Abstract
One of the most striking examples of plant developmental plasticity to changing environmental conditions is the modulation of root system architecture (RSA) in response to nitrate supply. Despite the fundamental and applied significance of understanding this process, the molecular mechanisms behind nitrate-regulated changes in developmental programs are still largely unknown. Small RNAs (sRNAs) have emerged as master regulators of gene expression in plants and other organisms. To evaluate the role of sRNAs in the nitrate response, we sequenced sRNAs from control and nitrate-treated Arabidopsis seedlings using the 454 sequencing technology. miR393 was induced by nitrate in these experiments. miR393 targets transcripts that code for a basic helix-loop-helix (bHLH) transcription factor and for the auxin receptors TIR1, AFB1, AFB2, and AFB3. However, only AFB3 was regulated by nitrate in roots under our experimental conditions. Analysis of the expression of this miR393/AFB3 module, revealed an incoherent feed-forward mechanism that is induced by nitrate and repressed by N metabolites generated by nitrate reduction and assimilation. To understand the functional role of this N-regulatory module for plant development, we analyzed the RSA response to nitrate in AFB3 insertional mutant plants and in miR393 overexpressors. RSA analysis in these plants revealed that both primary and lateral root growth responses to nitrate were altered. Interestingly, regulation of RSA by nitrate was specifically mediated by AFB3, indicating that miR393/AFB3 is a unique N-responsive module that controls root system architecture in response to external and internal N availability in Arabidopsis.
Keywords: nitrogen, microRNA, auxin, feed-forward mechanism
Nitrate is one of the major forms of inorganic nitrogen in the biosphere and nitrate availability is the most limiting factor for plant growth and agricultural productivity. Besides its role as a nutrient, nitrate (as well as other N metabolites) has been shown to act as a signal that regulates global gene expression (1–5). Although genomic data show that the nitrate response is global, little is known about the molecular basis of nitrate sensing and signaling and how the transcriptomic changes can result in developmental responses. In the last years, microRNAs (miRNAs) have emerged as master regulators of gene expression in plants and other systems (6–8). miRNAs are small ∼21–22 nt molecules, that play critical roles in various developmental, stress, and signaling responses (reviewed in refs. 9, 10). Microarray analysis showed that target transcripts of miRNAs are regulated by nitrate and/or sucrose treatments in Arabidopsis roots (4), suggesting that posttranscriptional gene expression control by miRNAs can be a general mechanism integrating nitrate signals into developmental changes. miRNAs have been known for years to be important for phosphate and sulfate deprivation responses in plants (11–13). More recently, miR167 and its target ARF8 were shown to be part of an organic N-responsive regulatory network that controls lateral root initiation in Arabidopsis (14) and N- and P-limitation regulated miRNAs have been identified in Arabidopsis seedlings (15). In this work, we used 454 sequencing to detect N-regulated miRNAs and we identified a nitrate responsive miRNA/target regulatory module that integrates N and auxin signaling to control root system architecture (RSA) in response to changes in nitrate availability.
Results
Identification of Nitrate-Responsive sRNAs in Arabidopsis Roots.
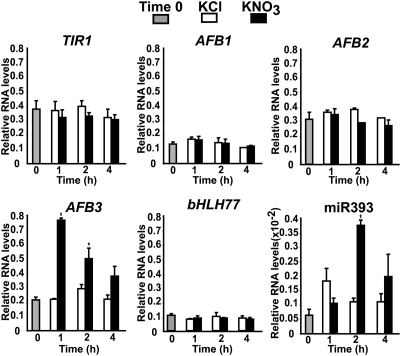
As a first approximation to evaluate the contribution of sRNAs to the nitrate response in Arabidopsis, we used the 454 sequencing technology to identify nitrate responsive sRNAs (16). Sequencing approaches have been shown to provide accurate estimates of transcript levels (17, 18) and to allow for the discovery of miRNAs [or other sRNAs such as small interfering RNAs (siRNAs) not previously identified (19–24)]. Previous genomic analyses of the nitrate and sucrose response in Arabidopsis roots have shown that most of the previously identified nitrate-responding genes are in fact regulated by some type of carbon (C)/nitrogen (N) interaction (4). Therefore, we chose to perform a combined nitrate/sucrose treatment to maximize N-responding sRNA discovery. Arabidopsis plants were grown hydroponically in basal MS media without N, supplemented with 1 mM ammonium as sole N source and 3 mM sucrose for 2 weeks, and were then treated with 5 mM KNO3 and 30 mM sucrose (treatment) or with 5 mM KCl and 30 mM mannitol (control) for 20 min and for 2 h. These experimental conditions have been shown to elicit a robust transcriptomic response in previous studies (1, 4). Total RNA