Abstract
The abnormally folded form of the prion protein (PrPSc) accumulating in nervous and lymphoid tissues of prion-infected individuals can be naturally cleaved to generate a N-terminal-truncated fragment called C2. Information about the identity of the cellular proteases involved in this process and its possible role in prion biology has remained limited and controversial. We investigated PrPSc N-terminal trimming in different cell lines and primary cultured nerve cells, and in the brain and spleen tissue from transgenic mice infected by ovine and mouse prions. We found the following: (i) the full-length to C2 ratio varies considerably depending on the infected cell or tissue. Thus, in primary neurons and brain tissue, PrPSc accumulated predominantly as untrimmed species, whereas efficient trimming occurred in Rov and MovS cells, and in spleen tissue. (ii) Although C2 is generally considered to be the counterpart of the PrPSc proteinase K-resistant core, the N termini of the fragments cleaved in vivo and in vitro can actually differ, as evidenced by a different reactivity toward the Pc248 anti-octarepeat antibody. (iii) In lysosome-impaired cells, the ratio of full-length versus C2 species dramatically increased, yet efficient prion propagation could occur. Moreover, cathepsin but not calpain inhibitors markedly inhibited C2 formation, and in vitro cleavage by cathepsins B and L produced PrPSc fragments lacking the Pc248 epitope, strongly arguing for the primary involvement of acidic hydrolases of the endolysosomal compartment. These findings have implications on the molecular analysis of PrPSc and cell pathogenesis of prion infection.
Keywords: Metals/Copper, Neurobiology/Neuroscience, Prions, Protease/Inhibitor, Protein/Processing, Subcellular Organelles/Lysosomes, Tissue/Organ Systems/Spleen
Introduction
Prions are the infectious agent of transmissible spongiform encephalopathies (TSE),4 a group of fatal neurodegenerative disorders that include scrapie in sheep, bovine spongiform encephalopathie in cattle, and Creutzfeldt-Jakob disease in humans. The pathogenesis of these diseases is crucially linked to the cellular prion protein (PrPC) (1), a host-encoded glycoprotein attached to the membrane by a glycosylphosphatidylinositol (GPI) anchor, whose normal function is uncertain (2). Infected individuals accumulate an abnormal form of this protein (PrPSc), principally in their nervous and lymphoid tissues (3). Conversion of PrPC into PrPSc seems to take place at the cell surface or along the endocytic pathway. It involves a profound conformational change, leading to the acquisition of new properties such as insolubility in nondenaturing detergent, a strong tendency to aggregate, and an increased resistance to protease digestion, properties that are commonly used to distinguish the two PrP isoforms. Incubation of TSE-infected tissue homogenate with proteinase K (PK) in conditions that completely degrade PrPC generates N-terminal-truncated fragments of PrPSc, referred to as PrPres. Molecular analysis of the relative mass and glycoform ratio of these fragments allows the categorization of the clinicopathological heterogeneous TSE affecting humans and animals (4, 5). This phenotypic diversity is due to the existence of multiple strains of prion in conjunction with host genetic factors including prnp gene polymorphism or mutation. Variation in structural organization of PrPSc within multimers is thought to underlie prion strain diversity. These strain-specific, conformational differences in turn lead to exposure of distinct cleavage sites for PK.
Proteolytic processing of PrPSc has been shown to occur both in brain tissue and cultured cells. A well recognized event is N-terminal truncation leading to the production of PrPSc species commonly referred to as C2, possibly a step toward its complete degradation. Cleavage to produce C2 takes place within the unstructured region of the molecule, distal to the tandem array of octarepeats, and upstream of the physiological cleavage site of PrPC (position 111–112, human numbering) leading to a fragment called C1 (6). C2 is PK resistant and assumed to be the in vivo counterpart of the C-proximal fragment generated by PK digestion of full-length PrPSc. The presence of the N-terminal-truncated PrPSc species in infected brain tissue has been reported in naturally affected species, humans and sheep (6–9), as well as in mouse and hamster models (10–14). Immunohistochemical studies in sheep combined with PrP peptide mapping have demonstrated the intracellular accumulation in the brain and lymphoid tissues of the various N-terminal-truncated PrPSc species, some of which might correspond to C2 (15, 16). More recently, a region-specific deposition of C2 fragments was reported in sheep brain (9). Altogether these findings provide some evidence that the endocellular processing of PrPSc can be influenced by the agent strain but also possibly by the cell or tissue where it propagates.
Cell culture systems steadily infected with prions offer a convenient system in which PrPSc processing can be studied. N-terminal-truncated, C2-like fragments present before any PK digestion have been observed to accumulate in several mouse cell models, including N2a, GT1, and SMB cell lines (12, 13, 17). This trimming can occur within a few hours after PrPSc acquires its protease resistance, as revealed by the use of metabolically labeled or more recently, tetracysteine-tagged PrP (12, 18). Although matrix metalloproteases have been ascribed a role in the generation of the C1 PrPC fragment (19), the identity and relative contribution of the cellular proteases acting in PrPSc processing is less clear. Treatments of cultures by lysosomotropic compounds such as NH4Cl have been reported to inhibit the generation of C2 cleavage products (12, 17), thus potentially involving hydrolases from the acidic endosomal cell compartment, a recognized site of PrPSc accumulation (20–22). Cysteine protease inhibitors have also been shown to affect PrPSc clearance in cell culture (23). One detailed study has led to the proposal that endoproteolytic C2 cleavage of PrPSc, and prion propagation, are calpain-dependent processes (13). Although lysosome inhibition appeared to prevent PrPSc trimming without any major effect on its biosynthesis (12, 17), cysteine protease inhibitors were shown to either increase (23), reduce (24) the PrPSc steady-state level, or leave it unaffected (18) depending on the cell model, thus raising the possibility that cysteine proteases may indirectly control PrPSc propagation.
In this study, we investigated the endogenous processing of PrPSc in various cell cultures and mouse tissues infected by the same TSE agent. We found that the proportion of N-terminal-truncated versus full-length molecules varies considerably depending on the cellular environment. This process, in which hydrolases from the acidic cell compartment, not calpain, appeared to be primarily involved, did not or only marginally affected prion formation in the cell culture. We also show that the N terminus of naturally trimmed PrPSc molecules can differ from those produced by PK digestion. Our findings bring new information on the natural processing of PrPSc molecules, which is important for prion cell biology and molecular characterization or subtyping of TSE agents.
EXPERIMENTAL PROCEDURES
Cell Culture
Rov cells (clone Rov9) and Rom cells are derived from the RK13 epithelial cell line and express the ovine or mouse PrP, respectively, in a doxycycline-dependent manner (25, 26). They were grown in Opti-MEM medium supplemented with 10% fetal calf serum (FCS), penicillin, and streptomycin, and split at a ¼ dilution once a week. MovS cells (clone MovS6) are Schwann cell-derived, immortalized cells isolated from tg338 mice constitutively expressing the VRQ allele of ovine PrP (27). The cells were grown in Opti-MEM medium supplemented with 10% FCS plus antibiotics and split once a week at dilution. CAD cells are issued from a clone of the CathA cell line, originally derived from mouse brain neurons (28). The cells were cultivated in Opti-MEM (Invitrogen) supplemented with 10% FCS, and split after mechanical resuspension twice a week at a or dilution. Primary cultures of cerebellar granule neurons (CGN) and astrocytes (CAS) were isolated from 6-day-old tg338 or C57Bl/6 mice as previously described (29, 30). For neuron cultures, isolated cells were seeded on poly-d-lysine-coated plates and cultivated in Dulbecco's modified Eagle's medium-glutamax I high glucose (Invitrogen) with 10% FCS, antibiotics, with the addition of 20 mm KCl plus N2 and B27 supplements (Invitrogen). The cultures were complemented weekly with 1 mg/ml of glucose and antimitotic agents, uridine and fluorodeoxyuridine (Sigma) were added to inhibit astrocyte proliferation. Astrocyte cultures were prepared as described above and left for 8 days. To eliminate the neuronal population, the cells were shifted to Dulbecco's modified Eagle's medium containing 10% FCS and antibiotics. The medium was changed weekly for 3 weeks.
Drugs and Antibodies
NH4Cl was purchased from Prolabo and leupeptin hydrochloride from Sigma. Calpain inhibitor III (or MDL 28170), cathepsin L inhibitor I, and cathepsin B inhibitor I were from Calbiochem (Merck, Darmstadt, Germany). The anti-PrP monoclonal antibody Pc248 directed to the octarepeat domain (54–92, sheep PrP numbering) was generated in our laboratory using sheep brain PrPC (31, 32); its epitope sequence was determined by Pepscan analysis of sequential nonapeptides (see supplemental Fig. S2). Other anti-PrP monoclonal antibodies were the following: 12B2 (epitope 93–97) (33), Sha31 (epitope 148–159) (34), and ICSM4 that specifically recognizes the nonglycosylated form of PrP (35). The rabbit polyclonal antibody 179-CT was directed against the C-terminal end of the ovine PrP (epitope 218–230) (36).
