Abstract
Background
Lack of sequencing-validation and complexity of deletion testing hinder genetic diagnosis of SDH-associated paraganglioma/pheochromocytoma.
Methods
We developed sequencing assays and multiplex ligation-dependent probe amplification (MLPA) deletion-detection for SDHB, SDHC and SDHD. Clinical performance was validated on 141 blinded samples, previously tested at NIH.
Results
Sequencing and deletion-detection were highly reproducible and agreed with previous NIH results in 99.3% and 100%, respectively.
Conclusions
DNA sequencing combined with MLPA allows reliable and simplified genotyping of SDHB, SDHC and SDHD.
Keywords: SDHB, SDHC, SDHD, paraganglioma, pheochromocytoma, genotyping, DNA-sequencing, deletion detection, MLPA
Introduction
Succinate dehydrogenase (SDH) is a mitochondrial oxireductase complex consisting of four subunits: SDHA, SDHB, SDHC and SDHD (1, 2). Germline heterozygous mutations or deletions of SDHB, SDHC or SDHD cause an autosomal dominant tumor syndrome, manifesting when the second copy is somatically lost or mutated in sympathetic or parasymphatetic ganglia (3). SDHB mutations are observed most commonly and lead (usually) to catecholamine-secreting paragangliomas (PGL) or pheochromocytoma (PC), while the rarer SDHC or SDHD tumors cause chiefly nonfunctioning head and neck PGL (4).
Between 30–50% of apparently sporadic and >90% of clinically hereditary PGL are caused by SDH germline mutations or deletions; the corresponding figures for PC are 1–10% and 20–30% (1, 3). Consequently, mutation and deletion testing of SDHB, SDHC or SDHD has become indispensible in patients with PGL, or, seemingly, hereditary PC (3, 4, 5, 6)
When both full gene sequencing and deletion detection are performed, testing is highly accurate in identifying affected individuals. However, deletion detection by traditional methods, e.g. Southern blotting, is labor intensive, costly and slow. In addition, clinical validation of sequencing assays, in particular for SDHC and SDHD, has proven challenging, because of the low numbers of affected patients. Testing is sometimes only available through research laboratories, resulting in difficult access, unpredictable turn-around times, possible need for independent confirmation for patient care, and potential rejection of charges by insurance carriers.
To address these problems, we developed an easier deletion detection method, using multiplex ligation-dependent probe amplification (MLPA), performed an extensive analytical validation, including quantitative measurements of repeatability of sequencing quality and MLPA allelic ratios, and performed clinical validation of full gene sequencing and MLPA deletion detection of SDHB, SDHC and SDHD, using a large cohort of well characterized samples obtained from collaborators at the National Institute of Health (NIH; Bethesda, MD).
Methods
Samples
This study was approved by the Mayo Clinic Institutional Review Board. We purified DNA from 15 de-identified normal volunteer blood samples. Fifty archival, de-identified normal DNA samples and two samples with known SDHB deletions were used for deletion detection validation.
We obtained, and tested blindly, 141 de-identified, coded DNA samples, which had previously been mutation/deletion tested at NIH.
Sequencing of SDHB, SDHC, and SDHD
We sourced primers for PCR-amplification and cycle sequencing of all 18 exons of SDHB, SDHC and SDHD from IDT (Integrated DNA Technologies, Coralville, IO) (Supplemental table 1). Amplification reactions of 25μl contained: 100 ng of genomic DNA, 200 μmol/L dNTPs, 0.5U Platinum Taq Polymerase (both Invitrogen, Carlsbad, CA), 0.2 μmol/L primer mix and 1.5 mmol/L MgCl2. The cycling conditions were: 1 × 95 C (10 min), 12 × 95 C (30 s), 64 C (30s) and 72 C (1 min), 12 × 95 C (30 s), 62 C (30 s) and 72 C (1 min), 12x 95 C (30 s), 58 C (30 s) and 72 C (1 min), 1x 72 C (5 min).