was extracted from seedlings and the small RNA fraction was isolated for 454 sequencing (16, 25). In a pilot experiment, we pooled the two time points and obtained ∼16,000 sequences from the treatment and control samples. The raw sequence data were processed with custom made PERL scripts, mapped to the Arabidopsis genome, and a list of sRNAs was generated with the normalized frequency of occurrence in the control and treated samples. Using a fourfold difference cutoff between the normalized frequency in the treatment and control samples, we identified miR393 as the only sRNA induced by the treatment in these experiments. miR393 targets the transcripts that code for a basic helix-loop-helix (bHLH) transcription factor (bHLH77, ref. 26) and the auxin receptors TIR1, AFB1, AFB2, and AFB3 (13, 27, 28). Auxin is a key phytohormone, mediating growth and developmental responses in plants (29). Auxin has been proposed as a long-range signal from shoot to root mediating root developmental responses to nitrate (30, 31) and is clearly important based on network analysis of nitrate-regulated genes (4). Thus miR393 was an attractive candidate to mediate developmental plant responses to nitrate. According to our sequencing results, miR393 was induced by the treatment with a log2 (treatment normalized expression/control normalized expression) of 3.3. To corroborate our results, and to better define the timing and organ regulation of miR393, we used a modified Northern blot procedure to analyze the regulation of this miRNA in shoot and root tissue after 20 min and and 2 h of nitrate plus sucrose treatment. We found miR393 to be induced by the treatment specifically in root tissue, after 2 h of treatment (Fig. S1). To define whether miR393 was responding to nitrate or to a combined nitrate/sucrose effect, we subjected the plants to an N-only treatment (5 mM KNO3 or 5 mM KCl). miR393 was regulated similarly by nitrate treatments in the absence of sucrose, indicating that miR393 responds to nitrate independently of external sucrose levels (Fig. 1). These results prompted the hypothesis that nitrate regulation of miR393 in roots controls root auxin receptor levels, which is important for root morphological changes in response to nitrate.
Fig. 1.
Nitrate consistently regulates miR393 and its target AFB3 in Arabidopsis roots. Plants were grown hydroponically for 14 days with ammonium as the sole N source and were treated with 5 mM KNO3 or 5 mM KCl for the times indicated. Root transcript levels for bHLH77, AFB1, AFB2, AFB3, and mature miR393 were analyzed by real-time qPCR. We show the mean and standard error for three biological replicates. The asterisk indicates means that significantly differ between the control and treatment conditions (P < 0.01).
The miR393/AFB3 Module Is a Unique N-Regulatory Network Integrating External and Internal N Availability.
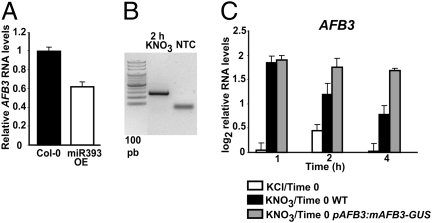
To evaluate the effect of miR393 regulation over the transcript levels of its targets, we analyzed the expression of bHLH77 and of the auxin receptors in Arabidopsis roots after nitrate treatments. Plants were grown for 2 weeks in ammonium as sole N source and were treated with 5 mM KNO3 or 5 mM KCl as control for 1, 2, and 4 h. Transcript levels for bHLH77, TIR1, AFB1, AFB2, and AFB3 were analyzed using real-time quantitative reverse transcription PCR (qRT-PCR). As shown in Fig. 1, nitrate treatment did not affect bHLH77, TIR1, AFB1, or AFB2 transcript levels in roots. However, AFB3 was induced in root organs with a peak 1 h after exposure to nitrate (Fig. 1). Interestingly, we found that AFB3 mRNA levels decreased rapidly over time, suggesting active transcript degradation. As expected, miR393 shows a peak of accumulation 2 hours after nitrate treatment, just as the transcript levels of AFB3 begin to decrease (Fig. 1). Analysis of AFB3 expression in a miR393 overexpressor line (28) showed that AFB3 levels are diminished in comparison with wild-type plants (Fig. 2A). In addition, we found a fragment corresponding to a miR393 AFB3 cleavage product (Fig. 2B) 2 h after KNO3 treatment using a modified RNA ligase-mediated 5′ rapid amplification of cDNA ends (RLM-RACE) procedure (28, 32). Moreover, we compared the kinetics of mRNA accumulation in afb3 mutant plants expressing a miR393-resistant version of AFB3 under the control of the endogenous AFB3 promoter (pAFB3:mAFB3-GUS) (33) (Fig. 2C). In contrast to the rapid decrease of AFB3 mRNA levels seen in wild-type plants, AFB3 levels did not decrease over time in pAFB3:mAFB3-GUS plants after nitrate induction (Fig. 2C). These results indicate that miR393 specifically cleaves AFB3 transcripts under our experimental conditions, controlling AFB3 mRNA accumulation in roots in response to nitrate exposure.
