Abstract
Vascular endothelial growth factors (VEGFs) regulate blood and lymph vessel formation through activation of three receptor tyrosine kinases, VEGFR-1, -2, and -3. The extracellular domain of VEGF receptors consists of seven immunoglobulin homology domains, which, upon ligand binding, promote receptor dimerization. Dimerization initiates transmembrane signaling, which activates the intracellular tyrosine kinase domain of the receptor. VEGF-C stimulates lymphangiogenesis and contributes to pathological angiogenesis via VEGFR-3. However, proteolytically processed VEGF-C also stimulates VEGFR-2, the predominant transducer of signals required for physiological and pathological angiogenesis. Here we present the crystal structure of VEGF-C bound to the VEGFR-2 high-affinity-binding site, which consists of immunoglobulin homology domains D2 and D3. This structure reveals a symmetrical 2∶2 complex, in which left-handed twisted receptor domains wrap around the 2-fold axis of VEGF-C. In the VEGFs, receptor specificity is determined by an N-terminal alpha helix and three peptide loops. Our structure shows that two of these loops in VEGF-C bind to VEGFR-2 subdomains D2 and D3, while one interacts primarily with D3. Additionally, the N-terminal helix of VEGF-C interacts with D2, and the groove separating the two VEGF-C monomers binds to the D2/D3 linker. VEGF-C, unlike VEGF-A, does not bind VEGFR-1. We therefore created VEGFR-1/VEGFR-2 chimeric proteins to further study receptor specificity. This biochemical analysis, together with our structural data, defined VEGFR-2 residues critical for the binding of VEGF-A and VEGF-C. Our results provide significant insights into the structural features that determine the high affinity and specificity of VEGF/VEGFR interactions.
Keywords: angiogenesis, lymphangiogenesis, vascular endothelial growth factor C, vascular endothelial growth factor receptor-2
Angiogenesis and lymphangiogenesis, the growth of new blood and lymphatic vessels from preexisting ones, are important biological processes during embryonic development, tissue growth, wound healing, and in the pathogenesis of various diseases. The mammalian vascular endothelial growth factors (VEGF-A, VEGF-B, VEGF-C, VEGF-D, and placenta growth factor, PlGF) and their tyrosine kinase receptors (VEGFR-1, VEGFR-2, and VEGFR-3) are the major mediators of angiogenesis. In addition, these receptors regulate vascular permeability and vessel dilation [reviewed in Lohela et al. (1)]. VEGF-A signaling through VEGFR-2 is the major pathway regulating endothelial cell sprouting, migration, proliferation, and survival (2), whereas VEGF-C signaling through VEGFR-3 is indispensable for the development of lymphatic vessels (3, 4). VEGFR-2 activation is responsible for the angiogenic properties of VEGF-C in many experimental conditions (5 –7), but angiogenic signaling also involves VEGFR-3 (8). In addition, VEGF-C promotes the formation of VEGFR-2/VEGFR-3 heterodimers whose signaling potential is not yet clear (9). VEGF signaling is modulated through interactions with distinct heparan sulfate proteoglycans and neuropilins, which act as coreceptors (10 –14). VEGFs exist in multiple isoforms that are generated by alternative splicing and posttranslational processing and display distinct receptor specificities (15).
All VEGFs are antiparallel, cystine-knot polypeptide dimers that are covalently linked by two intermolecular disulfide bonds (16 –19). In VEGF-C and VEGF-D, this VEGF homology domain is flanked by C- and N-terminal propeptides that are sequentially cleaved, giving rise to VEGF homologs with distinct functions. Interestingly, mature VEGF-C has been described as a mixture of covalently and noncovalently bound dimers (20, 21).
C-terminally cleaved VEGF-C and VEGF-D are high-affinity ligands for VEGFR-3 and, upon removal of both propeptides, they acquire binding affinity for VEGFR-2 (20, 22). All VEGF-A isoforms bind to VEGFR-1 and VEGFR-2, whereas PlGF and VEGF-B are specific for VEGFR-1. Furthermore, pox viruses encode VEGF variants collectively called VEGF-E that specifically bind to VEGFR-2 (23 –25).
Crystal structures have been published for VEGF-A (26), PlGF (27), VEGF-B (28), and VEGF-E (29). In addition, structures for VEGF-A (30) and PlGF (31) in complex with domain 2 of VEGFR-1 (VEGFR-1D2) are available. Analysis of VEGFR-1 and VEGFR-2 mutants showed that the second and third immunoglobulin homology domains are essential for high-affinity VEGF-A binding (32, 33), in agreement with the recently published EM structure of the VEGF-A/VEGFR-2 complex (34). According to our EM data, receptor dimers are held together by ligand interacting with immunoglobulin homology domains 2 and 3 and by homotypic receptor contacts mediated by the membrane-proximal domains (34). This rigid conformation of the extracellular domain may then instigate transmembrane signaling resulting in the activation and autophosphorylation of the intracellular kinase domain (12).
To obtain high-resolution structural information of VEGF/VEGFR interactions and to understand VEGF receptor specificity in molecular terms, we determined the crystal structure of VEGF-C in complex with immunoglobulin homology domains 2 and 3 of VEGFR-2. This structure, in combination with our mutational analysis, provides insights into the high affinity interactions of VEGFs with their receptors.
Results
Biochemical Analysis.
