Abstract
Toxin-antitoxin (TA) systems form a ubiquitous class of prokaryotic proteins with functional roles in plasmid inheritance, environmental stress response, and cell development. ParDE-family TA systems are broadly conserved on plasmids and bacterial chromosomes, and have been well characterized as genetic elements that promote stable plasmid inheritance. We present a crystal structure of a chromosomally-encoded ParD-ParE complex from Caulobacter crescentus at 2.6 Å resolution. This TA system forms an α2β2 heterotetramer in the crystal and in solution. The toxin-antitoxin binding interface reveals extensive polar and hydrophobic contacts of ParD antitoxin helices with a conserved recognition and binding groove on the ParE toxin. A cross-species comparison of this complex structure with related toxin structures identified an antitoxin recognition and binding sub-domain that is conserved between distantly-related members of the RelE/ParE toxin superfamily despite low overall primary sequence identity. We further demonstrate that ParD antitoxin is dimeric, stably folded, and largely helical when not bound to ParE toxin. Thus, the paradigmatic model in which antitoxin undergoes a disorder-to-order transition upon toxin binding does not apply to this chromosomal ParD-ParE TA system.
Two-gene operons encoding toxin-antitoxin (TA) systems are broadly-distributed components of plasmids, bacterial and archaeal chromosomes, and bacteriophage genomes (1–3). TA toxins kill or arrest growth of bacterial cells by inhibiting essential processes including DNA replication and translation (4–7). TA antitoxins bind and neutralize their cognate, co-transcribed toxin proteins, but are generally less structured than toxins and thus more susceptible to proteolysis (8–10). The differential protein stability between toxins and antitoxins underlies the function of TA systems as genetic stability elements. Specifically, plasmid and prophage TA systems increase stability of their encoding genes via post-segregational killing of daughter cells that fail to inherit the TA genes (11–15). The functions of chromosomally-encoded TA systems appear to be more diverse: these genes have been implicated in processes as varied as stress adaptation (4, 5, 16), persistence (17), maintenance of genome stability (18), and multicellular development (19). For some chromosomal TA systems, the association between toxin and antitoxin genes and a cellular phenotype remains under debate (20).
TA systems are typically classified by the primary sequence of their toxins, as antitoxins are poorly conserved and more difficult to classify phylogenetically. A broadly distributed superfamily of TA toxins is RelE/ParE, which encompasses several smaller toxin families including RelE, YoeB, and ParE (1). The ParE toxin, originally identified as a stabilizing element on plasmid RK2 (13, 15), inhibits DNA gyrase and thereby blocks DNA replication (21). RelE and YoeB, on the other hand, function to inhibit translation by inducing cleavage of mRNAs in the ribosomal A-site (22–25). Thus, phylogenetically related toxins can function to target distinct cellular processes: replication and translation. The molecular basis of how related toxins interact with their targets is not well understood, nor is the structural basis of how these toxins specifically recognize and bind their cognate antitoxin.
A current model of recognition and binding in TA systems invokes a disorder to order transition in the antitoxin. Specifically, a disordered region of the free antitoxin is organized into a well-defined secondary structure upon binding to its cognate toxin (26–30). The Phd-Doc system of prophage P1 is a well-characterized system that exhibits this behavior: binding of Doc toxin induces α-helix formation in an unstructured region of its cognate antitoxin, Phd (29). Analogously, RelE toxin of Escherichia coli induces secondary structure formation in the disordered C-terminus of its cognate antitoxin, RelB (30). The C-terminus of unbound ParD antitoxin from plasmid RK2 is also reported to be disordered in solution (28). However, the YoeB-YefM TA system of Mycobacterium tuberculosis presents an exception to this disorder-order binding model; the YefM antitoxin is not intrinsically disordered, but rather forms a stably-folded structure in the absence of toxin (31). Therefore, the disorder to order recognition model is not universally applicable to TA antitoxins.
The chromosome of the α-proteobacterium, Caulobacter crescentus, encodes four parDE TA operons (parDE1-4) (3). Of the C. crescentus parDE systems, the proteins encoded by parDE1 are most similar to the archetypal ParD (antitoxin)-ParE (toxin) system first characterized on plasmid RK2 (15). Therefore, in this study we have chosen to investigate the biochemical properties of the C. crescentus ParD1-ParE1 complex as a general model. Henceforth, we will refer to the C. crescentus proteins simply as ParD and ParE.
Prior to this study, no high resolution structural information has been presented on toxin-antitoxin interaction in a ParD-ParE family complex. We present the crystal structure of a heterotetrameric ParD-ParE complex at 2.6 Å resolution. The heterotetrameric state observed in the crystal is consistent with the oligomeric state in solution as assessed by sedimentation velocity analytical ultracentrifugation (AUC) and small angle X-ray scattering (SAXS). As in the RK2 system (32), we demonstrate that unbound C. crescentus ParD antitoxin is dimeric and that the N-terminus of ParD forms a ribbon-helix-helix DNA binding motif. Unlike RK2 ParD, in which the majority of the protein is natively unstructured (32), circular dichroism (CD) spectroscopy, CD thermal denaturation, and one-dimensional 1H nuclear magnetic resonance (NMR) spectroscopy reveal that unbound C. crescentus ParD antitoxin is largely α-helical and stably folded in solution.
Overall, the data presented herein elucidate the specific molecular contacts responsible for ParD-ParE binding, and identify general structural features that underlie toxin-antitoxin binding and specificity in the ParDE/RelBE superfamily of TA systems. Moreover, the data demonstrate that unbound ParD antitoxin encoded by this chromosomal TA locus forms a folded, primarily α-helical structure, and thus does not undergo a substantial disorder to order transition upon binding to ParE toxin.
Materials and Methods
Construction of ParD-ParE and ParD protein expression vectors
The C. crescentus parD gene (locus CC-0874) was amplified by PCR and cloned into the NdeI and XhoI sites of pETDuet1 (Novagen, Madison, WI) to generate a plasmid carrying wild-type parD under the control of an inducible T7 promoter. This plasmid was then transformed into E. coli DH5α amplified, purified, and digested with EcoRI and BamHI. The PCR-amplified parE gene (CC-0873) was then ligated into the EcoRI and BamHI sites to generate plasmid pAF178. pAF178 thus encodes an N-terminal polyhistidine-tagged ParE toxin and an untagged ParD antitoxin; each gene is under the control of separate T7 promoters. Additionally, parD was cloned into the EcoRI and BamHI sites of pETDuet1. This plasmid was sequenced and subsequently cut with NdeI and XhoI and the parE gene was ligated in to generate pAF202. However, sequence confirmation of pAF202 revealed that toxin parE was inserted incorrectly into the vector. Therefore, under inducing conditions, pAF202 generates only a polyhistidine tagged version of antitoxin ParD. All PCR amplifications were carried using whole C. crescentus cells to provide a chromosomal gene amplification template. PCR reactions for gene amplification were conducted in the presence of 5% DMSO using KOD Hot Start Polymerase (Novagen, Madison, WI).
