Abstract
Many well-characterized examples of antisense RNAs from prokaryotic systems involve hybridization of the looped regions of stem–loop RNAs, presumably due to the high thermodynamic stability of the resulting loop–loop and loop–linear interactions. In this study, the identification of RNA stem–loops that inhibit U1A protein binding to the hpII RNA through RNA–RNA interactions was attempted using a bacterial reporter system based on phage λ N-mediated antitermination. As a result, loop sequences possessing 7–8 base complementarity to the 5′ region of the boxA element important for functional antitermination complex formation, but not the U1 hpII loop, were identified. In vitro and in vivo mutational analysis strongly suggested that the selected loop sequences were binding to the boxA region, and that the structure of the antisense stem–loop was important for optimal inhibitory activity. Next, in an attempt to demonstrate the ability to inhibit the interaction between the U1A protein and the hpII RNA, the rational design of an RNA stem–loop that inhibits U1A-binding to a modified hpII was carried out. Moderate inhibitory activity was observed, showing that it is possible to design and select antisense RNA stem–loops that disrupt various types of RNA–protein interactions.
INTRODUCTION
RNA–protein interactions play important roles in gene regulation, in the assembly of functional RNA–protein complexes such as the ribosome, and in viral replication. Therefore, molecules that regulate specific RNA–protein interactions provide an attractive means to dissect molecular steps of various biological processes, and to establish the validity of targeting an RNA–protein interaction for future drug design.
Various strategies have been developed for the inhibition of RNA–protein interactions, and can be classified into two groups depending on whether the protein or the RNA is targeted. Methods for targeting the protein include the use of RNA decoys or in vitro selected DNA or RNA aptamers. In the case of the human immunodeficiency virus (HIV) regulatory proteins Tat and Rev, RNA decoys corresponding to the respective RNA sites, the trans-activating response region (TAR) and the Rev-responsive element (RRE), as well as aptamers have been shown to inhibit viral replication (1). In particular, several Rev aptamers with affinities significantly higher than the wild-type RRE that compete with the RRE for Rev-binding have been generated (2).
Approaches for targeting RNA range from the use of small molecules (3) and peptides (4) to nucleic acid-based agents such as antisense RNA/DNA(5), siRNA(6) and aptamers (7,8). Targeting RNA using small molecules is a particularly attractive approach because such molecules may directly lead to the development of therapeutic agents; however, the desired specificity has been difficult to achieve by such compounds (3). On the other hand, nucleic-acid-based agents, such as antisense RNA/DNA and siRNA, have been shown to be effective in regulating gene expression, and a useful tool in elucidating molecular mechanisms (9,10). However, stable RNA secondary structure formation has been known to be an obstacle for both antisense oligonucleotides (11) and siRNA (12).
In many prokaryotic antisense control systems, RNA stem–loops are used for initial recognition, resulting in hairpin loop–loop (‘kissing’) and loop–linear interactions (13–15). Loop–loop interactions are also observed in RNA folding (16–18) and in the dimerization of retroviral genomic RNAs (19–21). These interactions appear to have been optimized for rapid and stable intermolecular interactions which are essential for their function (22). While loop–loop interactions generally use only five to seven complementary base pairs to join the two hairpin loops, this short complementary region may be an advantage since increasing affinity by increasing complementarity may be a source of decreased specificity (23,24).
However, the rational design of novel loop–loop interactions is not straightforward because the factors governing stable loop–loop complex formation appear to be complex and diverse, and the stability of loop–loop interactions are difficult to predict (25). For example, the stability of the extensively studied loop–loop interaction derived from RNA I and RNA II from plasmid ColE1, which consists of seven bases in the loop, of which all seven form base pairs, has been shown to increase 350-fold by simply inverting the loop sequences of the hairpins 5′ to 3′ (26). In this case, the major determinant of complex stability was found to be the identity of the base at the first and seventh position in the loop (27). An in vitro selected antisense stem–loop targeting the HIV TAR with a six base-pair loop, has an eight base loop with a closing G-A base-pair that has been shown to be crucial for stable complex formation (28). In the case of the dimerization initiation site (DIS) of HIV, six of the nine loop bases participate in base-pair formation, while the remaining three purine bases are important for stacking interactions (29–32). Surprisingly, stable loop–loop complexes with only two intermolecular G-C base-pairs have also been found (33).
In this study, we have attempted to identify RNA stem–loops that inhibit RNA–protein interactions through the formation of loop–loop interactions between the antisense RNA stem–loop and the target RNA structure. The complex formed between hairpin II of U1 snRNA (U1 hpII) and U1A protein, which is a component of the U1 snRNP, was chosen as a target (34). U1 hpII RNA contains a 10-nt apical loop, which is recognized by the N-terminal RRM of U1A protein with high specificity and affinity (35), and was expected to be a potential target for kissing complex formation. As it is difficult in general to predict the stability of loop–loop interactions as described above, an RNA stem–loop library was screened for sequences that bind to the target U1 hpII loop and inhibit U1A protein binding. A bacterial two-plasmid system for detecting RNA–polypeptide interactions based on bacteriophage λ N protein-mediated antitermination was used (Figure 1A) (36). In this system, N protein is expressed from a pBR322-based N expressor plasmid and LacZ is expressed from a pACYC184-based reporter plasmid containing the nut site (boxA–boxB) and four terminators upstream of LacZ. Binding of the N-terminal RNA-binding domain of N (N peptide) to the nut site boxB stem–loop of the nascent RNA transcript nucleates the formation of an antitermination complex, which includes the bacterial host factors NusA, NusB, NusG and S10, thereby causing transcription antitermination by RNA polymerase and expression of LacZ. This system can be modified to study heterologous RNA–polypeptide interactions by replacing the pBR and pACYC plasmid DNA regions corresponding to the N peptide and boxB RNA with those of the peptide and RNA of interest (37).
Figure 1.
The strategy and design of a bacterial assay for detecting RNA loop–loop interactions that inhibit U1A–hpII-mediated antitermination complex formation. (A) A bacterial two-plasmid system based on phage λ N-mediated antitermination for detection of RNA–polypeptide interactions. (B) The secondary structure of the region of the RNA transcript containing the target U1 hpII and downstream RNA stem–loop library. The randomized nucleotides are indicated by N, and the regions corresponding to the PstI, BsrGI, and BamHI sites are shown in italic. (C) Possible mechanism for the disruption of antitermination complex formation by antisense RNA stem–loops.
In this study, the N/boxB interaction was replaced by that of the U1A protein and the U1 hpII RNA, and an RNA stem–loop library with a completely randomized 10-nt loop was placed 38 nt downstream of U1 hpII (Figure 1B). Individual clones showing repressed reporter gene expression were isolated and the sequences were analyzed for intramolecular RNA–RNA binding. Contrary to our expectations, it was found that the selected RNA stem–loop was most likely binding to the boxA region, which is an essential element in antitermination complex formation (Figure 1C, left) (38,39). We therefore used a rational approach to design RNA stem–loops that inhibit the binding of the U1A protein to a U1 hpII variant (Figure 1C, right). The result shows that it is possible to inhibit RNA–protein interaction in an efficient way using antisense RNA stem–loops.