We expressed the human VEGFR-2 immunoglobulin homology domains 2 and 3 (VEGFR-2D23) with a C-terminal Fc-tag and human VEGF-C (22) in insect cells. VEGF-C was used with a Cys137Ala transversion for increased protein stability (7) and is hereafter referred to as VEGF-C. The complex was purified by protein A affinity chromatography followed by size exclusion chromatography, resolving as a major peak consisting of the VEGF-C/VEGFR-2D23 complex (Fig. S1A). The molecular weight of the VEGF-C/VEGFR-2D23 complex was 78.0 kDa (Fig. S1B) as determined by multiangle laser light scattering (MALS), suggesting that the complex consists of two receptor molecules and one VEGF-C homodimer. To further characterize the complexes, we measured the binding affinity of VEGF-C for VEGFR-2D23 (D23; Fig. S1C) and VEGFR-2D2 alone (D2; Fig. S1D) using isothermal titration calorimetry (ITC). The data confirmed the 2∶2 ligand∶receptor stoichiometry and showed that binding is enthalpically and entropically favorable. VEGFR-2D2 (21) alone was sufficient for VEGF-C binding, but the presence of VEGFR-2D3 was essential for high-affinity binding.
VEGF-C/VEGFR-2D23 Complex Structure.
The crystal structure of VEGF-C in complex with VEGFR-2D23 (Fig. 1) in an orthorhombic crystal form was determined to 2.7 Å resolution by single isomorphous replacement with anomalous scattering phases, and it was refined to a crystallographic R value of 22.5% and an R free of 27.7% (Table S1). The asymmetric unit contains two crystallographically independent copies of 2∶2 VEGF-C/VEGFR-2D23 complexes. The VEGF-C/VEGFR-2D23 complex structure was solved also in a tetragonal spacegroup at 3.1 Å resolution with an R value of 25.7% and an R free of 34.6% (Table S1). Here, the asymmetric unit contains only one chain each of VEGF-C and VEGFR-2D23. The two structures are highly similar, but some loops and the VEGF-C extended N-terminal helix are differentially resolved (SI Materials and Methods). The two VEGF-C and the four VEGFR-2D23 N-linked glycans are not equally ordered in all chains and were only partially modeled.
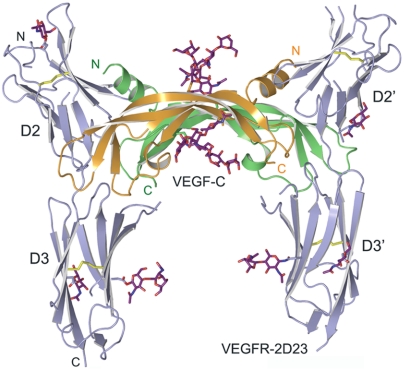
Fig. 1.
Structure of the VEGF-C/VEGFR-2D23 complex in a cartoon representation. The VEGF-C homodimer is shown in orange and green, and the two VEGFR-2 receptor chains are colored in light blue. The sugar moieties and the disulfide bonds are shown in purple and yellow sticks, respectively. VEGF-C binds to the VEGFR-2 interface between domains 2 and 3.
The crystal structure of human VEGF-C is an antiparallel homodimer, covalently linked by two disulfide bridges between Cys156 and Cys165 (Fig. S2A–C). The structure of the monomer is similar to that of other cystine-knot proteins with an antiparallel four-stranded β-sheet, three connecting loops (L1–L3), and an extended N-terminal α-helix (α1) that folds on top of the second monomer, providing several van der Waals and ionic interactions for the dimer interface.
VEGFR-2D2 (residues 120–218) and VEGFR-2D3 (residues 222–326) are immunoglobulin homology domains with two antiparallel β-sheets. They adopt the topology of an intermediate I-set domain (35), with part of the β-strand A (A′; I-set β-strand naming) moved to the opposite layer. D2 is a globular domain with relatively short β-strands. The N-terminal bulge (30) between strands A and A′ is disordered and was omitted from the model. D3 is an elongated domain with long β-strands in both sheets. Residues 265–269 between the C–D strands are partially disordered and the C′ strand is absent from all D3 domains. Both the D2 and the D3 domains have a disulfide bridge between the β-sheets that is buried in the hydrophobic core. They have an overall extended structure and are separated by a three-residue (Val-Gly-Tyr) linker peptide such that there are only a few interactions between the domains.
Consistent with the biochemical studies (Fig. S1), the independent complexes in the two crystal forms follow the approximate 2-fold symmetry of the VEGF-C dimers with 2∶2 stoichiometry (Fig. 1). VEGFR-2 immunoglobulin homology domains 2 and 3 are positioned perpendicular to the long axis of VEGF-C and D2 is approximately in the same plane as VEGF-C, wheras D3 is located below this plane (Fig. 1). The bending angle between D2 and D3 is 122–149° and results in a left-handed twisted domain arrangement about the VEGF-C 2-fold axis (Fig. S2D). The superpositions of the VEGF-C molecules in the independent complexes result in rmsd between 0.7 and 1.1 Å for 192 VEGF-C Cα atoms, and the whole complexes superimpose with an rmsd of about 3.5 Å for 567 Cα atoms. The differences in the superpositions result mainly from variation in D3 orientations relative to the rest of the structure (Fig. S2E). The variation in D3 orientation and the VEGF-C loops 1 and 3 result in differences in the surface area buried at the ligand–receptor interface, in particular between VEGF-C and D3 (Table S2), whereas the VEGF-C/D2 interfaces are essentially identical.
VEGF-C/VEGFR-2D23 Interface.
VEGF-C binds to the D2-D3 junction so that the D2 strand G and the linker between the receptor domains occupy the groove between the VEGF-C monomers. Both VEGF-C monomers interact with the VEGFR-2 domains D2 and D3, and the VEGF-C binding surface is continuous. To better describe the numerous interactions, we assigned two binding sites, 1 and 2, that mediate the VEGF-C monomer A and B interactions with VEGFR-2 (Fig. 2 A–C). The buried surface area at the interface varies in the independent complexes but can be divided into 1160–1410 Å 2 (48–58% of the total buried surface area) for site 1 and 1040–1250 Å 2 (42–52% of the total buried surface area) for site 2.