Expression and purification of the ParD-ParE complex and free ParD
Expression plasmids pAF178 and pAF202 were transformed into electrocompetent E. coli Rosetta2(DE3)pLysS (Novagen, Madison, WI). 10mL starter cultures of containing pAF202 in LB medium containing 100μg/ml ampicillin were allowed to grow to saturation overnight in a 30°C incubator. These were added to 1.5L of LB medium containing 100μg/ml ampicillin. The inoculated cultures of Rosetta 2(DE3)pLysS containing pAF202 were shaken in an incubator at 37°C, allowed to grow to an optical density of 0.4–0.5 OD600, and expression was induced with the addition of 0.5mM IPTG. Four hours after induction, cell cultures were pelleted by centrifugation and frozen in a −20°C freezer. Cultures of Rosetta 2(DE3)pLysS containing pAF178 were grown and induced as described by Johnson et al (33). Cells were then resuspended in buffer A (20mM Tris pH 8.0, 200 mM NaCl, 50mM imidazole), lysed by sonication, the cell debris was spun down at 18,000 rpm for 10 minutes, and the lysate was loaded onto GE Chelating Sepharose Fast Flow resin (GE Life Sciences, Piscataway, NJ) that had been pre-bound with Ni2+. The protein was eluted from the column over a linear gradient from 50 to 500 mM imidazole. The eluate was analyzed on 16% polyacrylamide gel using the tricine-SDS protocol of Schagger and von Jagow (34), and judged to contain stoichiometric quantities of His6-ParE and ParD. The protein was collected, dialyzed against 20 mM Tris pH 8.0, and loaded onto a column containing GE Q-Sepharose anion exchange resin. The protein was eluted from the ion exchange column using a linear NaCl gradient from 0 to 500 mM, and judged to be greater than 95% pure.
HPLC purification of ParD and His6-ParD antitoxin
Crude His6-ParD and was purified by the same affinity protocol described above for the ParDE complex. Reverse-phase HPLC was used to further purify His6-ParD expressed from pAF202 and to separate untagged ParD from the purified ParD-His6-ParE complex expressed from pAF178. Samples for HPLC purification were prepared by the addition of 0.1% trifluoroacetic acid, loaded onto a Grace Vydac 214TP C4 reversed-phase HPLC column, and eluted over a linear gradient from 10 to 70% acetonitrile. Fractions were collected and lyophilized and purity was assessed on 16% SDS-tricine gels. Molecular weights of these purified proteins were confirmed by electrospray ionization mass spectrometry.
Expression of selenomethionine-labeled protein for crystallography
ParD-His6-ParE protein complex was expressed from pAF178 in Rosetta2(DE3)pLysS in M9 minimal medium with 4% glucose, supplemented with 60mg of Se-Met and 90mg each of Lys, Phe, Thr, Ile, Leu, and Val per liter, and allowed to induce overnight; protocol according to Doublié (35). Selenomethionine labeled protein was purified by the same protocol as native protein with 5 mM BME added to the buffers to prevent oxidation of selenomethionine.
ParD-ParE complex crystal growth
Initial crystallization conditions were identified in a high throughput screen conducted by the Hauptman-Woodward Medical Research Institute (36). Diffraction quality crystals of Se-Met ParD-His6-ParE complex were grown via hanging drop vapor diffusion from a well buffer containing 100 mM MES pH 7.5, 100 mM (NH4)2HPO3, 10% PEG 20,000. Purified ParD-ParE protein complex in 20 mM Tris buffer pH 8.0 was diluted 1:1 with well buffer. Drops were set at 2, 4, and 6 μl total volume.
X-ray crystallography data collection, phasing, and model building
5 selenium atoms were located in the unit cell using SHELXD (37) from a single 3.0 Å resolution data set collected at an energy of 12.66 keV (0.9793 Å) at 100 K on beamline 21-ID-D (Advanced Photon Source, Argonne, IL). Heavy atom positions were refined in MLPHARE (38) and initial phases were determined using Phaser (39). A partial 3.0 Å resolution model of the ParD-ParE complex was built automatically using Buccaneer (40).
A second 2.6 Å resolution data set collected at an energy of 12.66 keV (0.9793 Å) at 100 K on beamline 24-ID-C (Advanced Photon Source, Argonne, IL) was phased by molecular replacement with the initial 3.0 Å structural model using Phaser (39). Manual model building and refinement was carried out iteratively in Coot (41). Refmac5 or Phenix.refine (42) were used to calculated 2FO-FC as well as FO-FC maps at each stage in the iterative refinement process. The agreement of the model with these maps was examined in Coot, and manual model correction was carried out between each cycle of refinement whenever necessary. Maximum likelihood model refinement was carried out initially in Refmac5 (43) using 2-fold non-crystallographic symmetry constraints, TLS and anisotropic scaling. Further model refinement and addition of solvent molecules was carried out using Phenix. Solvent placement was examined first by the “check waters” function in Coot first and secondly by hand to yield 46 water molecules in the final build. The final refinement preceding submission to the Protein Data Bank was carried out in phenix.refine using individual isotropic atomic displacement parameters for each atom, bulk solvent flattening, and anisotropic scaling. The data were strongly anisotropic based on spread in values of the three principle components (30.98 Å2), as assessed by the ellipsoidal truncation and anisotropic scaling routine of Eisenberg and colleagues (44). The statistics in Table 1 reflect this anisotropy. The coordinates of the ParD-ParE complex structure have been deposited in the Protein Data Bank (http://www.rcsb.org) under PDB ID 3KXE.
Table 1.
