Abstract
Most processes involving an organism’s genetic material, including replication, repair and recombination, require access to single stranded DNA as a template or reaction intermediate. To disrupt the hydrogen bonds between the two strands in double stranded DNA, organisms utilize proteins called DNA helicases. DNA helicases use duplex DNA as a substrate to create single stranded DNA in a reaction that requires ATP hydrolysis. Due to their critical role in cellular function, understanding the reaction catalyzed by helicases is essential to understanding DNA metabolism. Helicases are also important in many disease processes due to their role in DNA maintenance and replication. Here we discuss ways to rapidly purify helicases in sufficient quantity for biochemical analysis. We also briefly discuss potential substrates to use with helicases to establish some of their critical biochemical parameters. Through the use of methods that simplify the study of helicases, our understanding of these essential proteins can be accelerated.
Keywords: Helicase, Purification, DNA Substrate
INTRODUCTION
The double-stranded structure of DNA provides for an elegant mechanism to ensure accurate and faithful replication and repair of the DNA – the genetic information stored in one strand is also encoded in its complement. To take advantage of the properties of complementary base pairing, the two strands of the double helix must be separated or unwound to provide DNA polymerases, strand transferases, primases, RNA polymerases, and other proteins with access to single-stranded DNA (ssDNA). Enzymes able to catalyze the unwinding of double-stranded DNA (dsDNA) were first discovered over three decades ago [1, 2]. Referred to as DNA helicases, these enzymes disrupt the hydrogen bonds that hold the two strands of duplex DNA together to generate, at least transiently, the ssDNA required as a template or reaction intermediate in the various aspects of DNA metabolism [3, 4].
Studies conducted during the last 30 years have demonstrated that DNA helicases are ubiquitous in nature. Indeed, a multitude of distinct helicases have now been identified and characterized [5, 6]. Genetic studies have been useful in determining that DNA helicases are involved in every aspect of DNA metabolism. Specific helicases are required in DNA replication, in DNA repair, in recombination and in bacterial conjugation. A similar but distinct class of enzymes called RNA helicases serves an analogous role in processes involving RNA including transcription, translation and RNA processing [7–13]. Thus, helicases are involved in all aspects of nucleic acid metabolism and, as such, play a central role in promoting and controlling growth and duplication of the cell.
It is also clear that each cell contains multiple distinct helicases. For example, the bacterium Escherichia coli (E. coli) contain at least 11 DNA helicases [4, 14, 15]. Even the relatively simple bacteriophage T4 encodes (at least) two different DNA helicases [16]. Genetic and biochemical studies have revealed that each DNA helicase has a role in a specific pathway in the cell [4]. Presumably, each helicase makes specific protein-DNA and protein-protein contacts that direct it to the appropriate DNA substrate in the appropriate reaction pathway. For example, the DnaB protein in E. coli is the primary replicative helicase in the cell [17]. No other helicase found in E. coli can substitute for DnaB protein in this capacity. Similarly, DNA helicase II is involved in UvrABC-mediated excision repair of pyrimidine dimers [18] and methyl-directed mismatch repair [19], and no other helicase in the cell can substitute for helicase II in these roles [20]. Thus, it seems likely that the variety of DNA helicases found in the cell has evolved to provide helicase function in specific reaction pathways.
Although a great deal has been learned regarding the essential roles of DNA helicases in DNA metabolism, less is known regarding the mechanism of helicase-catalyzed duplex DNA unwinding. All helicases catalyze a DNA-stimulated hydrolysis of nucleoside 5′-triphosphates (NTPs), usually ATP. The energy liberated upon hydrolysis of the high energy phosphate bond between the beta and gamma phosphates is required for DNA unwinding. Precisely how the energy-providing ATPase activity and the energy-requiring helicase activity are coupled is an active area of investigation. It has been suggested that hydrolysis of the nucleotide serves to cycle the enzyme through conformational states with differing affinities for dsDNA and ssDNA [21, 22]. It has also been proposed that most helicases function as multimers, either dimers or hexamers [5, 21]. This arrangement provides for multiple DNA binding sites as well as multiple NTP binding sites on the active form of the enzyme. Two general mechanisms for DNA unwinding have been advanced; a passive mechanism and an active mechanism [21, 23]. The passive mechanism relies on thermal breathing of the duplex at the junction of ssDNA and dsDNA with the helicase passively occupying the thermally denatured region and preventing it from reannealing. The active mechanism, on the other hand, requires the helicase to actively disrupt the hydrogen bonds holding the duplex together. Helicases that are carefully examined to date appear to catalyze unwinding by the latter, active mechanism [24]. Lohman and colleagues have proposed a rolling model to account for unwinding by the E. coli Rep protein dimer [22]. In this model each Rep monomer (protomer) alternates between binding duplex DNA at the ssDNA-dsDNA junction and binding ssDNA just behind the ssDNA-dsDNA junction. Upon ATP hydrolysis the enzyme unwinds the duplex region to render it single-stranded leaving both protomers bound to ssDNA. The protomer originally bound to ssDNA then moves relative to the other protomer to occupy the region of duplex DNA at the ssDNA-dsDNA junction and the process is repeated. Thus, the enzyme can be envisioned to roll into and unwind the duplex region. A thorough review of the recent literature on helicase unwinding reaction mechanism has recently appeared [5] and we will not consider this topic further. However, it should be noted that this mechanism has not been rigorously proved although compelling evidence, obtained using the E. coli Rep helicase, has been obtained in support of this model. It is possible that other helicases unwind duplex DNA by other mechanisms. Specifically, the mechanism utilized by hexameric helicases remains unknown, although significant work in this area has appeared recently [25]. This represents an active and exciting area of continuing investigation.