Prion Infection of Cell Cultures and Preparation of Lysates
Rov and MovS cells were infected with the 127S scrapie strain as previously described (25, 27) using 1% (w/v) brain homogenate of terminally ill tg338 mice (37). CGN and CAS primary cultures from tg338 mice were exposed to 0.01% homogenate of 127S-infected tg338 mouse brain for 3 days and the accumulation of PrPSc was determined after 3 weeks as previously described (29). CGN from C57Bl/6 mice were exposed to 0.01% homogenate of 139A or 22L-infected C57Bl/6 mouse brain (30). CAD cells were incubated for 1 or 2 days with 0.5% brain homogenate of C57Bl/6 mice infected with 22L, 139A, or Chandler strains of mouse-adapted scrapie as described (38). Scrapie-infected cells were analyzed from passages 2 to 12 or more after infection.
Transgenic Mice, in Vivo Infections, and Tissue Homogenate Preparation
tg338 mice, expressing the VRQ allele of ovine PrP (37), were infected by intracranial inoculation with the 127S scrapie strain. tga20 mice overexpressing mouse PrP were infected with the 139A strain. Brains and spleens of infected animals were harvested at the terminal stage of the disease and homogenized with a ribolyzer (Hybaid, Middlsex, UK) and adjusted to 20 and 10% (w/v) in medium containing 5% glucose, respectively.
Cell and Tissue Lysate Preparation
The cells were washed twice in phosphate-buffered saline and whole cell lysates were prepared in TL1 buffer (50 mm Tris-HCl, pH 7.4, 0.5% deoxycholate, 0.5% Triton X-100) or TNT buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100) supplemented with protease inhibitors (Roche Applied Science). Lysates were clarified by centrifugation at 1000 × g for 2 min before use or freezing. For analysis, brain and spleen homogenates were diluted in TL1 or TNT buffers. The protein concentration of cells and tissue lysates was determined by BCA (Pierce). The tissues were frozen immediately after removal to minimize spontaneous proteolysis and precautions were taken during the preparation of lysates.
PrPSc Sedimentation Experiment
250 to 500 μg of protein from cell or tissue lysates of control or infected samples were sedimented at 22,000 × g for 30 min. Supernatants were recovered and pellets were washed once with TL1 buffer and resolubilized in Laemmli sample buffer for SDS-PAGE analysis. In some experiments, pellets were further resuspended in TL1 buffer and treated with PK (see below) before denaturation with Laemmli sample buffer.
Cu2+-IMAC Hi-Trap Chromatography
The AKTA Purifier100 FPLC chromatographic system was used (GE Healthcare). A 1-ml Hi-Trap IMAC column (GE Healthcare) was charged with 0.2 m CuSO4. The column was equilibrated with TNT buffer containing 3 mm imidazole. One milliliter of cell (1 mg of protein) or 0.5 ml of brain lysates (0.75 mg of protein) were injected into the column. The flow-through fraction was recovered, the column was washed, and a 10-min linear gradient of 3–200 mm imidazole in TNT buffer was applied to elute column-bound proteins at a flow rate of 1 ml/min; 0.5-ml fractions were collected. As a final step, 6 m urea was used to remove tightly bound proteins. Column fractions were analyzed by Western blotting for PrP detection before and after PK treatment. Individual columns were dedicated to each type of lysate.
Proteinase K Digestion
Aliquots of 50 μl of whole cell lysates were treated with 10 μg/ml (10 μg/mg of protein) of PK at 37 °C for 1 h and denatured with Laemmli sample buffer. Mouse brain and spleen lysates were either treated with 50 μg/ml (10 μg/mg of protein) of PK at 37 °C for 1 h or PK digested according to the Bio-Rad test protocol (39). To analyze cell lysates fractionated on IMAC columns, 400 μl of eluted fractions were treated with 5 μg/ml of PK and then precipitated with cold methanol after addition of 20 μg of bovine serum albumin as a protein carrier. Methanol-precipitated proteins were sedimented at 22,000 × g for 15 min and the pellets were solubilized in 40 μl of 1× sample buffer. To analyze brain lysates, 50 μl of IMAC column fractions were digested with 10 μg/ml of PK for 1 h at 37 °C and then denatured with 4× sample buffer.
Thermolysin Treatment
Cell and brain lysates were treated with thermolysin (Sigma) as previously described (40) with slight modifications. Thermolysin was used at a concentration of 10 μg/ml (10 μg/mg protein) for cell lines and CGN, and at 200 μg/ml for brain lysates. After incubation at 37 °C for 1 h, samples were denatured with an equal volume of ×2 sample buffer.
Cathepsins Treatment
Sedimentation pellets of infected Rov9 cell lysates were solubilized in cathepsin digestion buffer containing (i) dibasic sodium phosphate brought to pH 6 or 5 with citric acid; (ii) l-cysteine (18 or 195 mm) for redox conditions mimicking that in slightly acidic or acidic compartments, respectively (41). Human liver cathepsin B and L (Merck) were used at 50 μg/ml overnight at 37 °C. Reactions were terminated by addition of an equal volume of 2× sample buffer.
Quantification of Deglycosylated PrPSc Species
Samples were treated with PNGase F according to the manufacturer's instructions (New England Biolabs) and quantification of full-length and C2 PrPSc bands was determined by GeneTools software after acquisition of chemiluminescent signals with a GeneGnome digital imager (Syngene).
Removal of GPI Anchor
Removal of the GPI anchor with hydrofluoric acid was performed according to a published procedure (42) with one critical modification. Briefly, 1 volume of crude lysates from uninfected brain or cell materials was mixed directly with 6 volumes of aqueous 48% hydrofluoric acid (Merck), incubated for 24 h at 4 °C, and then vacuum-dried material was solubilized in 2× SDS-PAGE sample buffer. PrPC was immunodetected using ICSM4 mAb, specific for the unglycosylated form.
Immunoblotting
Either 12 or 4–12% NuPage gels (Invitrogen) were used for SDS-PAGE. Transfer of proteins on nitrocellulose filters was performed using a semi-dry transblot system (Bio-Rad). For detection of PrP by immunoblotting, an enhanced chemiluminescence (ECL) detection system (Pierce or Roche) was used with goat anti-mouse IgG coupled to peroxidase as secondary antibodies.
RESULTS
PrPSc Processing Greatly Varies Depending on Cell and Tissue Types
We looked for possible quantitative and qualitative differences in the natural processing of PrPSc depending on which cells support the propagation of the prion. We first examined the molecular profile of the immunoreactive PrP species in different cell systems and mouse tissues infected by the same prion strain, 127S, a well characterized strain of sheep scrapie agent. The cell systems studied comprised previously described Rov and MovS cell lines (27, 43), primary CGN and astrocytes (CAS) derived from tg338 mice (29), and the tissues were whole brain and spleen homogenates from tg338 mice. All these materials are genetically engineered to express solely the same ovine PrPVRQ allotype. On immunoblot analysis, a typical profile, including a well individualized band of 26 kDa corresponding to unglycosylated PrPC, was observed in uninfected cell cultures and healthy tissues. In scrapie-infected Rov and MovS cells, additional PrP fragments shorter than 26 kDa were present, whose size matched those of PK-resistant PrPSc fragments (Fig. 1A). Centrifugation of cell lysates corroborated the presence of detergent-insoluble, truncated PrPSc, which was quantitatively recovered in the pellets, whereas normal PrPC did not sediment in these conditions. As expected, sedimented PrPSc was also PK-resistant (data not shown). Therefore, the bulk of PrPSc accumulating in Rov and MovS cells consisted of naturally truncated molecules, shown below to be bona fide C2 (i.e. N-terminal-truncated) fragments. Upon quantification after deglycosylation by PNGase the full-length (FL) species were found to represent only 5 and 15% of PrPSc accumulated in Rov and MovS cells, respectively (see Fig. 12 for an overview of the quantification data). Rov cultures were maintained in serum-free medium and passed without using trypsin for splitting accumulated truncated PrPSc-like cultures kept in standard conditions, arguing that the observed PrPSc processing was actually endogenous to cells rather than caused by exogenous proteases (data not shown).
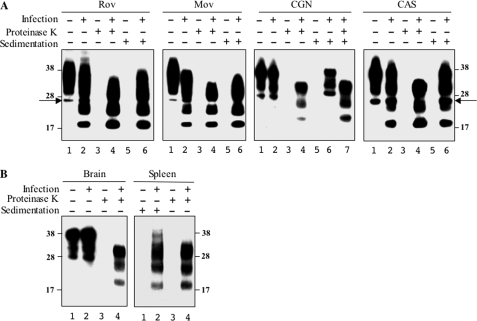
FIGURE 1.