Crystallographic Data and Refinement Statistics
| Data Collection Statistics | Se-Met (1) | Se-Met (2) |
|---|---|---|
| Energy (keV) | 12.66 | 12.66 |
| Resolution range (Å) | 20-3.00 (3.07-3.00) | 20-2.60 (2.64-2.60) |
| Unique Reflections | 7080 | 11164 |
| Rmergea | 0.10 (0.63) | 0.10 (0.58) |
| 〈 I 〉/〈 sI 〉 | 32.3 (3.2) | 22.2 (2.5) |
| Redundancy | 13.3 (12.0) | 5.2 (4.9) |
| Completeness | 99.7 (98.0) | 99.8 (99.3) |
| Phasing statisticsb | Resolution shells (Å) | |||||||
|---|---|---|---|---|---|---|---|---|
| 20-10.6 | 10.6-7.1 | 7.1-5.7 | 5.7-4.8 | 4.8-4.3 | 4.3.-3.9 | 3.9-3.6 | 3.6-3.4 | |
| Overall | ||||||||
| Figure of Merit 0.34 | 0.31 | 0.45 | 0.43 | 0.40 | 0.36 | 0.33 | 0.27 | 0.23 |
| Refinement Statistics | |
|---|---|
| Space group | P212121 |
| a, b, c (Å) | 65.7, 72.7, 76.8 |
| Rcrystc | 23.7 |
| Rfreed | 29.1 |
| 〈B〉 (Å2) | 41.5 |
| Rmsde bond lengths (Å) | 0.014 |
| Rmsd bond angles (°) | 1.532 |
| Ramachandran analysis | |
|---|---|
| Preferred (%) | 97.3 |
| Allowed (%) | 2.7 |
| Disallowed (%) | 0 |
Rmerge = ΣhklΣi |Ii-〈I〉|/ΣhklΣiIi, for all data I/σ(I) > −3
Phases were determined by selenium SAD from data set Se-Met(1).
Rcryst = Σhkl ||Fobs|−|Fcalc||/Σhkl|Fobs|, includes all data
Rfree uses 4.8% of the data for the test set.
rmsd, root-mean-square deviation
Calculation of protein physical and structural properties
Isoelectric points were estimated from primary sequence using the ProtParam application of the ExPASy Proteomics Server (45). Surface areas were estimated in AreaImol of the CCP4 Suite (38). Hydrodynamic radii were estimated for each of the gel-filtration standard proteins based on high-resolution structures: aprotinin (PDB ID 2ZJX), ribonuclease A (PDB ID 2AAS), carbonic anhydrase (PDB ID 1V9E), and ovalbumin (PDB ID 1OVA). Hydrodynamic radius estimates were calculated using the Simulaid software package (http://atlas.physbio.mssm.edu/~mezei/simulaid/).
Analytical ultracentrifugation
The multimeric state of the ParD-His6-ParE complex in solution was measured via sedimentation velocity ultracentrifugation on a Beckman Optima XL-A analytical ultracentrifuge (Beckman Coulter, Fullerton, CA). 215μL of protein complex solution (A280=0.6) was loaded into the cell in 20 mM Tris pH 8.0, 200mM NaCl and spun at 40,000 rpm. 300 successive scans were measured over a time course of 18 hours at a temperature of 20° C. Data were fit to the Lamm equation in Sedfit (46) and a differential molar mass distribution curve was generated.
Small Angle X-Ray Scattering
SAXS data were collected at the Advanced Photon Source beamline 18-ID. Se-Met ParD-His6-ParE in 20 mM Tris pH 8.0, 200 mM NaCl was suspended in a 1 mm capillary at final concentrations of 500 mM and 15 mM and small angle scattering was measured from this sample. Scattering data were recorded on an Aviex CCD detector (Aviex, Chicago, IL) and data analysis carried out using a custom SAXS analysis plugin implemented in Igor Pro (WaveMetrics, Lake Oswego, OR). A P(r) plot was calculated from the SAXS data using the indirect Fourier inversion algorithm of Svergun (47) in Gnom version 4.5a. The shell Rg of the ParD-ParE heteroteramer crystal structure was calculated using Crysol version 2.6 (48)
Circular dichroism spectroscopy and thermal denaturation
Lyophilized ParD was resuspended in 50 mM potassium phosphate pH 7.2. The CD spectrum of ParD antitoxin was measured on a Jasco J-715 spectropolarimeter at a 1 nm bandwidth with 1 nm step resolution from 200 nm to 250 nm. Data were averaged over 3 accumulations. The thermal denaturation curve of ParD was measured by monitoring the change in ellipticity at 222 nm in the spectropolarimeter using a Jasco PFD-425S Peltier temperature controller. Temperature was raised at a rate of 1°C/minute; data were acquired at a 2 nm bandwidth at 222 nm at a frequency of 0.25 hertz.
Nuclear Magnetic Resonance Spectroscopy
Lyophilized ParD was resuspended in 50 mM phosphate buffer pH 7.2 at a concentration of 500μM. 10% D2O was added to the solution. A 1H NMR spectrum was acquired on a 600MHz Varian Unity Spectrometer (Varian Inc., Palo Alto, CA) at 25°C.
Results
ParD-ParE forms a heterotetrameric protein complex in crystal and in solution
The structure of the ParD-ParE complex was initially solved at 3.0 Å resolution from a single seleno-methionine crystal phased by single-wavelength anomalous dispersion (49). A higher resolution data set from a second crystal, collected to 2.6 Å resolution, was phased by molecular replacement using the initial 3.0 Å model. We refined the 2.6 Å model to a final Rcryst of 23.7 and Rfree of 29.1 (see Table 1). Residues M1, G96, and the N-terminal polyhistidine tags of both ParE monomers are poorly resolved in the 2.6 Å electron density maps. M1-K4 of both ParD monomers, A79-E88 of one ParD monomer, and P80-E88 of the other are poorly resolved as well. These regions are either disordered or were proteolyzed during purification and crystallization.