Cycle-sequencing was performed using nested primers and BigDyeTerminatorsTM v1.1 (Applied Biosystems, ABI, Foster City, CA). Electropherograms were generated on ABI-3730xl automated DNA sequencers (ABI). The traces were analyzed with Mutation Surveyor® (Softgenetics, LLC State College, PA).
Detection of SDHB, SDHC, and SDHD deletions
A combination of MLPA and Luminex® xTAG (Luminex Molecular Diagnostics, Inc., Toronto, Ontario, Canada) technologies was used for deletion detection. DNA (400 ng) was denatured at 95 C (5 min), followed by addition of M13-tailed probes for each exon of SDHB, SDHC, SDHD and control genes at 25 C (see below). Samples were denatured at 95 C (5 min) before probe-hybridization at 60 C (>=16 h). Ligase-65 (MRC-Holland, Amsterdam, Netherlands) was then added for probe-ligation at 54 C for 15 min. The ligase was inactivated at 95 C (5 min) and the ligation-mixes were added to pre-made PCR-mixes containing universal M13-primers (forward primer 5′ biotin-labeled). PCR consisted of 23 cycles of 95 C (30 s), 60 C (30 s) and 72 C (60 s); final elongation at 72 C (20 min). PCR-products were mixed with xTAG beads, 2000 of each bead per sample, denatured at 96 C (2 min) and hybridized at 37 C (1 h). The hybridization buffer was 1x TMAC (300 ml 5M tetramethyl ammonium chloride, 2.5 ml of 20% Sarcosyl, 25 ml of 1M TRIS pH 8.0, 4 ml of 0.5M EDTA pH 8.0 and 168.5 ml of water). Hybridized products were incubated with 25 μl of a 1:125 mixture of Streptavidin R-phycoerythrin and 1x TMAC (15 min) at room temperature in the dark, and then applied to a Luminex 200 instrument (Luminex Molecular Diagnostics, Inc.). Two lasers with different wavelengths detect the bead identity and the reporter molecule signal. For each probe, deviations of >20% from a 1:1 ratio between signals from normal control samples and patient samples indicate possible deletions or insertions/duplications. Less than 4% of normal samples fall outside this ratio. Data was analyzed semi-automatically with Gene Marker® (Softgenetics), using CNNTB (3p22.1), DMD (Xp21.1), HIRA (22q11.2), NPC-E17 (18q11.2), and TRNSFR7 (12p13.31) as reference/control genes.
Analytical and Clinical Validation
Intra- and inter-assay precision for sequencing and deletion detection were assessed by running one DNA sample 3 times in the same run, and 10 times in different runs. We reviewed the traces for mutations and tracked the Mutation Surveyor quality scores for these samples, for the fresh normal samples, and the samples used in the clinical validation (see below), to calculate the coefficient of variation (CV) for the quality scores of each exon. The quality scores indicate the fraction of noise in a trace (e.g. a score of 20 corresponds to a noise of 1/20=5%).
The freshly collected 15 normal DNA and the 50 archival DNA samples were used to verify absence of false-positive sequencing and deletion testing, respectively, and to establish the signal cut-off ratios for deletion detection (see above). The two samples with known deletions were used for initial verification of these cut-offs. One of these patients had a complete SDHB deletion (Figure 1), while the other’s deletion encompassed part of the promotor and exon 1 of SDHB. Reproducibility of signal ratios for MLPA was assessed by calculating the CVs for the signal ratios for each tested exon and control gene.
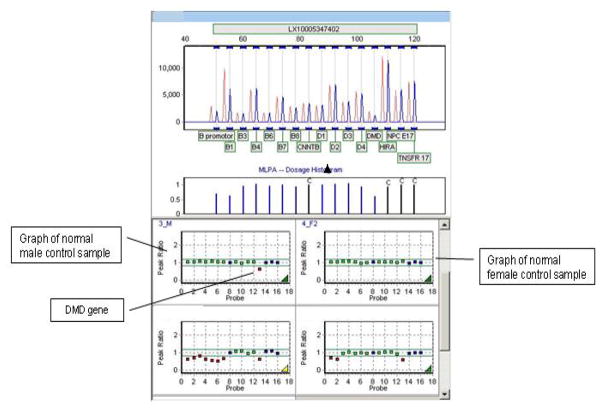
Figure 1. MLPA deletion detection.