Fig. 2.
AFB3 transcript is cleaved by miR393 in response to nitrate. (A) Wild-type Col-0 plants and miR393 overexpressor plants (28) were grown in 0.5× MS salts supplemented with 30 mM sucrose in Petri dishes for 14 days. AFB3 levels were analyzed in seedlings using qPCR. We show the mean and standard error for three biological replicates. (B) Plants were grown hydroponically for 14 days with ammonium as the sole N source and were treated with 5 mM KNO3 for 2 h. Poly(A)+ RNA was extracted from roots and a modified RLM-RACE procedure was used to amplify a miR393 cleavage product from AFB3 (28). NTC, no template control. (C) pAFB3:mAFB3-GUS plants (33) were grown as described in B and were treated with 5 mM KNO3 for the times indicated. AFB3 transcript levels in roots were analyzed by real-time qPCR. Values are presented as the log2 ratio between the treatment level and the time 0 levels. As a reference, we also present the AFB3 transcript levels in wild-type plants from Fig. 1.
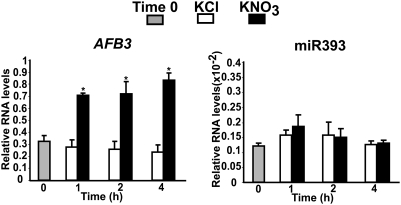
Nitrate in roots can be converted to other inorganic and organic N metabolites such as ammonium and the amino acids glutamate and glutamine. To test whether miR393 or AFB3 were responding directly to nitrate or to N metabolites produced by nitrate reduction and/or assimilation, we used a nitrate reductase (NR)-null mutant of Arabidopsis thaliana (34). This mutant plant is unable to reduce nitrate, therefore genes responding to nitrate treatments in the NR-null mutant are controlled by nitrate and not by N metabolites produced after nitrate reduction. In the NR-null mutant, AFB3 was induced by nitrate treatments with a peak of mRNA accumulation 1 h after the treatment (Fig. 3), similar to what was observed in wild-type plants (Fig. 1). However, AFB3 levels did not decline over time in the NR-null mutant (Fig. 3) as compared to wild type (Fig. 1). As expected, this lack of repression over time, correlated with the lack of induction of miR393 in the NR-null mutant (Fig. 3). These results indicate that the induction of AFB3 gene expression is caused by a nitrate signal (likely acting at the transcriptional level) but the downregulation seen at later times is caused by miR393 induction by an N metabolite downstream of nitrate reduction. To determine possible N signals controlling miR393, we tested the regulation of miR393 by ammonium and glutamate, N sources downstream of nitrate reduction and assimilation. Both N sources caused an increase in mature miR393 levels after 2 h of treatment (Fig. S2 A and B). These results indicate that miR393 responds to N signals produced after nitrate reduction and assimilation and acts as a negative feedback loop regulating AFB3 levels over time according to external and internal N availability.
Fig. 3.
AFB3 is directly induced by nitrate and is under posttranscriptional regulation by N metabolites produced after nitrate reduction by a pathway involving miR393. Nitrate reductase-null mutant plants (34) were grown hydroponically as described before and were treated with 5 mM KNO3 or 5 mM KCl for the times indicated. AFB3 transcript levels and mature miR393 levels were analyzed by real-time qPCR in roots. We show the mean and standard error for three biological replicates. The asterisk indicates means that significantly differ between control and treatment conditions (P < 0.01).
Nitrate Regulates Primary Root Growth by a Pathway Involving the AFB3 Auxin Receptor.
