hit, thus allowing them to rank order all of the hits according to their potencies. Compared to Kodadek’s microarray method, which still involves immobilized ligands, the method of Hintersteiner et al. has the advantage of being able to measure the KD values in solution. But the microarray method is faster and more sensitive.
The recent developments described above have made OBOC library screening a much more efficient process. Indeed, it is now realistic for a single person to synthesize and screen a 100 million-member library and obtain the most active compound in a matter of weeks, without the need of any elaborate robotic systems. Although the methodologies have been demonstrated with peptide and peptoid libraries, they should be readily applicable to any compound class, as long as the compound structure can be decoded by using the sample from a single bead. For peptides and peptoids, compound decoding has become trivial due to the advent of several powerful mass spectrometry-based techniques (Paulick et al. 2006, Thakkar et al. 2009). For small molecule libraries of 104 or lower diversity, several innovative encoding techniques have been developed (Ohlmeyer et al., 1993, Song et al., 2003). For larger small-molecule libraries (≥105 diversity), hit identification remains a significant challenge.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Astle JM, Simpson LS, Huang Y, Reddy MM, Wilson R, Connell S, Wilson J, Kodadek T. Chem Biol. 2010;17:xxx–xxx. doi: 10.1016/j.chembiol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tan PH, Zhang Y, Pei D. J Comb Chem. 2009;11:604–611. doi: 10.1021/cc9000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintersteiner M, Kimmerlin T, kalthoff F, Stoeckli M, Garavel G, Seifert JM, Meisner NC, Uhl V, Buehler C, Weidemann T, Auer M. Chem Biol. 2009;16:724–735. doi: 10.1016/j.chembiol.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- Ohlmeyer MHJ, Swanson RN, Dillard LW, Reader JC, Asouline G, Kobayashi R, Wigler M, Still WC. Proc Natl Acad Sci USA. 1993;90:10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulick MG, Hart KM, Brinner KM, Tjandra M, Charych DH, Zuckermann RN. J Comb Chem. 2006;8:417–426. doi: 10.1021/cc0501460. [DOI] [PubMed] [Google Scholar]
- Song AM, Zhang JH, Labrilla CB, Lam KS. J Am Chem Soc. 2003;125:6180–6188. doi: 10.1021/ja034539j. [DOI] [PubMed] [Google Scholar]
- Thakkar A, Cohen AS, Connolly MD, Zuckermann RN, Pei D. J Comb Chem. 2009;11:294–302. doi: 10.1021/cc8001734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Peng L, Liu R, Xu B, Lam KS. J Pep Res. 2005;65:130–138. doi: 10.1111/j.1399-3011.2005.00192.x. [DOI] [PubMed] [Google Scholar]