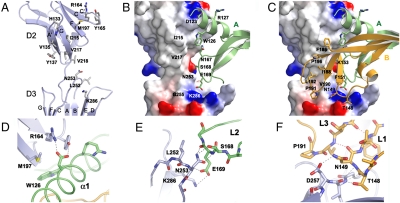
Fig. 2.
Interface between VEGF-C and VEGFR-2. (A) VEGF-C binding interface on VEGFR-2. VEGFR-2 is shown as a cartoon representation with the VEGF-C binding key residues highlighted in sticks and labeled. (B) An overview of the VEGF-C/VEGFR-2D23 site 1 interface, with VEGF-C monomer A colored in green and VEGF-C residues at the interface labeled. VEGFR-2 charge distribution shown as a surface potential model. (C) The same as in (B) for the site 2 interface with VEGF-C monomer B in orange and the monomer 2 key residues labeled. (D) VEGF-C Asp123 interactions with VEGFR-2. Hydrogen bonds and salt bridges are shown in gray dashed lines. (E) VEGF-C Glu169 interactions with VEGFR-2 as in (D). (F) VEGF-C Thr148 and Asn149 interactions with VEGFR-2 as in (D).
The VEGF-C site 1 interface (Fig. 2 B) consists of the N-terminal helix (residues 113–129) and loop L2 (residues 167–171). The VEGFR-2D2 hairpin turn C-C′ (residues 164–166) packs against the VEGF-C helix α1 (Fig. 2 D) while the connecting loop E–F and the beginning of strand F (residues 194–197) interact with the VEGF-C loop L2 and the N-terminal helix. The VEGFR-2D3 loop B–C (residues 250–257) and strand E also interact with loop L2. The major hydrophobic contacts of site 1 consist of VEGF-C Trp126 and Arg127 interacting with Gly196, Met197, and Tyr165 of VEGFR-2 (Fig. 2 and Table S2). Most of the site 1 interactions are hydrophilic and involve hydrogen bonds between VEGF-C N167 Nδ2 and the carbonyl oxygen of VEGFR-2 Tyr194, and between VEGFR-2 Asn253 Oδ1 and the main chain amide of VEGF-C Glu169 (Fig. 2 E). Moreover, Glu169, which is highly conserved in the VEGF family (29), forms a salt bridge with VEGFR-2 Lys286 and a hydrogen bond with the main chain amide of VEGFR-2 Asn253, and VEGF-C Asp123 is in contact with the D2 Arg164 and Tyr165.
The site 2 VEGF-C interface (Fig. 2 C) consists of loops L1 (residues 139–155) and L3 (residues 188–196) held together by hydrogen bonding (Fig. 2 F). On the receptor side, the D2 site 2 interface consists of strands A′ and G (Fig. 2 A) whereas VEGFR-2D3 interacts with the bottom face of VEGF-C with a surface made up of the strands C, D, and E, as well as the loops B–C and F–G (residues 311–314). The D2 strands A′ and G both contribute hydrophobic interactions (Val135, Ile215, Val217, and Val219) to the interface (Table S2). Together with Gly196 and Met197 of the neighboring site 1, they form a distinct hydrophobic patch on D2. VEGF-C loops L1 and L3 bear multiple hydrophobic residues providing a complementary surface. Several hydrophobic residues in the interface are beyond the van der Waals distance (> 4 Å) but the exclusion of water from around these residues upon binding may contribute to the favorable thermodynamic parameters (Fig. S1). The D3 interface in site 2 also involves hydrophilic interactions (Table S2) although the interactions vary between the four chains in the asymmetric unit.
VEGFR-2 Mutagenesis and Ligand Binding.
To better understand why VEGF-C binds to VEGFR-2 and VEGFR-3, but not to VEGFR-1 (20), we replaced five potential epitopes in VEGFR-2D23 with the corresponding sequences from VEGFR-1 (Fig. 3 A and B). Also, a VEGFR-2 Leu252Ala/Asn253Ala double mutant was prepared. The binding behavior of the double mutant and the five chimeric receptor proteins (called C1–C5) to VEGF-C and VEGF-A165 was assessed by BaF3/VEGFR-2 cell proliferation and by binding assays. The chimeric proteins C1, C4, and C5 showed close to wild-type activity, whereas C2 and C3 showed reduced binding in both VEGF-C (100 ng/mL) and VEGF-A165 (30 ng/mL) induced proliferation assays in comparison with the two (D23) and three domain (D1–3) VEGFR-2 constructs (Fig. 3 C).
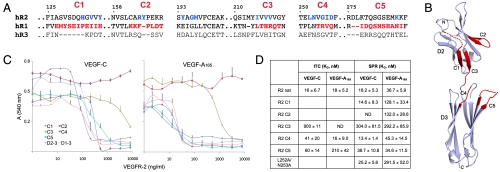
Fig. 3.
Characterization of VEGF-A and VEGF-C binding to the VEGFR-1/VEGFR-2 chimeric proteins. (A) Structure-based sequence alignment of human VEGFR-1, -2, and -3 (hR1–3) covering the VEGF-C binding interface in VEGFR-2 and the VEGFR-1/-2 chimera (C1–C5). The key VEGFR-2 residues (Table S2) and the VEGFR-1 residues inserted to the VEGFR-2 construct are highlighted in blue and red, respectively. The VEGFR-2 residues are numbered. (B) VEGFR-2D23 backbone with the swapped areas (C1–C5) highlighted in red. (C) The inhibition of VEGF-C (Left) and VEGF-A165 (Right) stimulated BaF3/VEGFR-2 cell proliferation by the VEGFR-1/-2 chimeric proteins C1–C5 together with the VEGFR-2 domains 2–3 and 1–3. (D) Summary of the VEGF-C and VEGF-A165 binding affinities (K d ± SD) to the VEGFR-2D23 constructs measured by ITC and SPR. ND, not determinable.