It should also be noted that each helicase appears to unwind duplex DNA with a specific polarity defined with respect to the strand of DNA on which the protein is presumed to be primarily bound. Unwinding polarity is empirically determined using a linear DNA substrate with a large region of ssDNA in the middle and duplex regions of differing lengths on each end. Since most helicases appear to bind and preferentially initiate unwinding from a ssDNA region, the enzyme is presumed to bind the ssDNA and then move toward one end of the linear DNA. Alternatively, the enzyme could bind preferentially to one or the other of the two available ssDNA-dsDNA junctions. In either case, one of the two duplex regions is preferentially unwound and this event is used to operationally define the macroscopic polarity of the unwinding reaction. Presumably the polarity of unwinding has relevance to the biological role of the enzyme although this has yet to be rigorously tested.
Interest in fully understanding this class of proteins has been heightened by the discovery that mutations in specific DNA helicases result in human genetic diseases [26–28]. For example, Xeroderma pigmentosum, a disease resulting in severe sensitivity to sunlight and the occurrence of skin cancer, results from a defect in the DNA excision repair pathway in human cells. Seven complementation groups have been identified; two encode DNA helicases [29–32]. The gene responsible for Bloom’s syndrome (BLM) was identified as a DNA helicase [33, 34] and patients with Bloom’s syndrome are predisposed to several different classes of neoplasm suggesting that this gene product plays a fundamental role in an important aspect of DNA metabolism that, when defective, leads to cancer in a wide range of tissues [35]. The Werner’s syndrome gene (WRN) has also been shown to encode a DNA helicase/exonuclease [36]. Patients with Werner’s syndrome appear to age prematurely [37, 38] suggesting that this gene product plays a fundamental role in nucleic acid metabolism. As a final example, the gene encoding another human RecQ helicase family member, RecQL4, is linked to Rothman-Thomson syndrome [39, 40]. There is no doubt that other diseases will be linked with defects in DNA helicases as more disease genes are identified. This places added emphasis on the importance of understanding this class of proteins.
Fully understanding the mechanism and role played by any helicase in the cell requires that the protein be purified to near homogeneity for biochemical study. This requires cloning of the gene encoding the protein in an appropriate expressions system followed by purification. Here we describe an E. coli expression system that allows rapid purification through the use of affinity tags. This system has proven useful for the routine purification of several helicases including eukaryotic helicases and large helicase proteins.
RAPID HELICASE PURIFICATION
Most proteins benefit from rapid purification which reduces susceptibility to proteolysis and denaturation. Affinity tags offer several advantages in rapid purification strategies but have limitations of their own. Though some tags, such as a poly-histidine (His-tag) or a HA-tag, are quite small and comprise of only a few amino acids they still raise the question of what effect those few extra amino acids have on the native activity of the protein. Because of this limitation we have modified the Impact System (New England Biolabs, Ipswich MA) which allows removal of the affinity tag in the last purification step and utilized this modification for rapid purification of helicase proteins. The pTYB4 plasmid contains a C-terminal tag that consists of an intein fused to a chitin binding domain (intein-CBD). The CBD is able to bind a chitin column very efficiently in the presence of high salt and, in a reducing environment (50 mM DTT), the intein cleaves itself from the C-terminus of the protein leaving only one or two extra amino acids depending on the cloning procedure. This tag system is very effective for rapid protein purification, but it is rare to achieve greater than 90% homogeneity with a single column.