The top half of the figure depicts the signals of a patient sample (blue peaks) compared to multi-run averaged female control signals (red peaks).
The bottom half of the figure shows the peak ratios of averaged control signal to the signal generated in a run by control samples (male, left upper and female, right upper) and the response ratios observed in the sample of a male patient with a complete deletion of the SDHB gene (left lower) and a male patient with a deletion of exon 1 of SDHB. The horizontal green lines and the green shaded area on each graph indicate the expected normal range for the signal ratios.
Of note is the difference between both normal control and patient sample with regards to the signal ratio of the DMD control gene. The DMD gene is located on the X-chromosome and is therefore only present in a single copy in male patients, thus serving as a convenient control gene for the expected deviation of a heterozygous deletion from the established normal signal ratio range.
Clinical validation utilized the 141 samples provided by NIH; 99 had been characterized for SDH mutations by DNA-sequencing, while 42 had been tested for deletions by quantitative multiplex PCR. After completion of assay validation, our blinded genotyping results were send back to NIH, where they were compared with the original results.
Results
Within and between run agreement of DNA sequencing and deletion detection was 100%. We did not detect any mutations or deletions in the normal samples and correctly identified both known SDHB deletions in the two unblinded samples. The average sequencing quality scores for bidirectional sequencing of SDHB, SDHC and SDHD ranged from 17–71, with intraassay and interassays CVs of 0–40.8% and 1.2–75.4%, respectively. For MLPA, the average signal ratios for control genes in normal samples and in samples of patients with deletions ranged from 0.980–1.025, with total CVs of 2.85–5.22%. For DMD the average ratios were 0.913–0.992 for females and 0.591–0599 for males, with respective CVs of 5.04–7.76% and 7.00–11.00%. For undeleted and deleted SDH exons the average ratios were 0.932–1.077 and 0.646–0.708, respectively, with corresponding CVs of 3.49–15.34% and 4.38–6.86% (Supplemental table 2).
The results of the blinded validation portion of the study are summarized in the Table. The 141 samples contained 60 disease-causing mutations or big deletions, which were all correctly identified in our study. We found one additional mutation (confirmed), not previously found by NIH, corresponding to an agreement of 61/60 (99.2%) for disease-causing DNA changes between NIH and us. We also found an additional NSV (normal sequence variant) in one sample. Overall agreement between methods for deletions, mutations and NSVs was 75/76 (99.3 %), for a total genotyping agreement of 98.6%.
Table.
Comparison of original genotyping with blinded re-testing in 141 patients with suspected SDH mutations/deletions
| Sequence variants | Original genotyping result (N of patients) | Blinded retesting result (N of patients) | Method agreement (%)* | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SDHB | SDHC | SDHD | Combination# | Total | SDHB | SDHC | SDHD | Combination# | Total | ||
| Total N of samples sequenced | - | - | - | - | 99 | 99 | - | ||||
| Matches Genebank ref-sequence | - | - | - | - | 44 | - | - | - | - | 43 | 98.9 |
| Normal sequence variants (NSV) | 3 | 3 | 4 | 5 | 15 | 3 | 3 | 3 | 6 | 15 | 100## |
| Disease-causing mutations, no NSV | 27 | 0 | 2 | 0 | 29 | 27 | 0 | 3 | 0 | 30 | 98.3 |
| Missense | 20 | 0 | 1 | 0 | 21 | 20 | 0 | 2 | 0 | 22 | 97.7 |
| Nonsense | 7 | 0 | 1 | 0 | 8 | 7 | 0 | 1 | 0 | 8 | 100 |
| Disease-Causing mutations + NSV | 2 | 0 | 0 | 9 | 11 | 2 | 0 | 0 | 9 | 11 | 100 |
| Missense + one NSV | 1 | 0 | 0 | 5 | 6 | 1 | 0 | 0 | 5 | 6 | 100 |
| Nonsense + one NSV | 1 | 0 | 0 | 4 | 5 | 1 | 0 | 0 | 4 | 5 | 100 |
| Total N of samples tested for deletions | 42 | 42 | 100 | ||||||||
| No deletions | 22 | 22 | 100 | ||||||||
| Deletions | 20 | 0 | 0 | 0 | 20 | 20 | 0 | 0 | 0 | 20 | 100 |
| Total N of disease-causing mutations and deletions detected | 49 | 0 | 2 | 9 | 60 | 49 | 0 | 3 | 9 | 61 | 99.2 |
| Total N of all sequence variants and deletions detected | 52 | 3 | 6 | 14 | 75 | 52 | 3 | 6 | 15 | 76 | 99.3 |
| Total genotyping** | 98.6** | ||||||||||
Calculated as follows for each row: 100 × total original/(0.5 × total original + 0.5 × total retesting).
