Abstract
The red light:far-red light ratio perceived by phytochromes controls plastic traits of plant architecture, including branching. Despite the significance of branching for plant fitness and productivity, there is little quantitative and mechanistic information concerning phytochrome control of branching responses in Arabidopsis (Arabidopsis thaliana). Here, we show that in Arabidopsis, the negative effects of the phytochrome B mutation and of low red light:far-red light ratio on branching were largely due to reduced bud outgrowth capacity and an increased degree of correlative inhibition acting on the buds rather than due to a reduced number of leaves and buds available for branching. Phytochrome effects on the degree of correlative inhibition required functional BRANCHED1 (BRC1), BRC2, AXR1, MORE AXILLARY GROWTH2 (MAX2), and MAX4. The analysis of gene expression in selected buds indicated that BRC1 and BRC2 are part of different gene networks. The BRC1 network is linked to the growth capacity of specific buds, while the BRC2 network is associated with coordination of growth among branches. We conclude that the branching integrators BRC1 and BRC2 are necessary for responses to phytochrome, but they contribute differentially to these responses, likely acting through divergent pathways.
Branching is an important parameter that contributes to plant architecture, affecting plant fitness in natural environments (Juenger and Bergelson, 2000; Lortie and Aarssen, 2000; Van Kluenen and Fischer, 2001; Bonser and Aarssen, 2003) and productivity in agricultural crops (Peng et al., 1994; García del Moral and García del Moral, 1995; Zhao et al., 2006; Boe and Beck, 2008) and pastures (Zarrough et al., 1983). Branching is the result of several interrelated developmental programs beginning with axillary meristem initiation, the formation of an axillary bud, the initiation of bud outgrowth, and then branch elongation. Elaboration of branching patterns can occur through the repetition of this process at higher order nodes, giving rise to secondary branches, tertiary branches, etc.
In Arabidopsis (Arabidopsis thaliana) grown under long days, axillary meristems become obvious following the reproductive transition of the shoot apical meristem (Hempel and Feldman, 1994). Bud outgrowth subsequently occurs in a basipetal wave beginning with activation of the uppermost cauline bud and progresses to sequentially lower buds, including buds formed in the axils of rosette leaves. In ecotypes Columbia (Col) and Landsberg erecta, all cauline buds produce branches but only a limited number of rosette buds are activated to outgrow.
Genes involved in axillary meristem initiation in eudicots include REVOLUTA (Otsuga et al., 2001), LATERAL SUPPRESSOR (Schumacher et al., 1999), and BLIND (Schmitz et al., 2002). While their loss of function leads to dramatic reductions in the frequency of axillary meristems formed, there is little evidence to suggest that meristem initiation is a plastic trait contributing to variations in branching. Arabidopsis branching is strongly regulated at the level of bud outgrowth, and teosinte branched1 (tb1) homologs are key players in this process. The tb1 gene of maize (Zea mays) was identified as a major contributor to the domestication process through its inhibitory effects on branching (Doebley et al., 1995, 1997). The ancestral tb1 gene has apparently radiated into three genes in the eudicots (Howarth and Donoghue, 2006) of which at least two, BRANCHED1 (BRC1 or TEOSINTE BRANCHED1-LIKE1 [TBL1]) and BRC2, function in Arabidopsis axillary buds as suppressors of outgrowth (Aguilar-Martínez et al., 2007; Finlayson, 2007). tb1 and its homologs including BRC1 and BRC2 belong to the TCP domain group of proteins, which may function as transcriptional regulators (Cubas et al., 1999). BRC1 and BRC2 have been proposed to act as integrators of a variety of branching signals, including signals related to high planting density (Aguilar-Martínez et al., 2007).
The inhibition of branch development by signals received from more or less remote parts of the plant is termed correlative inhibition. An example of this is apical dominance, which describes the inhibition of axillary bud outgrowth by signals generated by the main shoot apex. Decapitation of the main shoot generally results in increased initiation of outgrowth of axillary buds (Thimann and Skoog, 1934; Cline, 1996). Another specific type of correlative inhibition is apical control, which describes the inhibitory effects of the main shoot apex and/or other branches on the growth of branches after they have initiated outgrowth (Cline, 1997). Removing the main shoot or stronger branches typically results in increased growth of the remaining branches (Cline and Sadeski, 2002). While auxin plays an important role in regulating apical dominance (Thimann and Skoog, 1934; Cline, 1996), its role in apical control has been called into question while other possible signals have been implicated (Cline and Sadeski, 2002; Cline et al., 2009).
The roles of plant hormones in branching have been studied extensively. As described above, auxin contributes to apical dominance. Auxin transported basipetally from the shoot apex inhibits bud outgrowth by a cryptic mechanism whereby this auxin does not enter the bud (Ongaro and Leyser, 2008). Expression of the dormancy-associated DRM1 gene is responsive to decapitation in pea (Pisum sativum; Stafstrom et al., 1998) and Arabidopsis (Aguilar-Martínez et al., 2007), implying its utility as an indicator of bud dormancy status and potentially as an indirect probe of basipetal auxin transport stream function. Lesions in genes encoding auxin signaling components, such as AXR1, result in increased branching in Arabidopsis (Lincoln et al., 1990; Stirnberg et al., 1999). Conversely, auxin may be necessary within the bud to promote outgrowth in bean (Phaseolus vulgaris), because bud auxin levels increase coincident with outgrowth (Gocal et al., 1991). It has also been proposed that auxin transport out of the bud may be required for outgrowth in Arabidopsis (Bennett et al., 2006). Therefore, auxin can act as both a non-organ-autonomous signal to inhibit bud outgrowth (basipetal transport stream) and may act as an organ-autonomous signal to permit bud outgrowth.
A strigolactone-related hormone also inhibits branching through a non-organ-autonomous pathway that may originate in roots (Gomez-Roldan et al., 2008; Umehara et al., 2008). This pathway is associated with Arabidopsis MORE AXILLARY GROWTH (MAX) genes that correspond to homologs in other species, including pea, petunia (Petunia hybrida), and rice (Oryza sativa; Ongaro and Leyser, 2008). Loss of function of components potentially involved in biosynthesis (MAX1, MAX3, and MAX4) or in signaling (MAX2) of the hormone results in increased branching (Stirnberg et al., 2002; Sorefan et al., 2003; Booker et al., 2004, 2005).
