Abstract
The CHO1-encoded phosphatidylserine synthase from Saccharomyces cerevisiae is phosphorylated and inhibited by protein kinase A in vitro. CHO1 alleles bearing Ser46 → Ala and/or Ser47 → Ala mutations were constructed and expressed in a cho1Δ mutant lacking phosphatidylserine synthase. In vitro, the S46A/S47A mutation reduced the total amount of phosphorylation by 90% and abolished the inhibitory effect protein kinase A had on phosphatidylserine synthase activity. The enzyme phosphorylation by protein kinase A, which was time- and dose-dependent and dependent on the concentration of ATP, caused a electrophoretic mobility shift from a 27-kDa form to a 30-kDa form. The two electrophoretic forms of phosphatidylserine synthase were present in exponential phase cells, whereas only the 27-kDa form was present in stationary phase cells. In vivo labeling with 32Pi and immune complex analysis showed that the 30-kDa form was predominantly phosphorylated when compared with the 27-kDa form. However, the S46A/S47A mutations abolished the protein kinase A-mediated electrophoretic mobility shift. The S46A/S47A mutations also caused a 55% reduction in the total amount of phosphatidylserine synthase in exponential phase cells and a 66% reduction in the amount of enzyme in stationary phase cells. In phospholipid composition analysis, cells expressing the S46A/S47A mutant enzyme exhibited a 57% decrease in phosphatidylserine and a 40% increase in phosphatidylinositol. These results indicate that phosphatidylserine synthase is phosphorylated on Ser46 and Ser47 by protein kinase A, which results in a higher amount of enzyme for the net effect of stimulating the synthesis of phosphatidylserine.
Keywords: Membrane Enzymes, Membrane Lipids, Phosphatidylserine, Phospholipid, Protein Kinase A (PKA), Yeast
Introduction
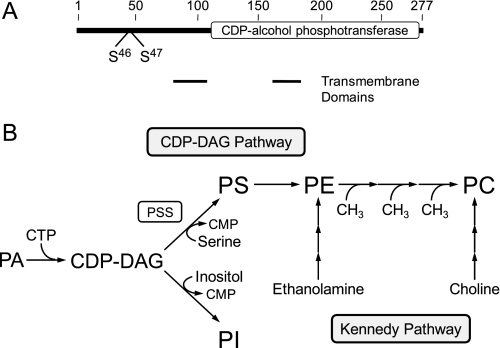
In the yeast Saccharomyces cerevisiae, the CHO1-encoded PS2 synthase (CDP-diacylglycerol:l-serine O-phosphatidyltransferase, EC 2.7.8.8)3 is an ER-associated enzyme that catalyzes the formation of PS by a Mn2+-dependent sequential reaction by displacing CMP from CDP-DAG with serine (Fig. 1) (1–5). The enzyme possesses a CDP-alcohol phosphotransferase motif, DGX2ARX7,8GX3DX3D (residues 130–152) within a larger domain that is common to phospholipid biosynthetic enzymes (e.g. PI synthase) that catalyze similar types of reactions (6) (Fig. 1). The reaction product PS is a major membrane phospholipid in yeast, accounting for up to 18% of the total membrane phospholipids (7–9). In addition, PS synthase catalyzes the committed step in the synthesis of PE and PC via the de novo CDP-DAG pathway (7–11) (Fig. 1). As such, PS synthase is one of the most highly regulated enzymes of phospholipid synthesis in this organism (10, 12, 13).
FIGURE 1.
Domain structure of PS synthase and pathways for the synthesis of the major phospholipids in S. cerevisiae. A, the diagram shows the positions of the CDP-alcohol phosphotransferase domain, the transmembrane-spanning domains, and the target sites of protein kinase A phosphorylation. B, the pathways shown for the synthesis of the major phospholipids include the relevant steps discussed in this work. The reaction catalyzed by PS synthase (PSS) is indicated in the figure. A more comprehensive figure that shows the individual steps in the CDP-DAG and Kennedy pathways may be found in Ref. 11.
Genetic and biochemical mechanisms regulate the expression and activity of PS synthase. The expression of CHO1 is regulated by water-soluble phospholipid precursors (e.g. inositol, choline, ethanolamine, and serine) (14–18), essential nutrients (e.g. zinc) (19), and growth phase (20, 21). These forms of regulation occur through a regulatory circuit involving a UASINO cis-acting element in the CHO1 promoter, the positive transcription factors Ino2p and Ino4p, and the transcriptional repressor Opi1p (10, 11, 22, 23). The expression of CHO1 is elevated in the exponential phase when cells are grown in the absence of the phospholipid precursors (14–18) and grown in the presence of the essential nutrient zinc (19). The elevated expression of the gene is mediated by an Ino2p-Ino4p complex that binds to the UASINO cis-acting element in the promoter to drive transcription (10, 22, 24, 25). This results in an increase in CHO1 mRNA abundance, PS synthase protein, and its activity (14–19). Overall, this regulation favors an increase in the amount of PS relative to PI (because both phospholipids are derived from CDP-DAG) and the synthesis of PE and PC via the CDP-DAG pathway (10, 23) (Fig. 1).
In contrast, the expression of CHO1 is reduced in the exponential phase by inositol supplementation, and this regulation is enhanced by the inclusion of choline, ethanolamine, or serine in the growth medium (14–18). CHO1 expression is also reduced in exponential phase cells when zinc is depleted from the growth medium (19) or when cells progress from the exponential to stationary phases of growth (20, 21). The regulations by zinc and growth phase occur in the absence of inositol supplementation (19–21). The reduction in CHO1 expression is mediated by the repressor Opi1p, which interacts with Ino2p to attenuate transcription for reduced abundance of mRNA abundance, protein, and enzymatic activity (10, 22, 24, 25). Consequently, CDP-DAG is favorably partitioned to PI at the expense of PS, and there is decrease in the synthesis of PE and PC via the CDP-DAG pathway (10, 23). The attenuation of the CDP-DAG pathway for PE and PC synthesis by CHO1 repression is compensated by the Kennedy pathway when cells are supplemented with ethanolamine or choline (10, 11, 23). In the Kennedy pathway, ethanolamine and choline undergo a series of reactions leading to the formation of PE and PC, respectively (10, 11) (Fig. 1). In fact, the Kennedy pathway is essential to cho1 mutants devoid of PS synthase activity, and as such, they are ethanolamine/choline auxotrophs (26, 27).
The expression of CHO1 is also controlled at the post-transcriptional level by mechanisms that do not involve its UASINO element and the transcription factors Ino2p, Ino4p, and Opi1p. For example, the level of the CHO1 transcript is controlled by its rate of decay (28, 29). The CHO1 transcript is primarily degraded through the general 5′–3′ mRNA decay pathway that involves deadenylation, mRNA decapping, and 5′–3′- exonuclease activities (29). In wild type cells, the CHO1 transcript is moderately stable with a half-life of 12 min (29). However, defects in mitochondrial respiration stabilize the CHO1 transcript to a half-life of >45 min (29). This regulation results in increases in the abundance of CHO1 mRNA and PS synthase protein, enzymatic activity, and the synthesis of PS in vivo (29).
With respect to its biochemical regulation, the activity of PS synthase is stimulated by the phospholipid precursor phosphatidate (30), whereas its activity is inhibited by the water-soluble phospholipid precursor inositol (31) and the nucleotide CTP (32). Stimulation of PS synthase favors phospholipid synthesis via the CDP-DAG pathway (12), whereas enzyme inhibition favors the synthesis of PI relative to PS and the synthesis of phospholipids via the Kennedy pathway (11, 12).