To achieve the purity required for biochemical analysis, we modified the pTYB4 plasmid by adding an eight-histidine tag on the C-terminus of the CBD (Fig 1). The his-tag was introduced into the plasmid via polymerase chain reaction (PCR) using the pTYB4 plasmid as template. The forward primer for this PCR was 5′-GGAATTGTGAGCGGATAACAATTCCCC-3′ and the reverse primer, containing the sequence for the histidine tag (underlined), was 5′-AATCTGCAGTCAGTGATGGTGATGGTGATGGTGATGTTGAAGCTGCCACAAGGC-3′. The PCR product and the original pTYB4 vector were digested with HindIII and PstI, and the PCR product was ligated into pTYB4 to create the new plasmid, pTYB4-His. The addition of the His-tag to pTYB4 allows for a rapid and efficient two-column purification that yields nearly homogeneous protein in a procedure that takes as little as a single day without the permanent addition of a tag to the protein of interest. Using this expression plasmid we have found that we get very efficient enrichment of the histidine-tagged fusion protein using cobalt based affinity columns such as TALON® (Clontech, Mountain View CA). We do not expect any problems with the use of a nickel based affinity column for this step in the procedure.
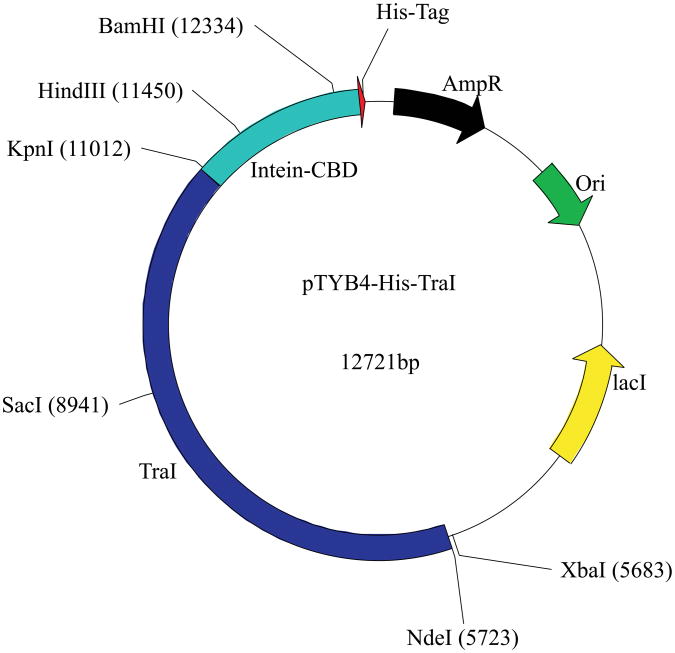
Figure 1. The pTYB4-His-TraI plasmid.
Schematic of the pTYB4-His-TraI plasmid with several unique restriction sites noted. The traI gene is shown in dark blue, the intein-CBD sequence is depicted in cyan, and the 8-His tag is depicted in red. In addition, the origin of replication (Ori), the ampicillin resistance gene (AmpR) and the lacI gene are shown.
In figure 2a we have delineated the basic purification protocol using the pTYB4-His expression plasmid. Briefly, the cells are lysed and the cleared lysate is passed over a TALON® column to allow binding of the target protein via the 8-his tag on the fusion protein. The column is washed extensively to remove unbound proteins, the target protein is eluted with imidazole and the eluted protein is applied directly to a chitin column. There is no need for a dialysis step since the high salt and imidazole concentrations do not interfere with binding to the chitin resin. The target protein is released from the chitin column by incubation with 50 mM DTT to allow intein-directed cleavage from the tag. The resulting eluted protein is ready for dialysis into storage buffer. Depending on the length of the incubation with DTT, this procedure can be completed in as little as 24 hours with excellent results.
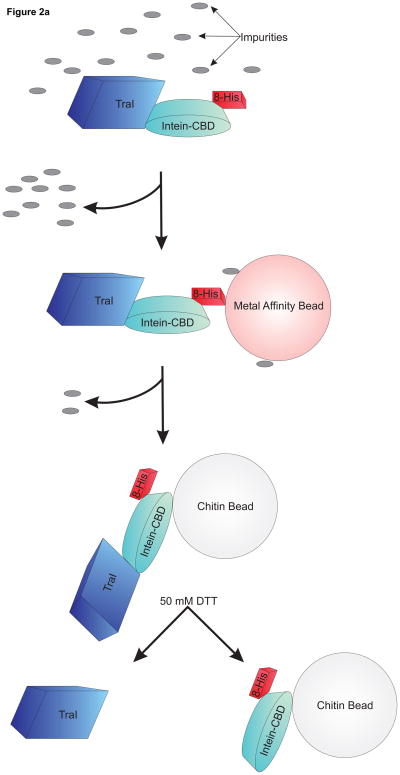
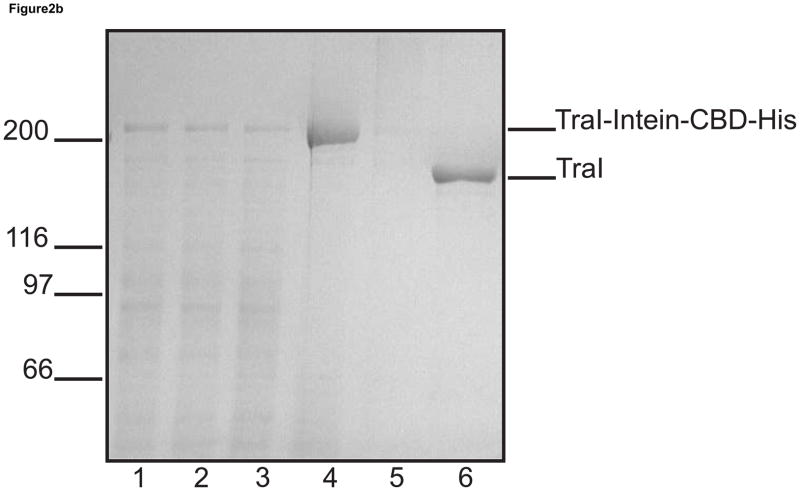
Figure 2. A schematic view of purification using pTYB4-His and results from TraI purification.