A variety of other hormones are also implicated in the regulation of branching, including cytokinins, which may act within the bud to promote outgrowth. Cytokinin levels increased within lupin (Lupinus albus) axillary buds coincident with outgrowth (Emery et al., 1998). Tobacco (Nicotiana tabacum) engineered to overproduce cytokinins showed increased branching (Medford et al., 1989), and application of cytokinins to axillary buds of plants such as sunflower (Helianthus annuus) and pea resulted in outgrowth (Sachs and Thimann, 1964, 1967). Cytokinin function has been probed by assessing the expression of the cytokinin-responsive ARABIDOPSIS RESPONSE REGULATOR5 (ARR5) gene (Taniguchi et al., 1998; D'Agostino et al., 2000).
Positive (SHOOT MERISTEMLESS [STM] and WUSCHEL [WUS; Long et al., 1996; Mayer et al., 1998]) and negative (CLAVATA1 [CLV1] and CLV3 [Clark et al., 1997; Fletcher et al., 1999]) regulators of stem cell populations determine meristem size and function (Tucker and Laux, 2007) and therefore may influence branching by altering axillary bud activity. Furthermore, bud outgrowth is anticipated to result, in part, from increased cell division in the meristem and derivative cells, which may be accompanied by changes in the expression of cell cycle-related genes, such as cyclins (de Jager et al., 2005) and PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA; Moldovan et al., 2007). Knowledge of the expression of these genes could offer insights into the mechanisms regulating bud outgrowth.
While intrinsic genetic programs are major determinants of architecture, environmental signals also modulate plant form. Among the environmental signals that may regulate branching is the red light:far-red light ratio (R:FR), perceived by the phytochrome family of photoreceptors. Reduced R:FR signals impending competition from neighboring plants, which absorb R and reflect or transmit FR (Ballaré, 1999). The phytochromes (phytochrome A [phyA] to phyE in Arabidopsis) sense and transduce light signaling parameters, including the R:FR, with phyB playing the major role. Reduced R:FR elicits a suite of responses collectively termed the shade avoidance response (Smith, 1995; Franklin and Whitelam, 2005). The resultant phenotype includes rapid elongation of the main shoot, reduced root growth, early flowering, reduced branching, and other effects, which may confer a competitive advantage to plants able to mount an early response. While the mechanisms contributing to shade avoidance downstream of phytochromes are poorly understood, expression of the homeobox gene A. THALIANA HOMEOBOX-2 (ATHB-2) is induced by low R:FR (Carabelli et al., 1993, 1996), thus providing a useful marker of shade responses.
The effect of phyB and R:FR on branching has been studied in both eudicots (Robin et al., 1994; Donohue and Schmitt, 1999) and monocots (Deregibus et al., 1983; Casal et al., 1985, 1986; Davis and Simmons, 1994; Wan and Sosebee, 1998), with a recent analysis conducted in sorghum (Sorghum bicolor; Kebrom et al., 2006). Loss of phyB function or low R:FR arrested bud outgrowth in sorghum seedlings and was associated with elevated expression of sorghum tb1. While loss of phyB function in Arabidopsis was anecdotally reported to result in decreased branching, the effect was not quantified, nor was the defective process(es) discovered (Reed et al., 1993). It is not known, for instance, whether in Arabidopsis the effects of low R:FR or phyB mutations on branching are the consequence of early flowering, which reduces leaf numbers, and therefore the number of axillary buds, or the result of more direct effects on bud initiation or outgrowth. Furthermore, although auxin biosynthesis via TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) is important during shade avoidance reactions (Tao et al., 2008) and auxin (Steindler et al., 1999) and strigolactone-related hormone genes (Shen et al., 2007) affect seedling photomorphogenesis, the link between these hormones and phytochrome-mediated control of branching has not been established.
The objective of this study was to investigate the interdependence between phytochrome signaling and the network controlling branching in Arabidopsis. A detailed analysis of a battery of architectural parameters was conducted to probe phytochrome-dependent variations in the complex processes contributing to the generation of the branching habit. It was hypothesized that phytochrome action would impact multiple branching processes and that the effects of phytochrome would be transduced through known branching regulators, including hormone pathways and the branching integrators BRC1 and BRC2.
RESULTS
Phytochrome Control of Shoot Architecture
The visual phenotypes at 10 DPA of various genotypes compromised in light sensing (phyA), branching-related hormone signaling and biosynthesis (axr1-12, max2, max4), integration of branching signals (brc1, brc2, brc1brc2), and their combinations with phyB are provided in Figure 1A. Since branching under long days occurs coincident with the reproductive transition, plants were evaluated a short time after anthesis to ensure that the duration of branch development was equivalent in all genotypes/treatments. In most cases, differences in the time to anthesis in phyB-sufficient and phyB-deficient genotypes were small; however, phyB deficiency substantially accelerated the time to anthesis in axr1-12 and max4 (Supplemental Fig. S1). Low R:FR reduced the time to anthesis in all cases. Primary rosette branches in all genotypes/treatments were actively growing at 10 DPA; therefore, collateral effects of senescence and fertility were minimized. The effect of R:FR on overall morphology of wild-type, brc1, brc2, and brc1brc2 plants is documented in Figure 1B. phyB deficiency and low R:FR promoted shoot elongation and appeared to reduce branching in most of the genotypes, but as rosette leaf numbers were also reduced, the specific basis for the branching defect was not revealed by visual observation alone and a quantitative analysis of the major architectural characteristics was required.
Figure 1.
Visual phenotypes of various Arabidopsis genotypes at 10 DPA. A, Plants were grown under high R:FR (R:FR of 2.08, PPFD of 180 μmol m−2 s−1, photoperiod of 18 h). B, Plants were grown under high (2.98) or low (0.05) R:FR (175 μmol m−2 s−1 PPFD, photoperiod of 18 h). WT, Wild type.
Compared with wild-type plants grown under high R:FR, phyB plants grown under high R:FR and wild-type plants grown under low R:FR showed a reduced number of primary rosette branches (Fig. 2A) and rosette leaves (Fig. 2B). Leaf number and branch number were highly correlated in most genotypes/treatments (Supplemental Fig. S2A). Although it cannot be concluded that increased leaf numbers caused increased branching, the correlation indicates that simple comparisons of branch numbers between genotypes/treatments with different numbers of leaves may provide an unsatisfactory estimate of the difference in branching that can be attributed to direct effects of phyB or R:FR on the process. To account for the association between leaf and branch numbers, the regressions of the phyB-sufficient genotypes (or high R:FR treatments) were used to derive branch numbers at the observed mean leaf values for the phyB-deficient genotypes (or low R:FR treatments). Standardization revealed the specific effects of phyB and low R:FR on branch numbers by eliminating the indirect effects caused by reductions in leaf number. A graphic explanation of the standardizing method and an example calculation are provided in Supplemental Figure S2B. Both loss of phyB function and low R:FR resulted in a significant decrease in standardized branch numbers in the wild-type background (Fig. 2C). A similar analysis was employed to assess the effects of phyB on bud initiation, since strong correlation was also evident between leaf and bud numbers (Supplemental Fig. S3). Consistent with the high branching potential of wild-type Col, buds initiated in most leaf axils (Figs. 2B and 3A). Loss of phyB function in wild-type plants resulted in a reduction in the number of standardized primary rosette buds (Fig. 3B); however, this effect did not account for the effects on branch numbers, because buds were always in excess of branches. These results demonstrate that phytochrome regulates branch numbers by modulating the bud outgrowth process. All primary cauline leaves produced buds, and all the buds gave rise to branches in every genotype and treatment (data not shown).