PrPSc is truncated in Rov and MovS cells but remains in a full-length form in tg338 CGN and the brain. A, cultures of Rov and MovS cell lines, and primary neurons (CGN) and astrocytes (CAS) derived from tg338 mice were infected (lanes 2, 4, and 6) or not (lanes 1, 3, and 5) by the ovine prion 127S strain. Crude lysates (lanes 1 and 2), PK-treated (lanes 3 and 4), or sedimented materials (lanes 5–7) were analyzed by immunoblotting. Lane 7 in the CGN panel corresponds to PK digestion of the sedimented material shown in lane 6. PrPSc sedimented as the truncated species from Rov and MovS cells and as FL species from CGN cells. Both truncated and FL PrPSc were detected in CAS cells. B, analysis of PrPSc in the brain and spleen homogenates from 127S-infected tg338 mouse (lanes 2 and 4) before (lanes 1 and 2) and after PK treatment (lanes 3 and 4). Mostly FL PrPSc is detected in the brain, whereas spleen PrPSc mainly contains the truncated species (lanes 2). All immunoblots were revealed with Sha31 anti-PrP mAb. Equivalent protein amounts were loaded per lane for each sample, except for the sedimented spleen lysate for which 10-fold more material was needed to detect PrPSc. The molecular profiles shown have been observed in multiple experiments on multiple blots. The positions of molecular mass markers (in kDa) are shown on both sides of the figure for all gels. The arrows designate the PrP 26-kDa aglycosylated species.
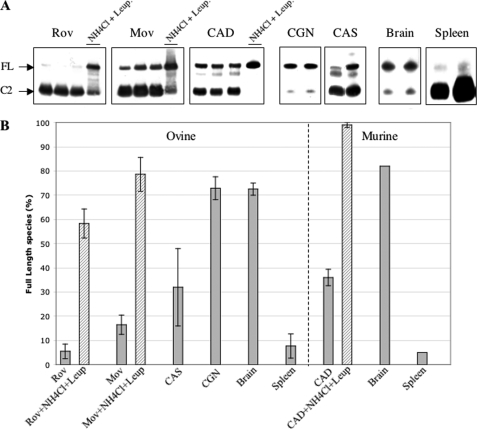
FIGURE 12.
Proportion of untrimmed PrPSc in infected cells and tissues. A, Western blot analysis (Sha31 mAb) of PNGase-treated samples: cell lines (Rov, Mov CAD), primary cultures of neurons (CGN) or astrocytes (CAS), and brain and spleen tissues. Each of the three left panels show three samples left untreated and one sample treated with lysosomal inhibitors (10 mm NH4Cl plus 10 μg/ml leupeptin); the four right panels each show two different samples. Sedimented material from lysates of cell lines and primary cultures were resuspended and treated with PNGase (similar results were obtained using samples that were digested with thermolysin before PNGase; not shown). Lysates from tg338 brain and spleen tissues were treated with thermolysin and further centrifuged in the case of spleen material to concentrate PrPSc. Arrows indicate deglycosylated full-length (FL) and N-terminal-truncated (C2) PrPSc species. B, the histogram shows the percentage of full-length species over total PrPSc as determined by quantification of samples such as seen in A. Average values and standard deviation were determined from four to seven different experiments.
In sharp contrast, in infected CGN cell cultures, fragments corresponding to the truncated PrPSc species were hardly detectable in whole cell lysates despite high PrPres levels (Fig. 1A). Only weak signals potentially corresponding to mono- and unglycosylated cleaved PrPSc were observed in overexposed blots (data not shown). Moreover, sedimentation pellets from infected CGN cells contained essentially FL PrPSc, which became truncated after PK digestion. The proportion of FL over total PrPSc averaged ∼75%, as determined after PNGase treatment (see Fig. 12). In CAS cell cultures, an intermediate situation was observed, in which both FL and truncated C2-like PrPSc molecules were detectable, with a predominance of the latter (Figs. 1B and 12). Immunoblot analysis of whole brain homogenates from terminally ill tg338 mice revealed that PrPSc accumulating in this tissue mostly consisted of FL species, i.e. a picture similar to that seen in CGN cells. This was not the case when spleen homogenates from the same mice were examined: indeed, the sedimented PrPSc material mainly consisted of the truncated PrP species (see Figs. 1B and 12). Collectively these data revealed major differences in terms of PrPSc processing both in different cultured cells and animal tissues.
PrPSc Molecules That Are N-terminal Truncated Predominate in Infected Cell Lines
As a means to further investigate the PrPSc processing occurring in our cell systems and confirm the sedimentation data, we first used thermolysin, a protease that allows the detection of intact PrPSc molecules while fully digesting PrPC (40). As shown in Fig. 2, the presence of thermolysin-resistant PrPSc with the same molecular mass as FL PrP was detected in both CGN cells and the tg338 mouse brain, whereas in Rov cells (and in MovS cells, not shown) PK- and thermolysin-resistant PrPSc displayed overlapping profiles.
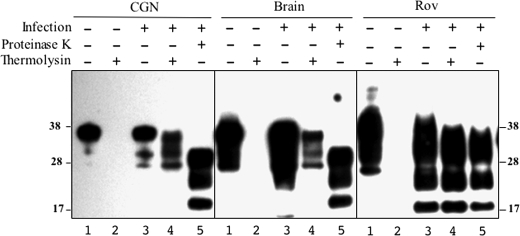
FIGURE 2.
CGN accumulate thermolysin-resistant, full-length PrPSc. Whole lysates from uninfected (lanes 1 and 2 of each panel) and 127S-infected (lanes 3–5) tg338 CGN, brain, and Rov cultures were digested with thermolysin (≥4 experiments) using conditions in which PrPC was completely proteolyzed (lanes 2 and 4). Immunoblots using the Sha31 mAb show that thermolysin-resistant PrPSc from CGN cells and the brain migrates with the mobility of FL species (lanes 4), not with PrPres generated by the PK treatment (lanes 5). In contrast, the profiles of thermolysin- and PK-resistant PrPSc from Rov cells are similar (compare lanes 4 and 5 in Rov panel). The positions of molecular mass markers are indicated.
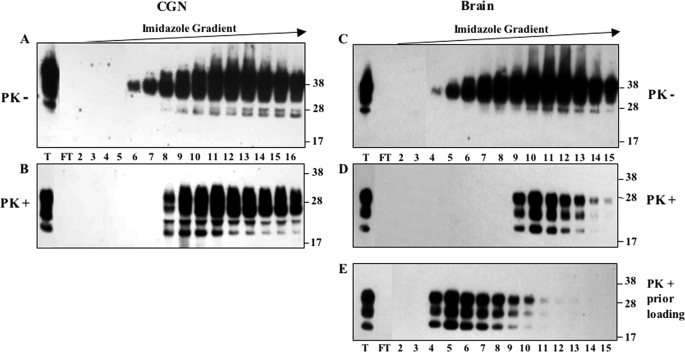
We next asked whether it would be feasible to physically separate the different PrPSc species, given the different binding activity of FL and 103–231 recombinant PrP molecules to immobilized copper ion (44). MovS cell PrPC was retained on an IMAC-Cu2+ column and recovered mainly in 60–80 mm imidazole-eluted fractions (Fig. 3A). In infected cells, PrP molecules including species with a molecular mass lower than 26 kDa were eluted at a lower imidazole concentration (Fig. 3B). Globally, these latter species were not recognized by an anti-octarepeat antibody (Fig. 3C) but were resistant to PK digestion (Fig. 3D). Similar results were obtained with lysates from infected Rov cells (supplemental Fig. S1). These results unambiguously demonstrate that most of the PrPSc accumulated in Rov and MovS cells was N-terminal-truncated before PK treatment. On the contrary, PrPSc produced in CGN cells was eluted from the IMAC-Cu2+ column mainly as FL PrP molecules, as assessed by PK digestion of the eluted fractions (Fig. 4, A and B).
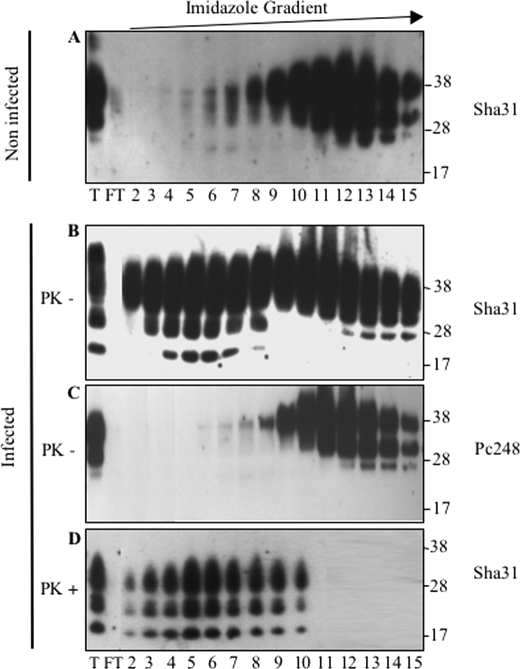
FIGURE 3.
Truncated PrPSc from MovS cells binds to the IMAC-Cu2+ column. Lysates of MovS cells (lane T) were loaded onto an IMAC-Cu2+ column. After recovery of unbound proteins (FT, flow-through), bound proteins were eluted by application of a 3–200 mm linear imidazole gradient and fractions (numbers 2–15) were analyzed by immunoblotting using either Sha31 mAb that recognizes the PrP core (A, B, and D) or anti-octarepeat Pc248 mAb (C). PrP elution profiles obtained with non-infected (A) and infected cells before (B and C) or after PK digestion of the eluted fractions (D) are shown. Pc248 mAb detects non-truncated PrP in infected cells (C), whereas Sha31 detects both truncated and FL molecules (B). PK digestion (D) proves that the population of truncated PrP corresponds to the bulk of PrPSc produced by MovS cells.