Phosphorylation is another mechanism by which PS synthase activity is regulated (33). PS synthase is a substrate for protein kinase A in vitro, and this phosphorylation results in an inhibition of enzyme activity (33). Protein kinase A is the principal mediator of signals transmitted through the RAS/cAMP pathway in S. cerevisiae (34, 35). The Ras-mediated stimulation of protein kinase A activity is associated with rapid cell growth and enhanced metabolic activity (34, 35). That phosphorylation inhibits PS synthase activity begs the question as to why the activity of an enzyme that is important to de novo membrane phospholipid synthesis is inhibited by a protein kinase whose function is to stimulate cell growth. To address this paradox, we identified Ser46 and Ser47 as the major sites of protein kinase A phosphorylation and constructed phosphorylation-deficient mutants for defined studies on the consequences of phosphorylation on phospholipid synthesis. Our studies with the mutant enzymes confirmed that protein kinase A phosphorylation inhibited PS synthase activity in vitro. However, these studies also indicated that the phosphorylated form of PS synthase was more abundant than the phosphorylation-deficient enzyme, which results in an elevated level of PS synthesis in vivo.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were reagent grade. Difco was the supplier of growth medium supplies. Restriction endonucleases, modifying enzymes, recombinant Vent DNA polymerase, and alkaline phosphatase were purchased from New England Biolabs. PCR and sequencing primers were prepared commercially by Genosys Biotechnologies. The plasmid DNA purification and DNA gel extraction kits were from Qiagen, Inc. The QuikChange site-directed mutagenesis and the Yeastmaker yeast transformation kits were purchased from Stratagene and Clontech, respectively. The DNA size ladder used for agarose gel electrophoresis, protein assay reagents, electrophoretic reagents, immunochemical reagents, protein molecular mass standards for SDS-PAGE, and acrylamide solutions were purchased from Bio-Rad. Polyvinylidene difluoride membranes and the enhanced chemifluorescence Western blotting detection kit were purchased from GE Healthcare. Ampicillin, aprotinin, ATP, benzamidine, bovine serum albumin, choline, leupeptin, pepstatin, protein A-Sepharose CL-4B, phenylmethylsulfonyl fluoride, phosphoamino acids, l-serine, sodium fluoride, l-1-tosylamido-2-phenylethyl chloromethyl ketone-trypsin, and Triton X-100 were purchased from Sigma. Mouse anti-phosphoglycerate kinase antibodies and alkaline phosphatase-conjugated goat anti-mouse IgG antibodies were from Invitrogen and Pierce, respectively. Protein kinase A catalytic subunit, protein kinase C, and casein kinase II were purchased from Promega. Radiochemicals were from PerkinElmer Life Sciences, and scintillation-counting supplies were from National Diagnostics. Lipids were from Avanti Polar Lipids, and thin layer chromatography plates (cellulose and silica gel 60) were from EM Science.
Strains and Growth Conditions
The bacterial and yeast strains used in this work are listed in Table 1. Standard methods were followed for the growth of yeast and bacteria (36, 37). Cultures were grown at 30 °C in YEPD medium (1% yeast extract, 2% peptone, 2% glucose) or in complete synthetic medium (38) containing 2% glucose. The appropriate amino acid of complete synthetic medium was omitted for selection purposes. Cells in liquid media were grown to the exponential phase (1–2 × 107 cells/ml), and cell numbers were determined spectrophotometrically at an absorbance of 600 nm. Escherichia coli strain DH5α was grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl (pH 7.4)) at 37 °C. Ampicillin (100 μg/ml) was added to cultures of DH5α carrying plasmids. Yeast and bacterial media were supplemented with 2 and 1.5% agar, respectively, for growth on plates.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source/Ref. |
|---|---|---|
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk−mk+) phoA supE44 l−thi-1 gyrA96 relA1 | Ref. 37 |
| S. cerevisiae | ||
| W303-1A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Ref. 63 |
| JSY94A | cho1Δ::TRP1 derivative of W303-1A | This study |
| W303-1A [rhoo] | rhoo derivative of W303-1A | Ref. 29 |
| Plasmid | ||
| pRS416 | Single copy vector containing the URA3 gene | Ref. 64 |
| pRS424 | Multicopy vector containing the TRP1 gene | Ref. 64 |
| pAS103 | Plasmid containing a 1.2-kb fragment of the CHO1 gene | Ref. 65 |
| pJS1 | Plasmid containing a 0.9-kb fragment of the CHO1 gene | This study |
| pJS2 | Plasmid containing the cho1Δ::TRP1 deletion cassette | This study |
| pHC101 | CHO1 gene from pAS103 ligated into the HindIII/SacI sites of pRS416 | This study |
| pHC102 | CHO1S46A derivative of pHC101 | This study |
| pHC103 | CHO1S47A derivative of pHC101 | This study |
| pHC104 | CHO1S46A/S47A derivative of pHC101 | This study |
| pHC105 | CHO1S140A derivative of pHC101 | This study |
| pHC106 | CHO1S141A derivative of pHC101 | This study |
| pHC107 | CHO1S140A/S141A derivative of pHC101 | This study |
| pHC108 | CHO1S46A/S47A/S140A/S141A derivative of pHC101 | This study |
| pHC109 | CHO1S46D derivative of pHC101 | This study |
| pHC110 | CHO1S47D derivative of pHC101 | This study |
| pHC111 | CHO1S46D/S47D derivative of pHC101 | This study |
Construction of Plasmids and PS Synthase Mutants
The plasmids used in this work are listed in Table 1. Standard methods were used for isolation and manipulation of DNA (37). Transformation of yeast (39, 40) and bacteria (37) with plasmids were performed as described previously. Plasmid pJS1 was constructed by insertion of the 0.9-kb CHO1 DNA from plasmid pAS103 into pBluescript II KS (−) at the EcoRI site. Plasmid pJS2 was constructed from pJS1 by replacing the 0.54-kb CHO1 DNA (NcoI/NdeI fragment) with the 0.94-kb TRP1 DNA that was released from pRS424 by digestion with SnaBI and SspI. For compatibility in ligation, the NcoI/NdeI-digested pJS1 was made blunt-ended using Klenow fragment. The 1.3-kb cho1Δ::TRP1 deletion cassette was released from plasmid pJS2 by digestion with EcoRI and used to transform W303-1A for one-step gene replacement. Transformants were selected on choline-supplemented complete synthetic medium lacking tryptophan. Of the yeast transformants, the cho1Δ mutant (strain JSY94A) was identified by its inability to grow in medium lacking choline. The cho1Δ mutant lacked the PS synthase protein and activity, and it did not synthesize PS in vivo. This mutant was used for the expression of wild type and phosphorylation site PS synthase mutants expressed on single copy plasmids (Table 1). Plasmid pHC101 was constructed by inserting CHO1 DNA (1.2 kb) derived from a HindIII/SacI digestion of plasmid pAS103 into plasmid pRS416 at the same restriction enzyme sites. The codons for the indicated serine residues in CHO1 were changed to alanine or aspartate codons by site-directed mutagenesis using plasmid pHC101 as the template and the primers listed in Table 2. The conditions for the amplification of DNA by PCR were optimized as described previously (41). All mutant alleles were completely sequenced to verify that no additional mutations were made.
TABLE 2.
Primers used in this work
| Primera | Sequence |
|---|---|
| S46A F | 5′-ACATTAAGCAGAAGGGCCGCTAGTATATTTTCTATAAA-3′ |
| S46A R | 5′-TATAGAAAATATACTAGCGGCCCTTCTGCT-3′ |
| S47A F | 5′-AGCAGAAGGGCCTCAGCTATATTTTCTATAAA-3′ |
| S47A R | 5′-TATAGAAAATATAGCTGAGGCCCTTCTGCT-3′ |
| S140A F | 5′-CTGAGAAATAGGGCTTCCTTAATGGGTCAA-3′ |
| S140A R | 5′-TTGACCCATTAAGGAAGCCCTATTTCTCAG-3′ |
| S141A F | 5′-CTGAGAAATAGGTCTGCTTTAATGGGTCAAGAA-3′ |
| S141A R | 5′-TTCTTGACCCATTAAAGCAGACCTATTTCTCAG-3′ |
| S46A/S47A F | 5′-ACATTAAGCAGAAGGGCCGCTGCTATATTTTCTATAAA-3′ |
| S46A/S47A R | 5′-TATAGAAAATATAGCAGCGGCCCTTCTGCT-3′ |
| S140A/S141A F | 5′-CTGAGAAATAGGGCTGCTTTAATGGGTCAAGAA-3 |
| S140A/S141A R | 5′-TTCTTGACCCATTAAAGCAGCCCTATTTCTCAG-3 |
| S46D F | 5′-ACATTAAGCAGAAGGGCCGATAGTATATTTTCTATAAA-3′ |
| S46D R | 5′-TATAGAAAATATACTATCGGCCCTTCTGCT-3′ |
| S47D F | 5′-AGCAGAAGGGCCTCAGATATATTTTCTATAAA-3′ |
| S47D R | 5′-TATAGAAAATATATCTGAGGCCCTTCTGCT-3′ |
| S46D/S47D F | 5′-ACATTAAGCAGAAGGGCCGATGATATATTTTCTATAAA-3′ |
| S46D/S47D R | 5′-TATAGAAAATATATCATCGGCCCTTCTGCT-3′ |
a F, forward orientation relative to sense strand; R, reverse relative to sense strand.