MATERIALS AND METHODS
Selection of RNA stem–loops that inhibit U1A-mediated antitermination complex formation using a bacterial reporter assay
The pAC hpII reporter plasmid containing a randomized stem–loop library downstream of the U1 hpII site was constructed in the following manner. A synthetic oligonucleotide cassette containing the boxA of nut, U1 hpII with a 9-bp stem, and a BsrGI site (denoted boxA–hpII; Tables S1 and S2) was cloned into the unique PstI and BamHI sites of pAC nut (40), to give pAC hpII. RNA stem–loop library dsDNA was prepared by annealing the two synthetic oligonucleotides, BsrGI-linker-1 and random-stem–loop (Tables S1 and S2) complementary at the 3′-end, and second-strand synthesis with Taq polymerase. The resulting dsDNA was introduced into the BsrGI and BamHI sites of the pAC hpII plasmid.
The procedure for the in vivo selection of library sequences that resulted in reduced antitermination activity was based on a previously described method for peptide selection of RNA binders (4,37). For the primary screen, the RNA stem–loop library plasmid was prepared by ligation of the library insert (7.5 ng) into pAC hpII reporter plasmid (250 ng), followed by phenol extraction and concentration to 10 µl using a filter unit (Montage PCR, Millipore). Plasmids were electroporated into N567/pBR U1A-N cells (80 µl) in 1-mm cuvettes at 2.0 kV using 1 µl of the above solution, Super Optimal Broth (SOC) medium (5 ml) was added immediately after electroporation, and cells were allowed to recover by incubating at 37°C; for 1 h. Transformants were spread onto tryptone plates (ϕ150 mm) containing ampicillin (100 µg/ml), chloramphenicol (20 µg/ml), isopropyl β-d-1-thiogalactopyranoside (IPTG; 0.05 mM) and X-gal (80 µg/ml) and incubated at 37°C; for 28 h. A total of 1.5 × 105 colonies were obtained, and the degree of colony color was visually scored by comparison with the blue intensity of a standardized set of controls that included the U1A–U1 hpII interaction (5+), as well that of the RSG-1.2 peptide and the HIV Rev-response element (RRE) (2+), the HIV Rev peptide and the RRE (1+), U1A and the RRE (0.5+) and the Rev peptide and U1 hpII (0). Individual light blue colonies (white to 3+; 1056) were then grown to saturation in 96-well plates containing tryptone and antibiotics, cultures were pooled and plasmid DNA was isolated. The library region of the selected pAC plasmid was amplified by polymerase chain reaction (PCR) using pAC forward primer (5′-GGCTTATCGAAATTAATACG-3′) and reverse primer (5′-ACGGTAAGAGTGCCAGTG-3′). The amplified fragments were then digested with BsrGI and BamHI, phenol-extracted and purified on a native 8% polyacrylamide gel.
In the secondary screen, library inserts from the primary screen (white to 3+ and >4+) were reintroduced into the pAC hpII reporter plasmid. Ligation mixtures were phenol-extracted and concentrated using Montage PCR (Millipore), and individually electroporated into N567/pBR U1A-N cells as described above. Transformants were spread onto X-gal plates, and plasmid DNA from individual colonies with various intensity of blue color were isolated and used to transform N567/pBR U1A-N cells by heat shock to confirm activity. The sequences of 25 clones with varying colony colors were determined.
Mutational analysis of intramolecular RNA stem–loop binding to boxA
Reporter plasmids with base substitutions in the boxA and antisense stem–loop region were prepared by ligating synthetic double-stranded inserts corresponding to the boxA–hpII (PstI–BsrGI fragments) and the antisense stem–loop (BsrGI–BamHI fragments) region into the PstI and BamHI sites of the pAC plasmid. The boxA–hpII inserts were prepared by annealing two complementary oligonucleotides listed in Supplementary Table S1 in the combinations indicated in Supplementary Table S2. BoxA–hpII and boxA–hpII A9 dsDNAs were digested with BsrG I, phenol-extracted and ethanol-precipitated. Inserts corresponding to the antisense stem–loops of clones 1-4, 1-2 and 1-8 were amplified by PCR using a pAC forward primer and reverse primer from selected plasmids, then digested with BsrGI and BamHI, phenol-extracted, and purified on a 10% polyacrylamide gel. antisense stem–loops with base substitutions were prepared by the same method as the preparation of library insert using the oligonucleotides listed in Supplementary Table S1 in the combinations indicated in Supplementary Table S2. The activities of these constructs were assessed using the LacZ colony color assay with N567/pBR U1A-N cells, and quantitated by using a β-galactosidase solution assay (41).
Native PAGE analysis of intra- and intermolecular RNA–RNA interactions between the boxA region and the selected antisense RNA stem–loops
RNA substrates (114-mer) for intramolecular RNA binding experiments and 18- or 34-mer RNAs for intermolecular RNA-binding experiments were transcribed in vitro by T7 RNA polymerase. DNA templates for 114-mer RNAs were PCR-amplified from the pAC constructs used in the in vivo antitermination assay using a T7 boxA primer and a reverse primer (Supplementary Table S3). 114-mer RNA substrates (a total of 48 pmol per experiment) in H2O (typically 24 µl) were heated at 95°C for 5 min and immediately cooled on ice for 5 min. To this solution, 4X PN buffer was added to give a final concentration of 10 mM sodium phosphate (pH 7.0) and 50 mM NaCl (1X PN buffer) and the mixture was incubated at 37°C; for 30 min, cooled on ice for 20 min and one-fourth of the volume of loading buffer containing 50% glycerol was added. One-fourth of this solution was analyzed separately by gel electrophoresis at 4°C; on 10% native polyacrylamide gels (acrylamide:methylenebisacrylamide = 20:1) in TBM (89 mM Tris, 89 mM borate, and 0.1 and 0.5 mM MgCl2). RNA bands were visualized by staining with ethidium bromide and irradiation with a UV trans-illuminator.
Gel mobility shift assays using 32P-labeled RNAs were carried out as follows. DNA templates for boxA (18mer) and antisense stem–loop (34mer) RNAs were prepared by annealing synthetic oligonucleotides (Supplementary Table S3) and T7 promoter DNA (5′-GTAATACGACTCACTATA-3′). Internally 32P-labeled RNAs (SL1-4 and SL1-4 GC) redissolved to 20 nM in 1X PN buffer were heated at 95°C and quickly cooled on ice. The RNAs were diluted to 5 nM with 1X PN buffer containing 50% glycerol. Unlabeled RNAs (boxA and boxA GC) diluted to 64 µM in 1X PN buffer were heated at 95°C and quickly cooled on ice, then diluted to 0.25–32 µM in 1X PN buffer. The 5 nM labeled RNAs (0.5 µl) were mixed with 4 vol of 1X PN buffer or the unlabeled RNAs (0.25–64 µM) and incubated at 37°C for 30 min, then chilled on ice for 20 min and subjected to 12% PAGE in TBM (0.1 mM MgCl2) at 4°C. The gels were dried up on 3MM Chromatography paper (Whatman) and analyzed with a fluorescence/radioisotope image analyzer (Fujifilm FLA-2000).
