Abstract
Proteins with Pumilio RNA binding domains (Puf proteins) are ubiquitous in eukaryotes. Some Puf proteins bind to the 3′-untranslated regions of mRNAs, acting to repress translation and promote degradation; others are involved in ribosomal RNA maturation. The genome of Trypanosoma brucei encodes eleven Puf proteins whose function cannot be predicted by sequence analysis. We show here that epitope-tagged TbPUF7 is located in the nucleolus, and associated with a nuclear cyclophilin-like protein, TbNCP1. RNAi targeting PUF7 reduced trypanosome growth and inhibited two steps in ribosomal RNA processing.
Keywords: Pumilio domain, Nucleolus, Cyclophilin, rRNA processing, Trypanosoma
1. Introduction
Proteins with domains of the Pumilio family (Puf) are ubiquitous in eukaryotes [1]; they are involved in various processes that require RNA binding. When several Puf domains are linked together, they form a crescent-like structure: the inner face binds to RNA while other surfaces are available for protein–protein interactions [2,3].
The genome of Saccharomyces cerevisiae encodes seven proteins with Puf domains. Five of them (ScPufs1-5) bind to, and regulate, specific subsets of mRNAs in the cytosol [4]. The remaining two, ScPuf6 and ScNop9, are predominantly nucleolar [5] (http://yeastgfp.ucsf.edu/). For Puf6, this is paradoxical because its only known function is in translational repression and localisation of the Ash1 mRNA [6,7]. The function of Nop9, in contrast, is consistent with the localisation since it plays a role rRNA maturation: it migrates with pre-ribosomes on a sucrose gradient, associates with pre-ribosomal particles and various ribosomal assembly factors, and is required for the first steps of cleavage of the rRNA precursor [8]. Puf6 and Nop9 associate with a variety of nucleolar proteins [9,10].
Trypanosoma brucei diverged from the animal/fungal branch early in eukaryotic evolution. It has eleven Puf proteins [11]. Phylogenetic analysis for TbPUF7 (Tb11.01.6600) gave conflicting information: using only the Puf domains placed TbPUF7 together with TbPUF8 (Tb927.3.2470), ScNop9 and ScPuf6 [11]. In contrast, when we used the whole TbPUF7 sequence, instead of the Puf domains alone, the protein sequence was grouped with Saccharomyces Pufs 3, 4 and 5. Since TbPUF7 also lacks a nuclear targeting signal, we therefore suspected that it might have a function in regulation of mRNA abundance or translation. We describe here results that indicate that TbPUF7 has a function in the nucleolus.
2. Methods
2.1. Plasmid constructs
A PUF7 RNAi construct was generated using p2T7 TAblu, with the Puf7 insert and oligonucleotide primers designed using RNAit [12]. This construct was transfected into T. brucei Lister 427 single marker (‘S16’) bloodstream-form cells expressing T7 RNA polymerase and the tetracycline repressor, and transfectants selected. The PUF7 construct used for RNAi in procyclic forms was based on p2T7 TAblu but targeted a different region of the gene than that described in [11]. Primers were cz3012 (gagaagatctgcatgcAAAATGTCTCCCAGCGAC) and cz3013 (cggaattcgtcgacCGAAGAGCGCTTTAC) (restriction sites are underlined and the hybridising parts of the primers are in upper case). The NMD3 (Tb927.7.970) RNAi construct was made using the stem-loop strategy [13] and RNAit; this and the PUF7 RNAi plasmid were transfected into procyclic trypanosomes expressing the tet repressor and T7 polymerase [14].
To express PUF7-TAP, pHD918 [15] was modified by addition of a polylinker (Avr I–Asc I–Xma I–Sph I) to give pHD1744. The PUF7 open reading frame was amplified and cloned into the Avr I–Asc I sites. For V5 tagging (pHD1911), the plasmid used was from [16] and the primers were: ORF–cz2992 (gacctcgagATGCCAAAAATGCGTTTAGA), cz2991 (gacgggcccGCCAAGGTAAGGGAGGAAAC); 5′-UTR–cz2994 (gacccgcggGAGTGGTGGCCTTCATTCAC), cz2993 (gactctagaTGCTCCCTTTAGTTCACTTCAA).