The binding constants were determined by ITC and surface plasmon resonance (SPR). According to the ITC measurements (Fig. S3), VEGF-C binding to the VEGFR-1/VEGFR-2 chimera C4 and C5 was only slightly affected, whereas the K d for the C3 protein was increased 50-fold (Fig. 3 D). VEGF-C and VEGF-A165 bind to VEGFR-2D23 with similar affinity and thermodynamic parameters (Figs. S1C and S3B). Furthermore, only C4 showed wild-type binding, whereas the K d of C5 for VEGF-A165 was increased about 10-fold (Fig. 3 D and Fig. S3C). No data fitting was possible for the VEGF-A/C3 calorimetric titrations. In the full set of binding assays carried out by SPR (Fig. S4), the binding affinities of C4 and C5 were not affected (Fig. 3 D), whereas C3 showed 17-fold and 8-fold increases in K d for VEGF-C and VEGF-A165 binding, respectively. The C1 and C2 showed about 3-fold increased K d for VEGF-A165 binding, whereas C1 binding to VEGF-C was not affected and C2 did not bind VEGF-C at all. The VEGFR-2 Leu252Ala/Asn253Ala double mutant showed wild-type VEGF-C binding but VEGF-A165 binding affinity was decreased 3-fold.
Discussion
Here we present the crystal structure of the ligand binding domain of VEGFR-2 in complex with VEGF-C. Our results reveal a unique view of a multidomain interaction of a ligand with a VEGF receptor. Our structure also gives important insights on the receptor specificity of VEGF family ligands.
The interaction surfaces 1 and 2 include contacts between VEGF-C and the two VEGFR-2 domains as well as the interdomain linker. D2 has been described as the major VEGF-C binding domain of VEGFR-2 (21). However, consistent with our binding studies using domain deletion mutants (Fig. S1C and D), VEGFR-2D3 also contributes a significant number of interactions to VEGF-C binding. These contacts involve site 1 interactions by the conserved Glu169 (Glu64 in VEGF-A) of VEGF-C with Asn253 and Lys286 of D3 (Table S2). D3 interacts with loops L1 and L3 of VEGF-C predominantly through site 1 (Table S2), suggesting that these interactions play an essential role in ligand binding.
VEGF-C binding to VEGFR-3 requires D1 and D2, but not D3 (21). Ligand binding to VEGFR-2 and Kit receptor D2 and D3 are structurally very similar (Fig. S5A and B) utilizing a similar interface at the  junction (Fig. S5C), despite the fact that the ligands are structurally dissimilar. In addition, comparison of the VEGFR-2 and the Kit (36) complexes suggests a role for VEGFR-3D1 in VEGF-C binding. The Kit D1 bends over its ligand, stem cell factor (SCF), making several interactions with it (36). Kit D1, and the related colony-stimulating factor 1 receptor D1 (37), together with the adjacent D2, forms a rigid two-domain D12 structure. The rigidity of D12, involving D2 strand A, which is only poorly resolved in our complex, suggests that the presence of D1 is important for the integrity of the major ligand binding epitope in D2. In analogy with Kit, D1 of VEGFR-3 may therefore directly interact with VEGF-C.
junction (Fig. S5C), despite the fact that the ligands are structurally dissimilar. In addition, comparison of the VEGFR-2 and the Kit (36) complexes suggests a role for VEGFR-3D1 in VEGF-C binding. The Kit D1 bends over its ligand, stem cell factor (SCF), making several interactions with it (36). Kit D1, and the related colony-stimulating factor 1 receptor D1 (37), together with the adjacent D2, forms a rigid two-domain D12 structure. The rigidity of D12, involving D2 strand A, which is only poorly resolved in our complex, suggests that the presence of D1 is important for the integrity of the major ligand binding epitope in D2. In analogy with Kit, D1 of VEGFR-3 may therefore directly interact with VEGF-C.
A comparison of VEGF-C/VEGFR-2D23 and the VEGF-A/VEGFR-1D2 (30) and PlGF/VEGFR-1D2 (31) structures suggests that VEGFR-1 and VEGFR-2 utilize the same interface for ligand binding. To test this and to understand VEGF-C receptor specificity, we constructed VEGFR-1/VEGFR-2 chimera. The exchange of nonconserved areas in the VEGF-C/VEGFR-2 binding interface was predicted to result in the loss of interactions revealed by our crystal structure. These constructs also include important residues for VEGF-A and PlGF binding to VEGFR-1D2 and presumably to VEGFR-1D3. The C1 chimera covering the D2 N-terminal bulge and strand A′ did not affect VEGF-C binding, whereas the C2 chimera did not bind VEGF-C, emphasizing the importance of Asp123 and Arg127 of VEGF-C for the interaction with Arg164 and Tyr165 of VEGFR-2 (Fig. 2 D). VEGF-C binding to the C3 chimera was also compromised, in line with the apparent loss of several hydrophobic interactions seen in the crystal structure. In C3, the hydrophobic residues (Val217, Val218, and Val219) in the G strand of D2 are replaced with hydrophilic and larger residues (His223, Arg224, and Gln225) from VEGFR-1. In the VEGF-A/VEGFR-1D2 complex Glu63 of VEGF-A, a determinant of VEGFR-1 specificity, is salt bridged to Arg224 of VEGFR-1 (26, 30). The corresponding VEGF-C residue, Ser168, is not capable to form this salt bridge. The C4 chimera has a Gly to Arg replacement and the corresponding VEGFR-1 DE loop in chimera C5 is shorter by two residues. The VEGF-C binding of these proteins is not changed, suggesting that the DE loop is not involved in VEGF-C binding.