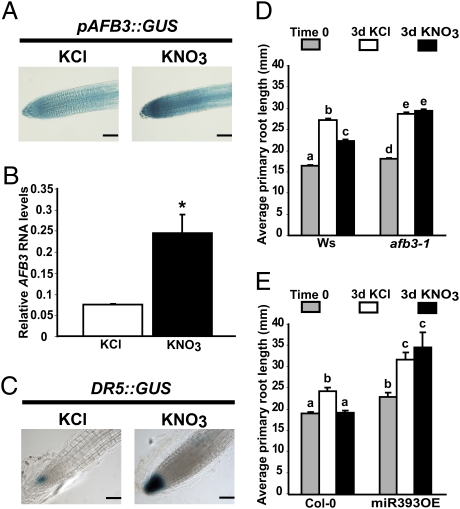
To understand the function of the miR393/AFB3 regulatory module in the nitrate response, we first analyzed the expression of this auxin receptor in roots after nitrate treatments. We used a previously described reporter line expressing the β-glucuronidase (GUS) reporter gene fused to a 1,800-bp sequence upstream of the AFB3 transcription initiation site, pAFB3::GUS (35). We treated the reporter lines with 5 mM KNO3 or 5 mM KCl for 1 h and we stained for GUS activity. We found that the AFB3 promoter is able to drive expression of GUS throughout the root in both KNO3- and KCl-treated roots, indicating root expression of the AFB3 gene as previously described (35). However, the nitrate treatment increased GUS activity preferentially in the root tip area (Fig. 4A), indicating that AFB3 mRNA accumulation by nitrate is due to transcriptional activation. To confirm these qualitative GUS results, we analyzed AFB3 RNA levels in the root tip using qRT-PCR. AFB3 was induced after 1 h of treatment in root tips (Fig. 4B). To evaluate whether this AFB3 induction correlated with increased auxin activity in the root tip, we analyzed GUS activity in the DR5::GUS reporter line (36). We found that GUS activity was also increased in the DR5::GUS line, indicating increased auxin response in the root tip in the nitrate-treated plants (Fig. 4C). Consistent with this result, we found that nitrate is able to regulate auxin-responsive genes in wild-type roots in our experimental conditions (Fig. S3A). In addition, we also found regulation of auxin-related genes that are not reported to be auxin responsive, such as the auxin response factors ARF9 and ARF18 and an auxin efflux carrier (At2g17500) (Fig. S3B). These results suggest that nitrate is able to modulate auxin signaling and responses at multiple levels as previously described (4).
Fig. 4.
Nitrate regulates primary root growth by a pathway involving AFB3. Plants were grown hydroponically as described before and were treated for the times indicated. (A) pAFB3::GUS plants (35) were treated for 1 h with 5 mM KNO3 or KCl and were then stained for GUS activity for 4 h. Qualitative GUS staining was analyzed using DIC optics. Photographs are representative of at least 15 stained plants. (Scale bar, 100 μm.) (B) Wild-type plants were treated for 1 h with 5 mM KNO3 or KCl. Root tips were excised from nitrate-treated or control-treated plants and AFB3 RNA levels were measured using qPCR. Bars represent SE. The asterisk represents means that significantly differ (P < 0.01). (C) Auxin reporter DR5::GUS plants (36) were treated for 1 h with 5 mM KNO3 or KCl and were then stained for GUS activity for 12 h. Qualitative GUS staining was analyzed using DIC optics. Photographs are representative of at least 15 stained plants. (Scale bar, 100 μm.) (D) Primary root length of Ws wild-type plants or afb3-1 mutant plants was measured using the ImageJ program after 3 days of 5 mM KNO3 or KCl treatment. Bars represent standard errors. Different letters represent significantly different means (P < 0.01). (E) Primary root length of Col-0 wild-type plants or 35S::miR393 overexpressor plants (28) was measured using the ImageJ program after 3 days of 5-mM KNO3 or KCl treatment. Bars represent standard errors. Different letters represent statistically different means (P < 0.01).
Auxin is known to control primary root growth in a concentration-dependent manner (37). Thus, the observed nitrate-induced auxin activity through AFB3 may lead to a repression in primary root growth in the nitrate condition. To test this hypothesis, we subjected wild-type plants and the AFB3 T-DNA insertional mutant afb3-1 (35) to a 3-day KNO3 or KCl treatment. We measured the primary root length of plants grown for 2 weeks on ammonium as sole N source and after 3 days of 5 mM KNO3 or KCl treatment. At the end of the 3-day treatment, we found that nitrate-treated wild-type plants have shorter primary roots as compared with control-treated plants (Fig. 4D), indicating that nitrate availability inhibits primary root elongation. However, the primary roots of afb3-1 plants were not inhibited by nitrate as observed in wild-type plants or in the other individual auxin receptor mutants tir1-1, afb1-1, and afb2-1 (Fig. 4D and Fig. S4 A and B). These results suggest that AFB3 plays a specific role in modulating primary root growth in response to nitrate. We also analyzed the response of the primary root to KNO3 treatments in the miR393 overexpressor line that shows reduced levels of AFB3 (Fig. 2A). Consistent with the results in the afb3-1 mutant line, the miR393 overexpressor line is completely insensitive to the KNO3 treatments as compared with the wild-type plants (Fig. 4E).
Our results show that AFB3 is involved in primary root growth inhibition in response to nitrate and that this effect is likely mediated by nitrate regulation of AFB3 levels in root tips.