Figure 2.
Primary rosette architectural parameters of various Arabidopsis genotypes at 10 DPA with or without functional phyB (left panel of each graph) or under high and low R:FR (shaded right panel of each graph). A and B, Statistical comparisons for primary rosette branch numbers (A) and primary rosette leaf numbers (B) were made within each genotype sufficient (PHYB) or deficient (phyB) for phyB or within each genotype grown under high versus low R:FR. Asterisks indicate significant differences between genotypes or light treatments at α = 0.05. Data are means ± se. C, Standardized primary rosette branch numbers employ regressions to account for correlation between leaf numbers and branch numbers (see text), and error bars represent 95% confidence intervals. For analyses comparing lines with or without functional phyB, n = 28 (phyBmax2) to 87 (wild type [WT]), average n = 51. For high and low R:FR, n = 26 (brc1) to 70 (wild type), average n = 37.
Figure 3.
Primary rosette bud parameters of various Arabidopsis genotypes at 10 DPA with or without functional phyB (left panel of each graph) or under high and low R:FR (shaded right panel of each graph). A, Statistical comparisons for primary rosette bud numbers were made within each genotype sufficient (PHYB) or deficient (phyB) for phyB or within each genotype grown under high versus low R:FR. Asterisks indicate significant differences between genotypes or light treatments at α = 0.05. Data are means ± se. B, Standardized primary rosette bud numbers employ regressions to account for correlation between leaf numbers and branch numbers (see text), and error bars represent 95% confidence intervals. For analyses comparing lines with or without functional phyB, n = 28 (phyBmax2) to 87 (wild type [WT]), average n = 51. For high and low R:FR, n = 26 (brc1) to 70 (wild type), average n = 37.
Low R:FR and the phyB mutation both reduced the number of secondary branches that arose in the axils of leaves produced on the uppermost primary rosette branch (branch n) compared with the wild type under high R:FR (Supplemental Fig. S4A) and increased (phyB) or reduced (low R:FR) secondary leaf numbers (Supplemental Fig. S4B). Similar results were obtained in the case of secondary cauline leaf numbers (Supplemental Fig. S5A) and secondary cauline branch numbers (Supplemental Fig. S5B). It is interesting that despite the presence of many nodes on each primary branch, the frequency of secondary branching from the uppermost rosette branch was about 0.62 in wild-type plants and was reduced in the wild type by low R:FR and by the phyB mutation. A comparable pattern was observed for secondary branching from the lowest cauline branch. These results are consistent with the above conclusion that low R:FR or loss of phyB function have specific effects on bud outgrowth rather than affecting branching solely by limiting the number of leaves or branching sites.
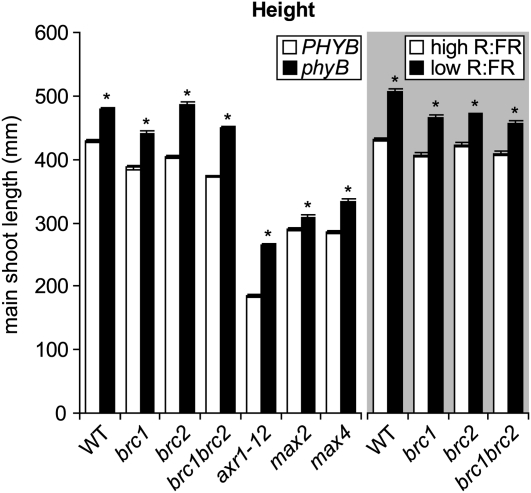
Loss of phyB function and low R:FR promoted the elongation of the main stem in all the genotypes tested (Fig. 4). Here and elsewhere, comparisons between phyA and phyAphyB grown under high R:FR provided results similar to those made between the wild type and phyB and have been omitted from the data set to improve clarity. phyA and phyAphyB data are provided in Supplemental Table S1.
Figure 4.
Overall height of the main shoot of various Arabidopsis genotypes at 10 DPA with or without functional phyB (left panel of the graph) or under high and low R:FR (shaded right panel of the graph). Statistical comparisons were made within each genotype sufficient (PHYB) or deficient (phyB) for phyB or within each genotype grown under high versus low R:FR. Asterisks indicate significant differences between genotypes or light treatments at α = 0.05. Data are means ± se. For analyses comparing lines with or without functional phyB, n = 28 (phyBmax2) to 87 (wild type [WT]), average n = 51. For high and low R:FR, n = 26 (brc1) to 70 (wild type), average n = 37.
The Effects of Phytochrome on Bud Outgrowth Kinetics
The end-point count of the number of branches per plant provides a static and incomplete picture of the effects of phytochrome on branching. phyB or wild-type plants grown under low R:FR produced two to three rosette branches. However, even these two to three branches showed indications of a reduced branching power in phyB or under low R:FR, which became obvious when the elongation kinetics of the top three buds/branches were monitored daily, beginning 2 d before anthesis until 10 d after anthesis. These branches were at similar positions in the bud activation sequence, since there was little variation in cauline node number (except axr1-12, which had approximately two more nodes than the other genotypes). Branches of wild-type plants grown under high R:FR began elongating earlier and at higher rates than phyB and also achieved a higher maximum sustained elongation rate (Fig. 5). The differences in branch elongation between the wild type and phyB were apparent in the uppermost branch (branch n), and these differences increased at sequentially lower nodes.
Figure 5.
Elongation of the top three primary rosette branches (A, branch n [uppermost branch]; B, branch n-1 [branch immediately below branch n]; C, branch n-2 [branch immediately below branch n-1]) of wild-type (WT) and phyB plants grown under high R:FR (left) and wild-type plants grown under high and low R:FR (right) with time. Insets show the elongation rates. The maximum sustained elongation rate (MSER [mm h−1]) was calculated by averaging the three greatest sequential daily elongation rates. Onset delay (Δ onset) is the delay in days between the onset of bud outgrowth in the wild type and phyB or between wild-type plants grown in high R:FR and low R:FR and was calculated by estimating the time at which a branch reached 3 mm in length. Asterisks indicate significant differences between genotypes or light treatments at α = 0.05. Data are means ± se; n = 12 to 14. Anthesis occurred on day 0.