VEGF-A binds to VEGFR-1 predominantly via D2, whereas both D2 and D3 are needed for VEGFR-2 binding. The deletion of D3 from VEGFR-1 and VEGFR-2 results in a 20-fold and 1,000-fold decreased VEGF-A affinity, respectively (30, 33). As expected, VEGF-A165 binding was retained in all VEGFR-1/VEGFR-2 chimeras. The C4 and C5 chimeras showed wild-type binding affinity and the K d was increased only 3-fold for C1 and C2. Interestingly, the C2 chimera did not bind to VEGF-C, consistent with the fact that the VEGF-A counterpart of Asp123 in VEGF-C is methionine. Hence, similar polar interactions with Arg164 and Tyr165 are not required for VEGFR-2 binding. Similar to VEGF-C, VEGF-A165 binding affinity was decreased for the C3 chimera (Fig. 3 C and D) suggesting that VEGF-A165 cannot utilize the same contacts as VEGFR-1, such as the Glu63-Arg224 interaction, in the context of VEGFR-2. Rather, it seems that VEGF-A utilizes the same interface and similar interactions as VEGF-C for VEGFR-2 binding. However, as indicated by the C2 chimera and the Leu252Ala/Asn253Ala double mutant, there are also differences between VEGF-A and VEGF-C binding to VEGFR-2. This interpretation is supported by the affinities and thermodynamic parameters determined for VEGF-A165 and VEGF-C binding to the VEGFR-2D23 construct.
When compared to other VEGFs, mature VEGF-C has an extra cysteine residue, Cys137 (Fig. 4 A). The Cys137Ala mutation improves dimer stability and increases biological activity, especially with respect to VEGFR-2, but does not change binding affinity for VEGFR-2 (Fig. S4) or VEGFR-3 (7). Residue 137 in the VEGF-C structure is solvent exposed and lies at the dimer interface close to the interchain Cys156-Cys165′ disulfide bridge (Fig. S2). The free Cys137 thiols are thus sensitive to changes in the redox environment, and because there are only few noncovalent interactions in the VEGF-C dimer interface, the stability of interchain disulfides may affect the monomer to dimer ratio as suggested for the Cys156Ser and Cys156Ala mutants (21, 38). In fact, the lack of these interchain disulfide bridges leads to an increased molar ratio of monomeric to dimeric molecules (21, 38). Interestingly, although the Cys156Ala mutant is a poor ligand for both VEGFR-2 and VEGFR-3, the VEGF-C Cys156Ser mutant is fairly selective for VEGFR-3 (38). Thus, the oligomerization state of mature VEGF-C, affected by the free thiol group, may provide additional control over receptor activity and specificity.
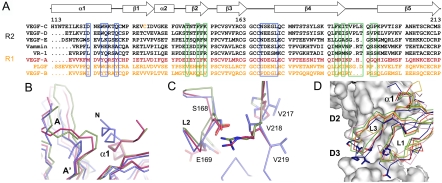
Fig. 4.
Comparison of the VEGF family ligand structures and the complexes. (A) Structure-based multiple sequence alignment of the VEGF-family members. Residues participating in the VEGF-C/VEGFR-2D23 interactions are boxed in blue (site 1) and green (site 2). VEGFR-1 and VEGFR-2 specific ligands are colored in yellow and in black, respectively. VEGF-A binds to both receptors and is highlighted in red. (B) Structural comparison of VEGF-C/VEGFR-2D23 (blue), VEGF-A/VEGFR1-D2 (red, 1FLT) and PlGF/VEGFR-1D2 (green, 1RV6) complex structures centered on the VEGF N-terminal helix (α1). VEGFR-2 D2 strands A and A′ are labeled. (C) Same as in (B) centered on VEGF-C loop L2 and VEGFR-2 D2 strand G. The centered residues are shown in sticks with VEGF-C residues labeled. (D) Conformational differences in the site 2 loops L1 and L3. The VEGFR-2 domains 2 and 3 and the VEGFs are in surface and ribbon representation, respectively. VEGF-C is in blue, VEGF-A (1FLT) in red, PlGF (1RV6) in green, VEGF-B (2VWE) in purple, and VEGF-E (2GNN) in yellow. VEGF-C mutants with reduced binding to VEGFR-2 (21) are shown in sticks.
VEGF-C and VEGF-D share 60% sequence identity including essentially all VEGF-C residues at the VEGFR-2 binding interface. Interestingly, VEGF-D binding to VEGFR-2 is not conserved between humans and mice because mVEGF-D binds hVEGFR-2 but fails to bind mVEGFR-2 (39). Humanizing the mVEGF-D sequence in the putative VEGFR-2 binding interface rescued binding only partially, indicating that residues outside this area also affect VEGF-D activity (39). Similarly to VEGF-C (7), substitution of the extra cysteine in VEGF-D with an aliphatic residue improved dimer stability but also increased VEGFR-2 binding and activation (40). Thus, consistent with the conclusions drawn from our VEGF-C/VEGFR-2 structure, dimer stability of VEGFs is likely essential for VEGFR-2 binding.