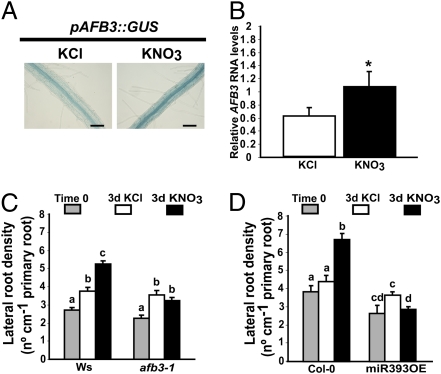
Nitrate Regulates Lateral Root Growth by a Pathway Involving the Auxin Receptor AFB3.
Analysis of GUS activity in the AFB3::GUS reporter lines indicated that AFB3 can be also induced by nitrate in the central vascular area, including the pericycle area (Fig. 5A). It is known that lateral roots are initiated from pericycle founder cells opposite to the xylem poles (38, 39). Therefore nitrate regulation of AFB3 in the pericycle may lead to changes in lateral root development. To quantify expression of AFB3 in the pericycle, we isolated KNO3 or KCl GFP-tagged pericycle cells using a fluorescence activated cell sorter (FACS) and extracted total RNA as described previously (9). KNO3 treatment induced AFB3 expression in the pericycle (Fig. 5B), suggesting that nitrate can also regulate auxin signaling in pericycle cells. Because lateral roots are produced from pericycle cells, we analyzed the number of initiating (stages I, II, III, IV, Va, Vb, VIa, VIb, and VII, after ref. 39) and emerging lateral roots using DIC microscopy in wild-type plants and in the afb3-1 mutant after 3 days of 5 mM KNO3 or 5 mM KCl treatment (Fig. 5C). In wild-type plants, nitrate treatments increased the density of lateral roots (both initiating and emerging) as compared with the KCl control condition. Most of the emerging laterals were short (<0.5 mm; Table S1), which can be related to an inhibitory effect of KNO3 on root elongation as described previously (40). On the other hand, the increased density of initiating laterals (Table S1) can be related to a positive KNO3 effect on root initiation, as described previously (14). The lateral root response was altered in the afb3-1 mutant, which showed decreased density of emerging and initiating lateral roots as compared to wild type (Fig. 5C). However, none of the other individual auxin receptor mutants showed altered lateral root response to nitrate treatments (Fig. S5 A and B). This result shows that, as we have seen for primary root, AFB3 has also a specific role in lateral root response to nitrate. We also analyzed lateral root density in the miR393 overexpressor line (Fig. 5D). Lateral root density after 3 days of KNO3 treatment was no different from lateral root density after 3 days of control treatment, indicating that this response was also absent in this line (Fig. 5D). These results show that nitrate regulates lateral root growth by a pathway involving AFB3.
Fig. 5.
Nitrate regulates lateral root growth by a pathway mediated by AFB3. Plants were grown hydroponically as described before and were treated for the times indicated. (A) pAFB3::GUS plants (35) were treated for 1 h with 5 mM KNO3 or KCl and were then stained for GUS activity for 4 h. Qualitative GUS staining was analyzed using DIC optics. Photographs are representative of at least 15 stained plants. (Scale bar, 100 μm.) (B) Pericycle marker line plants were treated for 1.5 h with 5 mM KNO3 or KCl. Protoplast were prepared from roots and pericycle cells expressing GFP were sorted by FACS. RNA levels for AFB3 were measured using qPCR. Bars represent standard errors. The asterisk represents statistically different means (P < 0.05). (C) The number of initiating and emerging lateral roots of afb3-1 mutants (35) or Ws wild-type plants treated for 3 days with 5 mM KNO3 or KCl was counted using DIC optics. Bars represent standard errors. Different letters indicate statistically different means (P < 0.01). (D) The number of initiating and emerging lateral roots of 35S::miR393 overexpressor plants (28) or Col-0 wild-type plants treated for 3 days with 5 mM KNO3 or KCl was counted using DIC optics. Bars represent SE. Different letters indicate statistically different means (P < 0.01).