Inhibition of U1A protein binding to a mutant U1 hpII RNA hairpin by designed antisense stem–loop RNAs
pAC reporter plasmids containing mutant U1 hpIIs were constructed by insertion of synthetic oligonucleotide cassettes prepared using the oligonucleotides in Supplementary Table S4 in the combinations shown in Supplementary Table S5 into the PstI and BamHI site of pAC nut− as described above. The U1A protein-binding activities of the resulting hpII RNA mutants were determined by transformation of N567/pBR U1A cells using the resulting pAC hpII mutant plasmids, and scoring antitermination activity as described above.
pAC reporter plasmids containing the U1 hpII Δ16/18A/10G (hpIIΔDIS) mutant and a downstream BsrGI site for subsequent cloning were prepared by annealing oligonucleotides shown in Supplementary Table S4, and inserting into the PstI and BamHI site of pAC nut− to yield pAC hpIIΔDIS. Oligonucleotide cassettes encoding the downstream antisense stem–loop structures, prepared by annealing oligonucleotide shown in Supplementary Table S4 in the combinations shown in Supplementary Table S5, second-strand synthesis, and digestion by BsrGI and BamHI, were inserted into the BsrGI and BamHI site of pAC hpIIΔDIS to give pAC hpIIΔDIS–aSL. Inhibition of the U1A–hpIIΔDIS interaction by the antisense stem–loops was determined by transformation of N567/pBR U1A cells using the resulting pAC hpIIΔDIS–aSL plasmids, and scoring antitermination activity as described above.
Native PAGE analysis of inhibition of the U1A-hpIIΔDIS interaction by the antisense stem–loops
RNAs were transcribed in vitro by T7 RNA polymerase from synthetic template DNAs (Supplementary Table S6) annealed with T7 promoter DNA. The plasmid expressing U1A with a C-terminal (His)6-tag was constructed as follows. The sequence of U1A1-102 followed by a glycine spacer was PCR-amplified from pBR U1A-N (40) and cloned into pBAD/Myc-His A (Invitrogen) at the NcoI and EcoRI sites. The generated plasmid was transformed into TOP 10 E. coli cells, and U1A-Myc-His was expressed by the induction of L-arabinose and purified using a Ni-NTA column (QIAGEN). The purified U1A-Myc-His protein was dialyzed against dialysis buffer containing 50 mM NaH2PO4, 300 mM NaCl and 20% glycerol and the concentration of the protein was determined by UV absorption at 280 nm using ε = 5120 M–1 cm–1(42). The protein was diluted to 163.84 µM with dialysis buffer and further diluted with 4 vol of TBS containing Triton X-100 [20 mM Tris–HCl (pH 7.6), 50 mM NaCl and 0.5 % Triton X-100]. The diluted protein was serially diluted to 0.125–16384 nM with the 1:4 mixture of dialysis buffer and TBS containing Triton X-100. Unlabeled RNAs (antisense stem-loop SL and antisense stem-loop SL 15C) diluted to 8192 nM in 1X PN buffer were heated at 95°C and quickly cooled on ice, then diluted to 8-4096 nM in 1X PN buffer. 32P-labeled RNAs (hpII and hpIIΔDIS) diluted to 2 nM in 1X PN buffer were heated at 95°C, quickly cooled on ice and diluted to 0.1 nM for hpII or 0.8 nM for hpIIΔDIS with 1X PN buffer. The labeled RNAs (2.5 µl) were mixed with the same volume of 4X binding buffer [40 mM Tris–HCl (pH 7.6) 850 mM NaCl, 1.2 mM MgCl2, 1.0 mg/ml tRNA and 40% glycerol]. To this mixture, 2.5 µl of the U1A protein solution (0 or 0.125–32768 nM) and 2.5 µl of the unlabeled RNA solution (0 or 8-8192 nM) were added and incubated at room temperature (22°C) for 20 min and further on ice for 20 min. Then, the mixtures (10 µl) were subjected to 12% PAGE in TBM (0.4 mM MgCl2) at 4°C. The gels were analyzed as described above.
RESULTS
Screening for RNA stem–loops that inhibit RNA–protein interactions using a bacterial reporter assay
The bacterial reporter system for detecting RNA–protein interactions described above was modified so that antisense RNA stem–loops that inhibit the U1A-hpII interaction through RNA–RNA interactions may be identified (Figure 1A and B). First, a DNA insert containing the boxA site followed by the wild-type U1 hpII stem–loop with an additional 4 bp in the stem to stabilize the structure and a BsrGI site was inserted into the PstI and BamHI sites of the pAC reporter plasmid, thereby yielding the pAC hpII reporter plasmid. Next, an RNA stem–loop library, consisting of a 9-bp stem with a 10-nt randomized loop encoding 410 = 1.0 × 106 sequences, was introduced 38 nt downstream of the hpII stem–loop using the BsrGI and BamHI sites (Figure 1B). N567/pBR U1A-N cells that express the U1A-N fusion protein were transformed using the pAC library plasmid and spread onto tryptone plates containing X-gal. Colonies exhibiting lighter blue intensities compared with that of the U1 hpII–U1A interaction, scored as 5+, were selected. Antitermination activities were visually scored by colony color intensities as the number of plusses, with more plusses indicating stronger intensities, by comparison with a set of standardized controls. We have previously shown that antitermination activities visually scored as the intensity of colony color correlate well with the stability of the RNA–peptide interaction replacing the boxB RNA and N peptide, and provide an accurate measure for the stability of the antitermination complex (37,43).
In the primary screen, ∼1.5 × 105 colonies were screened, of which ∼1.8% were white or light blue (white, 0.49%; 1+, 0.09%; 2+, 0.54%; and 3+, 0.65%) compared to the U1 hpII–U1A interaction (5+). A total of 1056 light blue colonies with varying blue intensity (white, 96; 1+, 125; and 2+∼3+, 835) and 96 blue colonies (>4+) as a control were individually pooled and plasmid DNA was isolated. In the secondary screen, to eliminate pAC reporter plasmid-related false-negative clones (37), the library region was PCR-amplified, reintroduced into pAC U1 hpII reporter plasmid and transformed into N567/pBR U1A cells yielding ∼9.7 × 103 colonies, of which ∼29% were white or light blue (white, 2.0%; +, 0.8%; 2+, 1.3%; and 3+∼4+, 24.9%) on X-gal plates. The library region of the control plasmid DNA isolated from the pool of blue colonies (>4+) was also reintroduced into pAC U1 hpII reporter plasmid, and transformed into N567/pBR U1A cells yielding ∼1.3 × 103 colonies with varying blue intensity (white, 1.8%; 1+, 0.2%; 2+, 1.3%; 3+∼4+, 55.4%; >5+, 27.9%; and 13.4% that could not be determined due to small colony size) on X-gal plates.