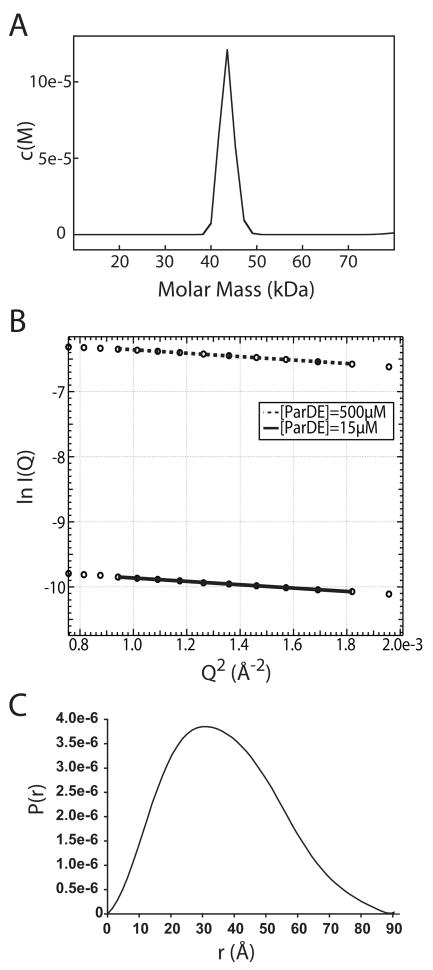
The quaternary structure of the complex in the asymmetric unit is an α2β2 heterotetramer (Figure 1). Two homodimer interfaces are evident in the ParD2-ParE2 tetramer. The ParD antitoxin in the crystal structure homodimerizes via an N-terminal ribbon-helix-helix DNA binding motif. The antitoxin C-termini extend outward from this dimerization domain sequestering the toxin homodimer between two largely helical domains (Figure 1). To test whether the oligomeric state of the ParD-ParE complex observed in the crystal structure is consistent with the oligomeric state in solution, we measured the sedimentation properties and solution scattering of the purified ParD-ParE complex. The results of both solution methods are consistent with a ParD2-ParE2 heterotetramer. The apparent molecular weight of the complex was measured to be 43.5 kDa by sedimentation velocity ultracentrifugation (Figure 2A); a tetrameric complex formed by the association of a ParD dimer with a ParE dimer has a predicted molecular weight of 44.6 kDa. Small angle X-ray scattering (SAXS) further confirmed the oligomeric state observed in the crystal. A Guinier fit of the SAXS data yielded an apparent solution state radius of gyration (Rg) for the complex of 28.1 ± 0.04 Å (at [ParD-ParE]= 500 mM) and 27.8 ± 0.5 Å (at [ParD-ParE]= 15 mM) (Figure 2B), while calculation of a P(r) plot yielded an Rg of 27.6 ± 0.02 Å (Figure 2C). These results are consistent with a hydrated shell Rg of 27.9 Å calculated from the heterotetramer crystal structure (Figure 1). Thus, we conclude that the ParD2-ParE2 heterotetrameric complex observed in the crystal structure is the same oligomer that we observe in solution.
Figure 1.

2.6 Å structure of the heterotetrameric ParD-ParE complex of C. crescentus. The ParE toxin is rendered in blue and the ParD antitoxin in red. The N-terminal ParD ribbon-helix-helix DNA binding motif (RHH) is labeled. The 4 secondary structural elements (®1, α1, α2, α3) of ParD are labeled in red on both monomers.
Figure 2.
A) Sedimentation velocity c(M) plot for the ParD-ParE complex. The predominant species, at 0.6 mg/ml, has a sedimentation coefficient that corresponds to a molecular weight of 43.5 kDa. B) Guinier analysis of small angle X-ray scattering data for the ParD-ParE complex. Fitted plots of protein solution at 500μM and 15μM yielded Rg values of 28.2 ± 0.04 and 27.8 ± 0.5 Å, respectively. C) P(r) plot of ParD-ParE calculated from the crystal structure yields a predicted hydration shell radius of gyration of 27.9 Å, consistent with the experimental SAXS data.
ParE is Structurally Homologous to E. coli YoeB and RelE Toxins
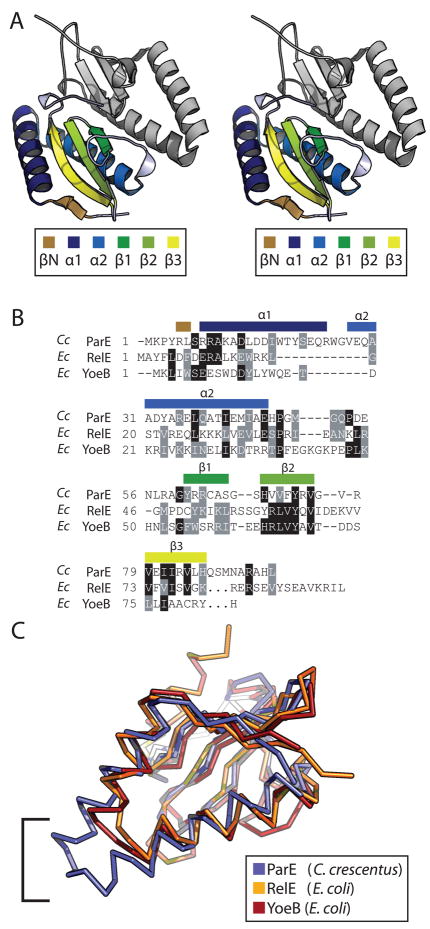
The ParE toxin is a basic (calculated isoelectric point = 8.9), 96 amino acid protein. The crystal structure of the complex reveals that ParE contains two N-terminal α-helices; α1 spans S7 to R24 and α2 spans G26 to E46. These helices are connected by a single tryptophan, W26, and run antiparallel forming a hairpin tertiary structure that packs against a three-stranded antiparallel β-sheet. β1 spans R62 to S66, β2 spans H69 to V75, and β3 spans V79 to L85. A short N-terminal β-strand, βN, hydrogen bonds in parallel to β3 of the antiparallel β-sheet (Figure 3A).
Figure 3.
A) Wall-eyed stereo rendering of the ParE toxin dimer. The lower toxin monomer is colored by secondary structure as indicated in the legend. The second monomer is rendered in grey for completeness. B) Structure-based sequence alignment of C. crescentus ParE, and E. coli RelE and YoeB, based on pairwise least-squares fitting of Ca for each structure. Sequence position of secondary structure elements are labeled above the alignment. C) An overlay of the Ca traces for C. crescentus ParE, E. coli RelE, and E. coli YoeB colored as indicated in the legend. The bracket indicates the area of C. crescentus ParE α1 + α2 that does not overlay with its structural homologs due to longer helices.
A search for structural homologs using the DALI server (50) revealed that C. crescentus ParE is most similar to the E. coli YoeB and RelE endoribonucleases, which are part of the YefM-YoeB and RelB-RelE TA systems, respectively (22, 51) (Figures 3B & 3C). YoeB has the highest structural homology with C. crescentus ParE with a backbone RMSD of 1.8 Å and a Dali Z-score of 10.8. The second highest hit to ParE, is endoribonuclease RelE which has a backbone RMSD of 2.2 Å and a Z-score of 8.4. The primary differences between C. crescentus ParE and its structural homologs from E. coli are localized to the N-terminal helices and extreme C-termini (Figure 3B & 3C). α1 of ParE is 5 residues longer than α1 of YoeB and 9 longer than α1 of RelE;α2 of ParE is 4 residues longer than the homologous helices of YoeB and RelE. The consequence of these extended helices in C. crescentus ParE is a substantially longer α1-α2 helix hairpin (Figure 3C). Furthermore, the toxin C-termini diverge: RelE contains an extended α-helix at its C-terminus, while the corresponding region of ParE is composed of random coil; in YoeB, the C-terminus is truncated.