Immunoprecipitation and Immunoblotting
The peptide sequences MVESDEDFAPQEFPH (residues 1–15) and FFIHGCGMISKSLKIPKP (residues 259–276) at the N-terminal and C-terminal portions of PS synthase, respectively, were synthesized and used to raise antibodies in New Zealand White rabbits by standard procedures (42) at Bio-Synthesis, Inc. The IgG fraction of the rabbit anti-PS synthase antibodies was purified from antisera by protein A-Sepharose chromatography (42) and used for immunoprecipitation and immunoblotting experiments. Cell extracts (0.5 mg of protein) were incubated for 1 h with 10 μg of anti-PS synthase antibodies in a total volume of 0.5 ml, followed by incubation with 100 μl of protein A-Sepharose CL-4B beads (10% slurry, w/v) for 1 h at 4 °C. Immune complexes were collected by centrifugation at 1500 × g for 30 s and washed three times with phosphorylation reaction buffer (40 mm Tris-HCl (pH 7.4), 60 mm β-mercaptoethanol, and 20 mm MgCl2). Following the washing steps, the immune complexes of the PS synthase were used as substrate for phosphorylation. For immunoblotting, cell extracts or phosphorylation reaction mixtures were separated by SDS-PAGE (43) and transferred to polyvinylidene difluoride membrane (44). The membrane was probed with anti-PS synthase antibodies (2 μg/ml) as described previously (42). Goat anti-rabbit IgG alkaline phosphatase conjugate was used as a secondary antibody at a dilution of 1:5000. For the detection of phosphoglycerate kinase, the membranes were probed with mouse anti-phosphoglycerate kinase antibodies (2 μg/ml) and then probed with a 1:5000 dilution of alkaline phosphatase-conjugated goat anti-mouse IgG antibodies. The PS synthase and phosphoglycerate kinase proteins were detected on immunoblots using the ECF Western blotting chemifluorescent detection kit as described by the manufacturer. Fluorescent signals on the immunoblots were acquired by FluorImaging and analyzed using ImageQuant software. Immunoblot signals were in the linear range of detectability.
Preparation of Cell Extracts and Membranes
All steps were performed at 4 °C. The cell extract and total membrane fraction were prepared as described previously (45). In brief, cells were disrupted by homogenization with glass beads in 50 mm Tris-maleate buffer (pH 7.0) containing 1 mm EDTA, 0.3 m sucrose, 10 mm 2-mercaptoethanol, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm benzamidine, and 5 μg/ml each of aprotinin, leupeptin, and pepstatin. The cell extract was obtained by centrifugation of the homogenate at 1500 × g for 10 min. The total membrane fraction was obtained from the cell extract by centrifugation at 100,000 × g for 1 h. Membranes were resuspended in 50 mm Tris-maleate (pH 7.0) buffer containing 10 mm MgCl2, 10 mm 2-mercaptoethanol, 20% glycerol (w/v), and 0.5 mm phenylmethylsulfonyl fluoride. Protein concentration was determined by the method of Bradford (46) using bovine serum albumin as the standard.
Phosphorylation and Dephosphorylation Reactions
Phosphorylation reactions were performed at 30 °C in a total volume of 25 μl for the indicated time intervals. Immunoprecipitated PS synthase or total membranes were incubated in 40 mm Tris-HCl (pH 7.4) buffer containing 60 mm β-mercaptoethanol, 20 mm MgCl2, 50 μm [γ-32P]ATP (5 μCi/nmol), and 5 pmol/min protein kinase A. The protein kinase C reaction buffer contained 50 mm Tris-HCl (pH 8.0), 10 mm MgCl2, 10 mm β-mercaptoethanol, 0.375 mm EDTA, 0.375 mm EGTA, 1.7 mm CaCl2, 20 μm diacylglycerol, 50 μm phosphatidylserine, 20 μm[γ-32P]ATP (5 μCi/nmol), and 50 nmol/min protein kinase C. The casein kinase II reaction buffer contained 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 10 mm dithiothreitol, 50 μm [γ-32P]ATP (5 μCi/nmol), and 25 pmol/min casein kinase II. For the dephosphorylation of PS synthase, immunoprecipitated enzyme was incubated for 60 min at 37 °C with 50 mm Tris-HCl (pH 8.0) buffer containing 100 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol, and 5 μmol/min alkaline phosphatase. At the end of the phosphorylation and dephosphorylation reactions, the samples were subjected to immunoblot analysis.
In Vivo Labeling and Analysis of PS Synthase
Cultures (5 ml) expressing the wild type and mutant forms of PS synthase were grown to the exponential phase in the presence of 32Pi (500 μCi). The cells were harvested, resuspended in 1 ml of 50 mm Tris-HCl (pH 7.5), and disrupted with glass beads (50 mg) using a vortex mixer. The samples were then centrifuged at 1500 × g for 5 min to obtain the cell extract. The 32P-labeled PS synthase was immunoprecipitated from the extract with 10 μg of anti-PS synthase antibodies, followed by immunoblot analysis as described above.
Analysis of Phosphoamino Acids and Phosphopeptides
For phosphoamino acid analysis, 32P-labeled PS synthase on pieces of polyvinylidene difluoride membrane was subjected to acid hydrolysis with 6 n HCl (47). Hydrolysates were dried in vacuo and applied to 0.1-mm cellulose thin layer chromatography plates with standard phosphoamino acids (2.5 μg of phosphoserine, 2.5 μg of phosphothreonine, and 5 μg of phosphotyrosine). After separation by two-dimensional electrophoresis (48), the plates were analyzed by phosphorimaging. The 32P-labeled phosphoamino acids were identified by comparison with standard phosphoamino acids that were visualized by staining with 0.25% ninhydrin in acetone. For phosphopeptide mapping (49), pieces of polyvinylidene difluoride membrane containing 32P-labeled PS synthase were subjected to digestion with l-1-tosylamido-2-phenylethyl chloromethyl ketone-trypsin, followed by electrophoresis and ascending chromatography on cellulose thin layer glass plates. Dried plates were then subjected to phosphorimaging analysis.
PS Synthase Assay
PS synthase activity was measured by following the incorporation of 0.5 mm [3-3H]serine (10,000 cpm/nmol) into PS in the presence of 50 mm Tris-HCl (pH 8.0), 0.6 mm MnCl2, 3.2 mm Triton X-100, and 0.2 mm CDP-diacylglycerol (50). The average S.D. value of the PS synthase assays (performed in triplicate) was ±5%. Enzyme reactions were linear with time and protein concentration. A unit of PS synthase activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of product/min. The relative amounts of PS synthase activity were normalized to the amount of PS synthase protein that was determined by ImageQuant analysis of immunoblot images.
Labeling and Analysis of Phospholipids
Labeling of phospholipids with 32Pi and [14C]serine was performed as described previously (26, 27). For the analysis of phospholipid composition, cells were labeled to steady state with 32Pi. For the analysis of PS synthesis, exponential phase cells were labeled for 30 min with [14C]serine. Phospholipids were extracted from labeled cells by the method of Bligh and Dyer (51). To aid in the extraction, the cell suspension in the extraction solvent was mixed with glass beads using a vortex mixer (52). Two-dimension TLC with chloroform/methanol/ammonium hydroxide/water (45:25:2:3) (dimension 1) and chloroform/methanol/acetic acid/water (63:8:10:2) (dimension 2) was used to separate phospholipids (28). The identity of the labeled phospholipids on TLC plates was confirmed by comparison with standards after exposure to iodine vapor. Radiolabeled phospholipids on the chromatography plates were visualized by phosphorimaging. The relative amounts of the 32P-labeled phospholipids were analyzed using ImageQuant software. Liquid scintillation counting was used to quantify the amount of [14C]PS.
Analyses of Data
Statistical analyses were performed with SigmaPlot software. Statistical significance was determined by performing Student's t test. p values of <0.05 were taken as a significant difference.
RESULTS
CHO1-encoded PS Synthase Exists in Two Forms That Differ in Electrophoretic Mobility
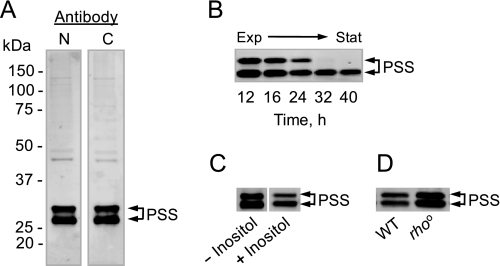
Antibodies generated against a peptide sequence found at the N-terminal portion of PS synthase have recognized proteins with molecular masses of 30 and 27 kDa (28). Based on the fact that the product of the CHO1 gene is predicted to be a 30.8-kDa protein and that previous studies indicated that PS synthase was subject to proteolysis (4, 53), it has been assumed that the 27-kDa protein was a proteolysis product of the full-length 30-kDa protein (28). In this work, immunoblot experiments were performed on cell extracts derived from exponential phase cells using antibodies generated against sequences found at both the N- and C-terminal ends of PS synthase. To our surprise, both antibodies recognized the 30- and 27-kDa proteins (Fig. 2A). The relative abundance of the proteins (∼45% for the 30-kDa form and ∼55% for the 27-kDa form) was about the same regardless of which antibody was used for the immunoblot analysis (Fig. 2A). Thus, the 27-kDa protein was not derived from the 30-kDa protein by proteolysis at either end of the protein.
FIGURE 2.