Plasmids were isolated from clones of varying blue intensity and reintroduced into N567/pBR U1A cells to confirm colony color. As a result, 13 unique sequences with colony colors 1 + ∼4+ were found, and the six clones exhibiting weak colony color (1 + ∼3+) possessed 7–8 base complementarity to the 5′ region of the boxA element, and not to U1 hpII loop (Table 1). This suggested that these stem–loops bind to the boxA region and inhibit binding of host factors (NusB and S10), which are important for antitermination (38,39,44). Interestingly, three clones exhibiting particularly low antitermination activity, clone 1-2, 1-4 and 1-8, all possessed the same 8 nt sequence complementary to the 5′-region of boxA at the 3′-side of 10-nt loop. The remaining seven clones showed reduced antitermination activity (3 + ∼4+), but little complementarity to both the hpII loop and the boxA region (Table 1). The RNA loop of 2-11 had 2-nt deletion in the linker region. Using an RNA folding algorithm, Mulfold, it was predicted that this nucleotide deletion caused incorrect folding, resulting in the linker region binding to boxA. The RNA loops of a number of the remaining six clones showed 3–4 nt complementary to the U1 hpII loop (Table 1). Although it is possible that these antisense RNA stem–loops bound to U1 hpII and destabilized the U1 hpII–U1A interaction, further analysis was not carried out since the inhibitory effect was small.
Table 1.
The nucleotide sequences of selected clones that exhibited low antitermination activity
 |
Bases in the boxA 5′-region and selected loop that were complementary to each other are underlined. Bases in the hpII loop and selected loop that were complementary to each other are shaded. The number of plusses indicates blue colony color scored by the colony color (X-gal) assay using the following controls: U1A/hpII, 5+; RSG-1.2/RRE, 2+; Rev/RRE, 1+; U1A/RRE, 0.5+; Rev/hpII, 0+.
In vivo mutational analysis of intramolecular RNA stem–loop binding to boxA
In order to confirm the presence of intramolecular binding of the selected loop sequences to the 5′-region of boxA, boxA and loop mutants based on 1-2, 1-4 and 1-8 that showed low antitermination activity (1+∼1.5+) were further examined in vivo using the antitermination assay (Table 2). LacZ expression scored by colony color correlated well with β-galactosidase activities quantitated using o-nitrophenyl β-d-galactopyranoside (ONPG). Clone 1-4 U107, having a C to U substitution in the selected RNA loop, showed high antitermination activity (Table 2, 4+), presumably due to the disruption of the RNA–RNA interaction; however, the activity was not as high as wild-type U1 hpII-U1A (5+). This weak disruption of inhibitory activity for the 1-4 U107 mutant could be the result of G-U base pair formation, and residual binding of the loop sequence and the boxA region. Conversely, clone 1-4 A9 mutant, having a G to A substitution upstream of boxA, showed antitermination activity (5+) similar to that of the wild-type U1 hpII-U1A (Table 2), suggesting that the intramolecular base pairing had been completely disrupted. Clone 1-4 A9/U107 mutant containing both the G to A substitution upstream of boxA and the C to U substitution in the original 1-4 RNA loop (Table 2), which restores complementarity, showed low antitermination activity (3+); however, the antitermination activity was not as low as the original clone 1-4 (Table 2, 1+). This weak restoration of inhibitory activity for clone 1-4 A9/U107 mutant could be the result of the substitution of a G-C pair in clone 1-4 to an A-U base pair in the double mutant. In an attempt to bring about more dynamic change in disruption and restoration of inhibitory activity, mutants containing double nucleotide substitutions were constructed. The mutants containing CG to GC substitutions either upstream of boxA or in the original 1-2, 1-4 and 1-8 RNA loop, denoted 1-2, 1-4 and 1-8 G8C9 or 1-4 G107C108, showed high antitermination activity (5+) comparable to wild-type U1 hpII-U1A (Table 2), suggesting that double nucleotide substitutions abolished the intramolecular RNA–RNA interactions. Conversely, CG to GC double nucleotide substitutions both upstream of boxA and in the original 1-4 RNA loop, denoted 1-4 G8C9/G107C108, restored complementarity and low antitermination activity (2+) comparable to the original clone 1-4 (Table 2). To confirm that the U1 hpII stem–loop was not involved in the inhibition of antitermination activity, the U1 hpII region was replaced by the boxB element (data not shown). As a result, the clone 1-4 SL repressed the antitermination activity, while the U107 and G107C108 mutants had no effect. Taken together, the above results strongly suggested that the selected loop sequences were binding to the boxA region and disrupting antitermination complex function.
Table 2.
Mutational analysis of intramolecular binding of the selected RNA loops to the boxA 5′-region
 |
Bases in the boxA 5′-region and selected loop that were complementary to each other are underlined. The boxA nucleotides are indicated in bold. The number of plusses indicates blue colony color scored by the colony color (X-gal) assay using the following controls: U1A/hpII, 5+; RSG-1.2/RRE, 2+; Rev/RRE, 1+; U1A/RRE, 0.5+; Rev/hpII, 0+.
In vitro mutational analysis of intramolecular RNA stem–loop binding to boxA
In order to further confirm that the selected loop sequences bound to the boxA region, the structures of RNA substrates derived from the mutants used in the in vivo assay described above were analyzed by native polyacrylamide gel electrophoresis. It was predicted that RNA substrates that form pseudoknot-like structures as a result of intramolecular RNA–RNA binding as shown in Figure 2A would be compactly folded and migrate faster on native gels compared to RNAs that cannot form pseudoknots.
Figure 2.
Native gel electrophoretic analysis of the intramolecular binding of the selected RNA stem–loop to boxA. (A) Schematic representation of the pseudoknot-like RNA folding by intramolecular RNA–RNA binding. (B) TBM gel (0.1 mM and 0.5 mM Mg2+) analysis of RNA substrates. All gels were run at 4°C;. Lane 1, 1-4; lane 2, 1-4 U107; lane 3, 1-4 A9; lane 4, 1-4 A9/U107; lane 5, 1-4 G107C108; lane 6, 1-4 G8C9; lane 7, 1-4 G8C9/G107C108; lane 8, 1-2; lane 9, 1-2 G8C9; lane 10, 1-8; lane 11, 1-8 G8C9.