FIGURE 4.
PrPSc from CGN and the brain binds to the IMAC-Cu2+ column and elutes as full-length PrP. Total lysates (lane T) of 127S-infected CGN (A) or the tg338 brain (C) were applied to the IMAC-Cu2+-charged column. The flow-through fraction (FT) was collected and bound proteins were eluted by application of a 3–200 mm linear imidazole gradient and analyzed by immunoblotting for the presence of PrP molecules before (A and C) and after PK treatment (B and D). In E, the lysate of the infected brain was first digested with PK prior to loading onto the column and the eluted fractions were analyzed. Immunoblots were revealed with Sha31 mAb. The binding and elution of FL PrPC and PrPSc produced in CGN and in the brain are similar (compare A to B and C to D). PK-digested brain PrPSc (E) is also retained on a copper column but elutes at lower imidazole concentrations than FL PrPSc (compare D to E). The positions of molecular mass markers are shown on the right of each immunoblot.
On analyzing infected brain homogenates, we found that FL PrPSc binds to copper with an avidity close to that of PrPC (Fig. 4, C and D). Brain samples treated with PK before loading also led to an efficient retention of the PrPSc PK-resistant core species on the column, although their elution occurred at a lower imidazole concentration than PrPC or FL PrPSc (Fig. 4E), as also shown for naturally truncated PrPSc accumulating in Rov and MovS cells (see Fig. 3 and supplemental S1). Altogether, these results fully corroborated the difference of processing PrPSc in cell lines versus primary nerve cells and brain tissue.
PrPSc Endogenous Trimming Involves Amino Acids Downstream of the Main PK Cleavage Sites
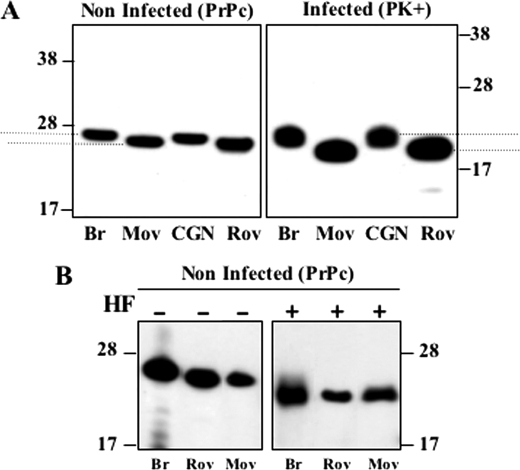
As noticeable on the immunoblot shown in Fig. 1, the electrophoretic mobility of PK-resistant fragments in cell culture and tissue appeared to differ, although the infecting prion was the same. To clarify this point, relevant samples were run on the same gels and immunoblots were performed using ICSM4 antibody specific for unglycosylated PrP. These experiments confirmed the slower mobility of the PK-resistant fragments from CGN cells and brain tissue compared with Rov and MovS cells (Fig. 5A). A similarly slower migration of PrPres fragments from the brain versus spleen tissue was also reproducibly observed (see Fig. 1B). These experiments also revealed a variation in the mobility of PrPC in corresponding, non-infected samples. To determine which cell-specific, post-translational modification of PrPC was involved, lysates from uninfected cultures or brain tissue were treated with hydrofluoric acid to remove the GPI moiety (Fig. 5B). Because after this treatment PrPC-unglycosylated bands migrated uniformly, it was concluded that a variation in the composition of the GPI anchor is mainly responsible for the observed mobility differences of PrPC.
FIGURE 5.
Higher electrophoretic mobility of PrPC and PrPres from MovS and Rov cells compared with CGN or brain tissue. A, the comparative mobility of PK-resistant PrPSc (right panel) and PrPC (left panel) from tg338 mouse brain, and MovS, CGN, and Rov lysates is shown on immunoblots revealed with ICSM4, an antibody specific for the non-glycosylated form of PrP. To compare the mobility differences accurately, in these experiments the runs were performed so that the migration distances of unglycosylated species PrPC and PrPSc were the same. B, treatment with hydrofluoric acid (HF) of non-infected brain tissue, Rov, and MovS cells generates a PrP species with identical mobility (immunodetection with ICSM4). The electrophoretic mobility differences shown were observed in three (B) or more (A) independent experiments.
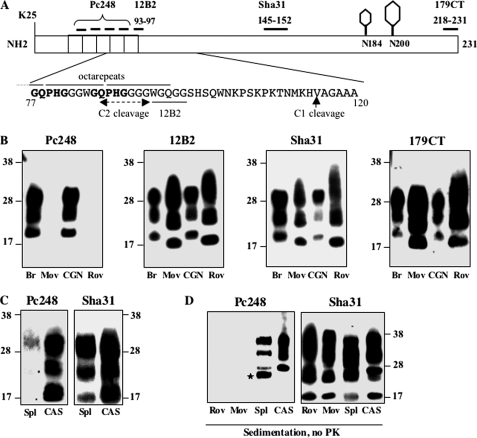
However, based on the relative mobility we calculated that the difference between PK-resistant fragments slightly exceeded that between PrPC species (i.e. 2.1 versus 1.0 kDa, respectively, data not shown), thus suggesting that the observed differential mobility was not ascribable solely to PrPC. Relevant monoclonal antibodies were used to probe epitopes of PrPSc present in the different cell cultures and tissues after PK digestion, and typical results are presented in Fig. 6. Whereas antibodies 12B2 (position 93–97 in sheep sequence) and CT179 (position 218–231) both produced strong signals in all cases, clear differences were observed with antibody Pc248. This antibody, previously described by our laboratory (32, 44), binds PrP within the octarepeat region with a very high avidity (epitope mapping and comparison with other anti-octarepeat mAbs are shown in supplemental Fig. S2). PK-resistant fragments from Rov and MovS cells were not recognized by Pc248, contrary to those from primary neurons or brain tissues (Fig. 6B), thus providing clear evidence that an octarepeat motif, still present in CGN cells and brain PrPSc after PK digestion, was lost in these cell lines. PK-resistant fragments from CAS exhibited some reactivity to Pc248 (Fig. 6C). When immunoprobing was performed on sedimented, non-PK-digested materials (Fig. 6D), CAS cell lysates were shown to contain a mixture of Pc248-negative and -positive (FL) PrPSc molecules, whereas in spleen homogenates FL PrPSc (Pc248-positive) was hardly detectable. Rov and MovS sedimented, undigested material failed to react with Pc248, confirming that a nearly exhaustive trimming of the PrPSc molecules rather than PK digestion accounted for loss of the Pc248 epitope in these cells. Altogether, these results established that N-terminal trimming of PrPSc molecules generated by ovine prion produces PrP fragments distinguishable from those resulting from the exogenous PK cleavage of FL PrPSc.
FIGURE 6.
N-terminal trimming of ovine PrPSc produced in Rov, MovS, CAS cells and in the spleen occurs downstream of the main PK cleavage sites. A, schematic diagram of ovine PrPC indicating the two glycosylation sites (N184 and N200, sheep numbering) and the location of epitopes used for mapping. The deduced region of cleavage to produce C2 (dashed line) is within the Pc248 epitope or just upstream of the 12B2 epitope (underlined), as indicated on the highlighted amino acid sequence. The Pc248 epitope is indicated in bold. B, PK-digested PrPSc from infected materials was tested for reactivity to the four antibodies indicated on the scheme. Only the anti-octarepeat mAb Pc248 discriminates PrPSc produced in CGN and the brain from PrPSc accumulated in Rov and MovS cells. C, PK-digested PrPSc from the spleen (Spl) of infected tg338 mice and from infected CAS was analyzed by immunoblotting with both Pc248 and Sha31 mAbs. D, sedimented, and non-PK-treated materials from infected Rov, MovS, spleen, and CAS were immunoblotted with Pc248 and Sha31 mAbs. The positions of molecular mass markers are indicated. The star on panel D points to a nonspecific band detected with Pc248 in the spleen-sedimented sample.
Differential PrPSc Trimming Occurs in Mouse Prion-infected Cells and Tissues
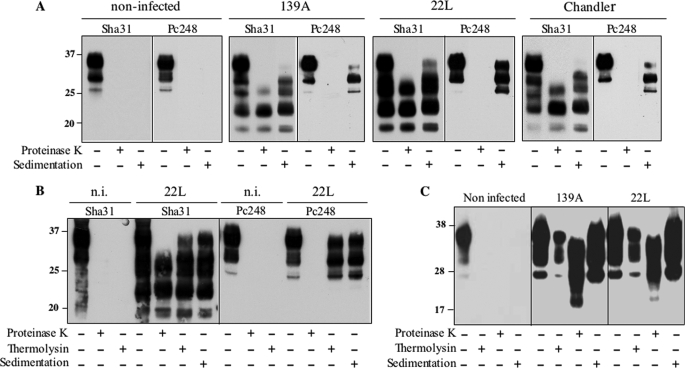
To see whether our findings would extend to other than ovine prions, we next focused our study on various models propagating mouse prions. The CAD cell line originating from brain tissue displays several features of neurons (28, 38). CAD cultures steadily infected by 139A, 22L, or Chandler prion strains were shown to produce naturally N-terminal-truncated, Pc248-negative PrPSc, which was predominantly monoglycosylated as is typically the case for PK-resistant PrPSc for these strains (Fig. 7A). Additional species of lower mobility were observed in sedimented material using Sha31 antibody (Fig. 7A), suggesting accumulation of FL PrPSc too. Indeed, sedimented, non-PK treated samples contained Pc248-reactive, FL PrPSc material, with predominant monoglycosylated forms, and Pc248-reactive species were resistant to thermolysin digestion (Fig. 7B). The full-length form represented around one-third of PrPSc in these cells (see Fig. 12). Infected cultures of Rom cells, similar to Rov but genetically engineered to express mouse instead of ovine PrP (26), were shown to accumulate PrPSc essentially under the C2, Pc248-negative form (supplemental Fig. 3), as did the Rov cells. In contrast, truncated species were hardly detectable in 139A- or 22L-infected, primary cultured CGN mouse neurons (Fig. 7C).