Discussion
RSA modulation in response to nutrients is a classical example of plant plasticity to changing environmental conditions (41–43). Given their importance in controlling growth and developmental programs, phytohormones have arisen as the missing links between nutrient availability and plant developmental responses. Because of its central role in root development (44, 45) a role for auxin has been proposed in the modulation of RSA in response to nutrients (46–51). Here, we showed that nitrate treatments modulate both primary and lateral root growth by a pathway involving the N-responsive miR393/AFB3 regulatory module. Recently, another auxin-related regulatory module, miR167/ARF8, has been identified in Arabidopsis to regulate the ratio between initiating and emerging lateral roots (14). miR393/AFB3 regulatory module is unique as nitrate can regulate auxin responses by direct regulation of an auxin receptor, modifying auxin perception in the roots and thus affecting both primary and lateral root growth. The regulatory mechanism of miR393/AFB3 regulation by nitrate involves at least transcriptional and posttranscriptional mechanisms. We showed that nitrate is able to transcriptionally induce expression of AFB3 in roots (Figs. 1, 4A, and 5A) and that N metabolites produced after nitrate reduction and assimilation lead to a downregulation of AFB3 levels due to the induction of miR393 (Figs. 1–3). This mechanism (Fig. S6) is consistent with the type I incoherent feed-forward loop (FFL) motif described for transcriptional networks in yeast, bacteria, and mammals (52–54). In this model, a transcription factor A can activate both a target gene Z and also a repressor of Z in response to a signal, leading to a transient activation of Z. This regulatory design allows for Z to be rapidly responsive to the input signal (55) and can also produce a nonmonotonic response, where the output of Z is first increased with the input signal but decreases when the signal is high (56–58). The observed regulation of AFB3 expression by nitrate and metabolites produced downstream of nitrate reduction might constitute a mechanism to rapidly and precisely adjust root growth depending on external and internal nitrate availability. Incoherent and coherent FFL involving miRNA-target pairs are recurrent motifs in mammalian gene regulatory networks (54, 59–62). Because most miRNA targets encode transcription factors in plants, incoherent FFLs are probably also a common feature of plant gene networks. Besides the FFL reported here, a coherent feed-forward loop involving miR164 and its target ORE1/NAC2 has been recently described in Arabidopsis that regulates age-dependent cell death (63).
AFB3 is part of the ubiquitin protein ligase SCFTIR1/AFB complex that targets and mediates the polyubiquitination and proteasomal degradation of the Aux/IAA transcriptional repressors to promote transcription of auxin-responsive genes (64–66). The finding that both primary and lateral root responses to nitrate are altered in the afb3-1 mutant (Fig. 4D and Fig. 5C) suggests that for this response to occur, a functional AFB3 is required. Moreover, AFB3 is induced by nitrate in primary root tips (Fig. 4C) and in pericycle cells (Fig. 5B). Therefore, we propose that the increase in the expression of AFB3 is responsible for the changes observed in RSA in response to nitrate.
We found that nitrate treatments were able to regulate the levels of auxin-responsive and auxin-related genes in Arabidopsis roots (Fig. S3), as previously demonstrated by network analysis of C- and N-regulated genes (4). However, we did not find misregulation of any of these genes in the afb3-1 mutant, suggesting that nitrate can modulate auxin signaling and responses at multiple levels and in an AFB3-dependent and AFB3-independent manner.
RSA modulation by auxin can depend on three main factors: changes in auxin homeostasis, auxin transport, and auxin signaling. Here we show that N regulation of a hormone receptor can lead to changes in hormonal signaling causing RSA changes. Recently, the auxin receptor TIR1 has been involved in lateral root formation in response to phosphate starvation in Arabidopsis; however, the other family members might also play a role in this response (49). Our evidence shows that RSA modulation in response to nitrate is a specific function of AFB3. Moreover, AFB3 has a dual role in primary and in lateral root development in response to nitrate whereas TIR1 seems to have a role only in lateral root formation (49).
N effects on RSA are complex and have been shown to depend on the N source and concentration available for plants as well as on other environmental conditions. In Arabidopsis, these effects can include changes in primary root growth (67), lateral root initiation (14, 68, 69), and elongation (40, 70). Under our experimental conditions, we found that supplementing 5 mM KNO3 to wild-type ammonium-fed plants causes an inhibition of primary root growth (Fig. 4 D and E) and an increase in initiating and emerging lateral root density as compared with the control KCl condition (Fig. 5 C and D). This observation contrasts the previous observation by Walch-Liu et al. (67), where it was seen that a 5-mM KNO3 treatment stimulated primary root growth when compared with a 5-mM KCl control. This discrepancy may arise from a higher apparent nitrate availability in our hydroponic condition, in which roots are surrounded by media, compared to the vertical plate condition in which roots are superimposed over the media-containing agar. This assumption is supported by findings by Tian et al. (71) where incubation of maize in solutions with KNO3 concentrations of 5 mM or more caused inhibition of root elongation. The inhibitory effect of KNO3 on primary root length was also seen after 18 days of continuous growth in vertical plates containing high nitrate concentrations in Arabidopsis (72). Our results are also similar to the ones reported by Gifford et al. (14), where the same KNO3 treatment caused an increase in the number of initiating lateral roots relative to the number of emerging lateral roots (14) although there is no mention of the effect of nitrate on primary root growth.