When compared with other family members, major structural differences are observed in the VEGF-C N-terminal helix α1 (Fig. 4 B), loops L1 and L3. In VEGFR-2, the D2 N-terminal bulge, the A′–B, the C-C′ hairpin, the E–F, and the F–G′ loops (Fig. S6) differ from VEGFR-1 both in sequence and in structure. Of the 21 receptor residues that form the interface with VEGF-C, only six are conserved between VEGFR-1 and VEGFR-2. The orientations of the ligands toward receptor domain D2 differ between the VEGFR-1 and VEGFR-2 complexes. When compared to VEGF-A and PlGF, VEGF-C is tilted by 15° and twisted by 9° from the interface to D2, which makes an overall comparison of the interfaces difficult. We therefore analyzed the local superpositions of the VEGF-C/VEGFR-2D23 structure with the VEGF-A and PlGF structures in complex with VEGFR-1D2 at critical sites of the interface. At site 1, Glu169 of VEGF-C loop L2 forms multiple interactions with VEGFR-2D3 providing an explanation for the importance of this glutamate residue (Asp in VEGF-B) that is highly conserved in the VEGF family. Of the two Glu169 counterparts in VEGFR-2, Asn253 is conserved in human VEGFRs, whereas Lys286 is not. As discussed above, Ser168 of VEGF-C and Val218 of VEGFR-2 form a weak hydrophobic contact in place of the Asp63-Arg224 salt bridge observed in the VEGFR-1D2 complexes with VEGF-A and PlGF (Fig. 4 C). However, as Asp63 of VEGF-A is conserved in all other VEGFs, except for VEGF-C and VEGF-D, this interaction alone cannot account for receptor specificity. In addition, our VEGFR-1/-2 chimeric protein C3 affecting also VEGF-A165 binding suggests that VEGF-A binding to VEGFR-1 and VEGFR-2 occurs via different interactions.
In comparison to VEGFR-1, D2 of VEGFR-2 has a one residue insertion in the C-C′ hairpin at site 1, whereas the side chains of Tyr165 and Pro166 occupy the same space as Phe172 and Pro173 in VEGFR-1. The side chain of the flanking residue Arg164 (salt bridged to VEGF-C Asp123) protrudes into an otherwise empty space in VEGFR-1. Of the complementary VEGF-C surface (Asp123, Trp126, Arg127, Gln130, Lys153, and Asn167), only Trp126 is conserved between VEGF-C and the VEGFR-1 specific PlGF. When compared to the other known VEGFs, the N-terminal helix α1 of VEGF-C extends from the complex interface by about two more turns. The proximity of the VEGF-C N terminus to the N-terminal bulge, which is disordered in the VEGFR-2 chains, may have implications for VEGFR-1 and VEGFR-3 binding specificity, possibly involving additional domains such as D1.
Besides the N-terminal helix, the largest variations in sequence and conformation among the ligands are observed at site 2 in loops L1 and L3. Consistent with this, we have reported earlier that alanine mutants in the segments Thr148-Phe151 and Thr189-Leu192 of VEGF-C, and a double deletion mutant Leu192Ser193 with a shorter loop L3, show reduced binding affinity for VEGFR-2 (21). The majority of these residues are in contact with VEGFR-2D3 and the linker region. This, together with the structural variation in loops L1 and L3 (Fig. 4 D), may explain the altered receptor specificity observed in VEGF loop L1 and L3 chimera between VEGF-E and PlGF (41), VEGF-E and VEGF-A (29), and VEGF-C and VEGF-A (21).
We have previously shown by negative stain EM how VEGF-A induces dimerization of the extracellular domain of VEGFR-2 (34). More recently, using membrane-bound receptor kinase constructs, we also have shown that dimerization is required but not sufficient for receptor activation (42). To compare the crystal structure of the ligand binding domain described here with the EM data, we calculated volumes filtered at 25 Å resolution from the two VEGF-C/VEGFR-2D23 complexes observed in the asymmetric unit of the orthorhombic crystal form. Domain projections derived from these volumes are virtually identical to the corresponding domains in the EM structure 34 (Fig. S7) and fit the model observed in the EM structure with additional homotypic interactions mediated by the membrane-proximal domains (34). The biological significance of such homotypic receptor–receptor interactions is further supported by recent findings for the Kit and PDGF receptors, where D4-mediated homotypic receptor interactions are mandatory for receptor activation (36, 43). As discussed above, the crystal structure of the complex between SCF and the extracellular domain of Kit (36), shares many features with the structure described here. To further unravel the molecular mechanism of transmembrane signaling by class III (Kit) and V (VEGFR) receptor tyrosine kinases, additional structural data for membrane-bound receptors is required. Combining low-resolution techniques such as EM and small angle solution scattering with x-ray crystallography or NMR spectroscopy should then lead to a model of receptor activation, as illustrated recently for a cytokine (44).
Signaling by VEGFR-2 represents the major pathway for the transduction of angiogenic signals and VEGF receptors are therefore prominent targets in the development of angiogenesis inhibitors (1, 45). Despite this, the structure of VEGFR-2 has only now been elucidated, more than a decade after the determination of the structure of the VEGF-A/VEGFR-1D2 complex (30). Our structural and functional analysis provides important insights into receptor/ligand interactions that explain previously published data and these are essential for the understanding of receptor/ligand specificity. Furthermore, using the structure described here, the binding sites of receptor blocking antibodies currently in clinical trials as inhibitors of tumor angiogenesis can now be mapped to the VEGFR-2 ligand binding epitopes. These results should further boost the rational design of new inhibitors of angiogenesis targeting the VEGF receptors.
Methods
Protein Expression and Purification.
Human VEGF-C, VEGF-C Cys137Ala mutant, VEGF-A165 (7), VEGFR-2 domains 2, 2–3, and 1–3 fused to IgGFc (21), and the VEGFR-1/-2 chimera were expressed in Sf9 insect cells and purified as described in SI Materials and Methods.
Cell Survival Assay and Binding Assays.
Cell survival assays with BaF3 cells expressing VEGFR-2/EpoR chimera (7), calorimetric titrations using a VP-ITC calorimeter (MicroCal), and SPR analysis with a Biacore 2000 biosensor (GE) were carried out as described in SI Materials and Methods.
Crystallization and Structure Determination.