PS synthase exists in two forms that differ in electrophoretic mobility. Cell extracts were prepared from wild type cells grown to the exponential (A) and stationary (B) phases of growth, from exponential phase wild type cells grown without and with 50 μm inositol (C), and from exponential phase wild type (WT) and rhoo mutant cells (D). For the experiment shown in B, cells were harvested at the indicated time points as the culture progressed from the exponential (Exp) to stationary (Stat) phases of growth. Samples (12.5 μg of protein) were subjected to immunoblot analysis using 2 μg/ml anti-PS synthase antibodies raised against the N-terminal (N) portion of the PS synthase (PSS) protein. Antibodies to the C-terminal (C) portion of the protein were also used in the experiment shown in A. The lanes shown in C were not adjacent to each other on the original immunoblot and are positioned side by side for comparison. The immunoblots shown are representative of three independent experiments. The positions of the 30- and 27-kDa PS synthase proteins are indicated.
The expression of CHO1 is reduced when wild type cells progress from the exponential to the stationary phases of growth (21) or when wild type exponential phase cells are supplemented with inositol (16, 21). On the other hand, the CHO1 transcript is stabilized by respiratory deficiency (29). These forms of regulation govern the amount of PS synthase activity (16, 29). We questioned if the regulated expression of CHO1 affected the presence of the different forms of PS synthase. Wild type cells were grown to the stationary phase (reached by 30 h of growth), and samples were taken at various time intervals for immunoblot analysis. Although both forms of PS synthase diminished as cells progressed from the exponential to stationary phases of growth, the 27-kDa form was essentially the only form present in stationary phase cells (Fig. 2B, 32 and 40 h time points). In the next experiment, wild type cells were grown to the exponential phase in complete synthetic medium without and with 50 μm inositol. Immunoblot analysis of cell extracts derived from these cultures showed that inositol repressed the expression of both forms of PS synthase, but it did not affect the relative amounts of the two forms (Fig. 2C). Finally, an immunoblot experiment was performed on cell extracts derived from exponential phase wild type and respiration-deficient rhoo mutant cells. Although the amounts of the 30- and 27-kDa forms of PS synthase proteins were more abundant in rhoo mutant cells, their relative amounts were similar to that found in wild type cells (Fig. 2C). Regardless of which antibodies (e.g. N- or C-terminal) were used, the results of these experiments were the same.
Protein Kinase A Phosphorylation Causes an Electrophoretic Mobility Shift of PS Synthase
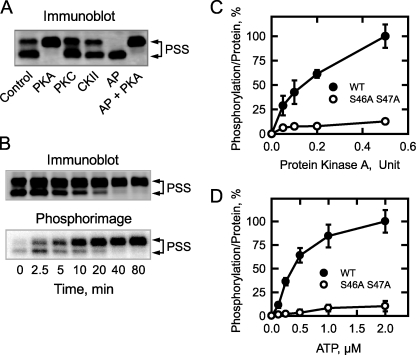
Previous studies have shown that purified PS synthase is a substrate for protein kinase A (33). Accordingly, we questioned if phosphorylation affected the relative amounts of the 30- and 27-kDa proteins. PS synthase was isolated from cell extracts by immunoprecipitation with anti-PS synthase (N-terminal) antibodies and then tested as a substrate for protein kinase A (Fig. 3A). The phosphorylation of PS synthase resulted in the electrophoretic mobility shift of the 27-kDa form to the 30-kDa form. By contrast, the incubation of PS synthase with alkaline phosphatase caused the electrophoretic mobility shift of the 30-kDa form to the 27-kDa form. Moreover, incubation of the alkaline phosphatase-treated PS synthase with protein kinase A resulted in the conversion of the 27-kDa form back to the 30-kDa form of the enzyme (Fig. 3A).
FIGURE 3.
The phosphorylation of PS synthase by protein kinase A causes an electrophoretic mobility shift of the 27-kDa protein to the 30-kDa protein; the phosphorylation of PS synthase is dependent on time and on the concentrations of protein kinase A and ATP. The cell extract was prepared from exponential phase cho1Δ mutant cells bearing CHO1 on a single copy plasmid. PS synthase (PSS) was immunoprecipitated from the cell extract (0.5 mg) with 10 μg of anti-PS synthase antibodies (N-terminal). A, immunoprecipitated PS synthase was incubated for 40 min with 50 μm ATP and 5 pmol/min protein kinase A (PKA), 50 nmol/min protein kinase C (PKC), 25 pmol/min casein kinase II (CKII), 5 μmol/min alkaline phosphatase (AP), or an alkaline phosphatase-treated sample was incubated with 5 pmol/min protein kinase A (AP + PKA). B, immunoprecipitated PS synthase was incubated for the indicated time intervals with 50 μm [γ-32P]ATP and 1 pmol/min protein kinase A. C, immunoprecipitated wild type (WT) and S46A/S47A mutant PS synthase were incubated for 40 min with 50 μm [γ-32P]ATP and the indicated amounts of protein kinase A. D, immunoprecipitated wild type and S46A S47A mutant PS synthase were incubated for 40 min with 1 pmol/min protein kinase A and the indicated concentrations of [γ-32P]ATP. After the reactions, samples were subjected to immunoblot analysis using anti-PS synthase antibodies (N-terminal). Phosphorimaging analysis was performed on the polyvinylidene difluoride membranes with 32P-labeled PS synthase (B–D). The data shown in A and B are representative of three independent experiments. For C and D, the relative amounts of phosphate incorporated into PS synthase were quantified using ImageQuant software. The maximum extent of PS synthase phosphorylation was set at 100%. The data were normalized to the amount of PS synthase protein as determined by immunoblot analysis. The values reported were the average of three separate experiments ± S.D. The positions of the 30- and 27-kDa PS synthase proteins are indicated.
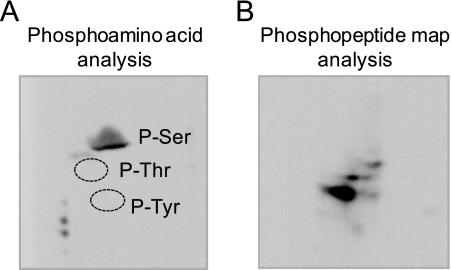
The conversion of the 27-kDa form of PS synthase to its 30-kDa form was time-dependent and coincident with the incorporation of the γ phosphate of 32P-labeled ATP into the 27-kDa form that shifted to the 30-kDa form of PS synthase (Fig. 3B). In addition, the phosphorylation of PS synthase (i.e. the γ-phosphate incorporation of ATP into the enzyme and the electrophoretic shift) was dependent on the amount of protein kinase A in the reaction (Fig. 3C) and the concentration of ATP (Fig. 3D). As described previously (33), phosphoamino acid analysis of the 32P-labeled PS synthase showed that protein kinase A phosphorylated the enzyme on a serine residue (Fig. 4A), and phosphopeptide mapping analysis of the 32P-labeled enzyme showed one major phosphopeptide (Fig. 4B). Additional minor radioactive spots were present in the phosphopeptide map.
FIGURE 4.
Phosphoamino acid and phosphopeptide mapping analyses of PS synthase phosphorylated by protein kinase A. The cell extract was prepared from exponential phase cho1Δ mutant cells bearing CHO1 on a single copy plasmid. PS synthase was immunoprecipitated from the cell extract (0.5 mg) with 10 μg of anti-PS synthase antibodies (N-terminal). Immunoprecipitated PS synthase was phosphorylated for 40 min with 50 μm [γ-32P]ATP and 1 pmol/min protein kinase A, followed by SDS-PAGE and transfer to polyvinylidene difluoride membrane. A, a piece of polyvinylidene difluoride membrane containing 32P-labeled PS synthase was hydrolyzed with 6 n HCl for 90 min at 110 °C, and the hydrolysate was separated by two-dimensional electrophoresis. The positions of the standard phosphoamino acids phosphoserine (P-Ser), phosphothreonine (P-Thr), and phosphotyrosine (P-Tyr) are indicated. B, a piece of polyvinylidene difluoride membrane containing 32P-labeled PS synthase was digested with trypsin. The resulting peptides were separated on cellulose thin layer plates by electrophoresis (from left to right) in the first dimension and by chromatography (from bottom to top) in the second dimension. The data shown in the two panels were representative of two independent experiments.
Although PS synthase has putative target sites for protein kinase C and casein kinase II, it was not a substrate for these protein kinases. The incubation of the immunoprecipitated enzyme with protein kinase C or with casein kinase II did not affect the electrophoretic mobility of PS synthase (Fig. 3A). In addition, these protein kinases did not catalyze the transfer of the γ phosphate from 32P-labeled ATP into the PS synthase protein (data not shown).