Our results are consistent with a model in which a high nitrate provision would prevent further rooting (as it would not significantly enhance N acquisition) by an interaction with the auxin signaling pathway mediated by miR393/AFB3, adjusting plant growth and development to external and internal N availability (Fig. S6).
Materials and Methods
Plant Material.
Arabidopsis (A. thaliana) plants were of Columbia (Col-0) ecotype or Wassilewskija (Ws) ecotype as indicated. tir1-1, afb1-1, afb2-1, afb3-1, pAFB3::GUS, and pAFB3:mAFB3-GUS lines were kindly donated by Mark Estelle, University of California San Diego, La Jolla, CA (33, 35). miR393 overexpressor lines were kindly donated by Jonathan Jones, the Sainsbury Laboratory, John Innes Centre, Norwich, UK (28). Nitrate reductase-NULL mutant lines were kindly provided by Nigel Crawford, University of California San Diego, La Jolla, CA (34). The GFP line that marks the pericycle (E374) was obtained from http://enhancertraps.bio.upenn.edu.
Growth and Treatment Conditions.
Approximately 1,500 Arabidopsis seedlings were grown hydroponically on Phytatrays on MS-modified basal salt media without N (Phytotechnology Laboratories, M531) supplemented with 0.5 mM ammonium succinate and 3 mM sucrose under a photoperiod of 16 h of light and 8 h of darkness and a temperature of 22 °C using a plant growth incubator (Percival Scientific, Inc.). After 2 weeks, plants were treated with 5 mM KNO3 with or without 30 mM sucrose or 5 mM KCl with or without 30 mM mannitol as control for different time periods as indicated. For the phenotypic analysis of the root response to nitrate treatment, seedlings were grown as described above and were treated with 5 mM KNO3 or 5 mM KCl for 3 days.
Histochemical Analysis of GUS Activity.
For histochemical analysis of GUS activity, Arabidopsis seedlings were incubated at 37 °C in a GUS reaction buffer (100 mM sodium phosphate buffer, pH 7.0, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 0.1% (vol/vol) Triton X-100, 0.1% (wt/vol) sodium lauroyl sarcosine) plus 1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-Gluc). The seedlings were cleared according to the protocol described in ref. 39 and were imaged using DIC optics on a Nikon Eclipse 80i microscope. For each marker line and treatment, at least 15 plants were analyzed.
Analysis of Root Architecture Traits.
Initiating and emerging lateral roots (stages I, II, III, IV, Va, Vb, VIa, VIb, and VII, according to ref. 39) were counted using DIC optics on a Nikon Eclipse 80i microscope. For primary root measures, plants were scanned using an Epson Perfection V700 Photo scanner, and roots were measured using the National Institutes of Health program ImageJ. The data were statistically analyzed in the Graph Pad Prism 5 Program.
Supplementary Material
Acknowledgments
We thank Dr. Mark Estelle for kindly providing the tir1-1, afb1-1, afb2-1, afb3-1, pAFB3::GUS, and pAFB3:mAFB3-GUS lines; Dr. Nigel Crawford for providing the NR-null mutant line; and Dr. Jonathan Jones for providing the miR393 overexpressor lines. We thank Dr. Miriam L. Gifford for sharing her experience in cell-specific analysis. This work was funded by the Fondo Nacional de Desarrollo Científico y Tecnológico(1060457), National Institutes of Health-Fogarty International Research Collaboration Award (F6414-01), International Centre for Genetic Engineering and Biotechnology (CRPCHI0501), and Millennium Nucleus for Plant Functional Genomics (P06-009-F) to R.A.G and PhD fellowship from Comisión Nacional de Investigación Científica y Tecnológica (AT-24080114) to E.A.V.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909571107/DCSupplemental.