Although ParE is highly homologous to RelE toxin at the level of primary sequence and tertiary structure, it does not contain any of the 3 critical catalytic residues required for mRNA cleavage on the ribosome (52). Thus, like its RK2 ortholog (21), C. crescentus ParE likely acts as an inhibitor of DNA gyrase.
ParD contains a N-terminal RHH motif and extended C-terminal helices
The ParD antitoxin protein observed in the crystal structure dimerizes via an N-terminal ribbon-helix-helix DNA binding domain. The dimer interface exhibits the characteristic intermolecular two-stranded antiparallel β-sheet of an RHH motif (Figure 1) (53). The four N-terminal residues of the antitoxin are not present in the electron density. As such, the N-terminal ParD β-sheet appears truncated when compared to other structures with RHH motifs (53). β1 of the ParD antitoxin is connected by a 3 amino acid coil to the 12 residue α1 helix. α1 is connected by a 5 amino acid coil to the helix α2, which packs against the N-terminal sheet and extends outward to the toxin-antitoxin interface. The tertiary arrangement of the ParD RHH motif is similar to the RelB antitoxin dimerization domain (30), but differs from the YoeB antitoxin, YefM, which has an unrelated N-terminal domain that is homologous to the Phd antitoxin of phage P1 (31). Helix α2 is 48 Å long and spans ParD residues S29 through S61. The C-terminal end of this helix interacts with a hydrophobic patch on the surface of ParE. Residues G62-F67 pack against the ParE toxin as a coil that connects α2 to the C-terminal helix α3. Helix α3 further interacts with another hydrophobic patch on the surface of ParE.
ParD binds ParE via conserved hydrophobic grooves
The interaction between the ParD antitoxin and ParE toxin proteins is largely mediated by conserved hydrophobic residues that line the bottom of two antitoxin binding grooves on the surface of the toxin dimer. An alignment of a broad phylogenetic range of ParE proteins by Anantharaman and Aravind (1) identified a number of residues that are greater than 80% conserved across the ParE/RelE superfamily. Coloring these highly conserved residues on a surface rendered model of ParE shows that they are largely restricted to two hydrophobic grooves on the toxin dimer, which form the critical structural motifs involved in ParD antitoxin binding (Figure 4A and 4B). If the toxin dimer is considered without the antitoxin, these conserved hydrophobic residues account for only 4.5% of its total surface area. However, these residues are overrepresented at the toxin-antitoxin interface where they account for 14% of the toxin surface area occluded by antitoxin. Of the 4.5% surface area that is highly conserved on the toxin dimer, 81% is covered by the antitoxin in the complex crystal structure.
Figure 4.
Surface representations of the C. crescentus ParE toxin monomer bound to the ParD antitoxin. The ParD antitoxin is shown as a stick model rendered in red; only Ala46 through Ala79 are shown. A) Hydrophobic surface plot of ParE based on the Protscale script for PyMOL. Blue indicates hydrophobic surface; white indicates hydrophilic surface. B) The ParE monomer is rendered in grey with yellow indicating residues identified by Anantharaman and Aravind (1) as greater than 80% conserved across the ParE/RelE superfamily. C) Surface representation of E. coli RelE toxin bound to RelB (red). D) E. coli YoeB surface bound to YefM (red).
Each ParD antitoxin interacts with a binding groove presented by a single toxin monomer. Within each binding groove, there are two hydrophobic subdomains where the antitoxin adopts an α-helical secondary structure. At both positions, ParD antitoxin buries multiple hydrophobic residues into the surface of the toxin. The first of the ParE hydrophobic subdomains is formed by conserved residues within α1 and α2, and interacts with the C-terminal ParD α3 helix (Figure 5A). Toxin residues Y4 and L6 from βN, A10 and L14 of α1, and L37 and I41 of α2 are the key conserved residues involved in subdomain 1 formation. ParD antitoxin buries several hydrophobic residues from its α3 into hydrophobic subdomain 1 including three phenylalanine side chains (F67, F69, F72) and I73 (Figure 5A).
Figure 5.
View of the two hydrophobic subdomains of the ParD-ParE binding groove. ParE is rendered as a hydrophobic surface based on the Protscale script for PyMOL. Blue represent hydrophobic surface while white indicates hydrophilic. The ParD antitoxin backbone is rendered in red with key hydrophobic sidechains rendered as ball and stick models. A) At hydrophobic subdomain 1, the helical antitoxin inserts F67, F69, F72, & I73 into a cavity on the toxin surface. The region N-terminal to this on ParD adopts a random coil conformation stabilized by F63 and I64. B) α2 of ParD buries L48, L51, L55, & I56 against the toxin surface.
ParE hydrophobic subdomain 1 is connected to hydrophobic subdomain 2 by a highly basic region, in which ParD adopts a random coil conformation that is stabilized by two hydrophobic residues (Figure 6B). ParD residues F63 and I64 (Figure 5A), which are contained within this region, alone account for 13% of the antitoxin surface area buried at the ParD-ParE interface. They are therefore, the key residues which stabilize the random coil conformation in the groove. Hydrophobic patch 2 on ParE toxin is formed principally by residues from the antiparallel C-terminal β2 and β3 strands. The conserved F72 of ParE β2 forms the bottom of this groove and contacts four residues in the antitoxins α2 helix: L48, L51, L55, and I56 (Figure 5B).
Figure 6.
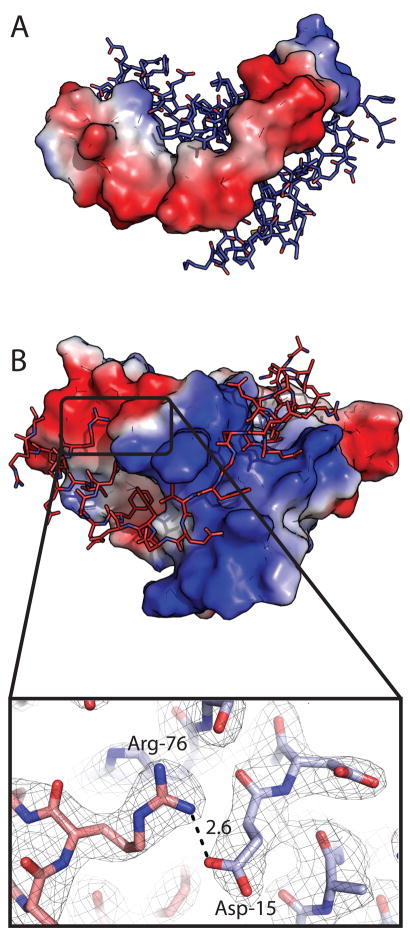
A) ParD electrostatic contact potential map (red = acidic, blue = basic) modeled with ParE as ball-and-stick. B) Electrostatic contact potential of ParE with ParD modeled as ball-and stick. ParD contacts an intensely basic binding groove in the toxin surface potential map. Inset: Salt-bridge between ParD R76 and ParE D15. Carbon atoms of ParD are represented in red, carbon atoms of ParE are represented in blue. Simulated annealing omit map was generated by omitting R76 and D15 prior to refinement; map contoured at 1 sigma. The electrostatic potential surfaces were calculated in PyMOL using the generate-vacuum electrostatics function.