Complete datasets to 2.7 and 3.1 Å resolution were collected from single orthorhombic and tetragonal VEGF-C/VEGFR2-D23 complex crystals, respectively (Table S1). For phasing, anomalous data on a Pt-derivative crystal was collected to 3.6 Å resolution (Table S1). Full crystallographic details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
The authors thank The Sigrid Juselius Foundation, The Louis Jeantet Foundation, the Finnish Cancer Research Organizations, and the European Union integrated projects on Lymphangiogenomics (LSHG-CT-2004 2004-503573), Tumor-Host Genomics (LSHC-CT-2005-518198), and MicroEnviMet (FP7/2007-2011, No. 201279) for financial assistance. Swiss Light Source is acknowledged for provision of synchrotron radiation resources. A.E.P. and K.B.-H. thank Swiss National Science Foundation (Grants 31003A-112455 and 3100A-116507, respectively) and Oncosuisse (Grant OC2 01200-08-2007) for support of their work.
Note Added in Proof.
Recent work by Y. Yang, P. Xie, Y. Opatowsky, and J. Schlessinger (PNAS, published online before print January 11, 2010) shows that salt bridges and van der Waals interactions between Arg726 of one VEGFR-2 D7 protomer and Asp731 of the other protomer are required for ligand-induced VEGFR-2 activation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2x1x and 2x1w).
This article contains supporting information online at www.pnas.org/cgi/content/full/0914318107/DCSupplemental.
References
- 1.Lohela M, Bry M, Tammela T, Alitalo K. VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr Opin Cell Biol. 2009;21:154–165. doi: 10.1016/j.ceb.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Shibuya M, Claesson-Welsh L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Exp Cell Res. 2006;312:549–560. doi: 10.1016/j.yexcr.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Mäkinen T, et al. Isolated lymphatic endothelial cells transduce growth, survival and migratory signals via the VEGF-C/D receptor VEGFR-3. EMBO J. 2001;20:4762–4773. doi: 10.1093/emboj/20.17.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kärkkäinen MJ, et al. Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat Immunol. 2004;5:74–80. doi: 10.1038/ni1013. [DOI] [PubMed] [Google Scholar]
- 5.Benest AV, Harper SJ, Ylä-Herttuala SY, Alitalo K, Bates DO. VEGF-C induced angiogenesis preferentially occurs at a distance from lymphangiogenesis. Cardiovasc Res. 2008;78:315–323. doi: 10.1093/cvr/cvm094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohela M, Heloterä H, Haiko P, Dumont DJ, Alitalo K. Transgenic induction of vascular endothelial growth factor-C is strongly angiogenic in mouse embryos but leads to persistent lymphatic hyperplasia in adult tissues. Am J Pathol. 2008;173:1891–1901. doi: 10.2353/ajpath.2008.080378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anisimov A, et al. Activated forms of VEGF-C and VEGF-D provide improved vascular function in skeletal muscle. Circ Res. 2009;104:1302–1312. doi: 10.1161/CIRCRESAHA.109.197830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tammela T, et al. Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature. 2008;454:656–660. doi: 10.1038/nature07083. [DOI] [PubMed] [Google Scholar]
- 9.Dixelius J, et al. Ligand-induced vascular endothelial growth factor receptor-3 (VEGFR-3) heterodimerization with VEGFR-2 in primary lymphatic endothelial cells regulates tyrosine phosphorylation sites. J Biol Chem. 2003;278:40973–40979. doi: 10.1074/jbc.M304499200. [DOI] [PubMed] [Google Scholar]
- 10.Kärpänen T, et al. Functional interaction of VEGF-C and VEGF-D with neuropilin receptors. FASEB J. 2006;20:1462–1472. doi: 10.1096/fj.05-5646com. [DOI] [PubMed] [Google Scholar]
- 11.Cebe-Suarez S, et al. A VEGF-A splice variant defective for heparan sulfate and neuropilin-1 binding shows attenuated signaling through VEGFR-2. Cell Mol Life Sci. 2006;63:2067–2077. doi: 10.1007/s00018-006-6254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cebe-Suarez S, Zehnder-Fjallman A, Ballmer-Hofer K. The role of VEGF receptors in angiogenesis; complex partnerships. Cell Mol Life Sci. 2006;63:601–615. doi: 10.1007/s00018-005-5426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawamura H, et al. Neuropilin-1 in regulation of VEGF-induced activation of p38MAPK and endothelial cell organization. Blood. 2008;112:3638–3649. doi: 10.1182/blood-2007-12-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cebe-Suarez S, et al. Orf virus VEGF-E NZ2 promotes paracellular NRP-1/VEGFR-2 coreceptor assembly via the peptide RPPR. FASEB J. 2008;22:3078–3086. doi: 10.1096/fj.08-107219. [DOI] [PubMed] [Google Scholar]
- 15.Robinson CJ, Stringer SE. The splice variants of vascular endothelial growth factor (VEGF) and their receptors. J Cell Sci. 2001;114:853–865. doi: 10.1242/jcs.114.5.853. [DOI] [PubMed] [Google Scholar]
- 16.Vitt UA, Hsu SY, Hsueh AJ. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol Endocrinol. 2001;15:681–694. doi: 10.1210/mend.15.5.0639. [DOI] [PubMed] [Google Scholar]
- 17.Oefner C, D’Arcy A, Winkler FK, Eggimann B, Hosang M. Crystal structure of human platelet-derived growth factor BB. EMBO J. 1992;11:3921–3926. doi: 10.1002/j.1460-2075.1992.tb05485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reigstad LJ, et al. Platelet-derived growth factor (PDGF)-C, a PDGF family member with a vascular endothelial growth factor-like structure. J Biol Chem. 2003;278:17114–17120. doi: 10.1074/jbc.M301728200. [DOI] [PubMed] [Google Scholar]
- 19.Muller YA, Heiring C, Misselwitz R, Welfle K, Welfle H. The cystine knot promotes folding and not thermodynamic stability in vascular endothelial growth factor. J Biol Chem. 2002;277:43410–43416. doi: 10.1074/jbc.M206438200. [DOI] [PubMed] [Google Scholar]