FIGURE 7.
Mouse PrPSc is truncated in CAD cells but remains full-length in CGN. A, CAD cells were infected with mouse-adapted scrapie prion (139A, 22L, or Chandler strain) or mock-infected and analyzed for their PrPC and PrPSc content. Immunoreactive PrP species were revealed in whole cell extracts, PK-digested and sedimented materials, as indicated, using either Sha31 or Pc248 mAbs. Note that both truncated and FL, Pc248-positive PrP species are recovered in sedimented material from infected samples, whatever the strain. B, whole cell lysate of 22L- or mock-infected CAD cells and thermolysin- or PK-digested samples were analyzed using Sha31 or Pc248 antibody. Note the Pc248-positive bands in thermolysin-resistant and sedimented materials, reflecting the presence of FL PrPSc. C, CGN from C57Bl/6 mice were mock-infected or infected with 139A or 22L scrapie strains. Cell lysates performed 24 days post-infection were centrifuged or digested either with thermolysin or PK as indicated, and analyzed by Western blot using Sha31 mAb. The molecular profiles have been observed in multiple experiments on multiple blots. n.i., non-infected.
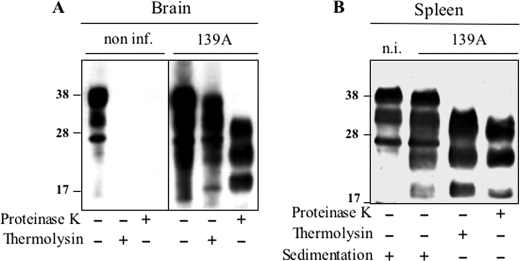
In tissues of 139A-infected tga20 mice, PrPSc was shown to accumulate as a FL species in a great majority in the brain (∼80%), whereas it appeared to be extensively trimmed in the spleen (Figs. 8 and 12), i.e. a situation similar to that seen in tg338 mice. Altogether these results indicated that the phenomenon of differential trimming is not a unique feature of ovine prions.
FIGURE 8.
Mouse PrPSc is efficiently truncated in the spleen but not the brain. Analysis of PrPSc in brain (A) and spleen (B) homogenates from non-infected or 139A-infected tga20 mice. Tissue lysates (from at least three animals) were centrifuged (spleen only) or digested either by thermolysin or PK as indicated, and analyzed by Western blot using Sha31 mAb. PrPSc detected in the spleen is essentially truncated prior to PK digestion (lane 3). Note the presence of sedimentable PrPC in both infected and non-infected tga20 mouse spleen, which was consistently observed with mouse PrPC in these conditions. n.i., non-infected.
Inhibition of Lysosomal Hydrolases but Not of Calpains Restores Accumulation of Full-length PrPSc
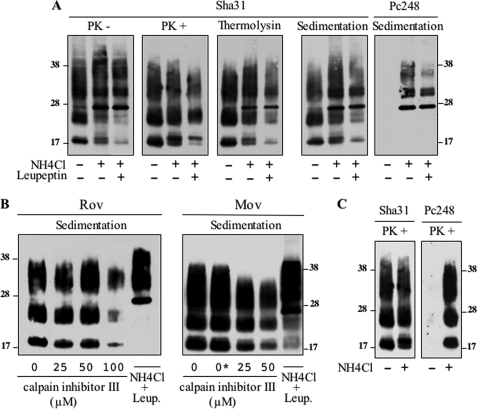
To test whether the endolysosomal compartment was involved in trimming of PrPSc in the cell systems studied here, we used NH4Cl, a compound that inhibits protease activity by raising the pH in this compartment, and/or leupeptin, a drug commonly used to impair lysosomal hydrolases. Treatment of infected Rov cultures modified the PrP immunoreactive profile, with a decrease of the C2 species and a concomitant increase of the FL PrP species, obvious for the unglycosylated band; overall, the accumulation of PrPSc did not decrease or decreased only slightly (Fig. 9A, left two panels). Accumulation of FL PrPSc in the lysosome-impaired culture was further substantiated by detection of thermolysin-resistant, detergent-insoluble, and Pc248-reactive PrP species (Fig. 9A, right three panels). Under lysosome inhibition the proportion of FL PrPSc increased from 5 to 58% and from 15 to 78% in Rov and MovS cells, respectively (see Fig. 12). Intriguingly, in a recent study, PrPSc trimming was reported to be impaired following inhibition of calpains, cysteine proteases that are not associated with the endolysosomal compartment, whereas inhibition of lysosomal proteases was ineffective (13). However, in neither Rov nor MovS cell cultures did treatment with ≥50 μm calpain inhibitor III (MDL28170) (13) affect PrPSc N-terminal trimming; at the highest dose, the inhibitor decreased the accumulation of PrPSc, yet the C2/FL ratio, as measured on sedimented material, remained unchanged (Fig. 9B).
FIGURE 9.
N-terminal trimming of PrPSc is inhibited in lysosome-impaired Rov and MovS cells but is not calpain-dependent. A, infected Rov cells were left untreated or cultivated for 7 days in the presence of 10 mm NH4Cl or 10 mm NH4Cl + 100 μg/ml of leupeptin, as indicated. The PrP profiles of undigested, PK-digested, and thermolysin-digested samples are shown after Sha31 mAb immunoblotting. Lysates were also sedimented without protease treatment and the pellets were revealed with either Sha31 or Pc248 mAbs (right two panels). B, infected Rov and MovS cells were treated with various concentrations of calpain inhibitor III or NH4Cl plus leupeptin, as indicated (0*: vehicle only). Sedimented materials from cell lysates were analyzed by Western blotting with Sha31 mAb. C, infected Rov cells treated or not with NH4Cl were digested with PK before immunoblot analysis using Sha31 or Pc248 mAbs. The above experiments have been repeated three times with consistent results.
Importantly, PrPres fragments generated by PK digestion of lysates from NH4Cl-treated cultures were recognized by Pc248 antibody, unlike that in control, untreated cultures (Fig. 9C). Therefore, PK-resistant fragments produced by digestion of FL PrPSc generated in endolysosome-impaired cells contained an octarepeat motif.
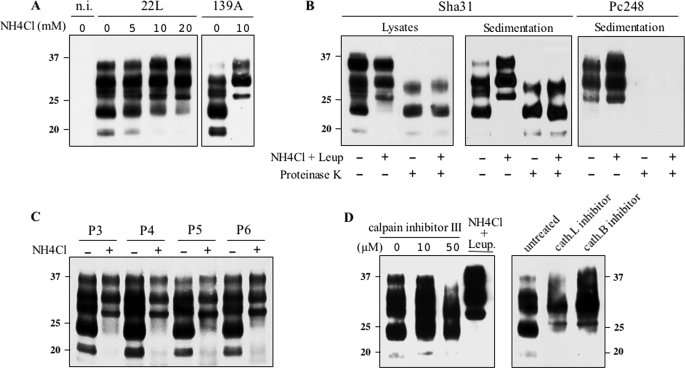
Similar experiments were performed in mouse prion-infected cells. Following treatment with NH4Cl, the proportion of FL PrPSc in 22L-infected cultures increased in a dose-dependent manner, and a 10 mm treatment led to the accumulation of an exclusively FL species in 139A-infected cells (Fig. 10A). When 22L-infected cells were cultivated for one passage in the presence of NH4Cl plus leupeptin, truncated PrPSc disappeared almost completely (Figs. 10, B and D, and 12); such cultures accumulated as much PK-resistant PrP as untreated cultures, implying that FL and truncated PrPSc had a similar resistance to PK digestion (Fig. 10B). Remarkably, blockade of N-terminal trimming was compatible with serial propagation of prion infection because the level of PrPSc remained fairly stable under sustained treatment over at least 6 passages (Fig. 10C). Upon discontinuation of the treatment, production of truncated forms resumed to its original level within one passage (supplemental Fig. S4). In CAD cell cultures treated with calpain inhibitor III the accumulation of C2 fragments was diminished, but again without an increase of the FL versus C2 species ratio (Fig. 10D, left panel). In contrast, inhibition of cathepsin L or B led to an increase of the FL species at the expense of trimmed fragments (Fig. 10D, right panel). Altogether, these results led us to conclude that the N-terminal trimming of PrPSc in ovine and mouse prion-infected cells primarily involves proteases other than calpains.