The ParD and ParE dimers contribute a total of 6319 Å2 to the toxin-antitoxin interfaces; this comprises 27.7% of the total surface area of the ParE2 and ParD2 dimers considered in isolation. If the four monomers in the crystal structure are considered as individual chains, their totoal surface area is 27,084 Å2 with 10,698 Å2 buried at the two homodimer interfaces and at the toxin-antitoxin interfaces. Thus approximately 40% of the total surface area of the four monomers is buried during oligomerization.
Extensive charge complementarity is observed at the ParD-ParE interface
Extensive Coulomb charge interactions are evident in the structure of the ParD-ParE complex in the form of interfacial salt bridges. The ParE surface recognized by ParD antitoxin is highly basic in the region between the two hydrophobic subdomains (Figure 6B). The corresponding region of ParD is commensurately acidic (Figure 6A). The C-terminal region of ParD, which contacts the surface of the toxin dimer (A46 to A79) has a theoretical isoelectric point (pI) of 4.25. Limiting the calculation to the residue range spanning the basic region of the toxin groove yields a theoretical pI of 3.68. In this region of ParD antitoxin, from E57 to D65, three interfacial salt bridges are observed between each ParD-ParE monomer (Figure 7).
Figure 7.

Interfacial salt bridges. (A) Ring of salt bridges linking residues R9, D13, R83 of ParE and E57 of ParD. (B) Chain of salt bridges linking residues R58, R74, E79, and R5 of ParE and E65 and E59 of ParD. A water molecule binding to the carboxyl group of ParD E65 and ParE R5 is shown as a red sphere. Side chains are rendered in red for ParD and blue for ParE. Labeled distances are in Å. Simulated annealing omit maps were generated by omitting the labeled residues; maps contoured at 1.0 sigma.
ParD E75 contacts both ParE R9 and R83. These two positively charged toxin residues are flanked by ParE D13, forming an interfacial charge network involving four residues (Figure 7A). Additionally, E59 and E65 of ParD contact R58 and R5 of ParE toxin, respectively, to form two additional interfacial salt bridges (Figures 7B and 7C). These salt bridges have the expected polarity given the acidic isoelectric point of ParD and basic isoelectric point of ParE. We note one well-ordered salt-bridge of atypical polarity in the ParD-ParE complex: at the ParD C-terminus, R76 of the antitoxin contacts an acidic patch on the surface of the toxin and interacts specifically with ParE D15 (Figure 6B, inset).
Unbound ParD antitoxin is helical, stably folded and dimeric in solution
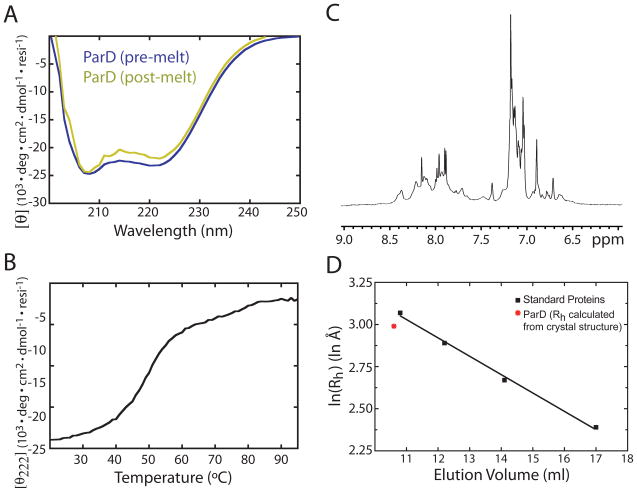
The secondary structure, stability, and oligomeric state of unbound C. crescentus ParD antitoxin were probed in solution using a variety of biophysical methods. The CD spectrum of ParD measured at 25°C has distinct minima at 207 nm and 222 nm (Figure 8A), indicating a protein that is largely α-helical at physiological temperatures. The normalized value for ellipticity (θ) at 222 nm for the ParD antitoxin is −23.1 deg·103·cm2·dmol−1·residue−1, which evidences a helical content well in excess of the expected value for the homologous ParD antitoxin of plasmid RK2 (28). We assessed the stability of the folded state of ParD by monitoring the loss of α-helical CD signal at 222 nm during thermal denaturation. The thermal denaturation curve of ParD is sigmoidal with a Tm of 49°C (Figure 8B) demonstrating that ParD undergoes a single, cooperative transition. Spectra of ParD before and after denaturation are congruent (Figure 8A), indicating that denaturation of the antitoxin is reversible. Thus, we conclude that unbound ParD antitoxin is a stable and cooperatively folded protein with a high degree of helical secondary structure. A one-dimensional 1H nuclear magnetic resonance spectrum of ParD provides additional evidence that the unbound antitoxin is folded. We generally observe sharp peaks in the N-H proton region of the spectrum with the expected amount of peak dispersion for an α-helical polypeptide of approximately 100 residues (Figure 8C). Some peak broadening is evident in this spectrum, which we attribute either to oligomer association/dissociation or dynamic conformational change in certain regions of the protein on an intermediate time scale. Size exclusion chromatography on purified ParD yielded a single peak at an elution volume of 10.2 ml on a Superdex 75 10/30 column, which corresponds to a protein with a solution hydrodynamic radius that matches the ParD dimer as defined in the ParD-ParE heterotetramer crystal structure (Figure 8D).
Figure 8.
A) Normalized circular dichroism spectrum of ParD. Two spectra are plotted, pre- and post-thermal denaturation. B) Melting curve of ParD over a temperature gradient of 20 to 95°C at 1°C/min. Helical signal at 222nm is plotted on the ordinate. C) The amide proton region of a 1H nuclear magnetic resonance spectrum of 500μM ParD. D) Size exclusion chromatography of ParD on Superdex 75. Elution volume is plotted versus the logarithm of the calculated hydrodynamic radii of ParD (red) and four standards (black) (see Materials and Methods). Standard proteins are indicated with black squares. ParD is indicated with a red asterisk. Linear regression of the gel filtration standard curve is indicated with a black line.