- 20.Joukov V, et al. A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. EMBO J. 1996;15:290–298. [PMC free article] [PubMed] [Google Scholar]
- 21.Jeltsch M, et al. Vascular endothelial growth factor (VEGF)/VEGF-C mosaic molecules reveal specificity determinants and feature novel receptor binding patterns. J Biol Chem. 2006;281:12187–12195. doi: 10.1074/jbc.M511593200. [DOI] [PubMed] [Google Scholar]
- 22.Joukov V, et al. Proteolytic processing regulates receptor specificity and activity of VEGF-C. EMBO J. 1997;16:3898–3911. doi: 10.1093/emboj/16.13.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyttle DJ, Fraser KM, Fleming SB, Mercer AA, Robinson AJ. Homologs of vascular endothelial growth factor are encoded by the poxvirus orf virus. J Virol. 1994;68:84–92. doi: 10.1128/jvi.68.1.84-92.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wise LM, et al. Viral vascular endothelial growth factors vary extensively in amino acid sequence, receptor-binding specificities, and the ability to induce vascular permeability yet are uniformly active mitogens. J Biol Chem. 2003;278:38004–38014. doi: 10.1074/jbc.M301194200. [DOI] [PubMed] [Google Scholar]
- 25.Mercer AA, et al. Vascular endothelial growth factors encoded by Orf virus show surprising sequence variation but have a conserved, functionally relevant structure. J Gen Virol. 2002;83:2845–2855. doi: 10.1099/0022-1317-83-11-2845. [DOI] [PubMed] [Google Scholar]
- 26.Muller YA, et al. Vascular endothelial growth factor: Crystal structure and functional mapping of the kinase domain receptor binding site. Proc Natl Acad Sci USA. 1997;94:7192–7197. doi: 10.1073/pnas.94.14.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer S, et al. The crystal structure of human placenta growth factor-1 (PlGF-1), an angiogenic protein, at 2.0 A resolution. J Biol Chem. 2001;276:12153–12161. doi: 10.1074/jbc.M008055200. [DOI] [PubMed] [Google Scholar]
- 28.Iyer S, Scotney PD, Nash AD, Ravi Acharya K. Crystal structure of human vascular endothelial growth factor-B: Identification of amino acids important for receptor binding. J Mol Biol. 2006;359:76–85. doi: 10.1016/j.jmb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Pieren M, et al. Crystal structure of the Orf virus NZ2 variant of vascular endothelial growth factor-E. Implications for receptor specificity. J Biol Chem. 2006;281:19578–19587. doi: 10.1074/jbc.M601842200. [DOI] [PubMed] [Google Scholar]
- 30.Wiesmann C, et al. Crystal structure at 1.7 Å resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 31.Christinger HW, Fuh G, de Vos AM, Wiesmann C. The crystal structure of placental growth factor in complex with domain 2 of vascular endothelial growth factor receptor-1. J Biol Chem. 2004;279:10382–10388. doi: 10.1074/jbc.M313237200. [DOI] [PubMed] [Google Scholar]
- 32.Davis-Smyth T, Presta LG, Ferrara N. Mapping the charged residues in the second immunoglobulin-like domain of the vascular endothelial growth factor/placenta growth factor receptor Flt-1 required for binding and structural stability. J Biol Chem. 1998;273:3216–3222. doi: 10.1074/jbc.273.6.3216. [DOI] [PubMed] [Google Scholar]
- 33.Fuh G, Li B, Crowley C, Cunningham B, Wells JA. Requirements for binding and signaling of the kinase domain receptor for vascular endothelial growth factor. J Biol Chem. 1998;273:11197–11204. doi: 10.1074/jbc.273.18.11197. [DOI] [PubMed] [Google Scholar]
- 34.Ruch C, Skiniotis G, Steinmetz MO, Walz T, Ballmer-Hofer K. Structure of a VEGF-VEGF receptor complex determined by electron microscopy. Nat Struct Mol Biol. 2007;14:249–250. doi: 10.1038/nsmb1202. [DOI] [PubMed] [Google Scholar]
- 35.Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 36.Yuzawa S, et al. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: Diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci USA. 2008;105:18267–18272. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joukov V, et al. A recombinant mutant vascular endothelial growth factor-C that has lost vascular endothelial growth factor receptor-2 binding, activation, and vascular permeability activities. J Biol Chem. 1998;273:6599–6602. doi: 10.1074/jbc.273.12.6599. [DOI] [PubMed] [Google Scholar]
- 39.Baldwin ME, et al. The specificity of receptor binding by vascular endothelial growth factor-d is different in mouse and man. J Biol Chem. 2001;276:19166–19171. doi: 10.1074/jbc.M100097200. [DOI] [PubMed] [Google Scholar]
- 40.Toivanen PI, et al. Novel vascular endothelial growth factor D variants with increased biological activity. J Biol Chem. 2009;284:16037–16048. doi: 10.1074/jbc.M109.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiba A, Yabana N, Shibuya M. A set of loop-1 and -3 structures in the novel vascular endothelial growth factor (VEGF) family member, VEGF-ENZ-7, is essential for the activation of VEGFR-2 signaling. J Biol Chem. 2003;278:13453–13461. doi: 10.1074/jbc.M210931200. [DOI] [PubMed] [Google Scholar]
- 42.Dell’Era Dosch D, Ballmer-Hofer K. Transmembrane domain-mediated orientation of receptor monomers in active VEGFR-2 dimers. FASEB J. 2010;24:32–38. doi: 10.1096/fj.09-132670. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci USA. 2008;105:7681–7686. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skiniotis G, Lupardus PJ, Martick M, Walz T, Garcia KC. Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol Cell. 2008;31:737–748. doi: 10.1016/j.molcel.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alitalo K, Adams RH. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.