Abstract
How instructive cues present on the cell surface have their precise effects on the actin cytoskeleton is poorly understood. Semaphorins are one of the largest families of these instructive cues and are widely studied for their effects on cell movement, navigation, angiogenesis, immunology and cancer1. Semaphorins/collapsins were characterized in part on the basis of their ability to drastically alter actin cytoskeletal dynamics in neuronal processes2, but despite considerable progress in the identification of semaphorin receptors and their signalling pathways3, the molecules linking them to the precise control of cytoskeletal elements remain unknown. Recently, highly unusual proteins of the Mical family of enzymes have been found to associate with the cytoplasmic portion of plexins, which are large cell-surface semaphorin receptors, and to mediate axon guidance, synaptogenesis, dendritic pruning and other cell morphological changes4–7. Mical enzymes perform reduction–oxidation (redox) enzymatic reactions4,5,8–10 and also contain domains found in proteins that regulate cell morphology4,11. However, nothing is known of the role of Mical or its redox activity in mediating morphological changes. Here we report that Mical directly links semaphorins and their plexin receptors to the precise control of actin filament (F-actin) dynamics. We found that Mical is both necessary and sufficient for semaphorin–plexin-mediated F-actin reorganization in vivo. Likewise, we purified Mical protein and found that it directly binds F-actin and disassembles both individual and bundled actin filaments. We also found that Mical utilizes its redox activity to alter F-actin dynamics in vivo and in vitro, indicating a previously unknown role for specific redox signalling events in actin cytoskeletal regulation. Mical therefore is a novel F-actin-disassembly factor that provides a molecular conduit through which actin reorganization—a hallmark of cell morphological changes including axon navigation—can be precisely achieved spatiotemporally in response to semaphorins.
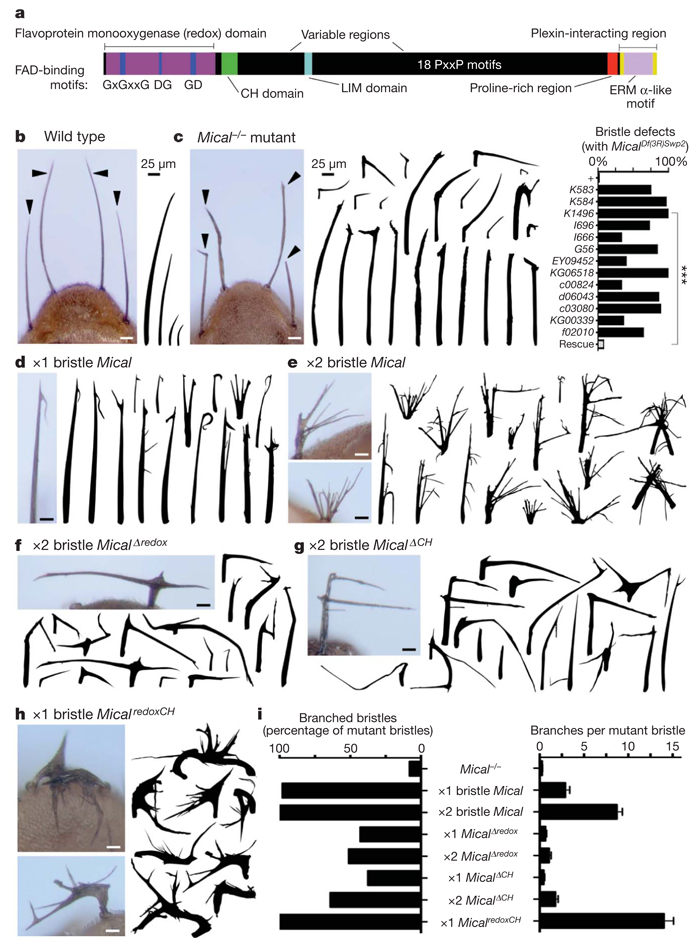
Multidomain cytosolic proteins of the Mical family (Fig. 1a) mediate semaphorin–plexin repulsive axon guidance and cell morphological changes4,5, but their role in controlling these events is unknown. While characterizing new hypomorphic Mical alleles, we found that surviving homozygous (Mical−/−) mutant adult flies had abnormally shaped bristle cell processes that were variously straight, thick, bent, twisted and/or had abnormal ‘club-like’ or blunt tips (Fig. 1b, c and Supplementary Figs 1–3). These morphological defects were rescued by expressing Mical specifically in Mical−/− mutant bristles (Fig. 1c), which showed that Mical is required for normal bristle process morphology. Bristle process elongation, like neuronal process extension, is actin dependent such that bristles are formed during pupal development when a bristle cell extends an unbranched, slightly curved, actin-rich cellular process (Supplementary Fig. 2). As development continues, a chitin-cuticle external skeleton forms around the membrane of the elongated bristle, preserving a record in the adult fly of the actin organization in the developing bristle (Fig. 1b and Supplementary Fig. 2; ref. 12). Consequently, the bristle process provides a simple, high-resolution model for characterizing the molecules and mechanisms that regulate actin filament dynamics12,13. We therefore considered whether the increased expression of Mical was sufficient to induce changes to bristle process morphology. Notably, Mical overexpression specifically in wild-type bristles generated branched bristles (Fig. 1d, e, i and Supplementary Figs 4, 5 and 7). To our knowledge, these bristle branching defects are more severe than any previously reported, and we did not find any defects in bristle cell numbers or positioning, indicating that Mical has specific effects on cell morphology.