For myc tagging, the TbNCP1 open reading frame (CZ2989 (gacaagcttATGCTAAAGAGCCCGCAAAATTTTCG) and CZ2990 (gacggatccTTTCTCTTCCGCCTGGGC)) was cloned into pHD1700 [17].
Trypanosome transfection and growth analysis were as described previously [13,18].
2.2. Northern blots and immunofluorescence
RNA was prepared using TRIzol, denatured with formamide and formaldehyde, and separated on denaturing formaldehyde-agarose or urea-acrylamide gels. RNA was blotted onto Nytran and hybridised. The NMD3 probe was made from a plasmid by random priming with 32P label. Oligonucleotide probes were labeled with 32P using polynucleotide kinase. These were: 3′ of the mature SSU rRNA, CZ3252 (ATTTTTGGTTGCATACTGTG); pre 5.8s, CZ1427 (GTTTTTATATTCGACACTG); mature 5.8S, CZ1193 (ACTTTGCTGCGTTCTTCAAC); 7SL, CZ1478 (CAACACCGACACGCAACC). Hybridisation with oligonucleotides was as described [15] except that for the SSU rRNA, washing was at 30 °C. Probes were detected by phosphorimager.
For immunofluorescence, Cells were prepared, labeled with primary antibodies to the V5 tag (Invitrogen), the TAP tag (peroxidase-anti-peroxidase, GE Healthcare) or RNA polymerase I (kind gift from Miguel Navarro, Granada, Spain), and secondary antibodies coupled to Alexa594, 488 or 568 (Molecular Probes), as in [19].
2.3. Tandem affinity purification (TAP) and co-immunoprecipitation
PUF7-TAP was purified, and proteins identified, as described [15,20]. For immunoprecipitation, the cell lysate was obtained as for TAP, and bound to myc-(Bethyl) or V5-(Sigma) coupled beads. After washing, bound protein was eluted by boiling with reducing SDS loading buffer and analysed by Western blotting. Blots were probed with antibodies to the myc tag (Santa Cruz Biotechnology), V5 tag (Invitrogen), or, for the TAP tag, with the ECL secondary antibody (GE Healthcare).
RNA associated with PUF7-TAP, after UV cross-linking, was obtained as previously described [21,22].
3. Results and discussion
3.1. Sequence alignments
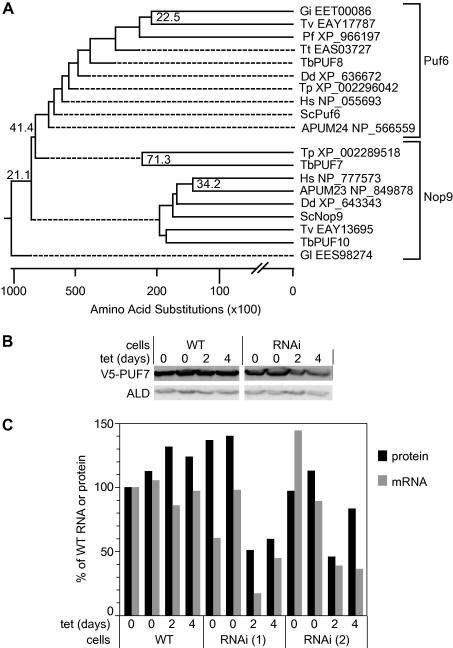
To investigate the phylogeny of S. cerevisiae Nop9 and Puf6, and of TbPUF7 and TbPUF8 in more detail, we used the S. cerevisiae genes to perform BLASTp searches on selected genomes from all major eukaryotic groups, then made a phylogenetic tree. Sequences with E values of less than 10−6 were checked by performing a BLASTp search back onto the S. cerevisiae genome, and sequences that gave the original input sequence as the best match, and had Puf domains, were included in subsequent analyses. Clear homologues of S. cerevisiae Puf6 were found in all groups, but several organisms appeared to lack Nop9. We constructed a phylogenetic tree with these sequences and all of the T. brucei and S. cerevisiae Puf proteins, and discovered that TbPUF8 was clustered with the Puf6 homologues, while TbPUF7, TbPUF10 and sometimes, TbPUF11, grouped with Nop9 proteins (not shown). A phylogenetic tree including fewer yeast and trypanosome Puf proteins is shown in Fig. 1A. Most branches were extremely poorly supported with bootstrap values below 20. The Puf6 group remained constant independent of which sequences were included, and TbPUF8 was always found within