Protein Kinase A Phosphorylation Site Mutations Result in the Loss of the 30-kDa Form of PS Synthase and a Reduced Amount of Total PS Synthase Protein
PS synthase has potential serine (Ser46, Ser47, Ser140, and Ser141) phosphorylation sites within a protein kinase A sequence motif. Mutagenesis of these sites was performed to examine the hypothesis that they are targets for the protein kinase A phosphorylation. PS synthase with serine-to-alanine (S46A, S47A, S46A/S47A, S140A, S141A, and S140A/S141A) mutations were constructed and expressed on a single copy plasmid in the cho1Δ mutant. Cells bearing the wild type and mutant alleles of the CHO1 gene complemented the ethanolamine/choline auxotrophy of the cho1Δ mutant. Although no morphological differences were observed in cells bearing the mutant enzymes, the growth rate of cells expressing the S46A/S47A mutant enzyme was slightly less than cells expressing the wild type enzyme (data not shown).
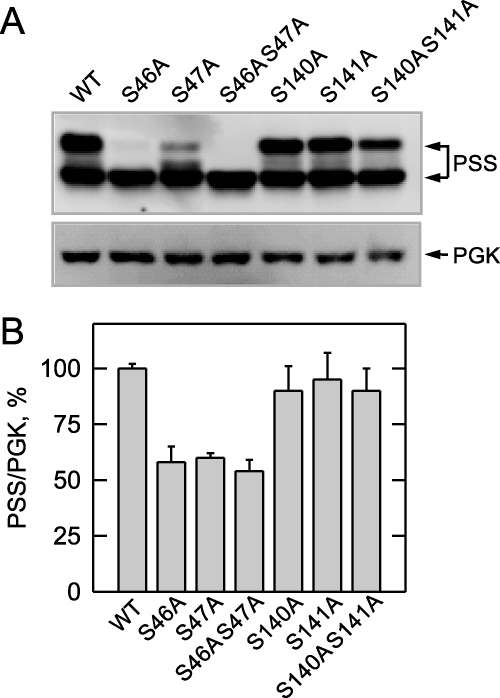
The cells expressing the S46A or S47A mutant enzyme primarily exhibited the 27-kDa form of PS synthase, whereas cells expressing the S46A/S47A mutant enzyme exhibited only the 27-kDa protein (Fig. 5A). In addition, the total amount of PS synthase (27-kDa plus 30-kDa forms) in cells expressing the S46A/S47A mutant enzyme was reduced by ∼50% (Fig. 5B). The S140A and S141A mutations had little effect on the relative amounts of the 30- and 27-kDa forms of PS synthase as well as the total amount of enzyme in the cell (Fig. 5, A and B).
FIGURE 5.
PS synthase protein levels are reduced in cho1Δ cells bearing the S46A and S47A mutations. cho1Δ cells expressing wild type (WT) and the indicated PS synthase (PSS) mutant enzymes were grown to the exponential phase of growth in complete synthetic medium. Cell extracts were prepared and used for immunoblot analysis (12.5 μg of protein samples) using anti-PS synthase and anti-phosphoglycerate kinase (PGK) antibodies (A). The relative amounts of PS synthase (30- plus 27-kDa forms)/phosphoglycerate kinase proteins from wild type and mutant cells were determined by ImageQuant analysis (B). Representative immunoblots of PS synthase and phosphoglycerate kinase are shown in A, whereas the quantitation data shown in B are from three independent experiments ± S.D. The positions of the 30- and 27-kDa PS synthase proteins are indicated.
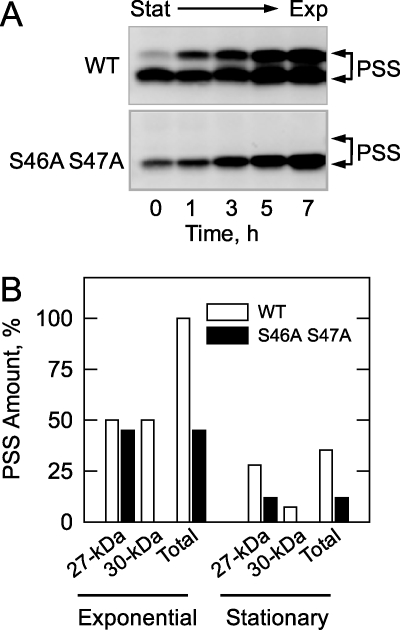
The effects of the S46A/S47A mutations on the amounts of the 27- and 30-kDa forms of PS synthase were examined during growth. For this experiment, wild type and mutant cells were first grown to stationary phase. The stationary phase cells were then resuspended in fresh growth medium for the resumption of growth into the exponential phase. As previously shown in Fig. 2B, the 27-kDa form of PS synthase was present in the exponential and stationary phases of wild type cells, but the 30-kDa form of the enzyme was hardly present in stationary phase cells (Fig. 6A, time zero). The amount of the 30-kDa form in stationary phase was reduced by 85% when compared with that in exponential phase (Fig. 6B). The total amount (both forms) of the wild type PS synthase was reduced by 65% in stationary phase when compared with exponential phase (Fig. 6B). The 27-kDa protein was the only form of PS synthase present in the exponential and stationary phases of cells expressing the S46A/S47A mutant enzyme (Fig. 6A). The amount of the 27-kDa protein in stationary phase cells was reduced by 75% when compared with the exponential phase (Fig. 6B). The total amount of PS synthase in cells expressing the S46A/S47A mutant enzyme was reduced by 55% in the exponential phase and by 66% in the stationary phase when compared with cells expressing the wild type enzyme (Fig. 6B).
FIGURE 6.
Effects of the S46A/S47A mutations on the amounts of the PS synthase proteins during growth. A, cultures (200 ml) of cho1Δ cells expressing wild type (WT) and the S46A/S47A mutant PS synthase (PSS) enzymes were grown to the stationary (Stat) phase of growth (32 h) in complete synthetic medium. The cells were harvested by centrifugation, resuspended in 200 ml of fresh growth medium, and allowed to resume growth back into the exponential (Exp) phase. At the indicated time intervals, cells (20 ml) were taken from the cultures, cell extracts were prepared, and samples (25 μg of protein) were subjected to immunoblot analysis using 2 μg/ml anti-PS synthase antibodies (N-terminal). The immunoblot data for the wild type and mutant enzymes are positioned vertically for comparison, and the positions of the 30- and 27-kDa forms of PS synthase are indicated. The immunoblot is representative of two independent experiments. B, the relative amounts of the PS synthase proteins were determined by ImageQuant analysis of the images in A. The amounts of the 27- and 30-kDa forms of the wild type and S46A/S47A mutant proteins were normalized to the total amount of the wild type enzyme (both forms) in the exponential phase of growth (7 h time point). The positions of the 30- and 27-kDa forms of PS synthase are indicated.
We also constructed and expressed PS synthase with S46D, S47D, and S46D/S47D mutations to examine the effects of a phosphorylation mimic on the enzyme. These mutations did not have major mimicking effects on the electrophoretic mobility of PS synthase.
Effects of Phosphorylation Site Mutations on the Phosphorylation of PS Synthase by Protein Kinase A
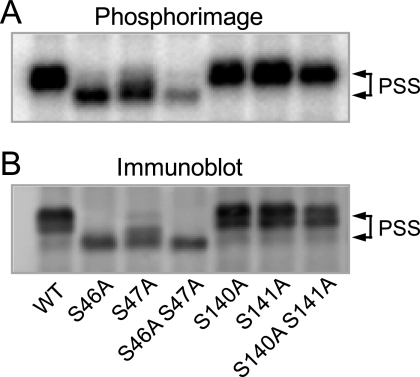
The effects of the phosphorylation site mutations on the phosphorylation of PS synthase by protein kinase A were examined. The immunoprecipitated wild type and mutant enzymes were incubated with protein kinase A and 32P-labeled ATP. After the phosphorylation reactions, samples were subjected to SDS-PAGE, followed by transfer to polyvinylidene difluoride membrane and phosphorimaging analysis. This analysis showed that the S46A and S47A mutant enzymes could be phosphorylated by protein kinase A, but the phosphorylation was reduced by ∼50% (Fig. 7A). Also, the phosphorylation did not result in the mobility shift of the 27-kDa form to the 30-kDa form of PS synthase (Fig. 7, A and B). The S46A/S47A double mutation caused >90% reduction in protein kinase A phosphorylation when compared with the control wild type enzyme, and there was no shift in the electrophoretic mobility of the enzyme (Fig. 3, C and D, and Fig. 7, A and B). This result indicated that phosphorylations of both Ser46 and Ser47 were required for the shift in the electrophoretic mobility of PS synthase.
FIGURE 7.