FIGURE 10.
Lysosome inhibition blocks PrPSc N-terminal trimming but not prion propagation in CAD cells. A, non-infected, 22L- and 139A-infected cells were either left untreated or treated with NH4Cl as indicated, and cell lysates were centrifuged. The pellets were analyzed for their PrPSc content by immunoblotting using Sha31 mAb. B, 22L-infected CAD cells cultivated in the absence or presence of 10 mm NH4Cl plus 10 μg/ml of leupeptin for two consecutive passages. Whole cell lysates and sedimented materials (right two panels) were PK-digested or not before immunoblotting using Sha31 or Pc248 mAb. C, 22L-infected CAD cells were cultivated in the absence or presence of 10 mm NH4Cl for 4 days and cells were passaged twice a week in the absence or presence of the lysosomal inhibitor. Cell lysates were immunoblotted using Sha31 mAb. Molecular mass markers in kDa are shown. D, infected CAD cells were treated with various concentrations of calpain inhibitor III, NH4Cl plus leupeptin, cathepsin L inhibitor I (cath.L), or cathepsin B inhibitor I (cath.B), as indicated. Sedimented materials from cell lysates were analyzed by Western blotting with Sha31 mAb. These experiments have been repeated three times with consistent results. n.i., non-infected.
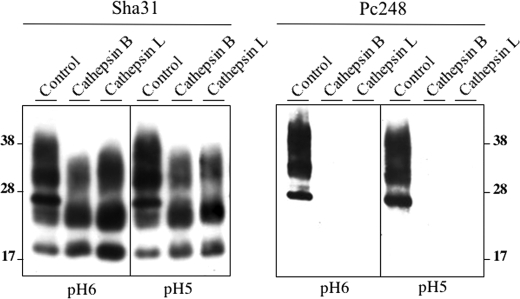
To further substantiate the nature of the enzymes involved in generation of the C2 fragment, FL PrPSc obtained following NH4Cl plus leupeptin treatment of infected Rov cells was subjected to in vitro digestion by cathepsin B or L, under two different buffer conditions; one mimicking the slightly acidic endosomal compartment (pH 6) and the other the acidic lysosomal compartment (pH 5) (Fig. 11) according to Jordans et al. (41). In both cases cleaved PrPSc fragments were obtained that showed a typical profile but lacked Pc248 reactivity (Fig. 11), as in Rov cell cultures whose endolysosomal compartment was unimpaired (see above).
FIGURE 11.
In vitro cleavage of full-length PrPSc by cathepsin B and L remove the Pc248 epitope. PrPSc from a lysate of infected Rov cells treated with NH4Cl and leupeptin was purified by sedimentation, resuspended in two different cathepsin buffers (pH 6 or 5), and digested or not (control lanes) with the indicated proteases as described under “Experimental Procedures.” The same samples were revealed with either Sha31 or Pc248 mAbs.
In the mouse prion cell systems, CAD (Fig. 10B) and Rom cells (supplemental Fig. S3), endogenously trimmed, and PK-cleaved PrPSc fragments exhibited the same mobility. Indeed, contrary to that seen in ovine prion-infected cells, PrPres fragments produced by PK digestion of FL PrPSc from lysosome-impaired cells showed no reactivity toward Pc248 antibody (Fig 10B). Similarly, PrPres in brain tissues from mice infected by 22L or other mouse prions, which mostly accumulate FL PrPSc, lacked Pc248 reactivity (data not shown). From these results it appears that, even though the main PK cleavage sites of sheep and mouse PrPSc differed, endogenous proteolytic cleavage generated C2 fragments with similar N termini, despite different cell types and PrP sequences.
DISCUSSION
The present study was focused on the natural processing of PrPSc leading to N-terminal-trimmed fragments commonly called C2. Several new findings emerged that will be discussed successively below.
Our study first revealed that the efficiency of trimming varies dramatically depending on which cell or tissue supported replication of the infecting prion (see Fig. 12). Thus, in cell systems infectible by sheep prion, the epithelial Rov and Schwann MovS cell lines, the bulk of PrPSc consisted of molecules that were N-terminal-truncated prior to PK digestion. In primary cultured cells, PrPSc produced by neurons was essentially full-length, whereas in astrocytes an important proportion was trimmed. A contrasted situation was also observed in vivo (tg338 mouse model), where intact PrPSc molecules largely predominated in the brain, although they were minimally represented in the spleen. The above mentioned materials all expressed the same ovine prnp allele and were infected by the same, biologically cloned sheep prion, therefore excluding any sequence or strain-dependent conformational effect. Examination of models propagating mouse-adapted prions led to similar conclusions, indicating that this may be a general phenomenon. Although in CAD neuronal cells, the proportion of C2 over FL molecules averaged two-thirds whatever the infecting prion strain, limited trimming occurred in primary cultured neurons. In the brain and spleen tissues of tga20 mice the FL/C2 ratios were roughly inverse, as seen in ovine PrP mice. It is reasonable to anticipate that such a markedly tissue-dependent, differential PrPSc trimming also exists in species naturally infected by prions.
Conceivably the endogenous cleavage of PrPSc, and more specifically the generation of C2 fragments, might differ quantitatively between different nerve cell categories within the brain. Accumulation of C2 fragments in the brain tissue has been reported to occur in rodent models other than mice (13, 14), as well as in naturally affected host sheep (9) and humans (6, 45). Moreover, several studies have offered evidence potentially linking the C2 accumulation level in the prion-affected mouse, sheep, and human brains or its neuroanatomical deposition (7, 9, 14, 45) to TSE strain variation. Our observation that PrPSc trimming efficiency in primary cultured astrocytes and neurons markedly differed, in particular, supports the view that certain nerve cell subpopulations might be more prone to endogenous proteolysis than others. Thus, the ratio of C2/FL PrPSc species in the brain tissue could also reflect the cellular tropism or “targeting properties” of a prion, rather than the solely conformational specificities of PrPSc. In future studies, it would be interesting to determine whether generation of the C2 fragments occurs preferentially in specific cell types within the central nervous system, and whether or not “high C2-producing” strains share common biological features.
It is generally considered that the C2 fragments are the in vivo counterparts of the protease-resistant fragments produced by PK digestion in vitro (6, 11, 13). However, the findings presented here do not support this view. As revealed by antibody mapping, C2 fragments consistently failed to react with the potent anti-octarepeat antibody Pc248, whatever the infecting prion. To be underlined, all the agents studied here exhibited molecular features of type 1 human prions (4, 5), i.e. PK-resistant PrP fragments around 21 kDa (unglycosylated band), being typically strongly reactive toward the 12B2 antibody (33). The observed lack of Pc248 reactivity of fragments generated by PK cleavage of FL PrPSc appears to be a specific feature of mouse prions. Indeed, in other prion-infected species, PK cleavage of “type 1-like” PrPSc tends to preserve C-proximal octarepeat residues. This is the case for sheep prions, as shown in this study by the strong Pc248 reactivity of PK-resistant fragments generated from FL PrPSc (e.g. in brain tissue or primary neurons (CGN)). Consistent with our findings, the Trp-84 residue, located upstream of the Pc248 epitope sequence delineated here, was recently identified by mass spectrometry analysis as one of the major PK cleavage sites in brains of natural or experimental scrapie-affected sheep (46), whereas residues within or downstream of the Pc248 epitope were identified as the main PK cleavage sites in scrapie-infected mice (11, 47, 48). Recently, PK-resistant PrPSc accumulating in the brain of human patients infected by type 1 prions (5, 49) was reported to be strongly reactive toward two newly introduced anti-octarepeat antibodies (50). We surmised that these antibodies (Pom2 and Pom12) have properties very similar to those of Pc248, which is characterized by high avidity for brain PrPres, whereas commonly used anti-octarepeat antibodies generally exhibit weak reactivity (for more information on Pc248 antibody, see supplemental Fig. S2). PK-resistant fragments retaining an octarepeat motif are produced upon infection by the 21K type of hamster prions also (51).5 In conclusion, the equivalence between C2 and PK-cleaved fragments commonly observed with mouse prions is far from the general rule. Endogenous processing events that precede in vitro proteolytic digestion determine the molecular features of PrPres to a variable extent. This notion might be worth accounting for while interpreting PrPSc molecular profiles in biopsies from brain or other tissues that might markedly differ in their trimming capability.