Discussion
Interfacial hydrophobic interactions are conserved across the ParDE/RelBE superfamily
The bulk of the energy in toxin-antitoxin binding in this superfamily likely arises from close packing interactions between hydrophobic residues on antitoxin C-termini and conserved hydrophobic residues located in a groove on the toxin surface. The conserved hydrophobic residues of the toxin groove organize into two hydrophobic subdomains. This motif appears to be conserved across the superfamily, and interestingly, is recognized by unrelated antitoxins.
In the case of E. coli RelB antitoxin, interaction with the two hydrophobic subdomains in the RelE toxin monomer has been shown to induce secondary structure formation (30), facilitating toxin-antitoxin binding (Figure 4C). RelE residues Leu5 and Phe7 of the N-terminal β-strand, Trp15 of α1, Leu16 and Leu30 of α2, as well as Leu62, Val73, and Val78 of the β sheet form hydrophobic subdomain 1. Involvement of homologous residues from the β-sheet in C. crescentus ParE was not observed; this is a consequence of lengthened N-terminal helices in ParE. The smaller helix hairpin in RelE forms a deeper hydrophobic subdomain 1 with greater sheet involvement relative to what we observe in the ParE toxin. Residues Val63 and Ile76 from the top of the RelE β-sheet form hydrophobic subdomain 2; in RelE this cavity is shallower than the homologous subdomain in ParE (Figure 4). Overall, we observe structural homology in the relative position of hydrophobic subdomains in the RelE and ParE toxins.
The YefM antitoxin, which is unrelated at the level of primary sequence and structure to the ParD and RelB antitoxins, also recognizes and binds a conserved hydrophobic motif (Figure 4D). However, YefM binds the YoeB toxin in an inhibited heterotrimer, with two molecules of YefM and one of YoeB (25, 54), presenting a case of a structurally dissimilar antitoxin recognizing conserved hydrophobic structural features of its toxin. We believe this example highlights the pervasiveness toxin-antitoxin recognition via a conserved two hydrophobic subdomain motif. Given the observed specificity of antitoxins for their cognate toxins in this superfamily, we were surprised to note the conservation of this interaction hot spot.
The role of hydrophilic contacts in TA recognition and binding
The structure of the ParD-ParE complex has revealed conserved features of toxin-antitoxin interactions in the ParDE/RelBE TA superfamily. Most notably, hydrophobic residues in toxins from this family serve to form the conserved antitoxin-binding groove (see Figures 4 and 5). However, hydrophobic residues account for approxiately 50% of the surface area buried at the ParD-ParE interface; hydrophilic and charged residues account for the other 50% of the surface area (see Figure 7). Notably, buried surface area formed between two residues which are canonically charged at biological pH comprise 11.7% of the interfacial surface area. The observation of multiple interfacial salt bridges is not surprising considering the disparity between the theoretical pI of ParD and ParE, but only 6 of the 8 observed interfacial salt bridges assumed the expected polarity. We propose that charge complementarity plays a substantial role in specific recognition between cognate toxins and antitoxins. Owing to the striking conservation of hydrophobic residues in the binding grooves of the toxin surface, hydrophobic interactions are unlikely to be highly specific. Electrostatic complementarity likely serves to guide specific antitoxins into the conserved hydrophobic groove on their cognate toxins.
Structural modes of toxin inhibition are not conserved across the superfamily
The mode of RelB-mediated inhibition appears to be considerably different from that of ParD. Inhibition of the RelE toxin proceeds by displacement of the C-terminal toxin α-helix (30). Specifically, the RelB antitoxin binds the surface of the three-stranded antiparallel β-sheet on RelE where the C-terminal RelE helix is located in the free toxin. In the complex, the RelE C-terminus is displaced and disordered. The displacement leaves RelB free to interact with numerous conserved hydrophobic residues in the β-sheet where the C-terminal helix normally packs. By contrast, we see limited interaction with conserved residues at this site in the ParD-ParE complex structure. ParD does not displace the ParE C-terminus, as ParE has a truncated C-terminus relative to RelE. Rather, ParD packs against the C-terminal region of ParE (SeMet89-Leu95), limiting access to hydrophobic residues located in the β-sheet. Structures of the RelB-RelE and YefM-YoeB family TA systems provide evidence that these antitoxins function by perturbing the toxin structure so as to distort an enzymatic active site (25, 30). Based on the heterotetramer complex structure of ParD-ParE presented here, we propose a mode of toxin inhibition for the ParDE family in which ParD functions to inhibit ParE interaction with its gyrase target without inducing large conformational changes in the toxin. Future experiments will determine if the variable modes of toxin inhibition across this large TA superfamily reflect the cellular targets with which they interact.
An example of a structured, stable ParD antitoxin
We have presented data demonstrating that the ParD antitoxin is largely α-helical in the absence of toxin. Furthermore, we demonstrate a single cooperative transition during thermal denaturation of ParD. From these observations, we conclude that the ParD antitoxin is more stable and structured than ParD from plasmid RK2 (28). A possible structural feature that may help to stabilize unbound ParD is a coiled-coil motif at the C-terminal end of the ribbon-helix-helix domain. Using a 14 amino acid sliding window, the Coils program (55) predicts a region of 16 residues spanning R39 to L55 of ParD with a considerable likelihood of coil formation. The residues in this range all have a >0.8 probability of forming a coiled-coil. The corresponding residue range within ParD-ParE complex structure is confined to the antitoxin helix α3 which mediates interaction with ParE subdomain 2 (Figure 9). We hypothesize that, in the absence of the toxin, the C-terminal helices can swing in to form a stable coiled-coil in solution.
Figure 9.
Output of the coiled-coil prediction algorithm, Coils (38), for the ParD antitoxin. ParD antitoxin is rendered as sticks; ParE toxin is rendered as a transparent blue surface. ParD residues with a greater than 80% probability of forming a coiled-coil are shown in white.
Conclusions
This first example of a ParD-ParE TA complex structure not only defines the molecular details of toxin-antitoxin interaction, but also reveals a conserved mode of antitoxin binding that is shared across the toxin superfamily. Additionally, this study presents evidence for a model in which interfacial charge-charge interactions are important for toxin-antitoxin interaction, and may serve as specificity determinants that “guide” the antitoxin into the conserved hydrophobic groove on the toxin. Finally, we provide evidence that the unbound ParD antitoxin is not natively unstructured like the ParD protein from plasmid RK2. Rather, ParD adopts a structured, helical conformation and thus does not likely undergo a significant disorder-to-order transition upon binding its cognate ParE toxin.