Figure 1. Mical regulates actin-rich cellular process morphology.
a, Mical protein organization. FAD, flavin adenine dinucleotide; CH, calponin homology. b, Adult Drosophila bristles (arrowheads, drawings) are of varying length, unbranched and slightly curved. c–h, Mical is necessary for normal bristle morphology and is also sufficient to alter morphology when one (×1) or two (×2) copies of different Mical transgenes are expressed specifically within bristles. Mutant, MicalDf(3R)Swp2 (ref. 4). n ≥ 25 animals per genotype; chi-squared test; ***P < 0.0001. i, Bristles resulting from Mical−/− and Mical overexpression are quantitatively distinct and the redox and calponin homology domains of Mical are both required and together are sufficient for Mical-like bristle branching. For wild type, transgene or bristle-specific driver only, there is 0% branching. n ≥ 25 bristles per genotype; data shown, mean ± s.e.m.; scale bars, 25 µm.
Mical proteins are unusual in that they each contain a redox enzymatic domain, and we wondered whether their redox activity was necessary for their specific effects on bristle shape. Point mutations that selectively disrupted the redox domain as well as bristle-specific expression of a MicalΔredox protein revealed that redox activity of Mical was required for both normal bristle morphology and Mical-dependent bristle branching (Fig. 1f, i andSupplementary Figs 6 and 7). In contrast, bristle-specific expression of the Mical redox domain alone (Micalredox) was not sufficient for bristle branching (Supplementary Fig. 7), showing that Mical also requires at least some of its other domains to mediate its full effect. Immediately carboxy-terminal to its redox domain, Mical contains a protein motif that is found in a number of cytoskeleton-associated proteins, namely a calponin homology domain (Fig. 1a). Notably, bristle-specific expression of both the Mical redox and calponin homology domains alone (MicalredoxCH) further increased bristle branching (Fig. 1h, i and Supplementary Fig. 7), indicating that MicalredoxCH is constitutively active. These results, in combination with observations that bristle-specific expression of a MicalΔCH protein acted dominant negatively (Fig. 1g, i and Supplementary Fig. 7), demonstrate that the redox and calponin homology domains of Mical are both necessary and sufficient to orchestrate specific cell morphological changes.