Low R:FR promoted the outgrowth of branch n compared with high R:FR, as evidenced by the earlier onset of elongation and elevated maximum sustained elongation rates (Fig. 5). This effect is opposite that observed for the elongation of branch n in phyB and is likely due to the effects of other phytochromes that are also inactivated by low R:FR but that remain active in phyB. It is possible that the reduced outgrowth of branch n in phyB compared with low R:FR is due in part to increased numbers of active primary and secondary cauline apices (11.43 in phyB versus 8.32 in the low R:FR wild type). These apices may collectively contribute to repress lower branches, such as branch n, by correlative inhibition. The promotion of branch n outgrowth in low versus high R:FR may actually reflect the often described functionality of phyB as an inhibitor of shoot elongation. Therefore, branch elongation may result from antagonistic mechanisms mediated by phyB: inhibition through a typical phyB-mediated pathway, and promotion by a reduction in cauline branching. Trends in the next uppermost branch (branch n-1) elongation were complicated, since some of the low R:FR buds/branches were highly arrested while others elongated at higher rates. Overall, branch n-1 elongation was reduced in plants grown under low R:FR compared with high R:FR, and this inhibition was due to reduced average elongation rates. The elongation of most branches at position n-2 was greatly reduced by low R:FR. Suppression by low R:FR at this position resulted from a delay in the onset of elongation and reduced average elongation rates throughout the sampling period.
Phytochrome Control of the Degree of Correlative Inhibition
Overall architectural profiles presenting the lengths of all primary branches and the main inflorescence are provided in Supplemental Figure S6. Low R:FR and the phyB mutation increased the final length of the main stem compared with the wild type under high R:FR. While the main stem was taller in the phyB mutant or as a result of low R:FR, the branches placed at lower positions were increasingly shorter under these treatments. As a result of these contrasting effects, loss of phyB function and low R:FR resulted in a larger gradient in the lengths of primary rosette branches in all backgrounds tested.
Variation in the relative growth of branches at different nodes is due, at least in part, to correlative inhibition resulting from the inhibitory influence of the main shoot and other branches (Thimann and Skoog, 1934). This correlative inhibition may include potentially disparate modes of action associated with apical dominance and apical control (see introduction) and possibly other modes of action as well. To obtain an estimate of the degree of correlative inhibition acting on the primary rosette branches, the lengths of the longest three rosette branches were plotted against their nominal positions (position 1 = branch n, position 2 = branch n-1, and position 3 = branch n-2; Fig. 6). The slope of the resulting curve provided an index of correlative inhibition, with more negative slopes indicating greater inhibition. Other ongoing studies have shown that decapitation, blocking main shoot auxin transport, and removal of superior branches results in a decrease of this correlative inhibition index (S.R. Krishnareddy and S.A. Finlayson, unpublished data). Our results here demonstrated the dramatic increase in correlative inhibition associated with phyB loss of function and low R:FR. In spite of the differential effects of the phyB mutation and low R:FR on the outgrowth of the uppermost bud, the relative growth of branches in the top three positions remained coordinated, as evidenced by the linear correlative index slopes.
Figure 6.
The lengths of the top three primary rosette branches at 10 DPA plotted against nominal positions of various Arabidopsis genotypes with or without functional phyB and under high and low R:FR. The slope of each line provides a primary correlative inhibition index, with more negative values indicating greater correlative inhibition. Asterisks indicate significant differences between the slopes plotted for the various genotypes or light treatments at α = 0.05. Data are means ± se. For analyses comparing lines with or without functional phyB, n = 28 (phyBmax2) to 87 (wild type [WT]), average n = 51. For high and low R:FR, n = 26 (brc1) to 70 (wild type), average n = 37.
Selected Components of the Control of Shoot Architecture by Phytochrome Are Reduced in brc1, brc2, brc1brc2, axr1-12, max2, and max4
As in the wild type, the number of standardized primary rosette branches in brc2 was significantly reduced with loss of phyB function (Fig. 2C). phyB loss of function did not significantly reduce the number of standardized primary rosette branches in any of the other genotypes. Standardized primary rosette branch numbers of brc1, brc2, and brc1brc2 were not significantly different under either high or low R:FR (Fig. 2C). axr1-12 and axr1-12phyB produced greater numbers of buds (and standardized buds) than other genotypes, as a consequence of greater numbers of primary rosette leaves in these genotypes (Fig. 2B). There were no statistically significant differences in standardized primary rosette bud numbers in any genotype/treatment combinations besides those noted for the wild type/phyB (Fig. 3B).
Secondary rosette branching responses to phyB presence or absence were complex, with promotion (brc1brc2, max4), inhibition (brc2), or no effect (brc1, axr1-12, max2) noted in the various genotypes (Supplemental Fig. S4A). Secondary leaf number responses were similarly complex (Supplemental Fig. S4B). Conversely, low R:FR resulted in reduced secondary rosette branching and leaf numbers in all genotypes tested. Secondary cauline branching and leaf number responses to phyB presence or absence were also complex, while low R:FR again decreased both branch and leaf numbers (Supplemental Fig. S5, A and B).
In the wild-type background, both the phyB mutation and low R:FR enhanced correlative inhibition. These effects were reduced in the brc1 background and actually inverted (i.e. phyB mutation and low R:FR reduced correlative inhibition) in the brc1brc2 double mutant background (Fig. 6). The effects of the phyB mutation and low R:FR in the single brc2 background were complex. The linear relationship between branch lengths observed in other genotypes did not hold in phyBbrc2 or in brc2 grown under low R:FR, indicating that correlative inhibition was affecting branch development at specific positions differentially. The enhanced correlative inhibition caused by the phyB mutation was also lost in the axr1-12, max2, and max4 mutant backgrounds (Fig. 6). In addition, while branch lengths were increased by phyB deficiency in axr1-12 and max4, branch lengths of max2 were unaffected. In summary, the full control of correlative inhibition by phytochrome requires BRC1, BRC2, AXR1, MAX2, and MAX4.