it (not shown), although the branching order was not at all robust. TbPUF7 and TbPUF10 did not branch with the Puf6’s, and their position relative to the putative Nop9 homologues changed every time the list of included proteins was altered. The Giardia protein with accession number EES98274 was consistently least related to all of the others, although the BLASTp analysis with Nop9 worked in both directions. From this analysis we could conclude that TbPUF8 could be a homologue of ScPUF6, but ScNop9 is poorly conserved in eukaryotic evolution. The status of TbPUF7 remained unresolved.
Fig. 1.
(A) Phylogenetic tree of possible homologues of S. cerevisiae Puf6 and Nop9. Accession numbers for all species apart from T. brucei and S. cerevisiae are shown. Each protein is designated according to the sequence sued for the BLASTp search – for example, TpPuf6 was the best match to ScPuf6. protein used for the Species abbreviations are: Tt – Tetrahymena thermophila, Pf – Plasmodium falciparum; Gi – Giardia intestinalis; Gl – Giardia lamblia; Dd – Dictyostelium discoideum; Tp – Thalassiosira pseudonana; A – Arabidopsis thaliana (official gene designations given); Hs – Homo sapiens; Tv – Trichomonas vaginalis (here there were several matches, only the best is shown). Note that the Dictyostelium Nop9 sequence gave the putative human Nop9 as the best match in the human genome, but did not give a significant match to S. cerevisiae. The tree was created in DNAStar using ClustalW; bootstrap values are from 1000 trials with a seed of 111. (B) Western blot for a procyclic trypanosome line with V5 tagged PUF7, with RNAi against PUF7. The blot was probed with antibody to the V5 tag, and to aldolase (ALD) as loading control. WT: trypanosomes expressing the tet repressor but without an RNAi construct. (C) Quantitation of PUF7 RNA and PUF7 protein after RNAi. Results are for trypanosomes without an RNAi plasmid (WT) and two experiments for the RNAi line.
3.2. TbPUF7 is required for growth and is located in the nucleolus
Previous trypanosome PUF7 RNA interference experiments revealed no effect on cell growth [11]. We here transfected two trypanosome life cycle stages, bloodstream forms and procyclic forms, with new RNAi plasmids. Several clones were obtained. Procyclic forms showed only negligible growth inhibition after RNAi (not shown). To check the level of PUF7 depletion, we tagged one PUF7 allele [16] to give expression of PUF7 with a V5 tag at the N-terminus, and subjected the cells to RNAi. In those cells after RNAi induction, PUF7 mRNA was decreased to 20–40% of normal while the level of V5-PUF7 was approximately halved (Fig. 1B and C). Although it is possible that the V5 tag affects protein stability, this suggests that substantial amounts of PUF7 protein remained after RNAi. In contrast, we observed severe growth inhibition in three different bloodstream-form lines. This was accompanied by an accumulation of cells blocked in cytokinesis: arrested cells increased to 20% in 24 h, with aberrant numbers of kinetoplasts and nuclei (not shown). The bloodstream-form cells rapidly lost the RNAi so we continued the work mainly with procyclic forms.