When the 11 RNA substrates shown in Table 2 were run on a Tris-boric acid (TB) gel containing 0.1 mM Mg2+ (TBM gel), most of the RNAs showed similar mobilities with the exception of the G8C9 mutants of 1-4, 1-2 and 1-8, which showed lower mobilities (Figure 2B). Since pseudoknot formation has been shown to be stabilized by divalent metal ions (25), the mobility of the pseudoknot-forming RNAs was expected to increase relative to the non-pseudoknot forming RNAs with increasing Mg2+ in the gel. Indeed, the original 1-4 RNA showed a significant increase in mobility relative to the 1-4 A9 mutant RNA at high Mg2+ concentrations, suggesting the formation of a compact pseudoknot conformation for 1-4 RNA as opposed to an open conformation for 1-4 A9 (Figure 2B, lanes 1 and 3). In contrast, the 1-4 U107 mutant only showed a slight difference in mobility compared to 1-4 (Figure 2B, lanes 1 and 2), presumably due to the retention of a G-U wobble base-pair, agreeing well with residual inhibitory activity observed in vivo (Table 2). On the other hand, the 1-4 A9/U107 double mutant, which was expected to restore pseudoknot formation, showed a high mobility comparable to that of 1-4 (Figure 2B, lanes 4). Similarly, the two mutants, 1-4 G107C108 and 1-4 G8C9, showed lower gel mobilities compared to the original 1-4, indicating that pseudoknot formation had been disrupted (Figure 2B, lanes 5 and 6). Conversely, the double mutant 1-4 G8C9/G107C108, in which pseudoknot formation was expected to be restored, a high gel mobility comparable to 1-4 was observed (Figure 2B, lane 7). The gel mobility of the G8C9 mutants of the RNAs corresponding to clones 1-2 and 1-8 also lead to a decrease in gel mobility relative to the original molecules (Figure 2B, lanes 8–11), indicative of destabilization of pseudoknot formation. These results correlate with the in vivo mutational analysis, strongly supporting the notion that the selected loop sequences are binding to the boxA region of the RNA transcript and disrupting antitermination complex formation.
In vitro mutational analysis of intermolecular RNA stem–loop binding to boxA
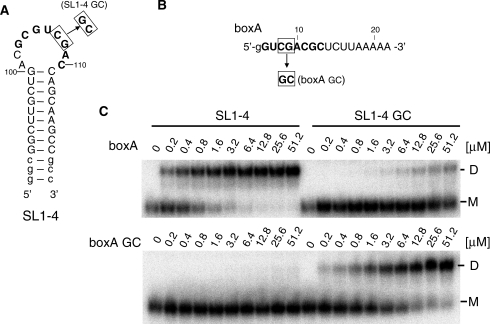
The binding affinity and specificity of the selected stem–loop RNAs toward the boxA RNA region was examined by a gel mobility shift assay (Figure 3). The boxA RNA and the mutant boxA containing the double nucleotide substitution, denoted as boxA and boxA GC, respectively, was mixed in different concentrations with internally 32P-labeled antisense stem–loop from clone 1-4 and the mutant with the double nucleotide substitution, denoted SL1-4 and SL1-4 GC, respectively, and analyzed on TBM gels containing 0.1 mM Mg2+. SL1-4 was found to bind to the boxA RNA with an apparent Kd of 0.50 µM, while no interaction with boxA GC could be detected up to a concentration of 50 µM. Similarly, only a very weak interaction (Kd > 50 µM) could be observed between the mismatched SL1-4 GC and wild-type boxA. On the other hand, restoration of complementarity in the case of SL1-4 GC and boxA GC resulted in a considerable recovery of binding affinity to 3.8 µM, which is somewhat weaker than that of SL1-4 and boxA, but correlates well with the slightly weaker inhibitory effect of the SL1-4 GC/boxA GC interaction in vivo (2+) (Table 2), compared to the wild-type SL1-4/boxA interaction (1+). Taken together, these results support the notion that the antisense stem–loop bound to the 5′-region of boxA and prevented the association of host factors important for antitermination complex formation.
Figure 3.
Gel mobility shift analysis of the intermolecular binding of the antisense stem–loop and boxA. (A) The secondary structure of the ‘inhibitor’ RNA stem–loop substrate. (B) The sequence of the boxA RNA substrates. (C) Gel shift analysis of the binding of 32P-labeled wild-type and mutant SL1-4 RNAs with boxA and mutant RNAs on a TBM gel (0.1 mM Mg2+) at 4°C.
In vivo analysis of the effect of the disruption of the stem region of antisense RNA stem–loops
The importance of the stem–loop structure for efficient inhibition of RNA–protein interaction by the antisense RNA stem–loops was investigated by disrupting the upper stem region and measuring antitermination activity using the in vivo reporter system (Figure 4). When the single and double mismatches (1 bp and 2 bp, respectively) were introduced into the upper stem of clones 1-4 and 1-4 G8C9/G107C108, a gradual increase in antitermination from 1+ to 3.5+ and from 1.5+ to 3+ was observed, respectively. On the other hand, constructs with mismatches in the loop region, clones 1-4 G107C108 and 1-4 G8C9, which do not inhibit RNA–protein interaction, showed no change. This suggested that the structure of the antisense RNA stem–loop, and not just the loop region is important for efficient inhibition of RNA–protein interaction.
Figure 4.
The effect of the disruption of 1 and 2 bp in the upper stem region of the antisense RNA stem–loop of clone 1-4 and mutant constructs. (A) The secondary structure of the boxA and antisense stem–loop region of clone 1-4 and base substitutions investigated. The effect of the disruption of 1 and 2 bp in the upper stem of the antisense RNA on the antitermination activity of (B) clone 1-4, (C) clone 1-4 G107C108, (D) clone 1-4 G8C9 and (E) clone 1-4 G8C9/G107C108.
Rational design of a modified U1 hpII RNA and cognate antisense RNA stem–loop
In order to show that an RNA–protein interaction resembling that of U1A–hpII could be disrupted by RNA loop–loop complex formation, rational design of a modified U1 hpII RNA and cognate antisense RNA stem–loop was carried out. The U1 hpII was modified so that it resembles the HIV dimerization initiation site (DIS) loop, which is known to dimerize through a loop–loop interaction, while still retaining significant affinity toward the U1A protein. Since the requirements for stable loop–loop complex formation by HIV DIS-like RNA stem–loops are fairly well understood both in vitro and in vivo (45–49), this would allow the rational design of an antisense RNA stem–loop that binds to the modified U1 hpII loop, which may inhibit binding to the U1A protein. Modification of the U1 hpII should also make possible the adjustment of the affinity toward U1A protein, since the wild-type interaction may be too strong for inhibition by RNA loop–loop complex formation.
The HIV DIS loop contains nine bases, of which positions 3 through 8 are involved in Watson–Crick base-pair formation, and the purine residues at positions 1, 2 and 9 are important for base stacking interactions (Figure 5A) (45,46). A sheared A–A base-pair was proposed to be important for positions 1 and 9 (46). The relative stability of the loop–loop complexes was found to correlate with the stability of the duplex region as estimated using nearest-neighbor parameters determined by Turner and co-workers (50), so that a higher G-C content leads to stable complex formation (47,48). On the other hand, the U1 hpII loop has a 10-nt loop (Figure 5B), which is one base larger than that of the DIS loop, of which the first 7 nt contact the U1A protein (34), and the remaining three nucleotides can be replaced by unnatural ethylene glycol linkers (51).
Figure 5.
Design of a modified U1 hpII RNA and cognate RNA stem–loop. The secondary structure of (A) HIV DIS, (B) U1 hpII, (C) U1 hpIIΔDIS and (D) the designed antisense stem–loop (aSL). (E) Antitermination activities of modified U1 hpII loop sequences.