The Effects of Phytochrome on the Expression of Branching-Related Genes in Unelongated Rosette Buds
Potential functional relationships between genes associated with branching (BRC1, BRC2), hormone physiology (TAA1, ARR5, DRM1), shade responses (ATHB-2), cell cycle regulation (PCNA1, CYCLINB1;1, CYCLIND2;1 [CYCD2;1], CYCD3;1), stem cell proliferation (CLV1, CLV3, STM, WUS), and branching phenotypes were examined by measuring expression in buds n and n-1 of the wild type and phytochrome mutants and of the wild type grown under high and low R:FR, just prior to the predicted onset of elongation of bud n (Fig. 7). Both buds n and n-1 typically formed branches in these genotypes/treatments, but the growth of bud n-1 was inhibited compared with bud n (see data above). Gene expression results from buds of phyA and phyAphyB were similar to those corresponding to the wild type and phyB, and the full data set is provided in Supplemental Figure S7.
Figure 7.
Abundance of various mRNAs in unelongated primary rosette bud n (uppermost bud) and bud n-1 (bud immediately below bud n) of wild-type (WT) and phyB plants grown under high R:FR (left panel of each graph) or wild-type plants grown under high and low R:FR (shaded right panel of each graph). Results are means of quantitative PCR analyses of four biological replicates ± se. Bars with different letters are significantly different at α = 0.05.
The mRNA abundances of ATHB-2 and BRC2 were elevated by both loss of phyB function and low R:FR, while WUS mRNA abundance was correspondingly reduced (Fig. 7). Therefore, the expression patterns of these three genes were consistent with regulation by R:FR transduced by phyB. The relationship between R:FR, phyB, and the expression of other genes was less clear. For instance, the expression of BRC1 was higher in phyB compared with the wild type but unaffected by R:FR, while CLV3 expression was increased by low R:FR but not by phyB deficiency. The differential expression patterns in response to R:FR and phyB status indicate complex regulation that in some cases may reflect bud outgrowth potential rather than phyB action per se (BRC1) and possible R:FR signal transduction by phytochrome(s) other than phyB (CLV3). In some instances, a logical relationship between the expression of genes with known or predicted functions and the growth characteristics of specific branches could not immediately be discerned. Correlation analysis, therefore, was used to further explore relationships between gene expression and branching.
The mRNA abundance of each gene tested was regressed with the mRNA abundance of all other genes, and correlation was determined (Supplemental Fig. S8). An interesting outcome of this analysis was that despite their sequence similarity, BRC1 and BRC2 grouped in different networks. The expression of BRC1 correlated (positively or negatively) with a set of genes including DRM1, TAA1, PCNA1, and WUS (Fig. 8). BRC2 expression correlated with a set of genes distinct from those that correlated with BRC1, including ARR5, CLV3, and ATHB-2 (Fig. 8). BRC1 and BRC2 expression were not correlated.
Figure 8.
Correlation analysis of the abundance of selected mRNA species regressed against the abundances of BRC1 and BRC2. Significant correlation at q = 0.10 is indicated by black squares.
Correlation analysis was also used to associate the expression of BRC1 and BRC2 from buds of all of the genotypes/treatments measured with two branching variables: one related to the specific bud fate (rosette branch length, which can be either promoted or reduced by the reduced phyB activity), and the other related to coordination of growth among branches (correlative inhibition). Since bud n and bud n-1 come from the same plants, the correlation with correlative inhibition was determined independently for each bud, because the data are not independent. BRC1 expression was significantly negatively correlated with branch elongation (Fig. 9). Other genes in the set that correlated with BRC1 expression (DRM1, TAA1, PCNA1, and WUS) also correlated with this parameter (Supplemental Fig. S9). On the other hand, BRC2 abundance showed significant negative correlation with the correlative inhibition index but not with branch elongation (Fig. 9). Genes in the set that correlated with BRC2 expression (ARR5, CLV3, and ATHB-2) likewise were correlated with the correlative inhibition index (Supplemental Fig. S9).
Figure 9.
Correlation analysis of the abundance of BRC1 and BRC2 mRNA regressed against primary rosette branch lengths (bud n [uppermost bud] and n-1 [bud immediately below bud n] data regressed together) and correlative inhibition (bud n and n-1 data regressed separately). Significant correlation at α = 0.01 is indicated by black squares.
DISCUSSION
Phytochrome Controls Bud Outgrowth in Arabidopsis
The presence of a reduced number of branches in phyB mutant Arabidopsis had already been reported (Reed et al., 1993). However, genetic or physiological reduction of phyB activity also accelerates the transition of the apex from the vegetative to the reproductive stage, interrupting the generation of leaves (Reed et al., 1993). Before trying to approach the molecular players in the control of branching by phytochrome, it was important to identify if this control was the direct consequence of flowering effects on leaf number (i.e. on the sites available for branch appearance) and/or whether phytochrome effects on branching resulted from effects on bud initiation or outgrowth. It was determined that low phytochrome activity caused some reduction in the number of buds produced (Fig. 3); however, it was concluded that the main pathway of phytochrome control of branching is via its promotion of bud outgrowth. This conclusion was supported by the observations that despite phytochrome effects on leaf and bud number, many buds with the potential to produce branches remain dormant in the wild type under low R:FR and in the phyB mutant under high R:FR, because there are far fewer branches than buds (Figs. 2 and 3). The analysis of the bud sites that did produce branches under high or low phytochrome activity conditions showed that phytochrome activity may promote bud outgrowth kinetics (Fig. 5). Finally, correlative inhibition (Thimann and Skoog, 1934), which regulates the sequential release and outgrowth of buds, was a major parameter regulated by R:FR perceived by phyB (Fig. 6). In sorghum, both loss of phyB function and low R:FR also resulted in strong repression of the outgrowth of the axillary buds (Kebrom et al., 2006). In fact, phyB-1 sorghum does not produce tillers until after flowering, which presumably occurs as a result of loss of the correlative inhibition imposed by the main shoot. Unlike the case with sorghum, Arabidopsis branching was not totally inhibited by loss of phyB function or low R:FR but instead was quantitatively repressed.
Non-Organ-Autonomous Hormone Pathways Transduce Phytochrome Branching Responses
Phytochrome activity affected leaf number, bud initiation, bud outgrowth, and stem elongation. Non-organ-autonomous hormone pathways defined by AXR1, MAX2, and MAX4 affected all four variables and were required for phytochrome effects on bud initiation and bud outgrowth parameters. For instance, correlative inhibition was not responsive to phyB status when any of these hormone components were nonfunctional. However, while rosette branches of axr1-12 and max4 maintained an elongation response to loss of phyB function, branches of max2 did not (Fig. 6). A study investigating the role of MAX2 in light signal transduction implicated MAX2 as contributing to the transduction of blue, R, and FR signals into the suppression of hypocotyl elongation (Shen et al., 2007), suggesting that MAX2 has a more general role in the control of elongation responses to phytochrome status.