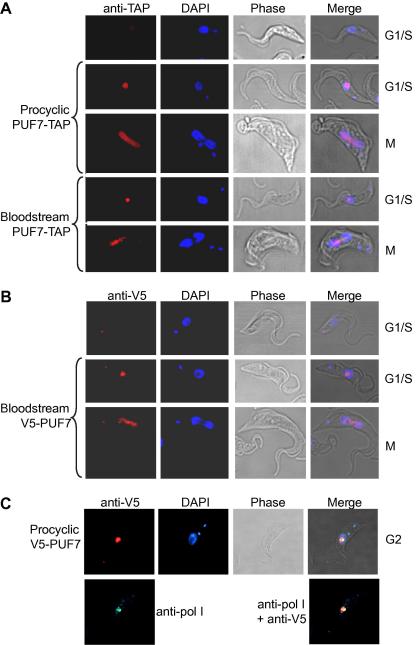
We inducibly expressed PUF7 with a C-terminal tandem affinity (TAP) tag. There was no growth effect and PUF7-TAP was located in a poorly DAPI-stained region of the nucleus (Fig. 2A), the nucleolus [23]. In dividing cells PUF7-TAP was distributed between the nuclei, the region expected to contain the mitotic spindle (Fig. 2A, bottom panel). To check that the localisation was not an artifact of tagging or over-expression, we examined V5-PUF7 in the in situ tagged procyclic forms. The localisation was the same as before (Fig. 2B) and the V5 tag colocalised with RNA polymerase I, confirming that it was in the nucleolus (Fig. 2C). Neither tag causes nuclear localisation in trypanosomes [11,16]. Bridges between nuclei were previously seen in dividing trypanosomes stained for other nuclear and nucleolar proteins [24–26]. Similar experiments with V5-tagged TbPUF8 confirmed the expected nucleolar location (C. Helbig, ZMBH, data not shown).
Fig. 2.
PUF7 is in the nucleolus. Trypanosomes expressing (A) PUF7-TAP or (B and C) V5-PUF7 were stained with antibodies and with DAPI as shown. Secondary antibodies were Alexa594 (A and B) and (C) Alexa 488 (pol I) and Alexa 568 (V5). The cell cycle stages of the parasites, judged from the numbers of nuclei and kinetoplasts, are indicated.
3.3. Depletion of TbPUF7 inhibits ribosomal RNA processing
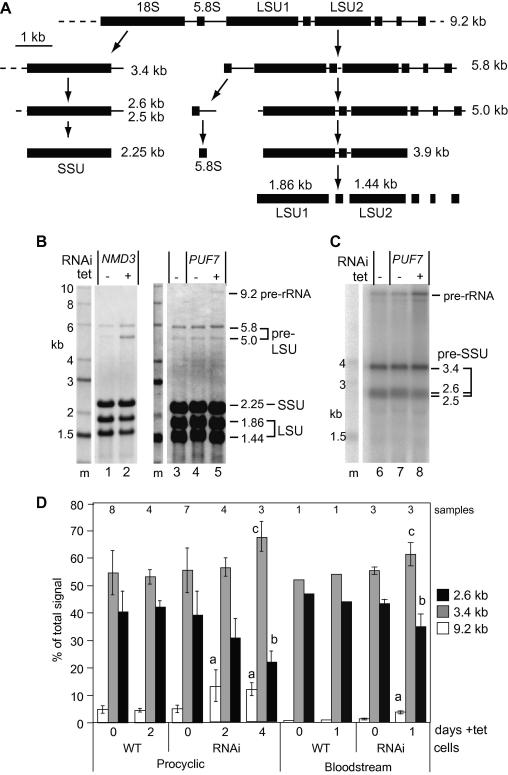
We next analysed the pattern of rRNAs after growth-inhibitory PUF7 RNAi (Fig. 3). As a control we used cells with RNAi against NMD3. Yeast NMD3 is involved in export of large ribosomal subunits: mutations cause feedback inhibition of rRNA processing [27,28]. Trypanosome NMD3 is associated with rRNA export factors [29].