In order to modify the U1 hpII loop so that it resembles the DIS loop, first, the eighth nucleotide in the U1 hpII loop, U16, was deleted, and C18 was substituted by an A to yield Δ16/18A, which resulting in a drop in antitermination activity of 5+ to 4+ (Figure 5E). Next, the second nucleotide in the loop, U10, was mutated to an A or G to improve base stacking interactions, and the third nucleotide, U11, was mutated to a C or G to increase the stability of complementary base pairing. However, a large decrease in antitermination activity was observed, except for the 10G mutation, which showed an activity of 3+ (Figure 5E). Therefore, the hpII triple mutant Δ16/18A/10G, designated hpIIΔDIS (Figure 5C), was chosen as a target for the design of an antisense stem–loop (Figure 5D).
Inhibition of U1A protein binding to a modified hpII loop by a designed antisense stem–loop
First, the antitermination system was used to determine whether the designed antisense stem–loop can indeed inhibit the interaction between the modified hpII (hpIIΔDIS) and the U1A protein (Figure 6). A synthetic DNA cassette encoding the antisense stem–loop was inserted into the parent reporter plasmid containing the hpIIΔDIS, so that the two stem–loops were separated by 37 nt. As a result, a considerable decrease in antitermination activity from 4+ for the parent plasmid containing the BsrGI site, which was somewhat higher than the activity of the hpIIΔDIS construct in Figure 5E (3+), to 1.5+ was observed, suggesting that antisense stem–loop was indeed inhibiting the interaction between hpIIΔDIS and the U1A protein. In order to show that the decrease in antitermination activity was due the specific interaction between the hpIIΔDIS and antisense stem–loop loop regions, the six bases within the antisense stem–loop loop (positions 12–17), which were anticipated to bind to the hpIIΔDIS, were individually substituted. As expected, an increase in antitermination activity to 2–3+ was observed, showing that a mismatch in this region leads to a decrease in the inhibitory activity of antisense stem–loop. Mutations at positions 15 and 16 lead to the largest decrease in inhibitory activity, and appeared to be the most important for the loop–loop interaction.
Figure 6.
In vivo analysis of the inhibition of U1A protein binding to a modified hpII loop by the designed antisense stem-loop (aSL) and the effect of loop mutations. (A) The secondary structure of modified hpII-aSL region. (B) Antitermination activities of the designed constructs consisting of the the designed hpII–aSL, and aSL mutants.
Next, the affinity of the modified hpII toward the U1A protein and the designed antisense stem–loop was determined by a gel shift assay (Figure 7) In the presence of 0.4 mM Mg2+, the apparent Kd of the interaction of the modified hpII and the U1A protein was between 1 and 5 µM, which was more than three orders of magnitude weaker than that of the wild-type hpII with the U1A protein (0.60 nM) (Figure 7A). The Kd value for the wild-type hpII/U1A interaction was similar to that determined under similar conditions (0.35 nM) (52). On the other hand, the interaction between the modified hpII and the designed antisense stem–loop was found to be 35 nM in the presence of 0.4 mM Mg2+ (Figure 7B). This indicated that it should be possible to inhibit the U1 hpIIΔDIS–U1A interaction by the antisense stem–loop RNA. Indeed, when a competition experiment was carried out, antisense stem–loop was found to effectively compete with the modified hpII-U1A complex with an apparent Ki of ∼160 nM, while antisense stem–loop 15C showed no inhibitory effect (Figure 7C). It should be noted that changes in the Mg2+ concentration showed opposite effects on the stability of the RNA loop–loop interaction and that of the RNA–protein interaction, so that stronger inhibitory effect by the antisense stem–loop would be expected to be observed at higher Mg2+ concentrations.
Figure 7.
Gel mobility shift analysis of the inhibition of U1A protein binding to the modified hpII RNA (hpIIΔDIS) by the designed antisense RNA stem–loop (aSL). (A) U1A protein binding to hpII and hpIIΔDIS. (B) Binding of aSL and a mutant aSL (aSL 15C) to hpIIΔDIS. (C) Inhibition of U1A–hpIIΔDIS binding by aSL and aSL 15C.
DISCUSSION
Inhibition of RNA–protein interactions by an antisense RNA stem–loop through long-range pseudoknot formation
In this study, a novel method for the identification of RNA stem–loops that disrupt the formation of RNA–protein interactions through RNA–RNA interactions is described. A bacterial reporter system for the detection of RNA–protein interactions, based on N-mediated antitermination (36), was reconstructed so that an stem–loop RNA library with 10 randomized loop nucleotides was positioned downstream of the target RNA, therefore enabling the selection of stem–loops that bind intramolecularly to the upstream target RNA and disrupt protein binding. While at least two mechanisms for the reduction of antitermination by the antisense stem–loop are conceivable, the selected RNA sequences of clones that suppressed antitermination activity were found to be complementary to the 5′-side of the boxA element, located upstream of the hpII stem–loop (Figure 1C). In vivo and in vitro mutational analysis strongly suggested that the selected sequences were binding to the boxA region to form a long-range pseudoknot structure (Table 2, Figures 2 and 3).
A pseudoknot is a structural motif commonly observed in functional RNA structures, where the loop nucleotides of an stem–loop forms base-pairs with adjacent 5′- or 3′-complementary single-stranded regions in a Mg2+-dependent manner, with the resulting double-stranded region generally forming a contiguous coaxially stacked helix with that of the stem–loop (53,54). The Mg2+-dependence of stable formation of RNA–RNA interactions, observed in the in vitro mutational analysis (Figure 2), support the formation of such a pseudoknot structure. Furthermore, in the loop sequences selected in this study, the nucleotides complementary to the boxA were adjacent to the 3′-stem nucleotides (Table 1), suggesting that binding of the RNA loop to the complementary boxA 5′-region resulted in similar coaxially stacked helices. The decrease in inhibitory activity upon disruption of the upper stem of the antisense stem–loop (Figure 4B and E) supports the importance of the putative co-axial stacking for stable pseudoknot formation.
The above results suggest that the selected antisense RNA stem–loops are disrupting the binding of host factors NusB and S10 to the boxA. Genetic analysis and cross-linking experiments have indicated that the NusB and S10 proteins bind as a heterodimer to the boxA site; however, the interaction is too weak to be detected, for example, by a gel shift assay (44,55). This implies that the assembly of NusB and S10 onto the boxA element requires the cooperative interaction of additional phage and host factors (39), and may explain why a relatively weak interaction between the antisense RNA stem–loop and the boxA site (Kd ∼ 0.5 µM) is sufficient for the disruption of the antitermination complex. While it is difficult to predict what the affinity required for efficient inhibition of antitermination by an RNA–RNA interaction at the boxA site is, the results demonstrate that it is possible to identify antisense stem–loops that disrupt the assembly of a functional ribonucleoprotein complex in a specific manner.