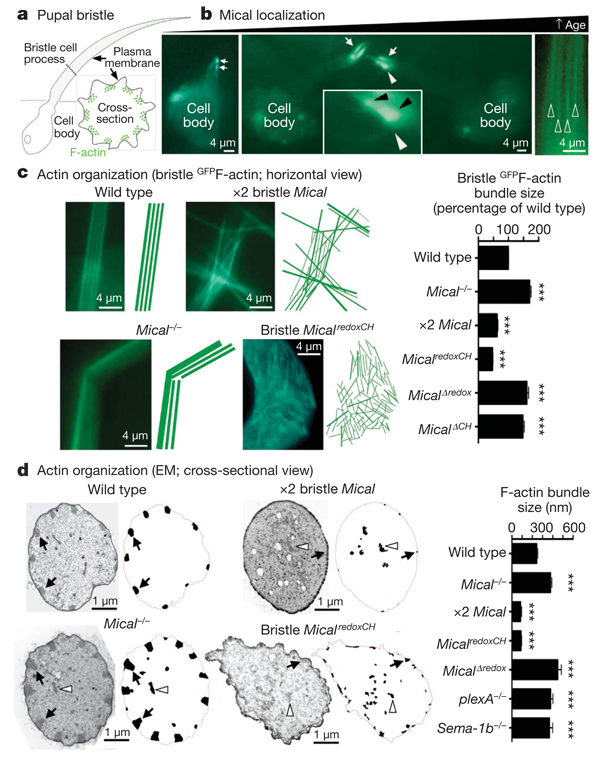
To better study the mechanisms underlying Mical-mediated morphological changes, we directly examined developing bristles. During bristle process elongation, Mical localized to growing bristle tips in close proximity to the bristle cell membrane and at sites of bristle branching and actin localization (Fig 2b and Fig 3A, B). Removing either the calponin homology domain or the C terminus of Mical altered this selective localization (Supplementary Fig. 7), indicating that Mical might be locally regulating the actin cytoskeletal network that specifies bristle morphology. In motile cells and neuronal growth cones, the majority of F-actin forms two array types: the parallel arrays of bundled F-actin found in filopodia and the meshwork F-actin arrays found in lamellipodia. Like filopodia, Drosophila bristle processes contain pillar-like bundles of F-actin comprising many individual actin filaments (Fig. 2a and Supplementary Fig. 2; ref. 12). A record of this F-actin organization was observed in the highly ordered, parallel arrangement of grooves in adult wild-type bristles but not in Mical−/− mutants, whose adult bristles indicated disorganized, intersecting and larger-than-normal F-actin bundles (Supplementary Fig. 3). Direct visualization of actin in developing bristles also showed that the F-actin bundles in Mical−/− mutants were significantly larger than wild-type F-actin bundles, were abnormally positioned and were intersecting (Fig. 2c, d and Supplementary Figs 2 and 3). These Mical−/− mutant bristles resembled those generated by overexpressing F-actin-stabilizing proteins such as actin-bundling/crosslinking proteins12, further indicating that Mical limits the size, abundance and bundling of F-actin.
Figure 2. Semaphorin, plexin and Mical control F-actin organization and bundling.
a, A pupal bristle cell extends a membranous process containing F-actin (green lines, and circles seen in cross-section) bundled together adjacent to the membrane. b, GFPMical (green) localizes to elongating pupal bristle tips (arrows) and adjacent (black arrowheads in a ×3 magnified view, inset) to filopodia-like extensions/branches, which were not seen in wild-type bristles. At older pupal ages, Mical localization forms an actin-like striped pattern (open arrowheads). The white arrowhead shows which region is seen at higher magnification in the inset. GFP, green fluorescent protein. c, d, Images and drawings of F-actin bundles. EM, electron microscopy. Membrane-associated (for example, arrows) and abnormally positioned (for example, arrowheads) bundles are drawn. n > 80 F-actin bundles per genotype; ***P < 0.0001 (compared with wild type); data shown, mean ± s.e.m.
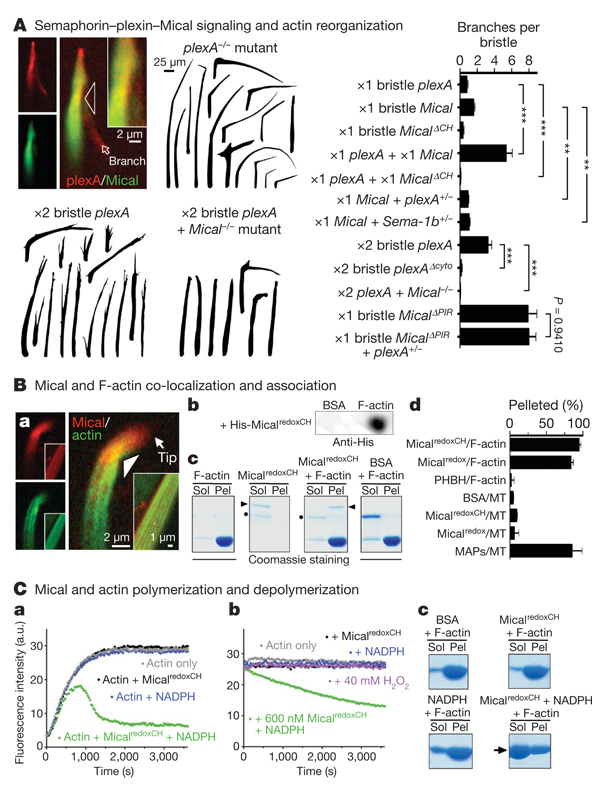
Figure 3. Semaphorin–plexin-mediated actin rearrangements require Mical, which binds and directly regulates actin dynamics.