Calculated as follows: 100 × N of sequence variants and deletions in agreement between both methods/(total N of sequenced samples + total N of deletion tested samples). I.e. in this case: 100 × 139/(99+42).
Combination category applies to NSV in >1 SDH gene. No combinations of mutations in more then one SDH gene were observed.
No additional patients with NSVs were found, but one additional NSV (in SDHD) was detected during blinded retesting in one patient with a SDHD NSV
Discussion
We have developed and comprehensively validated a method for complete genotyping of SDHB, SDHC and SDHD. The deletion detection component is less labor intensive and faster than many alternative approaches. Moreover, due to the high number of beads counted during Luminex® detection, the results are semi-quantitative, with control limits, highly reproducible, and easily amenable to standard quality control measures, thereby reducing the potential for mis-interpretation and facilitating quality assurance. With regards to DNA-sequencing, our collaboration with NIH allowed us to validate the performance for all three disease-causing SDH genes in actual patient samples, despite the relative rarity of this disorder, in a rigorous blinded fashion.
Reliable SDH genotyping, including deletion detection is, alongside the more widely available VHL and MEN2 testing, another step towards comprehensive genetic characterization of PGL and PC patients, conditions that are now recognized to have a much larger genetic component than previously thought (3,7). Identifying affected patients is important, as their tumors have an increased propensity for recurrence. Moreover, it allows screening of pre-symptomatic family members, enabling affected individuals to be entered in surveillance programs. Since the chances of metastatic spread and potentially fatal adrenergic storm increase in parallel with tumor size, this might reduce morbidity and mortality (1).
While we can offer clinicians a comprehensive SDH genotyping service, clinical judgment can be helpful in selecting the SDH gene that should be tested first. We suggest a tiered approach, based on the location of tumor, presence of malignancy, family history and tumor hormonal status (Figure 2). SDHB mutations/deletions are far more common than those of SDHC or SDHD and should be sought first in patients with hormonally active tumors, i.e. chest/abdominal/pelvic. SDHD mutations are mostly found in patients with head and neck PGL; PGL in chest (10), pelvis and abdomen, or seemingly sporadic PC are less prevalent. Finally, SDHC mutations are very rare and usually result in head and neck PC (1, 6, 8).
Figure 2. Decision tree and workflow for SDH genotyping.
The flow chart depicts a suggested decision tree and workflow for SDH genotyping in patients with suspected or known familial paraganglioma (PGL) or pheochromocytoma (PC). This includes most patients with PGL. With regards to PC, activating mutations in RET, and inactivating mutations or deletions in VHL and NF1 also have to be considered in suspected familial cases, and this is indicated at the top of the decision tree. However, an in-depth discussion and depiction of workflow for all familial variants of chromaffin tumors is beyond the scope of this article and the figure concentrates on the SDH genotyping.