The HIV DIS RNA loop as a framework for the rational design of antisense RNA stem–loops that target U1 hpII-like RNAs
There are a number of potential reasons why the U1 hpII stem–loop was not targeted in the initial screening for antisense stem–loops that inhibit U1 hpII–U1A-mediated antitermination. First, the affinity of the U1 hpII–U1A interaction may be too high to disrupt competitively by RNA loop–loop interactions. The dissociation constant of the U1 hpII–U1A protein complex has been determined by a gel mobility shift assay to be ∼0.35 nM (52). Conversely, the dissociation constants of RNA loop–loop interactions are typically higher (Kd = 10–8–10–9 M) than that of U1 hpII–U1A interaction (19,28,47,56). Another possibility that the hpII loop was not targeted is that the loop sequence may simply not be adequate for targeting by loop–loop interactions, which require a high percentage of G-C base pairs to support high-affinity binding. In fact, a reporter construct containing an RNA stem–loop with the loop sequence 5′-GGAGUGCAAU-3′, which is fully complementary to the U1 hpII loop did not show a decrease in antitermination activity (data not shown). In addition, the possibility that further extensive screening may lead to identification of stem–loops targeting hpII cannot be excluded, since we were only able to screen up to 15% of the total possible combinations of sequences (410 = 1.0 × 106).
In order to demonstrate that it is possible to inhibit the interaction of an RNA–protein interaction resembling that of the U1 hpII–U1A interaction, the hpII loop sequence was modified so as to resemble that of the HIV DIS, thereby facilitating the rational design of an antisense RNA stem–loop. This was also expected to result in a decrease in the affinity of the modified hpII toward the U1A protein, compared with the wild-type loop sequence. The designed antisense RNA stem–loop was able to decrease antitermination activity mediated by the modified U1 hpII and the U1A protein by up to 2.5 colony color units (Figure 6). Based on previous studies using the bacterial reporter system, in this case, one colony color unit is expected to correspond to a 20–30-fold difference in the affinity of the RNA–peptide interaction. This implies that the presence of the antisense stem–loop resulted in a two to three order of magnitude decrease in the proportion of the modified U1 hpII–U1A protein complex.
CONCLUSIONS
We have shown that it is possible to disrupt RNA–protein interactions using antisense RNA stem–loops through both loop–linear and loop–loop RNA–RNA interactions. Since it is still difficult to predict the stability of the resulting RNA–RNA interactions, the bacterial selection procedure provides a simple and powerful method for directly screening for stem–loop sequences that interfere with RNA–protein interactions. Other potential targets include internal loop regions such as in the case of the well-characterized tetraloop–tetraloop receptor interactions, possibly utilizing non-Watson–Crick interactions (57).
Attempts to disrupt the interaction between the U1A protein and hpII RNA has suggested the presence of an upper limit for the affinities of the RNA–protein interactions that may be inhibited. However, in contrast, it was also shown that antitermination complex formation could be inhibited presumably through the disruption of the weak interaction between NusB/S10 and the boxA element. This suggests that relatively weak RNA–RNA interactions may be sufficient for the inhibition of the assembly of cooperative multicomponent complexes such as the antitermination complex.
In this study, we were also able to show that the HIV DIS provides an attractive framework for the rational design of novel RNA–RNA interactions that may be used to target functional RNA structures, since the relative stability of loop–loop interactions by DIS variants has been shown to correspond fairly well with the predicted stabilities of hexanucleotide duplexes using nearest-neighbor parameters (47,48). Analysis of the resulting RNA–RNA interactions, as in this study, could lead to the identification of novel modes of interaction and an understanding of the structural requirements for the formation of stable loop–linear and loop–loop interactions. Antisense RNA stem–loops identified in this manner may potentially be used as a tool to understand RNA–protein interactions and RNA function, as well as drugs targeted against critical RNA–protein interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT); Ministry of Health, Labour and Welfare of Japan (to K.H.). Funding for open access charge: A Grant-in-Aid for Scientific Research from the Ministry of Health, Labour and Welfare of Japan (to K.H.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Midori Ohtsuki, Misa Mizuguchi, Mayumi Kunimoto, and Shoki Michi for their experimental assistance.
REFERENCES
- 1.Held DM, Kissel JD, Patterson JT, Nickens DG, Burke DH. HIV-1 inactivation by nucleic acid aptamers. Front Biosci. 2006;11:89–112. doi: 10.2741/1782. [DOI] [PubMed] [Google Scholar]
- 2.Symensma TL, Giver L, Zapp M, Takle GB, Ellington AD. RNA aptamers selected to bind human immunodeficiency virus type 1 Rev in vitro are Rev responsive in vivo. J. Virol. 1996;70:179–187. doi: 10.1128/jvi.70.1.179-187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 4.Harada K, Martin SS, Tan R, Frankel AD. Molding a peptide into an RNA site by in vivo peptide evolution. Proc. Natl Acad. Sci. USA. 1997;94:11887–11892. doi: 10.1073/pnas.94.22.11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crooke ST. Molecular mechanisms of action of antisense drugs. Biochim. Biophys. Acta. 1999;1489:31–44. doi: 10.1016/s0167-4781(99)00148-7. [DOI] [PubMed] [Google Scholar]
- 6.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Boiziau C, Dausse E, Yurchenko L, Toulme JJ. DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA–DNA kissing complexes. J. Biol. Chem. 1999;274:12730–12737. doi: 10.1074/jbc.274.18.12730. [DOI] [PubMed] [Google Scholar]
- 8.Aldaz-Carroll L, Tallet B, Dausse E, Yurchenko L, Toulme JJ. Apical loop-internal loop interactions: a new RNA–RNA recognition motif identified through in vitro selection against RNA hairpins of the hepatitis C virus mRNA. Biochemistry. 2002;41:5883–5893. doi: 10.1021/bi0121508. [DOI] [PubMed] [Google Scholar]
- 9.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 10.Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 11.Vickers TA, Wyatt JR, Freier SM. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 13.Eguchi Y, Itoh T, Tomizawa J. Antisense RNA. Annu. Rev. Biochem. 1991;60:631–652. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
- 14.Wagner EG, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 15.Brunel C, Marquet R, Romby P, Ehresmann C. RNA loop-loop interactions as dynamic functional motifs. Biochimie. 2002;84:925–944. doi: 10.1016/s0300-9084(02)01401-3. [DOI] [PubMed] [Google Scholar]
