Abstract
Neurons in the central nervous system lose their intrinsic capacity for axon regeneration as they mature, and it is widely hypothesized that changes in gene expression are responsible. Testing this hypothesis and identifying the relevant genes has been challenging because hundreds to thousands of genes are developmentally regulated in CNS neurons, but only a small subset are likely relevant to axon growth. Here we used automated high content analysis (HCA) methods to functionally test 743 plasmids encoding developmentally regulated genes in neurite outgrowth assays using postnatal cortical neurons. We identified both growth inhibitors (Ephexin, Aldolase A, Solute Carrier 2A3, and Chimerin), and growth enhancers (Doublecortin, Doublecortin-like, Kruppel-like Factor 6, and CaM-Kinase II gamma), some of which regulate established growth mechanisms like microtubule dynamics and small GTPase signaling. Interestingly, with only one exception the growth-suppressing genes were developmentally upregulated, and the growth-enhancing genes downregulated. These data provide important support for the hypothesis that developmental changes in gene expression control neurite outgrowth, and identify potential new gene targets to promote neurite outgrowth.
Introduction
After injury to the mature central nervous system (CNS), axons largely fail to regenerate. Besides the well-established presence of growth-inhibitory molecules in the CNS environment (Case and Tessier-Lavigne, 2005; Yiu and He, 2006), another major reason for this regeneration failure is that mature CNS neurons have an intrinsically low capacity for axon growth. This is particularly apparent in contrast to the developing CNS. Indeed, many axons in the embryonic and early postnatal CNS are capable of regeneration, including those in the brachium of the tectum (So et al., 1981), the nigral-striatal tract (Kawano et al., 2005), and the spinal cord (Bregman et al., 1989; Hasan et al., 1993; Saunders et al., 1998). Following a rapid developmental transition, neurons largely lose the capacity for regeneration. Importantly, experiments with age-mismatched co-cultures have shown consistently that the age of the neuron, and not the age of the recipient tissue, largely determines the success of regeneration (Chen et al., 1995; Senut et al., 1995; Dusart et al., 1997; Hafidi et al., 2004; Blackmore and Letourneau, 2006). Furthermore, retinal ganglion cells that are cultured at low density in defined, serum-free media, still show an age-dependent decrease in the rate of axon growth (Goldberg et al., 2002). Thus, changes intrinsic to developing neurons render them less able to regenerate as they mature. However, the precise nature of these neuron-intrinsic changes remains unclear.
One likely possibility is that developing neurons alter the expression of genes involved in axon growth, either by decreasing the expression of growth-promoting genes, increasing the expression of growth-suppressive genes, or some combination of the two (Moore et al, 2009). We tested the hypothesis that developmentally regulated genes control axon growth in this fashion by high content analysis of primary postnatal cortical neurons. Previous work used microarray analysis to profile gene expression in FACS-purified CST neurons during early postnatal periods (Arlotta et al., 2005). Starting from this publicly available microarray data, we tested 428 developmentally regulated genes and identified eight that consistently affected neurite length. Among these eight genes, we found that all 4 growth suppressing genes were developmentally upregulated, and 3 of 4 growth enhancing genes were developmentally downregulated. This screen is an important step in identifying developmentally regulated genes that control neurite outgrowth.
Methods
Microarray analysis
Microarray data, series GSE2039 (Arlotta et al., 2005), were downloaded from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). RMA normalization was performed as described in (Irizarry et al., 2003) using the software package RMA Express (http://rmaexpress.bmbolstad.com). Data were exported to Microsoft Excel, and probe values were averaged across replicate chips for comparison across age. A three-fold change in average probe intensity in either direction was used as a cutoff to select genes for screening.
Plasmid preparation
A glycerol stock library from the NIH Mammalian Genome Collection (Gerhard et al., 2004) was purchased from Open Biosystems (Thermofisher, Huntsville, Alabama), which included human (IRAT) and mouse (IRAV) clones in a pCMV-SPORT6 backbone. Glycerol stocks were used to inoculate 5ml of LB media containing 100µg/ml ampicillin, and miniprep spin columns (Qiagen) were used to prepare plasmid DNA. A concentration of ≥200ng/µl and an OD ratio (260/280) > 1.8 were minimal quality control standards. A protein considered inactive, mCherry (Shaner et al., 2004), was cloned into pCMV-SPORT6 plasmid to serve as control.
Preparation of 96-well plates and coverslips
96-well plates (Perkin Elmer #6005182, Waltham, MA) were coated with 10 µg/ml Poly-D-Lysine (PDL, Sigma #P7886) for at least 24 hours, rinsed five times with water, coated overnight at 4°C with 10 µg/ml laminin (Sigma #L2020), and rinsed before addition of 200µl neuronal media (see below). For experiments involving hand tracing of neurons, glass coverslips were prepared by dipping in 95% ethanol followed by brief flaming. Coverslips were coated with PDL and laminin as described above.