Fig. 3.
RNAi targeting PUF7 inhibits rRNA processing. (A) Schematic overview of rRNA processing in trypanosomes [30,31]. (B) Total RNA was prepared from cells with no RNAi (lane 3), RNAi targeting PUF7 (lanes 4, 5, 7 and 8) or RNAi targeting NMD3 (lanes 1 and 2). To induce RNAi the cells were grown for 48 h with tetracycline (lanes 2, 5 and 8). After denaturing agarose gel electrophoresis, RNA was blotted and stained with methylene blue. LSU: large subunit rRNA; SSU: small subunit rRNA; M: markers. (C) The blot was hybridised with a probe located 3′ of the mature SSU rRNA. (D) The intensities for the pre-rRNA, 3.4 kb, and 2.6 + 2.5 kb bands are expressed as mean ± standard deviation. Sample numbers are shown above the columns. The results shown are for procyclic cells without V5-tagged PUF7; in an experiment with the V5-tagged line, in which V5-PUF7 was decreased only two-fold, an increase in the 9.2 kb band was also seen. Using a Students t-test, the induced RNAi lines were statistically significant from the uninduced lines at the following levels: (a) <0.01; (b) <0.05; (c) <0.1.
A summary of rRNA processing in trypanosomes is shown in Fig. 3A [30,31]. A 9.2 kb precursor is cleaved to give a 3.4 kb small subunit (SSU) precursor, and a 5.8 kb precursor for the large subunit (LSU) and smaller rRNAs. The 3.4 kb RNA is processed via 2.6 kb and 2.5 kb intermediates; the 5.8 kb LSU precursor is processed via 5.0 kb and 3.9 kb intermediates. The 5.8 kb large subunit precursor is visible by total RNA staining (Fig. 3B). NMD3 RNAi inhibited growth (not shown) and large subunit processing, causing accumulation of the 5.0 kb fragment (Fig. 3B). After PUF7 RNAi, the initial processing step was inhibited such that the relative abundance of the 9.2 kb RNA increased two-to-four fold (Fig. 3B, C and D), while the 2.6 kb intermediate decreased (Fig. 3C and D). Neither RNAi affected maturation of the 5.8S rRNA, the 7SL (SRP) RNA, or the overall staining pattern of mature rRNA and small RNAs (not shown). In three bloodstream form RNAi lines, the baseline level of the 9.2 kb precursor was lower but PUF7 RNAi had the same effect as in procyclics (Fig. 3D). The results therefore indicated that PUF7 is required, either directly or indirectly, for efficient cleavage of the 9.2 kb precursor, and perhaps also for processing of the 3.4 kb pre-SSU RNA to 2.6 kb. The PUF7 RNAi did not inhibit these processes sufficiently for either to become rate-limiting in overall rRNA maturation, since neither the pre-LSU intermediates, nor steady-state SSU rRNA were affected.
In S. cerevisiae, the role of Nop9 in rRNA biogenesis was analysed by conditional expression under a Gal promoter. 12 h after repression, Nop9 was no longer detectable. A defect in processing of pre-rRNA to give 18S rRNA was evident from pulse-chase labeling, while production of the 25S rRNA was relatively unaffected. Steady-state levels of the 35S precursor (equivalent to the trypanosome 9.2 kb rRNA precursor) were strongly increased while the amounts of all smaller small subunit rRNA precursors, as well as the small subunit rRNA itself, were decreased [8]. The effects seen after PUF7 depletion were, in some ways, similar, but not as strong, perhaps because PUF7 down-regulation by RNAi was inefficient. Thus although our data suggest that PUF7 has a role in rRNA maturation, we cannot conclude that it is a functional homologue of yeast Nop9.
3.4. TbPUF7 is associated with a cyclophilin-like protein
S. cerevisiae Nop9 was found to be associated with a multitude of rRNA processing factors and with rRNA and pre-rRNA [8]. To find out if PUF7 was stably associated in an rRNA-processing complex, PUF7-TAP was purified. When RNA was purified from the preparations [21,22] we obtained a very low RNA yield, and reverse-transcription PCR revealed no specific association with pre-rRNA (not shown).