a. Overview of the purification procedure: The fusion protein is bound to a cobalt affinity column (TALON®) depicted in pink. The protein, including both tags, is eluted from the cobalt affinity column using an imidazole wash and directly bound to a chitin column depicted in light grey. After addition of the reducing agent DTT to promote intein-directed cleavage of the target protein from the affinity tags and incubation at 4°C, the final product is eluted from the chitin column leaving the intein-CBD-His tag bound to the beads. b. Coomassie blue stained SDS polyacrylamide gel from a TraI rapid purification. In each lane 5 μg of total protein has been loaded. Lane 1, crude lysate; lane 2, cleared lysate; lane 3, TALON® flow through; lane 4, TALON® column elution; lane 5, chitin column flow through; and lane 6, chitin elution containing purified TraI.
A critical issue for effective protein purification using the pTYB4-His system (or any system for that matter) is the ability to express significant amounts of soluble protein in E. coli. A key to effective expression in E. coli is determining the appropriate media and temperature for the specific protein being expressed. Though many proteins are expressed successfully in Luria-Bertani (LB) media with isopropyl-β-D-thiogalactopyranoside (IPTG) induction at 37 °C, some proteins – especially large or non-prokaryotic proteins – may require modification of these conditions for the expression of significant amounts of soluble protein. One means of increasing the solubility of proteins is by reducing the expression temperature. Cells are grown to an OD600 of 0.8, IPTG is added, and the induction is carried out either at room temperature or at 16°C depending on the protein being expressed (Table 1). When the temperature of the induction incubation is decreased the length of the incubation is increased to as much as 24 hours in some cases. We determine the appropriate length of the induction empirically by evaluating soluble protein at various time points after the addition of IPTG. If reduction in temperature is not sufficient then the growth media may be changed. One media that works particularly well for expressing difficult proteins is the autoinduction media ZYM-5052, as described in [41]. Autoinductions may also be tried at different temperatures to improve expression or solubility. Generally the lower the temperature the lower the level of expression, however, it may also result in greater solubility of the protein being expressed. As with IPTG inductions, when the temperature of the autoinduction is reduced the length of the incubation must be increased. Autoinduction at 16°C can require as much as a 40 hour incubation (Table 1).
Table 1.
| Protein | Expression Strain | Media | Temperature (°C) | Induction Type | Time of Induction (hours) |
|---|---|---|---|---|---|
| TraI | HMS174 (DE3) | LB | 25 | IPTG | 16 |
| UvrD | BL21 (DE3)uvrD::Tn5, mutL::Tn10 | ZYM-5052 | 25 | Auto | 40 |
| Hmi1p | RDK1896 (DE3) | LB | 16 | IPTG | 24 |
Media and expression conditions for three DNA helicases.
A good example of a rapid purification involving a large protein is E. coli helicase I (TraI). TraI is a 192 kDa protein and, with the addition of the intein-CBD-His tag, the size of the expressed fusion protein is increased to over 250 kDa. We utilized a two step procedure to clone the traI gene into pTYB4-His due to its large size. The XbaI and SacI fragment of pET11c-TraI [42] was removed and ligated into the pTYB4-His digested with the same enzymes to complete cloning of the first half of the traI gene into pTYB4-His. The second half of the gene was amplified via PCR using pET11c-TraI as the template and the following primers 5′-CGATGAGAGCTCAATGG-3′ (forward primer) and 5′-GTCTCCACCCAGGGTTTTC-3′ (reverse primer). The reverse primer amplified traI up to and including the last sense codon but did not include the stop codon so that the C-terminal tag could be fused to the traI gene allowing expression of the appropriate fusion protein. This PCR product was digested with SacI and the 3′ end of the amplicon was left blunt; an appropriately phosphorylated primer was used for this PCR. The pTYB4-His plasmid containing the first half of the traI gene was digested with SacI and SmaI to facilitate the directional cloning of the PCR amplicon creating the final desired construct – pTYB4-His-TraI (Fig 1). Alternatively, a SmaI site could be placed at the 3′ end of the gene being amplified. However, the creation of a full SmaI site in the final pTYB4-His construct will result in the addition of two extra amino acids at the C-terminus of the cleaved protein. The procedure used here results in the addition of a single glycine residue at the C-terminus of the purified protein product.