PrPres fragments in mouse and sheep prion-infected cell lines typically have a higher mobility than their counterparts in the brain tissue of wild type or ovine transgenic mice, respectively (25, 27, 52, 53). Re-inoculation to mice of the cell culture-propagated agent revealed no permanent change of its molecular profile and strain properties (26, 27, 52–55). The phenomenon has been proposed to simply reflect post-translational differences of PrPC molecules when expressed either in culture or tissue (52, 53). Thus, unglycosylated PrPC in mouse cell lines migrate faster than mouse brain PrPC, putatively reflecting different compositions of the GPI moieties (52). New findings of the present study offer clarification of the cause of these observations. Our results established that, whereas in mouse prion-infected cells the N termini of endogenously and PK-cleaved PrPSc are similar (i.e. both Pc248-negative and 12B2-positive), a more complex situation prevails in sheep prion-infected cells; indeed, two factors appeared to account for the molecular mass difference observed between brain and cell PrPres fragments: (i) a differential mobility of PrPC due to the variable composition of the GPI moiety, which was abolished after removal of the latter by hydrofluoric acid treatment, information lacking in the former studies; (ii) the production of distinguishable N termini produced by in vivo (endogenous) and in vitro (PK) cleavages. Likewise, not only PrPC tissue-specific differences, such as those reported in the human and sheep species (7, 31, 35), but also different trimming capabilities, as observed here in two mouse models (tg338 and tga20), might account for PrPres molecular mass discrepancies in neural and lymphoid tissues in naturally infected hosts.
Another unexpected finding in this study was the retention of nondenatured PrPSc by the copper ion, because failure to bind to immobilized copper ion has been reported by others to be a distinctive trait of nondenatured PrPSc (56, 57). In our experimental conditions, the binding of both FL PrPSc and PrPres was reproducible, and plainly ion-dependent (data not shown). It was observed irrespective of whether cell lysate or brain homogenate was used as a starting material. A strain-specific effect is implausible, because efficient binding was observed with hamster PrPSc also,6 the prion agent used in one previous study (56). Although we still have no clear explanation to offer for these discrepant data, we suggest reconsidering copper ion binding as a potentially useful means to prepare full-length or naturally trimmed PrPSc molecules in the absence of any treatment that would denature or cleave the PrPSc species. From a theoretical viewpoint, our observation suggests that determinants of the N-terminal region are accessible within native PrPSc multimers, because FL PrPSc and PrPC exhibited close avidities for bound copper.
Studying the effect of lysosomotropic compounds and protease inhibitors on the generation of C2 fragments has led to somewhat controversial observations in the literature. In all three cell lines examined here (Rov, MovS, and CAD), the formation of C2 fragments was blocked efficiently by leupeptin treatment and NH4Cl-induced pH elevation, with little or no effect on short term PrPSc accumulation. To be noted, PrPSc in lysosome-impaired Rov and CAD cells consisted of true FL species, not partially protected species with a shorter size truncation (17). Our results, in line with earlier observations in two other cell lines (12, 17), thus provided robust evidence for the involvement of the endolysosomal compartment in the N-terminal trimming of PrPSc. Furthermore, we showed that in vitro cleavage of full-length PrPSc by cathepsins B and L produced fragments that lacked the Pc248 octarepeat epitope, as observed for the endogenously produced C2 fragments. Conversely, cathepsin inhibitors were found to diminish the formation of C2 fragments in infected cell cultures. These data bring direct evidence for the involvement of these hydrolases into the generation of C2 PrPSc.
The above findings are in sharp contrast with those of a recent study, in which inhibitors of lysosomal proteases had no effect on the production of C2, whereas calpain inhibition adversely affected C2 accumulation, and also prion propagation, thus raising the possibility that C2 generation was critical (13). In none of the cell systems studied here, however, did inhibition of calpain lead to accumulation of the FL species at the expense of the C2 species, as it was consistently observed in lysosome-impaired cultures, therefore arguing that endoproteolytic cleavage of PrPSc is not a calpain-dependent process. Prion propagation was stably maintained along the reiterated subpassages in NH4Cl-treated CAD cell cultures, in which PrPSc endoproteolysis was essentially blocked, both quantitatively and qualitatively. Altogether, our findings support the view that hydrolases from the endolysosomal compartment rather than calpain are primarily involved in PrPSc trimming and that C2 formation neither profoundly affects nor is critically required for efficient prion propagation in the cell culture.
The mechanisms underlying the neurotoxic effects engendered by prion infection remain largely elusive but most probably involve some form of PrPSc. Could the trimming of PrPSc influence its neurotoxic potential? The N terminus of PrPC appears to be important for anti-apoptosis and antioxidant functions (58, 59), but what about PrPSc? Transgenic mice that express a deleted form of PrP mimicking C2 fragments can be infected and develop a lethal neurological disease, but this does not preclude the existence in the truncated region of determinants that could modulate the cytotoxicity of PrPSc. Interestingly, the neuropathology in one such transgenic line was reported to be profoundly modified, i.e. no histopathological lesions could be detected in the brain or brainstem of terminally ill animals, and the cytopathic anomalies were essentially restricted to neurons of the cervical spinal cord (60). Altogether our ex vivo observations raise the question of whether trimming the FL PrPSc could modulate its neurotoxic potential and thus exert some protective effect in the target cells. Although immortalized cells fail to recapitulate the deleterious effects occurring in vivo, primary cultured neurons show a gradually compromised survival upon prion infection (29). It is intriguing that in the latter, the FL PrPSc form largely predominates over the C2 form, whereas the opposite is commonly observed in immortalized cells even of neural origin. In vivo, prion infection causes no overt cell death of astrocytes, which showed good trimming capacity in primary cultures, or of lymphoreticular cells, where PrPSc could be extensively endoproteolyzed, considering our results with splenic tissue. The processing of PrPSc depends on its access to the endocytic pathway. Thus, a plausible hypothesis would be that non-internalized PrPSc is most harmful to the host cell. It is noteworthy that histopathological studies performed on scrapie-affected sheep brain have pointed to a relationship between cell-membrane or extracellular PrP deposition and vacuolation (61). Approaches using cell culture systems, including blockade of PrPSc trimming without impairing prion multiplication, should make it feasible to further explore these aspects.
Supplementary Material
Acknowledgments
We acknowledge the excellent technical support of Fabienne Reine and Laetitia Herzog for brain homogenate preparations; Vincent Beringue for critical reading of the manuscript; Jean Michel Peyrin, Julie Carimalo, and Emilie Jaumain for help with primary neuron cultures. We also thank Jacques Grassi (CEA, Saclay, France), Jan P. Langeveld (Central Institute for Animal Disease Control, Lelystad, Netherlands), John Collinge (MRC Prion Unit, UCL Institute of Neurology, United Kingdom), and Jean Luc Gatti (INRA, Tours, France) for antibody supply, and W. Brand-Williams for revising the manuscript.
This work was supported by the Institut National de la Recherche Agronomique.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
M. Moudjou, unpublished data.
M. Moudjou, unpublished results.
- TSE
- transmissible spongiform encephalopathies
- PK
- proteinase K
- PrP
- prion protein
- PrPC
- normal PrP
- PrPSc
- scrapie-associated PrP
- PrPres
- PK-resistant PrPSc
- FL
- full-length
- IMAC
- immobilized metal affinity chromatography
- mAb
- monoclonal antibody
- CGN
- cerebellar granule neurons
- CAS
- cerebellar astrocytes
- GPI
- glycosylphosphatidylinositol
- FCS
- fetal calf serum
- PNGase F
- peptide:N-glycosidase F.