The gel picture of purification of PUF7-TAP was published as the control-TAP lane in Fig. 2 of [20]. Two strong bands were seen. One was tagged PUF7 (migrating at 75 kDa, MOWSE score 1323, 38 peptides, 30.7% coverage) and the other was a 34 kDa protein encoded by locus Tb927.8.2000 (MOWSE score 583, 16 peptides, 38.1% coverage). The association appeared stoichiometric by SyproRuby staining [20]. No other specifically-associated proteins were present. A few clusters of bands were nevertheless sequenced, and found to contain ribosomal proteins. This might be a genuine association, but we cannot be certain because we have found ribosomal proteins as contaminants in every protein purification we have analysed so far. No proteins involved in rRNA processing or assembly were detected, although very few of these are actually known in trypanosomes. Thus our evidence so far suggests that PUF7 is not stably associated with pre-ribosomal particles.
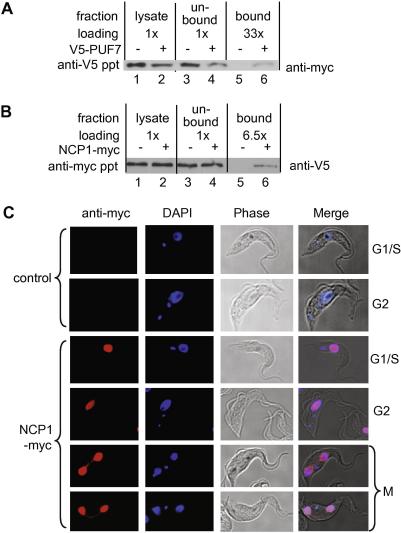
BLASTp searches with Tb927.8.2000 yielded cytoplasmic peptidyl-prolyl cis–trans isomerases so we designated it nuclear cyclophilin 1 (NCP1). We confirmed the interaction between PUF7 and NCP1 by expressing NCP1 with a C-terminal myc tag in cells expressing V5-PUF7. In cells expressing both tagged proteins, precipitation with anti-V5 antibody resulted in co-precipitation of NCP1-myc (Fig. 4A, lane 6). Precipitation with anti-V5 from extracts of cells expressing NCP1-myc alone did not result in co-immunoprecipitation (Fig. 4A, lane 5). In cells expressing both tagged proteins, precipitation with anti-myc antibody resulted in co-precipitation of V5-PUF7 (Fig. 4B, lane 6), with no co-precipitation in the control (Fig. 4B, lane 5).
Fig. 4.
PUF7 is associated with a cyclophilin-like protein. (A) NCP1-myc was expressed in trypanosomes with or without V5-tagged PUF7. Lysates were immunoprecipitated with anti-V5 antibody. The lysates, unbound and bound fractions were separated by SDS–PAGE, blotted and probed with an anti-myc antibody to detect NCP1-myc. The relative loading is indicated above the lanes. (B) As for (A), but using trypanosomes expressing V5-PUF7 with or without NCP1-myc, and precipitated with anti-myc antibody before probing to detect V5-PUF7. (C) NCP1 is in the nucleus. NCP1-myc was inducibly expressed and detected with anti-myc antibodies, with DAPI to stain DNA.
Inducibly expressed NCP1-myc was throughout the nucleus, and in thin inter-nuclear bridges during mitosis (Fig. 4C). In cells grown in the absence of tetracycline, a faint myc signal was still visible, again throughout the nucleus (not shown); thus the distribution was not due to over-expression. RNAi targeting NCP1 in procyclic trypanosomes had no effect on growth, but the extent of protein depletion was unknown. The distribution throughout the nucleus suggests that NCP1 may have several functions.