pTYB4-His-TraI was transformed into chemically competent HMS174 (DE3) cells for expression of the fusion protein. To increase the solubility of this large protein, cells were grown and induced using 0.3 mM IPTG at room temperature. Incubation at room temperature was continued for 16 hours (Table 1). Cells were harvested by centrifugation at 6000 × g for 10 minutes, suspended once using ice cold STE (100 mM NaCl, 10 mM Tris-HCl pH 8, 1 mM EDTA) and spun down again. The cell pellet was weighed and stored at −70°C until needed for protein purification.
To purify protein the cell pellet was thawed on ice and resuspended in a volume of lysis buffer (20 mM Tris-HCl pH 8, 100 mM NaCl, 10% glycerol, and 5 mM imidazole) equivalent to four times the cell pellet weight. The cells were lysed by the addition of lysozyme to a final concentration of 250 μg/ml with incubation at 4°C for 30 minutes followed by the addition of 0.05% sodium deoxycholate. Sodium chloride was added slowly to the lysate until the final salt concentration was at 500 mM. The lysate was sonicated to reduce viscosity and centrifuged at 21,000 rpm for 1 hour in a Sorvall SS34 rotor to prepare a cleared lysate. The cleared lysate, shown in figure 2b (lane 2), contains the soluble tagged fusion protein as the major band above the 200 kDa marker. The identity of the protein was confirmed with Western blotting (data not shown). Inductions at both 30°C and 37°C failed to produce significant amounts of soluble fusion protein.
The affinity tagged TraI was batch bound to cobalt affinity resin (TALON®) for 1 hour at 4°C. The protein-bound resin was then poured into a column and extensively washed with buffer (20 mM Tris-HCl (pH 8), 500 mM NaCl, 10% glycerol, and 5 mM imidazole) until the residual protein eluting from the column was less than 10 μg/ml. The TraI-intein-CBD-His fusion protein was eluted using wash buffer containing 250 mM imidazole. The TALON® elution was then directly batch bound to chitin beads (New England Biolabs, Ipswich MA) for 1 hour at 4°C. The protein-resin slurry was poured into a column and washed extensively with chitin wash buffer (20 mM Tris-HCl (pH 8), 750 mM NaCl, 10% glycerol, and 0.1 mM EDTA) until the residual protein eluting from the column was less than 10 μg/ml. Ten column volumes of wash buffer plus 50 mM dithiothreitol (DTT) were passed over the column and the column was plugged and incubated overnight at 4°C to allow intein-directed cleavage of the affinity tags from TraI. Purified TraI eluted from the chitin column at greater than 90% purity (Fig 2b, lane 6). This purification procedure requires a total of 24 hours from cell lysis to purified protein.
The example of TraI purification described above demonstrates two important advances in the purification of this protein. First is the successful expression of a very large fusion protein that is soluble in E. coli. This required reducing the temperature of induction. The second is that a purification scheme that used to take several days and multiple columns can now be completed in one 24 hour period, providing pure protein with high specific activity. We have also purified E. coli UvrD (unpublished) and S. cerevisiae Hmi1p [43] using this procedure demonstrating its utility for the purification of eukaryotic helicases.
SUBSTRATES FOR HELICASE ASSAYS
The availability of purified protein allows a detailed analysis of the biochemical activity of the helicase. There are many suitable assays and substrates available to measure helicase activity [44–46]. Here we will focus on simple assays that can be used to begin to assess the helicase activity of the purified protein. More sophisticated analyses are possible and are required to elucidate mechanism.
The first substrate we describe can be used to initiate the biochemical characterization of a helicase without prior knowledge of the polarity of unwinding. This substrate, shown in figure 3a, consists of M13 ssDNA to which an oligonucleotide is annealed to create a duplex region. Here we describe the construction of a substrate with a 93 base pair duplex region. However, the length of the duplex region can be extended or shortened by appropriate choice of oligonucleotide. In practice, substrates made with oligonucleotides less than about 25 bases in length are unstable and can undergo spontaneous melting under helicase assay conditions.