A, Mical (GFPMical) co-localizes (yellow) with plexA at sites of bristle branch formation (arrowhead and inset at a ×2 magnified view) and is activated and required for semaphorin–plexin-dependent branching. PIR, plexin-interacting region; cyto, cytoplasmic portion. n ≥ 28 bristles per genotype; t-test; **P < 0.001, ***P < 0.0001; data shown, mean ± s.e.m. B, Mical co-localizes (yellow) with F-actin during early and late (inset) stages of bristle elongation (a). Purified Mical robustly and selectively associates with F-actin as revealed by dot-blot (b) and actin and microtubule co-sedimentation/pelleting (c, d) assays. Arrowheads, MicalredoxCH; dots, Nus; MT, microtubules; BSA, bovine serum albumin; PHBH, p-hydroxybenzoate hydroxylase; MAPs, microtubule-associated proteins; Sol, G-actin (soluble); Pel, F-actin (pellet). n ≥ 2 per condition; data shown, mean ± s.e.m. C, Pyrene-actin assays, where the fluorescence of polymerized pyrene-actin is higher than monomeric pyrene-actin, reveal that purified MicalredoxCH + NADPH directly alters actin polymerization (a) and induces depolymerization (b), as do high-speed sedimentation/Coomassie staining assays (c; arrow). a.u., arbitrary units.
To further characterize Mical-mediated actin cytoskeletal regulation, we studied the transgenic flies expressing the different forms of Mical described above. In contrast to Mical−/− mutants, increasing the expression of full-length Mical or MicalredoxCH, but not MicalΔredox or MicalΔCH, in bristles generated bundles of F-actin that were significantly thinner than normal (Fig. 2c, d and Supplementary Fig. 4). Increasing Mical activity in bristles also induced a pronounced rearrangement of F-actin: a change from the normal, parallel, organization of bundled F-actin into a complex meshwork of short actin filaments (Fig. 2c and Supplementary Fig. 4). These Mical-mediated bristle alterations are similar to those seen when F-actin is destabilized in bristles using either cytochalasin treatment 14–17 or loss-of-function mutations in actin-bundling/crosslinking proteins12. Therefore, Mical is a critical regulator of F-actin instability that is sufficient to reorganize parallel F-actin networks into complex meshwork arrays.
Previous results have indicated that the generation of short bundles of F-actin is required for the characteristic, slightly curved morphology of Drosophila bristles12. Neuronal dendrites that extend alongside elongating bristles determine this actin arrangement and curved bristle shape (Supplementary Fig. 2), but little is known of the extra-cellular signals present on dendrites that direct these bristle alterations. Because Mical associates with the cytoplasmic portion of the semaphorin receptor plexA4, we wondered whether Mical-mediated actin destabilization in bristles occurred in response to semaphorin signals present on dendrites. Consistent with this hypothesis, we found that semaphorins localized to bristle-innervating dendrites whereas plexA co-localized with Mical within bristles (Fig. 3A and Supplementary Figs 8 and 10). Likewise, increasing or decreasing the expression of plexA or either of its transmembrane Sema-1a or Sema-1b ligands caused morphological and F-actin defects resembling those seen after similar manipulations of Mical (Fig 2d and Fig 3A and Supplementary Figs 8–13). Moreover, decreasing semaphorin–plexin signalling limited the actin-destabilizing activity of full-length Mical, but not that of constitutively active MicalredoxCH or Mical proteins missing their plexin-interacting regions (MicalΔPIR; Fig. 3A and Supplementary Fig. 13). Indeed, both loss-of-function and dominant-negative Mical mutants significantly suppressed or eliminated plexA-dependent bristle branching (Fig. 3A and Supplementary Fig. 7). These results indicate that Mical is both activated and required for semaphorin-induced F-actin destabilization.
Our genetic results, therefore, coupled with our observations that Mical and F-actin co-localize in vivo, indicate that Mical associates with F-actin or an F-actin regulatory protein and thereby targets the actin cytoskeleton for semaphorin-dependent reorganization. To further address this, we performed in vitro assays and found that different forms of purified Mical protein (Micalredox and MicalredoxCH), but not negative controls (bovine serum albumin and the Nus solubility tag), directly associated with in vitro-generated actin filaments (Fig. 3B and Supplementary Fig. 14). In contrast, neither of the purified Mical proteins associated with in vitro-generated microtubules (Fig. 3B and Supplementary Fig. 16), showing that Mical selectively associates with F-actin. Likewise, the Mical-related redox enzyme p-hydroxybenzoate hydroxylase did not associate with F-actin (Fig. 3B and Supplementary Fig. 14), indicating that the redox domain of Mical is a specific F-actin-binding module. In light of previous findings that Mical interacts with the cytoplasmic portion of the plexA receptor4,5, our results identify Mical as a direct physical link between the semaphorin-receptor plexin, which is present on the cell surface, and the F-actin cytoskeleton.