- 16.Lehnert V, Jaeger L, Michel F, Westhof E. New loop–loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 17.Ikawa Y, Ohta H, Shiraishi H, Inoue T. Long-range interaction between the P2.1 and P9.1 peripheral domains of the Tetrahymena ribozyme. Nucleic Acids Res. 1997;25:1761–1765. doi: 10.1093/nar/25.9.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen AA, Collins RA. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc. Natl Acad. Sci. USA. 2001;98:7730–7735. doi: 10.1073/pnas.141039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 20.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop–loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl Acad. Sci. USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muriaux D, Fosse P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 22.Zeiler BNaS, Robert W. In: RNA Structure and Function. Simons RW, G.-M. M, editors. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1998. pp. 437–464. [Google Scholar]
- 23.Herschlag D. Implications of ribozyme kinetics for targeting the cleavage of specific RNA molecules in vivo: more isn't; always better. Proc. Natl Acad. Sci. USA. 1991;88:6921–6925. doi: 10.1073/pnas.88.16.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larrouy B, Boiziau C, Sproat B, Toulme JJ. RNase H is responsible for the non-specific inhibition of in vitro translation by 2′-O-alkyl chimeric oligonucleotides: high affinity or selectivity, a dilemma to design antisense oligomers. Nucleic Acids Res. 1995;23:3434–3440. doi: 10.1093/nar/23.17.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 26.Eguchi Y, Tomizawa J. Complexes formed by complementary RNA stem–loops. Their formations, structures and interaction with ColE1 Rom protein. J. Mol. Biol. 1991;220:831–842. doi: 10.1016/0022-2836(91)90356-b. [DOI] [PubMed] [Google Scholar]
- 27.Gregorian RS, Jr, Crothers DM. Determinants of RNA hairpin loop–loop complex stability. J. Mol. Biol. 1995;248:968–984. doi: 10.1006/jmbi.1995.0275. [DOI] [PubMed] [Google Scholar]
- 28.Duconge F, Di Primo C, Toulme JJ. Is a closing “GA pair” a rule for stable loop–loop RNA complexes? J. Biol. Chem. 2000;275:21287–21294. doi: 10.1074/jbc.M002694200. [DOI] [PubMed] [Google Scholar]
- 29.Jossinet F, Paillart JC, Westhof E, Hermann T, Skripkin E, Lodmell JS, Ehresmann C, Ehresmann B, Marquet R. Dimerization of HIV-1 genomic RNA of subtypes A and B: RNA loop structure and magnesium binding. RNA. 1999;5:1222–1234. doi: 10.1017/s1355838299990982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dardel F, Marquet R, Ehresmann C, Ehresmann B, Blanquet S. Solution studies of the dimerization initiation site of HIV-1 genomic RNA. Nucleic Acids Res. 1998;26:3567–3571. doi: 10.1093/nar/26.15.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 32.Ennifar E, Dumas P. Polymorphism of bulged-out residues in HIV-1 RNA DIS kissing complex and structure comparison with solution studies. J. Mol. Biol. 2006;356:771–782. doi: 10.1016/j.jmb.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Kim CH, Tinoco I., Jr A retroviral RNA kissing complex containing only two G.C base pairs. Proc. Natl Acad. Sci. USA. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall KB. Interaction of RNA hairpins with the human U1A N-terminal RNA binding domain. Biochemistry. 1994;33:10076–10088. doi: 10.1021/bi00199a035. [DOI] [PubMed] [Google Scholar]
- 35.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 36.Franklin NC. Clustered arginine residues of bacteriophage lambda N protein are essential to antitermination of transcription, but their locale cannot compensate for boxB loop defects. J. Mol. Biol. 1993;231:343–360. doi: 10.1006/jmbi.1993.1287. [DOI] [PubMed] [Google Scholar]
- 37.Harada K, Martin SS, Frankel AD. Selection of RNA-binding peptides in vivo. Nature. 1996;380:175–179. doi: 10.1038/380175a0. [DOI] [PubMed] [Google Scholar]
- 38.Mogridge J, Mah TF, Greenblatt J. Involvement of boxA nucleotides in the formation of a stable ribonucleoprotein complex containing the bacteriophage lambda N protein. J. Biol. Chem. 1998;273:4143–4148. doi: 10.1074/jbc.273.7.4143. [DOI] [PubMed] [Google Scholar]
- 39.Mogridge J, Mah TF, Greenblatt J. A protein–RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 1995;9:2831–2845. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 40.Peled-Zehavi H, Horiya S, Das C, Harada K, Frankel AD. Selection of RRE RNA binding peptides using a kanamycin antitermination assay. RNA. 2003;9:252–261. doi: 10.1261/rna.2152303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peled-Zehavi H, Smith CA, Harada K, Frankel AD. Screening RNA-binding libraries by transcriptional antitermination in bacteria. Methods Enzymol. 2000;318:297–308. doi: 10.1016/s0076-6879(00)18059-0. [DOI] [PubMed] [Google Scholar]
- 42.Hall KB, Stump WT. Interaction of N-terminal domain of U1A protein with an RNA stem/loop. Nucleic Acids Res. 1992;20:4283–4290. doi: 10.1093/nar/20.16.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugaya M, Nishino N, Katoh A, Harada K. Amino acid requirement for the high affinity binding of a selected arginine-rich peptide with the HIV Rev-response element RNA. J. Pept. Sci. 2008;14:924–935. doi: 10.1002/psc.1027. [DOI] [PubMed] [Google Scholar]
- 44.Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, Urlaub H, Wahl MC. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell. 2008;32:791–802. doi: 10.1016/j.molcel.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lodmell JS, Ehresmann C, Ehresmann B, Marquet R. Convergence of natural and artificial evolution on an RNA loop–loop interaction: the HIV-1 dimerization initiation site. RNA. 2000;6:1267–1276. doi: 10.1017/s1355838200000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lodmell JS, Ehresmann C, Ehresmann B, Marquet R. Structure and dimerization of HIV-1 kissing loop aptamers. J. Mol. Biol. 2001;311:475–490. doi: 10.1006/jmbi.2001.4879. [DOI] [PubMed] [Google Scholar]
- 47.Horiya S, Li X, Kawai G, Saito R, Katoh A, Kobayashi K, Harada K. RNA LEGO: magnesium-dependent formation of specific RNA assemblies through kissing interactions. Chem. Biol. 2003;10:645–654. doi: 10.1016/s1074-5521(03)00146-7. [DOI] [PubMed] [Google Scholar]
- 48.Weixlbaumer A, Werner A, Flamm C, Westhof E, Schroeder R. Determination of thermodynamic parameters for HIV DIS type loop-loop kissing complexes. Nucleic Acids Res. 2004;32:5126–5133. doi: 10.1093/nar/gkh841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lorenz C, Piganeau N, Schroeder R. Stabilities of HIV-1 DIS type RNA loop–loop interactions in vitro and in vivo. Nucleic Acids Res. 2006;34:334–342. doi: 10.1093/nar/gkj435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 51.Williams DJ, Hall KB. RNA hairpins with non-nucleotide spacers bind efficiently to the human U1A protein. J. Mol. Biol. 1996;257:265–275. doi: 10.1006/jmbi.1996.0161. [DOI] [PubMed] [Google Scholar]
- 52.Laird-Offringa IA, Belasco JG. Analysis of RNA-binding proteins by in vitro genetic selection: identification of an amino acid residue important for locking U1A onto its RNA target. Proc. Natl. Acad. Sci. USA. 1995;92:11859–11863. doi: 10.1073/pnas.92.25.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyatt JR, Puglisi JD, Tinoco I., Jr RNA pseudoknots. Stability and loop size requirements. J. Mol. Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- 54.Batey RT, Rambo RP, Doudna JA. Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 55.Nodwell JR, Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- 56.Duconge F, Toulme JJ. In vitro selection identifies key determinants for loop–loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA. 1999;5:1605–1614. doi: 10.1017/s1355838299991318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.