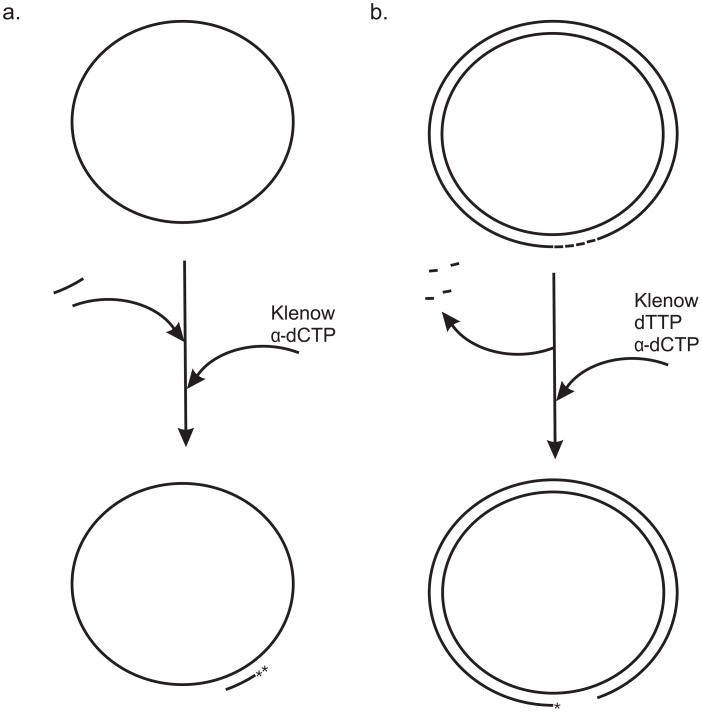
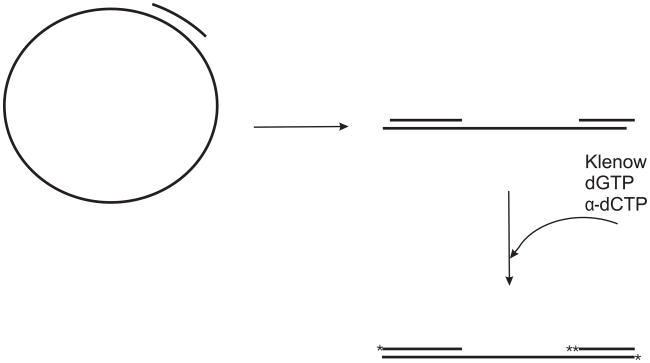
Figure 3. Two circular substrates for helicase assays.
a. A 91-mer oligonucleotide is annealed to M13 ssDNA as described in the text. The 3′-end is extended by two bases using [32P]dCTP and the Klenow fragment of DNA polymerase I to yield the 93 base pair partial duplex substrate. b. A plasmid containing several Nt.BbvCI sites is nicked to completion with Nt.BbvCI. The small oligonucleotide fragments are heat denatured and removed leaving a gapped plasmid. The gap-containing strand is labeled using [32P]dCTP, dTTP and the Klenow fragment of DNA polymerase I.
To prepare this substrate M13mp18 or mp19 ssDNA is annealed with a 91 base oligonucleotide with the sequence 5′-AGTAGCACCATTACCATTAGCAAGGCCGGAAACGTCACCAATGAAACCATCGATAGCAGCACCGTAATCAGTAGCGACAGAATCAAGTTTG-3′ in a 1:1 molar ratio by mixing ssDNA with the oligonucleotide in annealing buffer (50 mM Tris-HCl (pH 7.5), 10 mM Mg2Cl, 50 mM NaCl) followed by heating to 95°C and slow cooling to room temperature. The Klenow fragment of E. coli DNA polymerase I is then used to add two [32P]dCMP residues to the 3′ end of the duplex region to generate the 93 base pair partial duplex substrate (Fig 3a). In this case, the sequence of the oligonucleotide chosen allows the incorporation of two contiguous dCMP residues providing a substrate with higher specific activity. Alternatively, the oligonucleotide can be labeled on the 5′-end using [γ-32P]ATP and polynucleotide kinase. Because of the circular nature of this substrate, the polarity of the helicase is not a concern. This substrate can be processed further to study the polarity of a helicase as described below.
An alternative circular substrate with substantial duplex DNA is shown in figure 3b. This DNA substrate is particularly appropriate for biochemical analysis of processive DNA helicases. To construct this gapped duplex DNA we start with a plasmid of a desired size since the larger the plasmid the longer the unwinding tract for the helicase. Here we describe the construction using the pUC19 plasmid which yields a duplex region approximately 2.7 kbp in length. To construct a ssDNA gap of defined size we introduced a series of Nt.BbvCI recognition sequences (5′-CCTCAGC-3′) by cloning an appropriate duplex DNA fragment with multiple Nt.BbvCI recognition sites into the multiple cloning site in pUC19 using oligonucleotides. The Nt.BbvCI enzyme will introduce a nick after the second cytosine at each recognition site. The idea is to introduce several tandem Nt.BbvCI recognition sites, all on the same DNA strand, to allow the construction of a gapped molecule with a defined length gap. In the example mentioned we introduced four Nt.BbvCI recognition sites to produce a gap that is 31 nucleotides in length.