We next investigated whether Mical directly alters actin dynamics. Mical uses the pyridine nucleotide NADPH as a required coenzyme for its redox activity5,9, and our in vivo results show that it also requires its redox domain to alter the actin cytoskeleton. Activating purified Micalredox or MicalredoxCH protein using NADPH substantially decreased the rate, extent, and steady-state level of actin polymerization (Fig. 3C, a and Supplementary Fig. 14). These robust effects of Mical on actin dynamics were dependent on NADPH concentration, were substantially reduced in the presence of the related pyridine nucleotide coenzyme NADH and were eliminated in the presence of NADP+ (Supplementary Figs 14 and 15). Likewise, these actin alterations were observed using very low, substoichiometric, amounts of Mical (Supplementary Fig. 14), further indicating that Mical regulates actin dynamics through its enzymatic activity. In contrast, a Mical-related redox enzyme (p-hydroxybenzoate hydroxylase) and general redox reaction products (H2O2 and/or NADP+) had little to no effect on actin dynamics (Fig. 3C, b and Supplementary Figs 14 and 15), further indicating the specificity of Mical-mediated F-actin alterations in vivo and in vitro. Moreover, activated Mical does not affect tubulin polymerization (Supplementary Fig. 16), showing that Mical is a specific and selective regulator of F-actin dynamics.
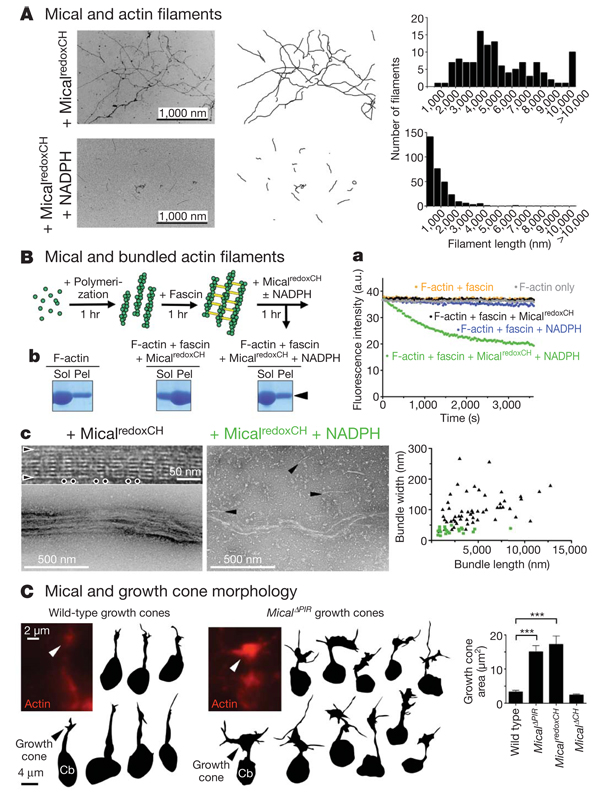
Semaphorins were identified in part on the basis of their ability to rapidly disassemble F-actin and ‘collapse’ elongating neuronal growth cones2,18, but the molecules directly mediating this effect remain unknown. Using actin depolymerization assays and electron microscopic analysis of purified proteins, we found that activated Mical directly induced actin depolymerization and significantly decreased actin filament length (Fig 3C, b, c and Fig 4A and Supplementary Fig. 14). These results are consistent with our in vivo observations and show that the critical semaphorin-signalling molecule Mical is a direct effector of F-actin disassembly. In contrast, we found no evidence that activated Mical directly induced actin branching or altered the bundling ability of actin-bundling/crosslinking proteins (Fig 3C and Fig 4A and Supplementary Figs 17 and 18), suggesting that Mical-dependent effects on branching and bundling in vivo are likely to be secondary to the ability of Mical to destabilize F-actin directly. To test this, we set up an in vitro assay using purified proteins that resembled the bundled organization of F-actin within our in vivo bristle model. We found that activated Mical also directly disassembled bundled actin filaments, decreasing both their length and their width (Fig. 4B and Supplementary Fig. 18). Therefore, Mical is an F-actin-disassembly factor that directly destabilizes both individual and bundled actin filaments.
Figure 4. Mical directly disassembles F-actin and regulates growth cone morphology.