Acknowledgments
We thank Sherwin Montaño and Xiaojing Yang for helpful advice on crystallographic data analysis, Spencer Anderson and Joe Brunzelle at APS sector 21 (LS-CAT) for guidance on data collection strategies, Shohei Koide and Josh Kurutz for assistance with NMR, Elena Solomaha for assistance with analytical ultracentrifugation, Team Sosnick for assistance with SAXS data collection, and Aretha Fiebig for expression plasmids pAF178 and pAF202 and for thoughtful criticism of this manuscript.
Abbreviations
- TA
toxin-antitoxin
- CD
circular dichroism
- NMR
nuclear magnetic resonance
- SAXS
small-angle X-ray scattering
- AUC
analytical ultracentrifugation
- RHH
ribbon-helix-helix
- pI
isoelectric point
Footnotes
K.M.D. is supported by an NIH Roadmap Physical and Chemical Biology training program grant (T90-DK070076). S.C. acknowledges support for this project from the Arnold and Mabel Beckman Foundation (BYI), the Mallinckrodt Foundation, and the NIH-NIAID Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research (RCE) Program (Region V ‘Great Lakes’ RCE; NIH award 1-U54-AI-057153). Advanced Photon Source is supported by the DOE Office of Basic Energy Sciences (Contract No. DE-AC02-06CH11357). LS-CAT is supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Funding for NE-CAT comes from the NIH National Center for Research Resources (NCRR). BioCAT is a NIH-supported Research Center (RR-08630).
References
- 1.Anantharaman V, Aravind L. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gronlund H, Gerdes K. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J Mol Biol. 1999;285:1401–1415. doi: 10.1006/jmbi.1998.2416. [DOI] [PubMed] [Google Scholar]
- 3.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 5.Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 7.Van Melderen L, De Bast MS. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehnherr H, Yarmolinsky MB. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuchimoto S, Nishimura Y, Ohtsubo E. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J Bacteriol. 1992;174:4205–4211. doi: 10.1128/jb.174.13.4205-4211.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Melderen L, Bernard P, Couturier M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol Microbiol. 1994;11:1151–1157. doi: 10.1111/j.1365-2958.1994.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaffe A, Ogura T, Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985;163:841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts RC, Helinski DR. Definition of a minimal plasmid stabilization system from the broad-host-range plasmid RK2. J Bacteriol. 1992;174:8119–8132. doi: 10.1128/jb.174.24.8119-8132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehnherr H, Maguin E, Jafri S, Yarmolinsky MB. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol. 1993;233:414–28. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- 15.Roberts RC, Strom AR, Helinski DR. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J Mol Biol. 1994;237:35–51. doi: 10.1006/jmbi.1994.1207. [DOI] [PubMed] [Google Scholar]
- 16.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol Microbiol. 2007;63:1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- 19.Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 20.Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J Bacteriol. 2007;189:6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Pogliano J, Helinski DR, Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 23.Christensen SK, Gerdes K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol. 2003;48:1389–400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- 24.Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51:1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 25.Kamada K, Hanaoka F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol Cell. 2005;19:497–509. doi: 10.1016/j.molcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Li GY, Zhang Y, Inouye M, Ikura M. Structural mechanism of transcriptional autorepression of the Escherichia coli RelB/RelE antitoxin/toxin module. J Mol Biol. 2008;380:107–119. doi: 10.1016/j.jmb.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 27.Madl T, Van Melderen L, Mine N, Respondek M, Oberer M, Keller W, Khatai L, Zangger K. Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA. J Mol Biol. 2006;364:170–185. doi: 10.1016/j.jmb.2006.08.082. [DOI] [PubMed] [Google Scholar]
- 28.Oberer M, Zangger K, Gruber K, Keller W. The solution structure of ParD, the antidote of the ParDE toxin-antitoxin module, provides the structural basis for DNA and toxin binding. Protein Sci. 2007;16:1676–1688. doi: 10.1110/ps.062680707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Pino A, Christensen-Dalsgaard M, Wyns L, Yarmolinsky M, Magnuson RD, Gerdes K, Loris R. Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J Biol Chem. 2008;283:30821–30827. doi: 10.1074/jbc.M805654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li GY, Zhang Y, Inouye M, Ikura M. Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site. J Biol Chem. 2009;284:14628–14636. doi: 10.1074/jbc.M809656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P, Issac B, Dodson EJ, Turkenburg JP, Mande SC. Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein. J Mol Biol. 2008;383:482–493. doi: 10.1016/j.jmb.2008.08.067. [DOI] [PubMed] [Google Scholar]
- 32.Oberer M, Zangger K, Prytulla S, Keller W. The anti-toxin ParD of plasmid RK2 consists of two structurally distinct moieties and belongs to the ribbon-helix-helix family of DNA-binding proteins. Biochem J. 2002;361:41–47. doi: 10.1042/0264-6021:3610041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson EP, Strom AR, Helinski DR. Plasmid RK2 toxin protein ParE: purification and interaction with the ParD antitoxin protein. J Bacteriol. 1996;178:1420–1429. doi: 10.1128/jb.178.5.1420-1429.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 35.Doublie S. Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol Biol. 2007;363:91–108. doi: 10.1007/978-1-59745-209-0_5. [DOI] [PubMed] [Google Scholar]
- 36.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 37.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 38.Project” CC. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowtan K. The Buccaneer software for automated model building. Acta Crystallogr D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; Totowa, NJ: 2005. pp. 571–607. [Google Scholar]
- 46.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 48.Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 49.Dauter Z. One-and-a-half wavelength approach. Acta Crystallogr D. 2002;58:1958–1967. doi: 10.1107/s0907444902016645. [DOI] [PubMed] [Google Scholar]
- 50.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Inouye M. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J Biol Chem. 2009;284:6627–6638. doi: 10.1074/jbc.M808779200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schreiter ER, Drennan CL. Ribbon-helix-helix transcription factors: variations on a theme. Nat Rev Microbiol. 2007;5:710–720. doi: 10.1038/nrmicro1717. [DOI] [PubMed] [Google Scholar]
- 54.Cherny I, Rockah L, Gazit E. The YoeB toxin is a folded protein that forms a physical complex with the unfolded YefM antitoxin: Implications for a structural-based differential stability of toxin-antitoxin systems. J Biol Chem. 2005;280:30063–30072. doi: 10.1074/jbc.M506220200. [DOI] [PubMed] [Google Scholar]
- 55.Lupas A, Van Dyke M, Stock J. Predicting Coiled Coils from Protein Sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]