In other organisms, several cyclophilin-like proteins have been reported to be located in the nucleus, or to shuttle between nucleus and cytoplasm, but none is a clear NCP1 homologue. S. cerevisiae cyclophilin A has been implicated in control of meiosis [32] and of nuclear protein trafficking [33]. Arabidopsis Cyp59 interacts with SR-domain proteins and RNA polymerase II [34]; the best trypanosome match is Tb927.5.3750. Yeast Pin1 has roles in mitotic chromosome condensation [35]; Tb927.8.690 is a likely homologue.
The location of PUF7, together with the effects of RNAi, indicated that PUF7 has a role in pre-rRNA processing; but we have no evidence for stable association with pre-ribosomal particles. We speculate, therefore, that the NCP1-PUF7 complex might assist in rearrangement of the rRNA processing complex, or in reorganisation of ribosomal proteins during small subunit assembly. Inhibition of this activity could cause a feedback inhibition on earlier steps, including rRNA processing.
Acknowledgements
Mass spectrometry was done at the ZMBH. This work was partially supported by the Wellcome Trust grant to C.C. and Mark Carrington (Cambridge) (salary of S.A. and P.D.; consumables costs). Katelyn Fenn was supported by a Wellcome Trust grant to K.R.M. We thank Miguel Navarro (López-Neyra Institute of Parasitology and Biomedicine, Granada, Spain) for the antibody to RNA polymerase I.
References
- 1.Wickens M., Bernstein D.S., Kimble J., Parker R. A PUF family portrait: 3′-UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 2.Edwards T.A., Pyle S.E., Wharton R.P., Aggarwal A.K. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 3.Wang X., Zamore P.D., Hall T.M. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 4.Gerber A., Herschlag D., Brown P. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:e79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huh W., Falvo J., Gerke L., Carroll A., Howson R., Weissman J., O’Shea E. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 6.Gu W., Deng Y., Zenklusen D., Singer R. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng Y., Singer R., Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomson E., Rappsilber J., Tollervey D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA. 2007;13:2165–2174. doi: 10.1261/rna.747607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark C., Breitkreutz B., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazbun T. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- 11.Luu V.D., Brems S., Hoheisel J., Burchmore R., Guilbride D., Clayton C. Functional analysis of Trypanosoma brucei PUF1. Mol. Biochem. Parasitol. 2006;150:340–349. doi: 10.1016/j.molbiopara.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Redmond S., Vadivelu J., Field M.C. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 2003;128:115–118. doi: 10.1016/s0166-6851(03)00045-8. [DOI] [PubMed] [Google Scholar]
- 13.Clayton C.E., Estévez A.M., Hartmann C., Alibu V.P., Field M., Horn D. Down-regulating gene expression by RNA interference in Trypanosoma brucei. In: Carmichael G., editor. RNA interference. Humana Press; 2005. [DOI] [PubMed] [Google Scholar]
- 14.Alibu V.P., Storm L., Haile S., Clayton C., Horn D. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 2004;139:75–82. doi: 10.1016/j.molbiopara.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Estévez A., Kempf T., Clayton C.E. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen S., Arhin G.K., Ullu E., Tschudi C. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 2001;113:171–173. doi: 10.1016/s0166-6851(00)00383-2. [DOI] [PubMed] [Google Scholar]