A, Negative-staining electron microscopy shows that MicalredoxCH + NADPH significantly decreases F-actin length. n > 120 per treatment; t-test; P < 0.0001. B, Actin (green) filaments bundled with fascin (yellow) are disassembled by MicalredoxCH + NADPH, as seen using pyrene-labelled actin (a), low-speed sedimentation/Coomassie staining (b;arrowhead) and electron microscopy (c). Electron microscopy shows that, similar to untreated controls, F-actin bundles treated with MicalredoxCH (black triangles in graph) are well organized, long and thick with ‘horizontally’ arranged individual actin filaments (arrowheads) and repeating ‘vertical stripes’ of fascin (dots). F-actin bundles treated with MicalredoxCH + NADPH (green squares in graph) are significantly shorter and thinner (n > 21 per treatment; t-test; P < 0.0001) and disassemble into single actin filaments (arrowheads). C, Measuring the area occupied by GFP-actin (red) shows that Mical significantly alters growth cone size. Cb, neuronal cell body. n > 40 growth cones per genotype; t-test; ***P < 0.0001; data shown, mean ± s.e.m.
Finally, we wondered whether the expression of Mical was also sufficient to reorganize the actin cytoskeleton of navigating axons. Semaphorins mediate axon repulsion in part by locally disassembling the actin cytoskeleton within neuronal growth cones. We found that Mical likewise strongly localized to growth cones in vivo and had a critical role in the repulsion of Sema-1a–plexA-responsive axons (Supplementary Figs 19 and 20; refs 4, 5). Furthermore, using GFP–actin to directly visualize actin cytoskeletal organization and growth cone complexity in vivo19, we found that individual growth cones became significantly more complex, with increased numbers of filopodia, when constitutively active (but not dominant-negative) forms of Mical were expressed within them (Fig. 4C). These observations indicate that the redistribution of actin we see in neuronal growth cones, like that which we observe in developing bristle processes and with purified Mical, is likely to result from direct Mical-mediated disassembly of actin filaments and F-actin bundles.
We have identified a previously unknown, redox-dependent actin-disassembly pathway that provides critical insights into the means by which semaphorins alter actin cytoskeletal dynamics and serve as navigational signals. Semaphorins have long been known to have localized destabilizing effects on F-actin2,18,20 that include a loss of F-actin18, a decreased ability to polymerize new F-actin18, a decrease in the number of F-actin bundles21 and the extension of new branches22–26. Our results are consistent with these observations and indicate that Mical is sufficient to trigger each of these semaphorin–plexin-dependent events. Specifically, we find that semaphorin–plexin–Mical signalling directly destabilizes F-actin, which triggers a secondary response that produces branched meshwork actin and actin-rich extensions (Supplementary Fig. 21).
Our observations also provide a more complete understanding of the roles of repulsive guidance cues in vivo. In particular, we propose that repellents such as semaphorins disassemble or ‘prune back’ the actin network in vivo, and that this pruning process initiates a cascade of secondary events that enhances cellular complexity/plasticity. These semaphorin–plexin–Mical-induced actin rearrangements, therefore, in combination with what are likely to be semaphorin–plexin-dependent, but Mical-independent, effects on microtubules and substrate adhesion1,3, would enable navigating cells/axons to identify new, more permissive substrates and could underlie the directional changes associated with semaphorin repulsive guidance. Indeed, neuronal growth cones develop complex morphologies with multiple extending filopodia when they encounter directional choice points in vivo27–30, and these new filopodia probe the environment and ultimately lead the growth cone to a more permissive substrate. Future work will seek to better understand these Mical-mediated events. These research directions are likely to be of significant biomedical importance given the roles of the more than twenty semaphorins in directing actin-dependent processes in neural connectivity, angiogenesis, immunity and cancer.