Media preparation
Enriched Neurobasal (ENB) media was modified from (Meyer-Franke et al., 1995) and contained Neurobasal medium (Invitrogen #12348017), 100 U/ml penicillin and 100µg/ml streptomycin (Invitrogen #15140122), 5µg/ml insulin (Sigma #I6634), 100µM sodium pyruvate (Invitrogen #11360070), 4µg/ml triiodothyronine (Sigma #T6397), 200µM L-glutamine (Invitrogen #25030149), 50µg/ml N-acetyl-L-cysteine (Sigma #A8199), B27 supplement (Invitrogen #17504044), 100µg/ml transferrin (Sigma #T1147), 100µg/ml bovine-serum albumen (Sigma #A4161), 63ng/ml progesterone (Sigma #P8783), 16µg/ml putrescine (Sigma #P7505) and 400ng/ml sodium selenite (Sigma #S5261). In experiments with cortical neurons, 20ml of this media was placed on confluent glial cultures in T-75 flasks (Nunc #156499) overnight for glial conditioned medium, which was removed and filtered prior to use.
Glial cell cultures
Astrocyte cultures were prepared as described in (De Hoop et al., 1998). Briefly, cortex from 3 P1 rat pups were dissected into ice-cold Hibernate E (Brainbits), minced with a razor blade, transferred to 10ml dissociation media (Hiberate E containing 0.25% trypsin (Gibco #15090 and 150µg/ml DNAse (Sigma #5025)), and incubated for 15 minutes at 37°C with constant shaking. Remaining cell clumps were allowed to settle for five minutes, and the supernatant was transferred to a collection tube containing 3ml horse serum (Gibco #26050-070). Cell clumps were incubated for an additional 15 minutes in Dissociation Media at 37°C with constant shaking. 3ml of horse serum was added, and cells were triturated 10 times in a 10 ml pipette. Cell suspensions were passed through 70 µm mesh (BD Falcon #352350), pelleted (180xg, 5 minutes), and resuspended in 10 ml culture media (MEM (Gibco #11095-080), Pen-Strep (Gibco #15145-014), 10% horse serum, 0.6% glucose (Sigma #67021). 1ml of cell suspension was placed in a T-75 flask (Nunc #156499) containing 20ml of culture media. Media were changed every 3 to 4 days, and cultures were used to condition neuronal culture media after two weeks.
Neuronal dissociation
Retinal ganglion cells (RGCs) were purified by immunopanning as previously described (Meyer-Franke et al., 1995). For cortical neurons we developed a method to maximize viability using sequential digestion with papain and trypsin followed by sequential trituration of cell clumps while periodically removing the overlying cell suspension to minimize mechanical stress on dissociated cells. Postnatal rats (P1 or P5) were sacrificed by decapitation and the brains placed in ice-cold Hibernate E (minus CaCl2) (Brainbits #HE-Ca 500). Meninges were removed. Frontal cortex was isolated, minced finely with a razor blade, and incubated in 10ml dissociation media (Hibernate E containing 20U/ml papain (Worthington #3126)) and 150µg/ml DNAse (Sigma #5025)), and incubated for 30 minutes at 37°C with constant shaking. Cell clumps were rinsed by pelleting (10Xg, 10min) and resuspended in Hibernate E + 2% B27, then pelleted, resuspended in 1.5ml Hibernate E + 2% B27, and gently triturated three times with a lightly fire polished pipette. Remaining cell clumps were pelleted, rinsed in 10ml of Hibernate E (no B27), then pelleted and resuspended in 10 ml Hibernate E containing 0.25% trypsin (Gibco #15090 and 150µg/ml DNAse (Sigma #5025)). Cells were incubated for 30 minutes at 37°C with constant shaking, then pelleted and resuspended in 5ml Hibernate E + 2% B27. Cells were then sequentially triturated by resuspending in 1.5ml Hibernate E + 2% B27, triturating three times with a lightly fire polished pipette, then letting cell clumps settle for two minutes. The supernatant was removed to a separate collection tube, and the process repeated until no cell clumps were visible. Typically a total of 12–14mls of cell suspension containing a total of 8–10 million cells was collected.