The phosphorylation of PS synthase by protein kinase A is reduced by the S46A and S47A mutations. The cell extract was prepared from exponential phase cho1Δ mutant cells expressing wild type (WT) and the indicated PS synthase (PSS) mutant enzymes. PS synthase was immunoprecipitated from the cell extract (0.5 mg) with 10 μg of anti-PS synthase antibodies (N-terminal). The immunoprecipitated PS synthase samples were incubated for 40 min with 50 μm [γ-32P]ATP and 1 pmol/min protein kinase A. After the incubation, the samples were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membrane. The polyvinylidene difluoride membrane was subjected to phosphorimaging (A) and immunoblot analysis using anti-PS synthase antibodies (N-terminal) (B). The data shown are representative of three independent experiments. The positions of the 30- and 27-kDa PS synthase proteins are indicated.
The major phosphopeptide of the phosphorylated wild type PS synthase (Fig. 4B) was still present in the phosphopeptide maps of the phosphorylated S46A and S47A mutant enzymes. As expected, this phosphopeptide had a reduced radioactive signal and migrated on the maps to slightly different positions (data not shown). This major peptide, however, was absent from the phosphopeptide map of the phosphorylated S46A/S47A double mutant enzyme (data not shown), indicating that Ser46 and Ser47 were contained in the major phosphopeptide of the phosphorylated wild type enzyme. The S140A, S141A, and S140A/S141A mutations did not affect the phosphorylation of PS synthase by protein kinase A as indicated by the accompanying conversion from the 27- to the 30-kDa form (Fig. 7, A and B). Moreover, a S46A/S47A/S140A/S141A quadruple mutant was indistinguishable from the S46A/S47A double mutant with respect to defects in protein kinase A phosphorylation (data not shown). Thus, the minor phosphorylation sites responsible for the residual ∼10% of protein kinase A phosphorylation of PS synthase were not due to the phosphorylation of Ser140 and Ser141. Additional studies will be required to identify the sites responsible for the residual phosphorylation.
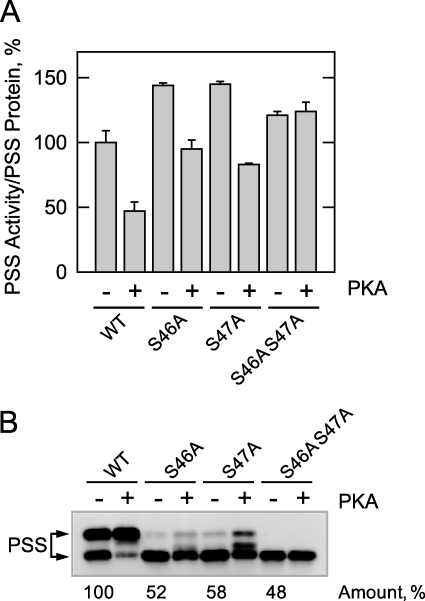
Effects of the Phosphorylation Site Mutations on the Protein Kinase A-mediated Inhibition of PS Synthase Activity
Membranes prepared from cho1Δ cells expressing the wild type and phosphorylation-deficient enzymes were incubated with protein kinase A and unlabeled ATP. Following incubation, the membrane samples were assayed for PS synthase activity and subjected to immunoblot analysis. As described previously (33), the protein kinase A phosphorylation of the wild type enzyme resulted in a decrease (53%) in PS synthase activity (Fig. 8A), and this inhibition correlated with a shift in the electrophoretic mobility of the 27- to the 30-kDa form of the enzyme (Fig. 8B). PS synthase activity, which was normalized to the total amount of PS synthase protein in the membrane samples, was elevated (25–50%) in the S46A, S47A, and S46A/S47A mutants (Fig. 8A). In addition, the S46A and S47A mutations caused a small attenuation of the inhibitory effect that the phosphorylation had on the PS synthase activity of the wild type enzyme (Fig. 8A), and a smaller portion of the 27-kDa form was converted to the 30-kDa form (Fig. 8B). Moreover, the protein kinase A-mediated inhibition of PS synthase activity was abolished by the S46A/S47A double mutation, and there was no shift in the electrophoretic mobility of the 27-kDa form of the enzyme (Fig. 8, A and B, respectively). Taken together, these data supported the conclusion that phosphorylation at Ser46 and Ser47 inhibited PS synthase activity. Consistent with the results from cell extracts (Fig. 5), the total amount of PS synthase (30-kDa plus 27-kDa forms) in the membrane fraction of the S46A, S47A, and S46A/S47A was reduced by about 50% when compared with the wild type control (Fig. 8B).
FIGURE 8.
The S46A and S47A mutations abolish the protein kinase A-mediated inhibition of PS synthase activity. The membrane fraction was prepared from exponential phase cho1Δ mutant cells expressing wild type (WT) and the indicated PS synthase (PSS) mutant enzymes. Samples (25 μg of protein) were incubated for 30 min in the phosphorylation reaction mixture containing 50 μm ATP without and with 5 pmol/min protein kinase A (PKA). After the incubation, one half of the sample was used for the measurement of PS synthase activity (A), and the other half of the sample was used for immunoblot analysis with anti-PS synthase antibodies (B). The PS synthase activity was normalized to the relative amount of PS synthase (30- plus 27-kDa forms) that was determined by ImageQuant analysis of the immunoblot. The PS synthase activity data are from three independent experiments ± S.D. A representative immunoblot of the three experiments is shown. The positions of the 30- and 27-kDa PS synthase proteins are indicated.
The Phosphorylation and Amount of PS Synthase and the Synthesis of PS Are Reduced by the S46A/S47A Mutations in Vivo
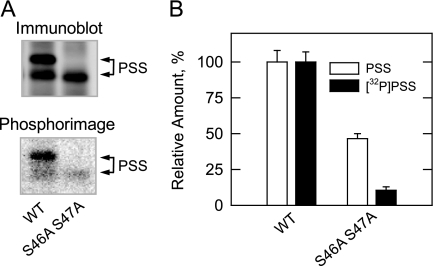
Cells expressing the wild type and S46A/S47A double mutant enzymes were labeled with 32Pi to detect the phosphorylated PS synthase in vivo. PS synthase was immunoprecipitated from cell extracts derived from exponential phase cells and subjected to immunoblot and phosphorimaging analyses (Fig. 9A). The immunoblot showed that both the 27- and 30-kDa forms of PS synthase were present in cells expressing the wild type enzyme. The phosphorimage showed that the 30-kDa form of the wild type PS synthase was the most heavily phosphorylated form in vivo. In contrast, the 27-kDa form was the only form of PS synthase found in the cells expressing the S46A/S47A double mutant (Fig. 9A). A barely detectable level of phosphorylated 27-kDa form PS synthase was observed in the double mutant. ImageQuant analysis of the data indicated that the S46A/S47A mutations caused a 54% decrease in total amount of PS synthase protein and a 90% decrease in the phosphorylation of the enzyme in vivo (Fig. 9B).
FIGURE 9.
Effects of the S46A/S47A mutations on the amount and the phosphorylation of the PS synthase in vivo. Cultures (5 ml) of cho1Δ mutant cells expressing wild type (WT) and the S46A/S47A mutant PS synthase (PSS) enzymes were grown to the exponential phase of growth in complete synthetic medium containing 32Pi (100 μCi/ml). Cell extracts were prepared and used for the immunoprecipitation of PS synthase with anti-PS synthase antibodies (N-terminal). A, the immunoprecipitated samples were subjected to immunoblot analysis (top) and phosphorimaging (bottom). B, the relative amounts of the PS synthase proteins and the extent of their phosphorylations were determined by ImageQuant analysis of the images in A. The data shown in A are representative of two independent experiments, whereas the data shown in B are the average of two independent experiments ± S.D. The positions of the 30- and 27-kDa forms of PS synthase are indicated.
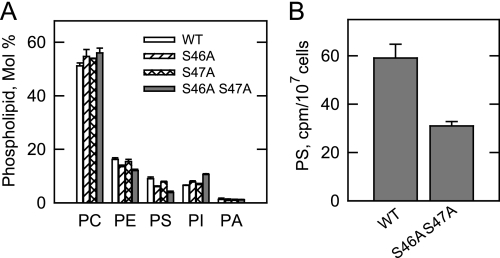
A phospholipid composition analysis was performed to further examine the in vivo consequences of the protein kinase A phosphorylation site mutations in PS synthase. The cho1Δ cells expressing the wild type and phosphorylation site mutant enzymes were labeled with 32Pi, followed by the extraction and analysis of phospholipids (Fig. 10A). 32Pi was incorporated into phospholipids that are synthesized by way of the CDP-DAG and the Kennedy pathways (11, 26). The phospholipids that were most affected by the phosphorylation site mutations were PS and PI, which are both derived from the common precursor CDP-DAG (Fig. 1) (11). Although the S46A and S47A mutations had relatively small effects on the relative amounts of PS and PI, the S46A/S47A double mutation resulted in a 57% decrease in PS and a 40% increase in PI (Fig. 10A). Cells expressing the wild type and the S46A/S47A mutant PS synthase enzymes were also subjected to pulse labeling with [14C]serine, followed by phospholipid analysis. As described previously (26), the major phospholipid identified after the 30-min labeling period was PS. In agreement with the 32Pi labeling experiment, the S46A/S47A mutations caused a 50% decrease in the synthesis of PS (Fig. 10B). The labeled serine was also incorporated into PE, phosphatidylmonomethylethanolamine, and phosphatidyldimethylethanolamine. However, the labeling of these lipids was not significantly affected by the mutations (data not shown).