To prepare the gapped DNA the plasmid is nicked to completion with Nt.BbvCI and then denatured at 80°C for 20 minutes to release the small oligonucleotides between each nick site. The gapped DNA is purified with the use of a PCR purification kit to remove the released oligonucleotides (Fig 3b). Finally, the DNA substrate is labeled using [α-32P]dCTP and the Klenow fragment of DNA Polymerase I. Unwinding of this molecule by a DNA helicase is evaluated using agarose gels.
Finally, as mentioned above, a modification of the 93 base pair partial duplex substrate shown in figure 3a can also be used to determine the directionality of a helicase. To modify this substrate to determine directionality, the 91 base oligonucleotide is annealed to the M13 ssDNA as described above. Then, prior to labeling with [32P]dCMP, the substrate is digested with BspDI or ClaI which cleave once in the duplex region to produce a linear DNA with duplex ends and a long internal region of ssDNA. Cleavage with BspDI or ClaI leaves a 5′-CG overhang at each end of the linear DNA molecule. The overhangs, along with the 3′-end of the oligonucleotide annealed to produce the duplex region, can then be filled in or extended using the Klenow fragment of DNA polymerase I in the presence of dGTP and [α-32P]dCTP to produce a molecule with a radioactive label on all three of the 3′-ends. The duplex region of the substrate is cut to produce a 52 bp duplex region at one end of the linear molecule and a 43 bp duplex region at the other end of the linear molecule (Fig 4). This substrate can be used to assess the directionality of the helicase – if the helicase has 3′ to 5′ directionality it will displace the 52 base oligonucleotide, and if it has 5′ to 3′ directionality it will displace the 43 base oligonucleotide.
Figure 4. Substrate for determining the directionality of a helicase.
A 91-mer oligonucleotide is annealed to M13 ssDNA as described in the text. This partial duplex DNA is digested with BspDI to generate a linear DNA molecule with double stranded regions of different length at either end as shown. The overhangs at the end of the DNA are not drawn to actual size. The resulting overhang ends and the 3′-end of the original oligonucleotide are filled in or extended using [32P]dCTP, dGTP and the Klenow fragment of DNA polymerase I to produce the labeled DNA molecule shown.
Using these simple DNA substrates it is possible to begin to determine many properties of a particular helicase. Directionality is a very important property of a helicase to determine. Furthermore, depending on the processivity of the helicase in question, the small 93 base pair partial duplex or the large plasmid based gapped substrate can help answer questions regarding the unwinding parameters of the helicase in question.
SUMMARY
The current dogma that large or eukaryotic proteins cannot be expressed in E. coli is an over generalization. By experimenting with different media conditions and temperatures we have successfully expressed and purified several prokaryotic and eukaryotic helicases in E. coli (Table 1). In addition, the use of the double affinity tag system we describe here has made it possible to obtain very pure protein quickly, compared to previous methods. The advantage of obtaining pure protein quickly goes beyond saving time spent on purification protocols, it also increases the specific activity of the purified protein.
Another significant and unique advantage of this double affinity tag system versus other systems is that it allows for almost complete removal of the affinity tags without the use of proteases. In other double affinity tag systems like the TAP tag system, not all tags are removed and the removal of the first tag requires protease treatment [47]. In our tandem affinity tag system the ability to remove both tags allows the study of the protein in near native form making this a novel approach to protein purification compared to other double affinity tag systems.
Once a pure and active helicase has been obtained there are a variety of DNA substrates that can be used to establish the polarity of unwinding and other DNA unwinding characteristics. Though not discussed here, synthetic oligonucleotides can also be used to create small substrates or substrates with unique structures such as forks, bubbles, and Holliday junctions.