Supplementary Material
Acknowledgments
We would like to thank Dr Mercedes Robledo from Human Cancer Genetics Program at the Spanish National Cancer Centre in Madrid, Spain for providing us with two SDHB samples with known deletion for our MLPA deletion detection initial validation step.
References
- 1.Timmers HJLM, Gimenez-Roqueplo AP, Mannelli M, Pacak K. Clinical Aspects of SDHx-related pheochromocytoma and paraganglioma. Endocrine-Related Cancer. 2009;16:391–400. doi: 10.1677/ERC-08-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lima J, Feijao T, da Silva AF, Pereira-Castro I, Fernandez-Ballester G, Maximo V, Herrero A, Serrano L, Sobrinho-Simones M, Garcia-Rostan G. High frequency of germline Succinate Dehydrogenase mutations in sporadic cervical paraganglioma in Northern Spain; Mitochondrial Succinate dehydrogenase structure-function relationships and clinical-pathological correlations. The Journal of Clinical Endocrinology and Metabolism. 2007;92(12):4853–4864. doi: 10.1210/jc.2007-0640. [DOI] [PubMed] [Google Scholar]
- 3.Bryant J, Farmer J, Kessler LJ, Townsend RR, Nathanson KL. Pheochromocytoma: The expanding Genetic Differential Diagnosis. Journal of the National Cancer Institute. 2003;95(16) doi: 10.1093/jnci/djg024. [DOI] [PubMed] [Google Scholar]
- 4.Gimenez-Roqueplo AP, Favier J, Rustin P, Rieubland C, Crespin M, Nau V, Khau Van Kien P, Corvol P, Plouin PF, Jeunemaitre X. Mutations in the SDHB gene are associated with extra- adrenal and/or malignant pheochromocytoma. Cancer Research. 2003;63:5615–5621. [PubMed] [Google Scholar]
- 5.Eisenhofer G, Bornstein SR, Brouwers FM, Cheung Nai-Kong V, Dahia PL, de Krijger RR, Giordano TJ, Greene LA, Goldstein DS, Lehnert H, Manger WM, Maris JM, Neumann HPH, Pacak K, Shulkin BL, Smith DI, Tischler AS, Young WF., jr Malignant pheochromocytoma: current status and initiatives for future progress. Endocrine – Related Cancer. 2004;11:423–436. doi: 10.1677/erc.1.00829. [DOI] [PubMed] [Google Scholar]
- 6.Young WF., jr Editorial: Paraganglioma-All in the Family. The Journal of Clinical Endocrinology & Metabolism. 2006;91(3):790–792. doi: 10.1210/jc.2005-2758. [DOI] [PubMed] [Google Scholar]
- 7.Lenders JWM, Eisenhofer G, Mannelli M, Pacak K. Pheochromocytoma. The Lancet. 2005:366. doi: 10.1016/S0140-6736(05)67139-5. [DOI] [PubMed] [Google Scholar]
- 8.Bayley JP, van Minderhout I, Weiss MM, Jansen JC, Oomen PHN, Menko FH, Pasini B, Ferrando B, Wong N, Alpert LC, Williams R, Blair E, Devilee P, Taschner Mutation analysis of SDHB and SDHC: novel germline mutations in sporadic head and neck paraganglioma and familial paraganglioma and/or pheochromocytoma. BMC Medical genetics. 2006;7:1. doi: 10.1186/1471-2350-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmut P, Neumann H, Eng C. The approach to the patient with Paraganglioma. The Journal of Clinical Endocrinology and Metabolism. 2009;94(8):2677–2683. doi: 10.1210/jc.2009-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghayee HK, Havekes B, Corssmit EP, Eisenhofer G, Hammes SR, Ahmad Z, Tessnow A, Lazúrová I, Adams KT, Fojo AT, Pacak K, Auchus RJ. Mediastinal paragangliomas: association with mutations in the succinate dehydrogenase genes and aggressive behavior. Endocr Relat Cancer. 2009 Mar;16(1):291–9. doi: 10.1677/ERC-08-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.