FIGURE 10.
Effects of the S46A, S47A, and S46A/S47A mutations in PS synthase on phospholipid composition and on the synthesis of PS. A, cultures (5 ml) of cho1Δ mutant cells expressing wild type (WT) and the indicated PS synthase mutant enzymes were grown to the exponential phase of growth in complete synthetic medium containing 32Pi (5 μCi/ml). B, cells expressing the wild type and the S46A/S47A mutant PS synthase enzymes were grown to the exponential phase and then labeled for 30 min with [14C]serine (10 μCi/ml). Phospholipids were extracted and separated by two-dimensional thin layer chromatography. The TLC plates were subjected to phosphorimaging. For the experiment shown in A, the images were subjected to ImageQuant analysis. The percentages shown for the individual phospholipids were normalized to the total 32P-labeled chloroform-soluble fraction that included minor phospholipids and sphingolipids. For the experiment shown in B, the amount of label in PS was determined by scintillation counting. The data shown in both panels are the average of three experiments ± S.D. PA, phosphatidate.
DISCUSSION
In this work, we addressed the regulation of PS synthase by protein kinase A phosphorylation. Using antibodies directed against the N-terminal and C-terminal portions of PS synthase, we discovered that the enzyme existed in two forms (30 and 27 kDa) whose abundance was dependent on growth phase but not on the regulated expression of CHO1 by inositol supplementation or respiratory deficiency. The 30- and 27-kDa forms were present in exponential phase cells, whereas the 27-kDa protein was primarily the only form of the enzyme present in stationary phase cells. In vitro experiments revealed that the 30- and 27-kDa forms were interchangeable by phosphorylation/dephosphorylation. The phosphorylation by protein kinase A, which was time- and dose-dependent and dependent on the concentration of ATP caused a shift in the electrophoretic mobility of the 27-kDa form to the 30-kDa form of the enzyme. That the doubly phosphorylated 30-kDa form was primarily present in exponential phase cells was consistent with the function of protein kinase A to stimulate cell growth (34, 35).
A combination of biochemical and molecular approaches was used to identify the protein kinase A phosphorylation sites in PS synthase. We examined the hypothesis that amino acid residues Ser46 and Ser47 contained in a protein kinase A sequence motif were target sites for phosphorylation. Ser46 → Ala and/or Ser47 → Ala mutations were constructed and used to support this hypothesis. These mutations alone and in combination did not affect the expression of the CHO1 transcript (data not shown), but they did affect the relative amounts of the 30- and 27-kDa forms of PS synthase. In exponential phase cells, the S46A, S47A, and S46A/S47A mutations caused loss of the 30-kDa form of PS synthase. This was consistent with the conclusion that phosphorylations at Ser46 and Ser47 were responsible for the shift in the electrophoretic mobility of the 27-kDa form to the 30-kDa form. Moreover, the in vitro phosphorylation experiments confirmed that the mutations reduced the protein kinase A-mediated incorporation of the γ-phosphate of ATP into the PS synthase enzyme. The S46A/S47A double mutation totally abolished the electrophoretic mobility shift in the enzyme and reduced the total amount of phosphorylation by over 90%. Importantly, the effects of the S46A/S47A mutations on PS synthase phosphorylation were reflected in vivo. In exponential phase cells expressing the wild type enzyme, the 30-kDa form was more heavily phosphorylated when compared with the 27-kDa form, and the 30-kDa form was absent in cells expressing the S46A/S47A mutant enzyme. Moreover, about half of the total amount of the PS synthase enzyme was missing in cells expressing the phosphorylation-deficient mutant.
As described previously (33), the protein kinase A phosphorylation resulted in a decrease in PS synthase activity. However, the protein kinase A-mediated inhibition of PS synthase activity was abolished by the S46A/S47A double mutation. Notwithstanding the inhibitory effect of phosphorylation on activity, cells expressing the S46A/S47A mutant enzyme showed a reduction in PS relative to PI and a decrease in PS synthesis in vivo. These effects on phospholipid synthesis were attributed to the reduction in the total amount of PS synthase observed in cells carrying the S46A/S47A mutant enzyme. Thus, on one hand, phosphorylation inhibited PS synthase activity, but on the other hand, it resulted in a higher amount of PS synthase protein for the net effect of stimulating the synthesis of PS. This regulation must be important to the function of PS synthase in exponential phase because the enzyme catalyzes the committed step of phospholipid synthesis via the CDP-DAG pathway (11). When the need for phospholipid synthesis is reduced in the stationary phase (54), the total amount of PS synthase is reduced because of a lack of phosphorylation, and at the same time, its expression is reduced by gene repression (21).
PS synthase was originally identified as a 23-kDa protein following its purification from stationary phase cells (1, 55). Subsequently, the enzyme was partially purified from exponential phase cells as both a 30- and 23-kDa protein with the 30-kDa protein being a predominant form (53). Some data have suggested that the 23-kDa enzyme is a proteolysis product of the 30-kDa enzyme (4). When PS synthase was first shown to be a substrate for protein kinase A, Ser46 was predicted to be the target site of phosphorylation (33). If so, the enzyme must be proteolyzed at the C terminus to generate the 23-kDa enzyme because proteolysis at the N terminus would remove the phosphorylation site. As discussed above, the antibodies generated against sequences found at each end of the full-length protein recognized the 30- and 27-kDa forms of PS synthase in exponential phase cells. An immunoreactive protein migrating at a molecular mass of 23 kDa was not observed in this work; nor did we find any evidence of proteolysis of either the 30- or 27-kDa proteins. It was difficult to reconcile this enigma because the 23-kDa enzyme originally purified by Bae-Lee and Carman (1) is no longer available to test with the antibodies generated here to the N-terminal and C-terminal portions of the full-length enzyme. The simplest explanation is that the 27-kDa form of PS synthase was misidentified as a 23-kDa protein due to differences in commercial preparations of molecular mass standards used in the present and previous studies.
Another difference between our present and previous studies (33) was that the phosphorylation of the 23-kDa protein did not result in the generation of the 30-kDa form of the enzyme. This might be explained if only one of the target sites was phosphorylated because both Ser46 and Ser47 must be phosphorylated for the shift in the electrophoretic mobility of the enzyme. In the first study (33), the amount of protein kinase A used for phosphorylation and the time of reaction were reduced by 50% and by 30 min, respectively, when compared with the conditions of phosphorylation used in the present study. It is also possible that the 23-kDa protein was derived from the 27-kDa protein and that an intact protein was required for the shift in electrophoretic mobility of the enzyme. Nonetheless, the work reported here clearly showed that the 27-kDa form of PS synthase was not a proteolysis product of the 30-kDa form but instead was the unphosphorylated form of the enzyme. That the 27-kDa form of PS synthase was primarily present in stationary phase cells provided a plausible explanation of why the enzyme was first identified as a 23-kDa protein (1). Moreover, although the primary translation product of CHO1 is predicted to be a 30.8-kDa protein (3, 4), the unphosphorylated form of the enzyme migrated on SDS-polyacrylamide gels as a 27-kDa protein.
It was unclear why about half of the PS synthase protein that was not phosphorylation by protein kinase A at Ser46/Ser47 was present and more active in the membrane. PS synthase is an ER-associated enzyme (5), and following its synthesis, it must properly insert and fold in the membrane bilayer. It is known that proper folding of membrane proteins may be governed by their phosphorylation and that the lack of phosphorylation may result in the loss of expressed protein in the membrane after its synthesis (56–58). It is also known that phosphorylation can influence the stability of some proteins by preventing proteasome-mediated degradation (59). In the case of PS synthase, we speculate that about half of the enzyme is properly folded in the membrane without protein kinase A phosphorylation, but about half of the enzyme is not properly folded and is degraded unless it is phosphorylated by protein kinase A. PS synthase has been identified as a candidate for ubiquitination (60), and computer algorithms predict that the most probable ubiquitination site (i.e. Lys66) is located near the protein kinase A phosphorylation sites. Thus, we envision that the phosphorylation at Ser46/Ser47 would prevent the ubiquitination and degradation of the enzyme. Whether PS synthase is subject to ubiquitination and proteasome-mediated degradation and, if so, whether protein kinase A phosphorylation prevents its degradation will be the subject of future studies.
Acknowledgment
We acknowledge Joseph Stukey for the construction of the cho1Δ mutant used in this work.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-50679.