BRC1 and BRC2 Control Phytochrome Branching Responses as Components of Different Signaling Networks
The teosinte tb1 gene, which suppresses axillary branching, was proposed to modulate plant architecture in response to environmental conditions through variations in its expression (Doebley et al., 1995). It was later shown that such changes in expression of the sorghum tb1 homolog were in fact regulated by R:FR perceived by phyB and that SbTB1 expression correlated with axillary bud outgrowth (Kebrom et al., 2006). The situation in Arabidopsis is complicated by the radiation of the ancestral tb1 homolog into three genes, TCP1, BRC1, and BRC2 (Howarth and Donoghue, 2006); therefore, the functional significance of BRC1 and BRC2 required further investigation. Functional BRC1 was essential for correlative inhibition responses to both phyB and low R:FR, whereas loss of BRC2 function disrupted, but did not eliminate, phyB and low R:FR correlative inhibition responses (Fig. 6). Previous experiments have shown that BRC1, but not BRC2, played a role in inhibiting branching at high planting density (Aguilar-Martínez et al., 2007). However, high-density planting is not directly comparable with low R:FR, since it can affect intercepted photosynthetic energy without appreciably altering the R:FR if common FR-poor fluorescent lamps are used as the light source. Additionally, the role of BRC2 is subtle and was only revealed by in-depth quantitative analysis.
BRC1 expression correlated with the specific fate of an individual bud, its growth capacity (rosette branch length), whereas BRC2 expression showed less variation at different bud positions with differential growth capacity. Instead, BRC2 expression correlated with growth coordination between different branches, the correlative inhibition index (Fig. 9). Furthermore, BRC1 and BRC2 are part of different gene expression networks (Fig. 8). It may be concluded that BRC1 and BRC2 are not functionally redundant, but each contributes to repress branching by unique pathways.
Phytochrome-Regulated Branching Pathways
The dissimilar elongation responses of the uppermost branches of phyB and the wild type under low R:FR (Fig. 5) indicate that phytochrome effects on branch outgrowth may integrate two opposing pathways. One pathway is defined by the promotion of branching by phytochrome-mediated inhibition of correlative inhibition, and the other is defined by the classical repression of stem elongation attributed to phytochrome, which manifests as increased shoot growth under shade. Regulation of stem elongation by the classical pathway is obvious in the case of main shoot lengths, where phyB deficiency and low R:FR result in increased plant height (Fig. 4). However, in the case of branch elongation, bud outgrowth is constrained by the correlative inhibition pathway and promoted by the classical pathway. Increased correlative inhibition could potentially result from increased apically derived branching inhibitor synthesis or transport from a more actively growing main shoot and cauline branches (phyB and the wild type under low R:FR) and also from elevated numbers of actively growing cauline branches (the wild type under low R:FR). Since both the correlative inhibition pathway and the classical pathway operate simultaneously, antagonism between these pathways ultimately determines the number of branches produced and their relative elongation.
In view of previous models (Morelli and Ruberti, 2002), analysis of shade avoidance mutants (Tao et al., 2008), and this study, it seems likely that phytochrome signals control plant architecture, at least in part, by modulating auxin physiology. The role of auxin in the control of plant architecture by phytochrome appears complex. Phytochrome has been shown to negatively regulate auxin biosynthesis via the TAA1-dependent pathway (Tao et al., 2008). However, in this study, phyB positively regulated TAA1 expression in the axillary bud (Fig. 7). TAA1 expression in the bud may reflect local auxin biosynthesis in the bud that promotes bud outgrowth, for instance by enabling vascularization of the bud by canalization (Sachs, 1969; Merks et al., 2007), in contrast to the bud-inhibiting basipetal auxin transport stream. Strigolactones may function downstream of the basipetal auxin transport stream (Brewer et al., 2009) to regulate the expression of the branching integrators BRC1 and BRC2 (Aguilar-Martínez et al., 2007; Finlayson, 2007). The exact targets of the integrators remain unknown; however, the expression of both BRC1 and BRC2 was demonstrated to correlate with the expression of distinct groups of known regulators of meristem function and also with distinct branching parameters (Figs. 8 and 9).
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Col ecotype of Arabidopsis (Arabidopsis thaliana) was used throughout. Wild-type Col-60000, axr1-12, and phyB (phyB-9) seeds were obtained from the Arabidopsis Biological Resource Center. brc1 (tbl1-1), brc2 (tcp12-1), and max2 (max2-336) mutant sources were described previously (Finlayson, 2007). max4 (max4-538) and phyA (phyA-575) were obtained by crossing SALK_147538 and SALK_014575 lines (Alonso et al., 2003), respectively, to the wild type and recovering individuals in the F2 that exhibited increased branching under white light or showed long hypocotyls under continuous FR.
Seeds were stratified for 3 d at 4°C and then sown in 36-cell trays (genotype comparisons) or 120-cm3 pots (low and high R:FR comparisons) filled with Metromix 200 soilless medium that was fertilized weekly with 4 mL of 1× Hoagland solution. Two different high R:FR light conditions were used in the study: one to grow all genotypes for comparison of PHYB versus phyB, and the other as a control for the low R:FR analyses. For PHYB/phyB comparisons between genotypes, plants were grown under 18-h photoperiods with 180 μmol m−2 s−1 photosynthetic photon flux density (PPFD) and a R:FR of 2.08 (Supplemental Fig. S10) and 24°C/18°C day/night temperatures. This light was provided by a mixture of T8 cool-white high output and incandescent lamps. For comparisons between wild-type, brc1, brc2, and brc1brc2 plants grown under low and high R:FR, plants were grown with 24°C/18°C day/night temperatures under 18-h photoperiods with 175 μmol m−2 s−1 PPFD and a R:FR of 2.98 (high R:FR) or grown for 7 d under these conditions following which the R:FR was reduced to 0.05 (low R:FR; Supplemental Fig. S10). This light was provided by a mixture of T12 very high output and compact fluorescent lamps with an overhead array of light-emitting diodes mounted in a clear acrylic sheet providing supplemental FR (735 nm) to the low R:FR plants. Light was measured with a Li-1800 spectroradiometer (Li-Cor). R:FR was calculated as the quantum flux density from 655 to 665 nm divided by the quantum flux density from 725 to 735 nm.
Architectural Analysis
Architectural characteristics were measured at 10 DPA. Leaf numbers were counted and the number of buds or meristems formed in each rosette axil was determined with a dissecting microscope. The lengths of the primary rosette and cauline branches (axis > 3 mm) and the main inflorescence were measured. The number of nodes of each primary rosette and cauline branch and the number of elongated secondary branches (axis > 3 mm) were recorded.