References
- 1.Wang R, et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136:2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang R, Okamoto M, Xing X, Crawford NM. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003;132:556–567. doi: 10.1104/pp.103.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheible WR, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infra-structure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutierrez RA, et al. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007;8:R7. doi: 10.1186/gb-2007-8-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palenchar PM, Kouranov A, Lejay LV, Coruzzi GM. Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biol. 2004;5:R91. doi: 10.1186/gb-2004-5-11-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 8.Voinnet O. Origin, biogenesis, and activity of plant micrornas. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 9.Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Curr Opin Plant Biol. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Sunkar R, Chinnusamy V, Zhu J, Zhu J-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Fujii H, Chiou TJ, Lin SI, Aung K, Zhu J-K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Bari R, Pant BD, Stitt M, Scheible W-R. PHO2, micro RNA399 and PHR1 define a phosphate signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pant BD, et al. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time PCR profiling and small RNA sequencing. Plant Physiol. 2009;150:1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloom J, Khan Z, Kruglyak L, Singh M, Caudy A. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics. 2009;10:221. doi: 10.1186/1471-2164-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu X, et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C, et al. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 20.Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 21.Lu C, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasschau KD, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fahlgren N, et al. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Heim MA, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 27.Sunkar R, Zhu J-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 29.Benjamins R, Scheres B. Auxin: The looping star in plant development. Annu Rev Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- 30.Walch-Liu P, et al. Nitrogen regulation of root branching. Ann Bot (Lond) 2006;97:875–881. doi: 10.1093/aob/mcj601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forde BG. Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol. 2002;53:203–224. doi: 10.1146/annurev.arplant.53.100301.135256. [DOI] [PubMed] [Google Scholar]
- 32.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 33.Parry G, et al. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, Xing X, Crawford N. Nitrite acts as transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol. 2007;145:1735–1745. doi: 10.1104/pp.107.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F-box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans ML, Ishikawa H, Estelle MA. Responses of Arabidopsis roots to auxin studied with high temporal resolution: comparison of wild type and auxin-response mutants. Planta. 1994;194:215–222. [Google Scholar]
- 38.Dolan L, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 39.Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Jennings A, Barlow PW, Forde BG. Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA. 1999;96:6529–6534. doi: 10.1073/pnas.96.11.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osmont KS, Sibout R, Hardtke CS. Hidden branches: Developments in root system architecture. Annu Rev Plant Biol. 2007;58:93–113. doi: 10.1146/annurev.arplant.58.032806.104006. [DOI] [PubMed] [Google Scholar]
- 42.Forde BG, Walch-Liu P. Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant Cell Environ. 2009;32:682–693. doi: 10.1111/j.1365-3040.2008.01927.x. [DOI] [PubMed] [Google Scholar]
- 43.Desnos T. Root branching responses to phosphate and nitrate. Curr Opin Plant Biol. 2008;11:82–87. doi: 10.1016/j.pbi.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Teale WD, Paponov IA, Palme K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 45.Benkova E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 46.Mishra BS, Singh M, Aggrawal P, Laxmi A. Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS One. 2009;4:e4502. doi: 10.1371/journal.pone.0004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Calderon L, et al. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:174–184. doi: 10.1093/pcp/pci011. [DOI] [PubMed] [Google Scholar]
- 48.Nacry P, et al. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 2005;138:2061–2074. doi: 10.1104/pp.105.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Torres CA, et al. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell. 2008;20:3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez-Bucio J, et al. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 2005;137:681–691. doi: 10.1104/pp.104.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franco-Zorrilla JM, Martin AC, Leyva A, Paz-Ares J. Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 2005;138:847–857. doi: 10.1104/pp.105.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 53.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mangan S, Itzkovitz S, Zaslaver A, Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol. 2006;356:1073–1081. doi: 10.1016/j.jmb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 56.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 57.Entus R, Aufderheide B, Sauro H. Design and implementation of three incoherent feed-forward motif based biological concentration sensors. Syst Synth Biol. 2007;1:119–128. doi: 10.1007/s11693-007-9008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaplan S, Bren A, Dekel E, Alon U. The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol Syst Biol. 2008;4:203. doi: 10.1038/msb.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLOS Comput Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Y, Ferguson J, Chang J, Kluger Y. Inter- and intra-combinatorial regu-lation by transcription factors and microRNAs. BMC Genomics. 2007;8:396. doi: 10.1186/1471-2164-8-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen EEW, et al. A feed-forward loop involving protein kinase C-alpha and microRNAs regulates tumor cell cycle. Cancer Res. 2009;69:65–74. doi: 10.1158/0008-5472.CAN-08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim JH, et al. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009;323:1053–1057. doi: 10.1126/science.1166386. [DOI] [PubMed] [Google Scholar]
- 64.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 65.Mockaitis K, Estelle M. Auxin receptors and plant development: A new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- 66.dos Santos Maraschin F, Memelink J, Offringa R. Auxin-induced, SCFTIR1-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 2009;59:100–109. doi: 10.1111/j.1365-313X.2009.03854.x. [DOI] [PubMed] [Google Scholar]
- 67.Walch-Liu P, Forde BG. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J. 2008;54:820–828. doi: 10.1111/j.1365-313X.2008.03443.x. [DOI] [PubMed] [Google Scholar]
- 68.Remans T, et al. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006;140:909–921. doi: 10.1104/pp.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Little DY, et al. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- 71.Tian Q, Chen F, Liu J, Zhang F, Mi G. Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J Plant Physiol. 2008;165:942–951. doi: 10.1016/j.jplph.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 72.Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002;29:751–760. doi: 10.1046/j.1365-313x.2002.01251.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.