Acknowledgments
ROLE OF FUNDING SOURCE: This investigation was supported by National Institutes of Health grants GM33476 and GM80599 to SWM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdel-Monem M, Hoffmann-Berling H. Eur J Biochem. 1976;65:431–440. doi: 10.1111/j.1432-1033.1976.tb10358.x. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Monem M, Durwald H, Hoffmann-Berling H. Eur J Biochem. 1976;65:441–449. doi: 10.1111/j.1432-1033.1976.tb10359.x. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn B, Abdel-Monem M, Krell H, Hoffmann-Berling H. J Biol Chem. 1979;254:11343–11350. [PubMed] [Google Scholar]
- 4.Matson SW, Bean DW, George JW. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 5.Lohman TM, Bjornson KP. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 6.Singleton MR, Dillingham MS, Wigley DB. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 7.Flores-Rozas H, Hurwitz J. J Biol Chem. 1993;268:21372–21383. [PubMed] [Google Scholar]
- 8.Deschavanne PJ, Harosh I. Mol Microbiol. 1993;7:831–835. doi: 10.1111/j.1365-2958.1993.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 9.Schmid SR, Linder P. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 10.Brennan CA, Dombroski AJ, Platt T. Cell. 1987;48:945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- 11.Ray BK, Lawson TG, Kramer JC, Cladaras MH, Grifo JA, Abramson RD, et al. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 12.Scheffner M, Knippers R, Stahl H. Eur J Biochem. 1991;195:49–54. doi: 10.1111/j.1432-1033.1991.tb15674.x. [DOI] [PubMed] [Google Scholar]
- 13.Jankowsky E, Fairman ME. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Matson SW, Kaiser-Rogers KA. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- 15.Matson SW. Prog Nucleic Acid Res Mol Biol. 1991;40:289–326. doi: 10.1016/s0079-6603(08)60845-4. [DOI] [PubMed] [Google Scholar]
- 16.Barry J, Alberts B. J Biol Chem. 1994;269:33063–33068. [PubMed] [Google Scholar]
- 17.LeBowitz JH, McMacken R. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 18.Husain I, Van Houten B, Thomas DC, Abdel-Monem M, Sancar A. Proc Natl Acad Sci USA. 1985;82:6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lahue RS, Au KG, Modrich P. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi M, Dao V, Modrich P. J Biol Chem. 1998;273:9197–9201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- 21.Lohman TM. Mol Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 22.Wong I, Chao KL, Bujalowski W, Lohman TM. J Biol Chem. 1992;267:7596–7610. [PubMed] [Google Scholar]
- 23.Lohman TM. J Biol Chem. 1993;268:2269–2272. [PubMed] [Google Scholar]
- 24.Amaratunga M, Lohman TM. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 25.Hingorani MM, Washington MT, Moore KC, Patel SS. Proc Natl Acad Sci USA. 1997;94:5012–5017. doi: 10.1073/pnas.94.10.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Brabant AJ, Stan R, Ellis NA. Annu Rev Genomics Hum Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 27.Brosh RM, Jr, Bohr VA. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, et al. DNA Repair (Amst) 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Sung P, Bailly V, Weber C, Thompson LH, Prakash L, Prakash S. Nature. 1993;365:852–855. doi: 10.1038/365852a0. [DOI] [PubMed] [Google Scholar]
- 30.Hwang JR, Moncollin V, Vermeulen W, Seroz T, van Vuuren H, Hoeijmakers JH, et al. J Biol Chem. 1996;271:15898–15904. doi: 10.1074/jbc.271.27.15898. [DOI] [PubMed] [Google Scholar]
- 31.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, et al. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, et al. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, et al. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 34.Ellis NA, German J. Hum Mol Genet. 1996;5:1457–1463. doi: 10.1093/hmg/5.supplement_1.1457. [DOI] [PubMed] [Google Scholar]
- 35.Cheok CF, Bachrati CZ, Chan KL, Ralf C, Wu L, Hickson ID. Biochem Soc Trans. 2005;33:1456–1459. doi: 10.1042/BST0331456. [DOI] [PubMed] [Google Scholar]
- 36.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 37.Martin GM. Birth Defects Orig Artic Ser. 1978;14:5–39. [PubMed] [Google Scholar]
- 38.Oshima J, Brown WT, Martin GM. Lancet. 1996;348:1106. doi: 10.1016/S0140-6736(05)64456-X. [DOI] [PubMed] [Google Scholar]
- 39.Larizza L, Magnani I, Roversi G. Cancer Lett. 2006;232:107–120. doi: 10.1016/j.canlet.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 40.Kellermayer R. Genet Med. 2006;8:213–216. doi: 10.1097/01.gim.0000214457.58378.1a. [DOI] [PubMed] [Google Scholar]
- 41.Studier FW. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Matson SW, Sampson JK, Byrd DR. J Biol Chem. 2001;276:2372–2379. doi: 10.1074/jbc.M008728200. [DOI] [PubMed] [Google Scholar]
- 43.Monroe DS, Jr, Leitzel AK, Klein HL, Matson SW. Yeast. 2005;22:1269–1286. doi: 10.1002/yea.1313. [DOI] [PubMed] [Google Scholar]
- 44.Sikora B, Eoff RL, Matson SW, Raney KD. J Biol Chem. 2006;281:36110–36116. doi: 10.1074/jbc.M604412200. [DOI] [PubMed] [Google Scholar]
- 45.Cadman CJ, Matson SW, McGlynn P. J Mol Biol. 2006;362:18–25. doi: 10.1016/j.jmb.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 46.Vindigni A. Mol Biosyst. 2007;3:266–274. doi: 10.1039/b616145f. [DOI] [PubMed] [Google Scholar]
- 47.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. Nat Biotech. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]