Analysis of Branch Elongation
The lengths of the three uppermost buds of selected genotypes were measured each day, beginning 2 d before anthesis. Branching rates were calculated by dividing the daily elongation increment by 24 h, and maximum sustained elongation rates were calculated by averaging the three greatest sequential daily elongation rates. The delay in the onset of elongation was determined by estimating the time that a branch achieved 3 mm of length using a second-order polynomial fitted to the early part of the elongation curve.
Analysis of Gene Expression
The uppermost (n) and next most uppermost (n-1) buds were collected separately just prior to the predicted onset of outgrowth of the uppermost bud. Bud n-1 was slightly less developed than bud n at the time of sampling, consistent with the slight delay in the onset of outgrowth of bud n-1 (calculated as 0.46 d in the wild type and 0.81 d in phyB). Total RNA was extracted using TRIzol (Invitrogen), the RNA samples were suspended to equal concentration, and subsamples were separated on glyoxal agarose gels (1%) to evaluate quality. Four micrograms of total RNA for each sample was digested with 3.5 units of DNaseI for 60 min and then reextracted with TRIzol (Invitrogen). cDNA was synthesized with the SuperScript III kit using random hexamers (Invitrogen). The cDNA was diluted 1:5, and 10-μL real-time PCRs were run in duplicate on an ABI 7900 SDS with corresponding minus-reverse transcriptase controls using the SYBR Green Jumpstart kit (Sigma) or the Power Sybr kit (ABI). A standard curve for each gene target was generated from a dilution series of known concentrations of amplicons (all except BRC1 and BRC2) or the cloned BRC1 and BRC2 genes in plasmid vectors and measured by the same quantitative PCR process. Target threshold cycle values were determined, converted to absolute transcript numbers per reaction, and normalized to 18 s rRNA transcript levels. Four biological replicates were measured for each genotype/treatment, and each replicate represented the RNA extracted from a pool of approximately 12 buds. Primer combinations are given in Supplemental Table S2.
Statistics
ANOVA followed by a Duncan's test (for more than two comparisons) or a t test (two comparisons) was used (SPSS 15.0) where differences between means were assessed and significance was determined at α = 0.05. Correlation significance was assessed using an F test (SPSS 15.0) with α = 0.05 and α = 0.01. Differences between correlative inhibition indices were determined by comparing the 95% confidence intervals of the slopes. Standardized branching parameter values and their 95% confidence intervals were derived using the regression curve and associated errors of the specific parameter versus leaf numbers to derive the parameter value at the corresponding phyB or low R:FR leaf number (Supplemental Fig. S1B). For correlation between gene expression levels, P values were transformed into q values (false discovery rate within the set of significant correlations) by the method of Storey and Tibshirani (2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF062581 (BRC1), AM408561 (BRC2), AF053746 (DRM1), NM_105724 (TAA1), NM_100611 (PCNA1), NM_117780 (ATHB-2), NM_001124926 (CLV3), NM_127349 (WUS), NM_114679 (ARR5), NM_119913 (CYCB1;1), NM_127815 (CYCD2;1), NM_119579 (CYCD3;1), NM_106232 (CLV1), NM_104916 (STM), and X16077 (18S).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Time to anthesis of various Arabidopsis genotypes with or without functional phyB (left panel of each graph) or under high and low R:FR (shaded right panel of each graph).
Supplemental Figure S2. Regressions of primary rosette branch numbers versus primary rosette leaf numbers of various Arabidopsis genotypes with or without functional phyB (top two rows) or under high and low R:FR (shaded bottom row).
Supplemental Figure S3. Regressions of primary rosette bud numbers versus primary rosette leaf numbers of various Arabidopsis genotypes with or without functional phyB (top two rows) or under high and low R:FR (shaded bottom row).
Supplemental Figure S4. Secondary branching parameters of primary rosette branch n (uppermost rosette branch) of various Arabidopsis genotypes at 10 DPA with or without functional phyB (left panel of each graph) or under high and low R:FR (shaded right panel of each graph).
Supplemental Figure S5. Secondary branching parameters of primary cauline branch n (lowest cauline branch) of various Arabidopsis genotypes at 10 DPA with or without functional phyB (left panel of each graph) or under high and low R:FR (shaded right panel of each graph).
Supplemental Figure S6. Main shoot height (longest bar), lengths of cauline branches (to left of longest bar, with lowest branch to immediate left of longest bar), and lengths of rosette branches (to right of longest bar) of various Arabidopsis genotypes at 10 DPA grown under high R:FR with or without functional phyB (left panels) or under high and low R:FR (shaded right panels).
Supplemental Figure S7. Abundance of various mRNAs in unelongated primary rosette bud n (uppermost bud) and bud n-1 (bud immediately below bud n) of wild-type, phyA, phyB, and phyAphyB plants grown under high R:FR (left panel of each graph) or wild-type plants grown under high and low R:FR (shaded right panel of each graph).
Supplemental Figure S8. Correlation matrix of the abundance of each measured mRNA species regressed against all others.
Supplemental Figure S9. Correlation analysis of the mRNA abundance of selected genes regressed against rosette branch lengths (bud n [uppermost bud] and bud n-1 [bud immediately below bud n] data regressed together) and correlative inhibition (bud n and n-1 data regressed separately).
Supplemental Figure S10. Spectra of light sources used in the experiments.
Supplemental Table S1. Selected architectural data for phyA and phyAphyB at 10 DPA grown under high R:FR.
Supplemental Table S2. Sequences of primers used for quantitative PCR.