The S. cerevisiae PS synthase enzyme should not be confused with the PS synthase from Gram-negative bacteria (e.g. E. coli), which catalyzes its CDP-DAG-dependent reaction via a ping-pong reaction mechanism (61) or the PS synthase enzyme from mammalian cells, which catalyzes an exchange reaction between PE or PC with serine (62).
- PS
- phosphatidylserine
- PI
- phosphatidylinositol
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- DAG
- diacylglycerol
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Bae-Lee M. S., Carman G. M. (1984) J. Biol. Chem. 259, 10857–10862 [PubMed] [Google Scholar]
- 2.Letts V. A., Klig L. S., Bae-Lee M., Carman G. M., Henry S. A. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 7279–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikawa J., Tsukagoshi Y., Kodaki T., Yamashita S. (1987) Eur. J. Biochem. 167, 7–12 [DOI] [PubMed] [Google Scholar]
- 4.Kiyono K., Miura K., Kushima Y., Hikiji T., Fukushima M., Shibuya I., Ohta A. (1987) J. Biochem. 102, 1089–1100 [DOI] [PubMed] [Google Scholar]
- 5.Natter K., Leitner P., Faschinger A., Wolinski H., McCraith S., Fields S., Kohlwein S. D. (2005) Mol. Cell. Proteomics 4, 662–672 [DOI] [PubMed] [Google Scholar]
- 6.Williams J. G., McMaster C. R. (1998) J. Biol. Chem. 273, 13482–13487 [DOI] [PubMed] [Google Scholar]
- 7.Rattray J. B., Schibeci A., Kidby D. K. (1975) Bacteriol. Rev. 39, 197–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henry S. A. (1982) in The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression (Strathern J. N., Jones E. W., Broach J. R. eds) pp. 101–158, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 9.Paltauf F., Kohlwein S. D., Henry S. A. (1992) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression (Jones E. W., Pringle J. R., Broach J. R. eds) pp. 415–500, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 10.Carman G. M., Henry S. A. (1999) Prog. Lipid Res. 38, 361–399 [DOI] [PubMed] [Google Scholar]
- 11.Carman G. M., Han G. S. (2009) J. Lipid Res. 50, S69–S73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carman G. M., Zeimetz G. M. (1996) J. Biol. Chem. 271, 13293–13296 [DOI] [PubMed] [Google Scholar]
- 13.Yamashita S., Nikawa J. (1997) Biochim. Biophys. Acta 1348, 228–235 [DOI] [PubMed] [Google Scholar]
- 14.Klig L. S., Homann M. J., Carman G. M., Henry S. A. (1985) J. Bacteriol. 162, 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poole M. A., Homann M. J., Bae-Lee M. S., Carman G. M. (1986) J. Bacteriol. 168, 668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailis A. M., Poole M. A., Carman G. M., Henry S. A. (1987) Mol. Cell. Biol. 7, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailis A. M., Lopes J. M., Kohlwein S. D., Henry S. A. (1992) Nucleic Acids Res. 20, 1411–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homann M. J., Bailis A. M., Henry S. A., Carman G. M. (1987) J. Bacteriol. 169, 3276–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwanyshyn W. M., Han G. S., Carman G. M. (2004) J. Biol. Chem. 279, 21976–21983 [DOI] [PubMed] [Google Scholar]
- 20.Homann M. J., Poole M. A., Gaynor P. M., Ho C. T., Carman G. M. (1987) J. Bacteriol. 169, 533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamping E., Lückl J., Paltauf F., Henry S. A., Kohlwein S. D. (1994) Genetics 137, 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg M. L., Lopes J. M. (1996) Microbiol. Rev. 60, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carman G. M., Henry S. A. (2007) J. Biol. Chem. 282, 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry S. A., Patton-Vogt J. L. (1998) Prog. Nucleic Acid Res. Mol. Biol. 61, 133–179 [DOI] [PubMed] [Google Scholar]
- 25.Chen M., Hancock L. C., Lopes J. M. (2007) Biochim. Biophys. Acta 1771, 310–321 [DOI] [PubMed] [Google Scholar]
- 26.Atkinson K., Fogel S., Henry S. A. (1980) J. Biol. Chem. 255, 6653–6661 [PubMed] [Google Scholar]
- 27.Atkinson K. D., Jensen B., Kolat A. I., Storm E. M., Henry S. A., Fogel S. (1980) J. Bacteriol. 141, 558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi H. S., Sreenivas A., Han G. S., Carman G. M. (2004) J. Biol. Chem. 279, 12081–12087 [DOI] [PubMed] [Google Scholar]
- 29.Choi H. S., Carman G. M. (2007) J. Biol. Chem. 282, 31217–31227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae-Lee M., Carman G. M. (1990) J. Biol. Chem. 265, 7221–7226 [PubMed] [Google Scholar]
- 31.Kelley M. J., Bailis A. M., Henry S. A., Carman G. M. (1988) J. Biol. Chem. 263, 18078–18085 [PubMed] [Google Scholar]
- 32.McDonough V. M., Buxeda R. J., Bruno M. E., Ozier-Kalogeropoulos O., Adeline M. T., McMaster C. R., Bell R. M., Carman G. M. (1995) J. Biol. Chem. 270, 18774–18780 [DOI] [PubMed] [Google Scholar]
- 33.Kinney A. J., Carman G. M. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 7962–7966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broach J. R., Deschenes R. J. (1990) Adv. Cancer Res. 54, 79–139 [DOI] [PubMed] [Google Scholar]
- 35.Thevelein J. M. (1994) Yeast 10, 1753–1790 [DOI] [PubMed] [Google Scholar]
- 36.Rose M. D., Winston F., Heiter P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 38.Culbertson M. R., Henry S. A. (1975) Genetics 80, 23–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito H., Fukuda Y., Murata K., Kimura A. (1983) J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiestl R. H., Gietz R. D. (1989) Curr. Genet. 16, 339–346 [DOI] [PubMed] [Google Scholar]
- 41.Innis M. A., Gelfand D. H. (1990) in PCR Protocols: A Guide to Methods and Applications (Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. eds) pp. 3–12, Academic Press, Inc., San Diego [Google Scholar]
- 42.Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 44.Haid A., Suissa M. (1983) Methods Enzymol. 96, 192–205 [DOI] [PubMed] [Google Scholar]
- 45.Oshiro J., Han G. S., Iwanyshyn W. M., Conover K., Carman G. M. (2003) J. Biol. Chem. 278, 31495–31503 [DOI] [PubMed] [Google Scholar]
- 46.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 47.Yang W. L., Carman G. M. (1996) J. Biol. Chem. 271, 28777–28783 [DOI] [PubMed] [Google Scholar]
- 48.Boyle W. J., van der Geer P., Hunter T. (1991) Methods Enzymol. 201, 110–149 [DOI] [PubMed] [Google Scholar]
- 49.MacDonald J. I., Kent C. (1994) J. Biol. Chem. 269, 10529–10537 [PubMed] [Google Scholar]
- 50.Carman G. M., Bae-Lee M. (1992) Methods Enzymol. 209, 298–305 [DOI] [PubMed] [Google Scholar]
- 51.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 52.Morlock K. R., Lin Y. P., Carman G. M. (1988) J. Bacteriol. 170, 3561–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohlwein S. D., Kuchler K., Sperka-Gottlieb C., Henry S. A., Paltauf F. (1988) J. Bacteriol. 170, 3778–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor F. R., Parks L. W. (1979) Biochim. Biophys. Acta 575, 204–214 [DOI] [PubMed] [Google Scholar]
- 55.Liberman I. (1956) J. Biol. Chem. 222, 765–775 [PubMed] [Google Scholar]
- 56.Odell A. F., Scott J. L., Van Helden D. F. (2005) J. Biol. Chem. 280, 37974–37987 [DOI] [PubMed] [Google Scholar]
- 57.Oh M. C., Derkach V. A., Guire E. S., Soderling T. R. (2006) J. Biol. Chem. 281, 752–758 [DOI] [PubMed] [Google Scholar]
- 58.Yang J. W., Vacher H., Park K. S., Clark E., Trimmer J. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20055–20060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q., Kennedy A., Das P., McIntosh P. B., Howell S. A., Isaacson E. R., Hinz S. A., Davy C., Doorbar J. (2009) J. Virol. 83, 3668–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 61.Larson T. J., Dowhan W. (1976) Biochemistry 15, 5215–5218 [DOI] [PubMed] [Google Scholar]
- 62.Vance J. E. (1998) Trends Biochem. Sci. 23, 423–428 [DOI] [PubMed] [Google Scholar]
- 63.Thomas B. J., Rothstein R. (1989) Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 64.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sperka-Gottlieb C., Fasch E. V., Kuchler K., Bailis A. M., Henry S. A., Paltauf F., Kohlwein S. D. (1990) Yeast 6, 331–343 [DOI] [PubMed] [Google Scholar]