METHODS SUMMARY
We performed complementation analysis, genetics, molecular biology, western blotting, immunostaining and generation of transgenic animals using standard techniques4. Multiple new lines of the full-length ‘short’ isoform of Mical, the MicalΔredox mutation (MicalG→W; ref. 4) and the other transgenic animals were generated and used for all experiments. Adult bristles were examined and quantified by crossing adults at 25 °C: adult offspring from these crosses were first sorted according to genotype and then examined under a dissecting microscope. We genotyped pupae using a Zeiss Discovery M2 Bio fluorescence stereomicroscope, and all preparation, staging and dissection of pupae were done using standard approaches. We imaged, drew and quantified the adult bristles with the aid of the Discovery M2 Bio stereomicroscope, a motorized focus and zoom, a Zeiss AxioCam HR camera and three-dimensional-reconstruction software (Zeiss AxioVision, version 4.6.3, and Extended Focus software). All other bright-field, dark-field, differential interference contrast and fluorescence visualization, and imaging of bristles, embryos and growth cones, was done using a Zeiss Axio Imager upright microscope with motorized focus and zoom and an ApoTome module, and images were captured and quantified using the AxioCam HR camera and AxioVision software. All electron microscopy of pupae and negative staining of purified proteins was done using a FEI Tecnai G2 Spirit BioTWIN transmission electron microscope. We purified recombinant Mical proteins10 and recombinant p-hydroxybenzoate hydroxylase using our previously developed approaches. Drosophila fascin (also known as singed) complementary DNA was inserted in a bacterial expression vector, and recombinant Drosophila fascin protein was purified. All F-actin and Mical co-sedimentation assays and G-actin/F-actin ratio experiments were performed using standard approaches, as were all pyrene-labelled actin polymerization and depolymerization assays, actin bundling assays, tubulin polymerization assays and microtubule co-sedimentation assays.
Supplementary Material
Acknowledgements
We thank C. Cowan, M. Rosen and T. Südhof for comments on drafts of our manuscript and M. Bailey, C. Gilpin, H. He, T. Januszewski, H. Krämer, W. Lin, T. Wedgeworth, X. Zhang and the University of Texas Southwestern Electron Microscopy Core Facility for discussions and assistance. We also thank the Bloomington, Harvard and Japanese stock centres for flies and H. Aberle, L. Cooley, C. Goodman, A. Kolodkin, B. Lee, L. Luo, J. Merriam and X. Zhang for flies and/or reagents. This work was supported by the US National Institute of Mental Health (MH085923) and a Basil O’Connor Starter Scholar Research Award to J.R.T. J.R.T. is a Klingenstein Fellow and the Rita C. and William P. Clements, Jr, Scholar in Medical Research.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions All authors contributed critical reagents, discussed the results and commented on the manuscript; R.-J.H., U.Y., J.Y., H.W., T.Y. and J.R.T. performed experiments, collected and analysed data, and prepared the manuscript; and J.R.T. oversaw all aspects of the project and wrote the paper.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Tran TS, Kolodkin AL, Bharadwaj R. Semaphorin regulation of cellular morphology. Annu. Rev. Cell Dev. Biol. 2007;23:263–292. doi: 10.1146/annurev.cellbio.22.010605.093554. [DOI] [PubMed] [Google Scholar]
- 2.Luo Y, Raible D, Raper JA. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell. 1993;75:217–227. doi: 10.1016/0092-8674(93)80064-l. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y, Gunput RA, Pasterkamp RJ. Semaphorin signaling: progress made and promises ahead. Trends Biochem. Sci. 2008;33:161–170. doi: 10.1016/j.tibs.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell. 2002;109:887–900. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt EF, Shim SO, Strittmatter SM. Release of MICAL autoinhibition by semaphorin-plexin signaling promotes interaction with collapsin response mediator protein. J. Neurosci. 2008;28:2287–2297. doi: 10.1523/JNEUROSCI.5646-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beuchle D, Schwarz H, Langegger M, Koch I, Aberle H. Drosophila MICAL regulates myofilament organization and synaptic structure. Mech. Dev. 2007;124:390–406. doi: 10.1016/j.mod.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Kirilly D, et al. A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nature Neurosci. 2009;12:1497–1505. doi: 10.1038/nn.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siebold C, et al. High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule. Proc. Natl Acad. Sci. USA. 2005;102:16836–16841. doi: 10.1073/pnas.0504997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadella M, Bianchet MA, Gabelli SB, Barrila J, Amzel LM. Structure and activity of the axon guidance protein MICAL. Proc. Natl Acad. Sci. USA. 2005;102:16830–16835. doi: 10.1073/pnas.0504838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta N, Terman JR. Flavins and Flavoproteins 2008. In: Frago S, Gómez-Moreno C, Medina M, editors. Proc. 16th Int. Symp. Flavins Flavoproteins; Prensas Univ. Zaragoza. 2008. pp. 345–350. [Google Scholar]