Supplementary Material
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ballaré CL. (1999) Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends Plant Sci 4: 97–102 [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563 [DOI] [PubMed] [Google Scholar]
- Boe A, Beck DL. (2008) Yield components of biomass in switchgrass. Crop Sci 48: 1306–1311 [Google Scholar]
- Bonser SP, Aarssen LW. (2003) Allometry and development in herbaceous plants: functional responses of meristem allocation to light and nutrient availability. Am J Bot 90: 404–412 [DOI] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I. (1996) Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA 93: 3530–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I. (1993) The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J 4: 469–479 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Deregibus VA, Sanchez RA. (1985) Variations in tiller dynamics and morphology in Lolium multiflorum Lam. vegetative and reproductive plants as affected by differences in red/far-red irradiation. Ann Bot (Lond) 56: 553–559 [Google Scholar]
- Casal JJ, Sanchez RA, Deregibus VA. (1986) The effect of plant density on tillering: the involvement of R/FR ratio and the proportion of radiation intercepted per plant. Environ Exp Bot 26: 365–371 [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Cline MG. (1996) Exogenous auxin effects on lateral bud outgrowth in decapitated shoots. Ann Bot (Lond) 78: 255–266 [Google Scholar]
- Cline MG. (1997) Concepts and terminology of apical dominance. Am J Bot 84: 1064–1069 [PubMed] [Google Scholar]
- Cline MG, Bhave N, Harrington CA. (2009) The possible roles of nutrient deprivation and auxin repression in apical control. Trees (Berl) 23: 489–500 [Google Scholar]
- Cline MG, Sadeski K. (2002) Is auxin the repressor signal of branch growth in apical control? Am J Bot 89: 1764–1771 [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Deruère J, Kieber JJ. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MH, Simmons SR. (1994) Tillering response of barley to shifts in light quality caused by neighboring plants. Crop Sci 34: 1604–1610 [Google Scholar]
- de Jager SM, Maughan S, Dewitte W, Scofield S, Murray JAH. (2005) The developmental context of cell-cycle control in plants. Semin Cell Dev Biol 16: 385–396 [DOI] [PubMed] [Google Scholar]
- Deregibus VA, Sanchez RA, Casal JJ. (1983) Effects of light quality on tiller production in Lolium spp. Plant Physiol 72: 900–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C. (1995) teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Donohue K, Schmitt J. (1999) The genetic architecture of plasticity to density in Impatiens capensis. EVol 53: 1377–1386 [DOI] [PubMed] [Google Scholar]
- Emery RJN, Longnecker NE, Atkins CA. (1998) Branch development in Lupinus angustifolius L. II. Relationship with endogenous ABA, IAA and cytokinins in axillary and main stem buds. J Exp Bot 49: 555–562 [Google Scholar]
- Finlayson SA. (2007) Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol 48: 667–677 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC. (2005) Phytochromes and shade-avoidance responses in plants. Ann Bot (Lond) 96: 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García del Moral MB, García del Moral LF. (1995) Tiller production and survival in relation to grain yield in winter and spring barley. Field Crops Res 44: 85–93 [Google Scholar]
- Gocal GFW, Pharis RP, Yeung EC, Pearce D. (1991) Changes after decapitation in concentrations of indole-3-acetic acid and abscisic acid in the larger axillary bud of Phaseolus vulgaris L. cv. Tender Green. Plant Physiol 95: 344–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 180–194 [DOI] [PubMed] [Google Scholar]
- Hempel FD, Feldman LJ. (1994) Bi-directional inflorescence development in Arabidopsis thaliana: acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192: 276–286 [Google Scholar]
- Howarth DG, Donoghue MJ. (2006) Phylogenetic analysis of the “ECE” (CYC_TB1) clade reveals duplications predating the core eudicots. Proc Natl Acad Sci USA 103: 9101–9106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T, Bergelson J. (2000) The evolution of compensation to herbivory in scarlet gilia, Ipomopsis aggregata: herbivore-imposed natural selection and the quantitative genetics of tolerance. EVol 54: 764–777 [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA. (2006) Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol 140: 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Lortie CJ, Aarssen LW. (2000) Fitness consequences of branching in Verbascum thapsus (Scrophulariaceae). Am J Bot 87: 1793–1796 [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Medford JI, Horgan R, El-Sawi Z, Klee HJ. (1989) Alterations of endogenous cytokinins in transgenic plants using a chimeric isopentenyl transferase gene. Plant Cell 1: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merks RMH, Van de Peer Y, Inzé D, Beemster GTS. (2007) Canalization without flux sensors: a traveling-wave hypothesis. Trends Plant Sci 12: 383–390 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Morelli G, Ruberti I. (2002) Light and shade in the photocontrol of Arabidopsis growth. Trends Plant Sci 7: 399–404 [DOI] [PubMed] [Google Scholar]
- Ongaro V, Leyser O. (2008) Hormonal control of shoot branching. J Exp Bot 59: 67–74 [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Peng S, Khush GS, Cassman KG. (1994) Evolution of the new plant ideotype for increased yield potential. Cassman KG, , Breaking the Yield Barrier: Proceedings of a Workshop on Rice Yield Potential in Favorable Environments. International Rice Research Institute, Los Banos, Philippines, pp 5–20 [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin CH, Hay MJM, Newton PCD, Greer DH. (1994) Effect of light quality (red:far-red ratio) at the apical bud of the main stolon on morphogenesis of Trifolium repens L. Ann Bot (Lond) 74: 119–123 [Google Scholar]
- Sachs T. (1969) Polarity and the induction of organized vascular tissues. Ann Bot (Lond) 33: 263–275 [Google Scholar]
- Sachs T, Thimann KV. (1964) Release of lateral buds from apical dominance. Nature 201: 939–940 [Google Scholar]
- Sachs T, Thimann KV. (1967) The role of auxins and cytokinins in the release of buds from dominance. Am J Bot 54: 136–144 [Google Scholar]
- Schmitz G, Tillman E, Carriero F, Fiore C, Cellini F, Theres K. (2002) The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci USA 99: 1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA 96: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E. (2007) The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol 145: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. (1995) Physiological and ecological function within the phytochrome family. Annu Rev Plant Physiol Plant Mol Biol 46: 289–315 [Google Scholar]
- Sorefan K, Booker J, Haurogne K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al. (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom JP, Ripley BD, Devitt ML, Drake B. (1998) Dormancy-associated gene expression in pea axillary buds. Planta 205: 547–552 [DOI] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Chatfield SP, Leyser HMO. (1999) AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol 121: 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HMO. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance of genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Kiba T, Sakakibara H, Ueguchi C, Mizuno T, Sugiyama T. (1998) Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett 429: 259–262 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer J, Ljung K, Pojer F, Hong F, Long J, Li L, Moreno J, Bowman M, Ivans L, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann KV, Skoog F. (1934) On the inhibition of bud development and other functions of growth substances in Vicia faba. Proc R Soc Lond 114: 317–339 [Google Scholar]
- Tucker MR, Laux T. (2007) Connecting the paths in plant stem cell regulation. Trends Cell Biol 17: 403–410 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormone. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Van Kluenen M, Fischer M. (2001) Adaptive evolution of plastic foraging responses in a clonal plant. Ecology 82: 3309–3319 [Google Scholar]
- Wan C, Sosebee RE. (1998) Tillering responses to red:far-red light ratio during different phenological stages in Eragrostis curvula. Environ Exp Bot 40: 247–254 [Google Scholar]
- Zarrough KM, Nelson CJ, Coutts JH. (1983) Relationship between tillering and forage yield of tall fescue. I. Yield. Crop Sci 23: 333–337 [Google Scholar]
- Zhao DL, Atlin GN, Bastiaans L, Spiertz JHJ. (2006) Developing selection protocols for weed competitiveness in aerobic rice. Field Crops Res 97: 272–285 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.