- 17.Colasante C., Alibu V.P., Kirchberger S., Tjaden J., Clayton C., Voncken F. Characterisation and developmentally regulated localisation of the mitochondrial carrier protein homologue MCP6 from Trypanosoma brucei. Eukaryot. Cell. 2006;5:1194–1205. doi: 10.1128/EC.00096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews K.R., Gull K. Evidence for an interplay between cell cycle progression and the initiation of differentiation between life cycle forms of African trypanosomes. J. Cell Biol. 1994:125. doi: 10.1083/jcb.125.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cristodero M., Clayton C. Trypanosome MTR4 in involved in ribosomal RNA processing. Nucleic Acids Res. 2007;35:7023–7030. doi: 10.1093/nar/gkm736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manful T., Cristodero M., Clayton C. DRBD1 is the Trypanosoma brucei homologue of the Spliceosome-Associated Protein 49. Mol. Biochem. Parasitol. 2009;166:186–189. doi: 10.1016/j.molbiopara.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Archer S.K., van Luu D., de Queiroz R., Brems S., Clayton C.E. Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog. 2009;5:e1000565. doi: 10.1371/journal.ppat.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archer S., Queiroz R., Stewart M., Clayton C.E. Trypanosomes as a model to investigate mRNA decay pathways. In: Maquat L.E., Kiledjian M., editors. RNA Turnover in Eukaryotes. Elsevier; San Diego, CA: 2008. pp. 359–377. [DOI] [PubMed] [Google Scholar]
- 23.Landeira D., Navarro M. Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. J. Cell Biol. 2007;176:133–139. doi: 10.1083/jcb.200607174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchetti M.A., Tschudi C., Kwon H., Wolin S.L., Ullu E. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J. Cell Sci. 2000;113:899–906. doi: 10.1242/jcs.113.5.899. [DOI] [PubMed] [Google Scholar]
- 25.Boucher N., Dacheux D., Giroud C., Baltz T. An essential cell cycle-regulated nucleolar protein relocates to the mitotic spindle where it is involved in mitotic progression in Trypanosoma brucei. J. Biol. Chem. 2007;282:13780–13790. doi: 10.1074/jbc.M700780200. [DOI] [PubMed] [Google Scholar]
- 26.Manger I.D., Boothroyd J.C. Identification of a nuclear protein in Trypanosoma brucei with homology to RNA-binding proteins from cis-splicing systems. Mol. Biochem. Parasitol. 1998;97:1–11. doi: 10.1016/s0166-6851(98)00118-2. [DOI] [PubMed] [Google Scholar]
- 27.West M., Hedges J., Lo K., Johnson A. Novel interaction of the 60S ribosomal subunit export adapter NMD3 at the nuclear pore complex. J. Biol. Chem. 2007;282:14028–14037. doi: 10.1074/jbc.M700256200. [DOI] [PubMed] [Google Scholar]
- 28.Ho J.H., Johnson A.W. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:2389–2399. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prohaska K., Williams N. Assembly of the Trypanosoma brucei 60S ribosomal subunit nuclear export complex requires trypanosome-specific proteins P34 and P37. Eukaryot. Cell. 2009;8:77–87. doi: 10.1128/EC.00234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White T., Rudenko G., Borst P. Three small RNAs within the 10kb rRNA transcription unit are analogous to Domain VII of other eukaryotic 28S rRNAs. Nucelic Acids Res. 1986;14:9471–9489. doi: 10.1093/nar/14.23.9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell D.A., Kubo K., Clark C.G., Boothroyd J.C. Precise identification of cleavage sites involved in the unusual processing of trypanosome ribosomal RNA. J. Mol. Biol. 1987;196:113–124. doi: 10.1016/0022-2836(87)90514-6. [DOI] [PubMed] [Google Scholar]
- 32.Arvalo-Rodriguez M., Heitman J. Cyclophilin A is localized to the nucleus and controls meiosis in Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:17–29. doi: 10.1128/EC.4.1.17-29.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu C. Cyclophilin A participates in the nuclear translocation of apoptosis-inducing factor in neurons after cerebral hypoxia-ischemia. J. Exp. Med. 2007;204:1741–1748. doi: 10.1084/jem.20070193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gullerova M., Barta A., Lorkovic Z. AtCyp59 is a multidomain cyclophilin from Arabidopsis thaliana that interacts with SR proteins and the C-terminal domain of the RNA polymerase II. RNA. 2006;12:631–643. doi: 10.1261/rna.2226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y., Manley J. The prolyl isomerase Pin1 functions in mitotic chromosome condensation. Mol. Cell. 2007;26:287–300. doi: 10.1016/j.molcel.2007.03.020. [DOI] [PubMed] [Google Scholar]