- 11.Suzuki T, et al. MICAL, a novel CasL interacting molecule, associates with vimentin. J. Biol. Chem. 2002;277:14933–14941. doi: 10.1074/jbc.M111842200. [DOI] [PubMed] [Google Scholar]
- 12.Tilney LG, DeRosier DJ. How to make a curved Drosophila bristle using straight actin bundles. Proc. Natl Acad. Sci. USA. 2005;102:18785–18792. doi: 10.1073/pnas.0509437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutherland JD, Witke W. Molecular genetic approaches to understanding the actin cytoskeleton. Curr. Opin. Cell Biol. 1999;11:142–151. doi: 10.1016/s0955-0674(99)80018-0. [DOI] [PubMed] [Google Scholar]
- 14.Turner CM, Adler PN. Distinct roles for the actin and microtubule cytoskeletons in the morphogenesis of epidermal hairs during wing development in Drosophila. Mech. Dev. 1998;70:181–192. doi: 10.1016/s0925-4773(97)00194-9. [DOI] [PubMed] [Google Scholar]
- 15.Tilney LG, Connelly PS, Vranich KA, Shaw MK, Guild GM. Actin filaments and microtubules play different roles during bristle elongation in Drosophila. J. Cell Sci. 2000;113:1255–1265. doi: 10.1242/jcs.113.7.1255. [DOI] [PubMed] [Google Scholar]
- 16.Guild GM, Connelly PS, Vranich KA, Shaw MK, Tilney LG. Actin filament turnover removes bundles from Drosophila bristle cells. J. Cell Sci. 2002;115:641–653. doi: 10.1242/jcs.115.3.641. [DOI] [PubMed] [Google Scholar]
- 17.Geng W, He B, Wang M, Adler PN. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics. 2000;156:1817–1828. doi: 10.1093/genetics/156.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan J, et al. The organization of F-actin and microtubules in growth cones exposed to a brain-derived collapsing factor. J. Cell Biol. 1993;121:867–878. doi: 10.1083/jcb.121.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez-Soriano N, Prokop A. The influence of pioneer neurons on a growing motor nerve in Drosophila requires the neural cell adhesion molecule homolog FasciclinII. J. Neurosci. 2005;25:78–87. doi: 10.1523/JNEUROSCI.2377-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan J, Raper JA. Localized collapsing cues can steer growth cones without inducing their full collapse. Neuron. 1995;14:263–274. doi: 10.1016/0896-6273(95)90284-8. [DOI] [PubMed] [Google Scholar]
- 21.Dent EW, Barnes AM, Tang F, Kalil K. Netrin-1 and semaphorin 3A promote or inhibit cortical axon branching, respectively, by reorganization of the cytoskeleton. J. Neurosci. 2004;24:3002–3012. doi: 10.1523/JNEUROSCI.4963-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapfhammer JP, Raper JA. Collapse of growth cone structure on contact with specific neurites in culture. J. Neurosci. 1987;7:201–212. doi: 10.1523/JNEUROSCI.07-01-00201.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell DS, et al. Semaphorin 3A elicits stage-dependent collapse, turning, and branching in Xenopus retinal growth cones. J. Neurosci. 2001;21:8538–8547. doi: 10.1523/JNEUROSCI.21-21-08538.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenstermaker V, Chen Y, Ghosh A, Yuste R. Regulation of dendritic length and branching by semaphorin 3A. J. Neurobiol. 2004;58:403–412. doi: 10.1002/neu.10304. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Halloran MC. Central and peripheral axon branches from one neuron are guided differentially by Semaphorin3D and transient axonal glycoprotein-1. J. Neurosci. 2005;25:10556–10563. doi: 10.1523/JNEUROSCI.2710-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai JA, Halloran MC. Semaphorin 3d guides laterality of retinal ganglion cell projections in zebrafish. Development. 2006;133:1035–1044. doi: 10.1242/dev.02272. [DOI] [PubMed] [Google Scholar]
- 27.Tosney KW, Landmesser LT. Growth cone morphology and trajectory in the lumbosacral region of the chick embryo. J. Neurosci. 1985;5:2345–2358. doi: 10.1523/JNEUROSCI.05-09-02345.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broadie K, et al. From growth cone to synapse: the life history of the RP3 motor neuron. Dev., Suppl. 1993:227–238. [PubMed] [Google Scholar]
- 29.Godement P, Wang LC, Mason CA. Retinal axon divergence in the optic chiasm: dynamics of growth cone behavior at the midline. J. Neurosci. 1994;14:7024–7039. doi: 10.1523/JNEUROSCI.14-11-07024.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray MJ, Merritt DJ, Brand AH, Whitington PM. In vivo dynamics of axon pathfinding in the Drosophila CNS: a time-lapse study of an identified motorneuron. J. Neurobiol. 1998;37:607–621. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.