REFERENCES
- 1.Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westergard L., Christensen H. M., Harris D. A. (2007) Biochim. Biophys. Acta 1772, 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguzzi A., Sigurdson C., Heikenwaelder M. (2008) Annu. Rev. Pathol. 3, 11–40 [DOI] [PubMed] [Google Scholar]
- 4.Gambetti P., Kong Q., Zou W., Parchi P., Chen S. G. (2003) Br. Med. Bull. 66, 213–239 [DOI] [PubMed] [Google Scholar]
- 5.Wadsworth J. D., Collinge J. (2007) Biochim. Biophys. Acta 1772, 598–609 [DOI] [PubMed] [Google Scholar]
- 6.Chen S. G., Teplow D. B., Parchi P., Teller J. K., Gambetti P., Autilio-Gambetti L. (1995) J. Biol. Chem. 270, 19173–19180 [DOI] [PubMed] [Google Scholar]
- 7.Jiménez-Huete A., Lievens P. M., Vidal R., Piccardo P., Ghetti B., Tagliavini F., Frangione B., Prelli F. (1998) Am. J. Pathol. 153, 1561–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanusso G., Righetti P. G., Ferrari S., Terrin L., Farinazzo A., Cardone F., Pocchiari M., Rizzuto N., Monaco S. (2002) Electrophoresis 23, 347–355 [DOI] [PubMed] [Google Scholar]
- 9.Owen J. P., Rees H. C., Maddison B. C., Terry L. A., Thorne L., Jackman R., Whitelam G. C., Gough K. C. (2007) J. Virol. 81, 10532–10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hope J., Morton L. J., Farquhar C. F., Multhaup G., Beyreuther K., Kimberlin R. H. (1986) EMBO J. 5, 2591–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hope J., Multhaup G., Reekie L. J., Kimberlin R. H., Beyreuther K. (1988) Eur. J. Biochem. 172, 271–277 [DOI] [PubMed] [Google Scholar]
- 12.Taraboulos A., Raeber A. J., Borchelt D. R., Serban D., Prusiner S. B. (1992) Mol. Biol. Cell. 3, 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadavalli R., Guttmann R. P., Seward T., Centers A. P., Williamson R. A., Telling G. C. (2004) J. Biol. Chem. 279, 21948–21956 [DOI] [PubMed] [Google Scholar]
- 14.Pan T., Wong P., Chang B., Li C., Li R., Kang S. C., Wisniewski T., Sy M. S. (2005) J. Virol. 79, 934–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffrey M., Martin S., González L. (2003) J. Gen. Virol. 84, 1033–1045 [DOI] [PubMed] [Google Scholar]
- 16.Jeffrey M., González L., Chong A., Foster J., Goldmann W., Hunter N., Martin S. (2006) J. Comp. Pathol. 134, 17–29 [DOI] [PubMed] [Google Scholar]
- 17.Caughey B., Raymond G. J., Ernst D., Race R. E. (1991) J. Virol. 65, 6597–6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taguchi Y., Shi Z. D., Ruddy B., Dorward D. W., Greene L., Baron G. S. (2009) Mol. Biol. Cell 20, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vincent B., Paitel E., Saftig P., Frobert Y., Hartmann D., De Strooper B., Grassi J., Lopez-Perez E., Checler F. (2001) J. Biol. Chem. 276, 37743–37746 [DOI] [PubMed] [Google Scholar]
- 20.McKinley M. P., Taraboulos A., Kenaga L., Serban D., Stieber A., DeArmond S. J., Prusiner S. B., Gonatas N. (1991) Lab. Invest. 65, 622–630 [PubMed] [Google Scholar]
- 21.Pimpinelli F., Lehmann S., Maridonneau-Parini I. (2005) Eur. J. Neurosci. 21, 2063–2072 [DOI] [PubMed] [Google Scholar]
- 22.Jeffrey M., McGovern G., Goodsir C. M., Síso S., González L. (2009) Brain Pathol. 19, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luhr K. M., Nordström E. K., Löw P., Kristensson K. (2004) Neuroreport 15, 1663–1667 [DOI] [PubMed] [Google Scholar]
- 24.Doh-Ura K., Iwaki T., Caughey B. (2000) J. Virol. 74, 4894–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilette D., Andreoletti O., Archer F., Madelaine M. F., Vilotte J. L., Lehmann S., Laude H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4055–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Courageot M. P., Daude N., Nonno R., Paquet S., Di Bari M. A., Le Dur A., Chapuis J., Hill A. F., Agrimi U., Laude H., Vilette D. (2008) J. Gen. Virol. 89, 341–347 [DOI] [PubMed] [Google Scholar]
- 27.Archer F., Bachelin C., Andreoletti O., Besnard N., Perrot G., Langevin C., Le Dur A., Vilette D., Baron-Van Evercooren A., Vilotte J. L., Laude H. (2004) J. Virol. 78, 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi Y., Wang J. K., McMillian M., Chikaraishi D. M. (1997) J. Neurosci. 15, 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cronier S., Laude H., Peyrin J. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12271–12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cronier S., Beringue V., Bellon A., Peyrin J. M., Laude H. (2007) J. Virol. 81, 13794–13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moudjou M., Frobert Y., Grassi J., La Bonnardière C. (2001) J. Gen. Virol. 82, 2017–2024 [DOI] [PubMed] [Google Scholar]
- 32.Moudjou M., Treguer E., Rezaei H., Sabuncu E., Neuendorf E., Groschup M. H., Grosclaude J., Laude H. (2004) J. Virol. 78, 9270–9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langeveld J. P., Jacobs J. G., Erkens J. H., Bossers A., van Zijderveld F. G., van Keulen L. J. (2006) BMC Vet. Res. 9, 2–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Féraudet C., Morel N., Simon S., Volland H., Frobert Y., Créminon C., Vilette D., Lehmann S., Grassi J. (2005) J. Biol. Chem. 280, 11247–11258 [DOI] [PubMed] [Google Scholar]
- 35.Beringue V., Mallinson G., Kaisar M., Tayebi M., Sattar Z., Jackson G., Anstee D., Collinge J., Hawke S. (2003) Brain 126, 2065–2073 [DOI] [PubMed] [Google Scholar]
- 36.Ecroyd H., Sarradin P., Dacheux J. L., Gatti J. L. (2004) Biol. Reprod. 71, 993–1001 [DOI] [PubMed] [Google Scholar]
- 37.Vilotte J. L., Soulier S., Essalmani R., Stinnakre M. G., Vaiman D., Lepourry L., Da Silva J. C., Besnard N., Dawson M., Buschmann A., Groschup M., Petit S., Madelaine M. F., Rakatobe S., Le Dur A., Vilette D., Laude H. (2001) J. Virol. 75, 5977–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dron M., Dandoy-Dron F., Farooq Salamat M. K., Laude H. (2009) J. Gen. Virol. 90, 2050–2060 [DOI] [PubMed] [Google Scholar]
- 39.Deslys J. P., Comoy E., Hawkins S., Simon S., Schimmel H., Wells G., Grassi J., Moynagh J. (2001) Nature 409, 476–478 [DOI] [PubMed] [Google Scholar]
- 40.Owen J. P., Maddison B. C., Whitelam G. C., Gough K. C. (2007) Mol. Biotechnol. 35, 161–170 [DOI] [PubMed] [Google Scholar]
- 41.Jordans S., Jenko-Kokalj S., Kühl N. M., Tedelind S., Sendt W., Brömme D., Turk D., Brix K. (2009) BMC Biochem. 22, 10–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borchelt D. R., Rogers M., Stahl N., Telling G., Prusiner S. B. (1993) Glycobiology 3, 319–329 [DOI] [PubMed] [Google Scholar]
- 43.Paquet S., Langevin C., Chapuis J., Jackson G. S., Laude H., Vilette D. (2007) J. Gen. Virol. 88, 706–713 [DOI] [PubMed] [Google Scholar]
- 44.Moudjou M., Bernard J., Sabuncu E., Langevin C., Laude H. (2007) Neurochem. Int. 50, 689–695 [DOI] [PubMed] [Google Scholar]
- 45.Pan T., Li R., Kang S. C., Pastore M., Wong B. S., Ironside J., Gambetti P., Sy M. S. (2005) J. Neurochem. 92, 132–142 [DOI] [PubMed] [Google Scholar]
- 46.Gielbert A., Davis L. A., Sayers A. R., Hope J., Gill A. C., Sauer M. J. (2009) J. Mass. Spectrom. 44, 384–396 [DOI] [PubMed] [Google Scholar]
- 47.Hayashi H. K., Yokoyama T., Takata M., Iwamaru Y., Imamura M., Ushiki Y. K., Shinagawa M. (2005) Biochem. Biophys. Res. Commun. 328, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 48.Howells L. C., Anderson S., Coldham N. G., Sauer M. J. (2008) Biomarkers 13, 393–412 [DOI] [PubMed] [Google Scholar]
- 49.Notari S., Capellari S., Langeveld J., Giese A., Strammiello R., Gambetti P., Kretzschmar H. A., Parchi P. (2007) Lab. Invest. 87, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 50.Polymenidou M., Stoeck K., Glatzel M., Vey M., Bellon A., Aguzzi A. (2005) Lancet Neurol. 4, 805–814 [DOI] [PubMed] [Google Scholar]
- 51.Stahl N., Baldwin M. A., Teplow D. B., Hood L., Gibson B. W., Burlingame A. L., Prusiner S. B. (1993) Biochemistry 32, 1991–2002 [DOI] [PubMed] [Google Scholar]
- 52.Arima K., Nishida N., Sakaguchi S., Shigematsu K., Atarashi R., Yamaguchi N, Yoshikawa D., Yoon J., Watanabe K., Kobayashi N., Mouillet-Richard S., Lehmann S., Katamine S. (2005) J. Virol. 79, 7104–71012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwamaru Y., Takenouchi T., Ogihara K., Hoshino M., Takata M., Imamura M., Tagawa Y., Hayashi-Kato H., Ushiki-Kaku Y., Shimizu Y., Okada H., Shinagawa M., Kitani H., Yokoyama T. (2007) J. Virol. 81, 1524–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arjona A., Simarro L., Islinger F., Nishida N., Manuelidis L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8768–8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birkett C. R., Hennion R. M., Bembridge D. A., Clarke M. C., Chree A., Bruce M. E., Bostock C. J. (2001) EMBO J. 20, 3351–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaked Y., Rosenmann H., Hijazi N., Halimi M., Gabizon R. (2001) J. Virol. 75, 7872–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Müller H., Strom A., Hunsmann G., Stuke A. W. (2005) Biochem. J. 388, 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng F., Watt N. T., Walmsley A. R., Hooper N. M. (2003) J. Neurochem. 84, 480–490 [DOI] [PubMed] [Google Scholar]
- 59.Sakudo A., Lee D. C., Nakamura I., Taniuchi Y., Saeki K., Matsumoto Y., Itohara S., Ikuta K., Onodera T. (2005) Biochem. Biophys. Res. Commun. 333, 448–454 [DOI] [PubMed] [Google Scholar]
- 60.Flechsig E., Shmerling D., Hegyi I., Raeber A. J., Fischer M., Cozzio A., von Mering C., Aguzzi A., Weissmann C. (2000) Neuron 27, 399–408 [DOI] [PubMed] [Google Scholar]
- 61.Jeffrey M., González L. (2007) Neuropathol. Appl. Neurobiol. 33, 373–394 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.