Transfection
For cortical neurons, transfection techniques were modified from (Buchser et al., 2006). Dissociated neurons were pelleted and resuspended in Internal Neuronal Buffer (INB) (KCL 135mM, CaCl2 0.2mM, MgCl2 2mM, HEPES 10mM, EGTA 5mM, pH 7.3) at a concentration of 2 ×106/ml. 25µl of this cell solution was placed in each well of a 96-well electroporation plate (Harvard Apparatus/BTX #45-0450) and mixed with 25µl of INB containing 0.5µg of mCherry reporter plasmid and 3ug of the test plasmid. The plate was placed in a plate handler (HT-200) attached to an ECM 830 square wave pulse generator (Harvard Apparatus/BTX), where each well received a single pulse of 350V for 300µs. 100µl of Hibernate E with 2% B27 supplement was added to each well to aid cell recovery, and cells were transferred to 96-well plates (1600 per well). For RGCs, 100,000 P4 postnatal RGCs were purified by immunopanning, resuspended in an electroporation solution containing 2µg of total DNA (GFP reporter and gene of interest; 1:6 ratio), placed in a cuvette for small cell numbers (Amaxa) and electroporated using Amaxa program SCN#1. Immediately following electroporation, growth media was added to the mixture and the whole solution placed into a 0.5ml Eppendorf tube. RGCs were centrifuged for 16 minutes at 1800 rpm prior to resuspension and plating. Transfection efficiencies averaged around 20% in RGC experiments.
Immunohistochemistry
Primary antibodies were: Rabbit anti-βIII tubulin (Sigma #T2200, 1:500 dilution), Mouse anti-Tau1 (A kind gift from Dr. Itzhak Fischer (Nothias et al., 1995) ), and Mouse anti-GAD67 (Chemicon #Mab 5046) . Cultures were fixed using 4% paraformaldehyde (Sigma #P1648) in PBS buffer at 37°C. Following rinses in PBS, cultures were blocked and permeabilized in 20% goat serum (Gibco #16210-72) )/0.02% Triton X-100 (Sigma) in antibody buffer (150mM NaCl, 50mM Tris base, 1% BSA, 100mM L-Lysine, 0.04% Na azide, pH 7.4) for 30 min to reduce non-specific binding. Cultures were incubated overnight at 4 deg C in antibody buffer containing primary antibodies, washed with PBS, incubated in antibody buffer containing secondary antibodies and 2µg/ml Hoechst 34580 (Invitrogen #H21846) for 4 hours at room temperature, washed with PBS, and left in PBS for imaging.
Quantification of neurite length
For automated image analysis, images of neurons were acquired using Cellomics KSR or VTI automated microscopes, using Hoechst dye to identify nuclei, βIII tubulin fluorescence to identify neuronal cell bodies and neurites, and mCherry fluorescence to identify transfected neurons. Neurites were traced using Neuronal Profiling v3.5. Data were exported to Spotfire DecisionSite software (Tibco) for further analysis. Untransfected neurons in a control well in each experiment were used to determine background fluorescence, and a threshold for mCherry+ cells was set such that <1% of untransfected cells were called mCherry +. Then, using only data from mCherry+ neurons, the average neurite length was determined for neurons transfected with each experimental plasmid. To calculate Z-scores, average neurite length was calculated for each plasmid, and these averages were then used to calculate an overall average and standard deviation within that set of 24 plasmids (no-plasmid and KLF4 treatments excluded). The overall average was subtracted from each individual average, and then divided by the standard deviation (Z-score). For hand tracing, neurons were imaged on an Olympus IX81 microscope and traced using Neurolucida software (MBF Bioscience).
Cell survival assay
Cortical neurons were cultured and transfected as described above, except the mCherry reporter was replaced with pMAX-GFP (Amaxa) to enable co-staining with Sytox orange. After three days, 150µl of culture media was removed and replaced with Hank’s Balanced Salt Solution (HBSS) containing 2µg/ml Hoechst 34580 (Invitrogen #H21846) and 5µM Sytox Orange (Invitrogen #S11368). After 15 minutes cells were scanned with the Cellomics KSR and percent of surviving cells (Hoechst+/Sytox–) was quantified for a minimum of 500 cells per treatment.
Results
Establishing a neurite outgrowth screen using high content analysis of primary postnatal cortical neurons
We hypothesized that changes in gene expression in postnatal cortical neurons reduce the intrinsic capacity for axon growth (Goldberg et al., 2002; Blackmore and Letourneau, 2006; Moore et al., 2009). To identify candidate genes we took advantage of publicly available microarray data comparing gene expression in purified corticospinal motor neurons (CSMNs) between embryonic day 18 (E18) and postnatal day 14 (P14) (Arlotta et al., 2005). This dataset was initially created to identify genes involved in cell specification, but fortuitously spans the time when CST neurons lose regenerative capacity in vivo (Bregman et al., 1989). Using RMA normalization (Irizarry et al., 2003) and a cut-off of three-fold or greater changes, we identified 237 genes that decreased and 834 genes that increased in expression between E18 and P14.
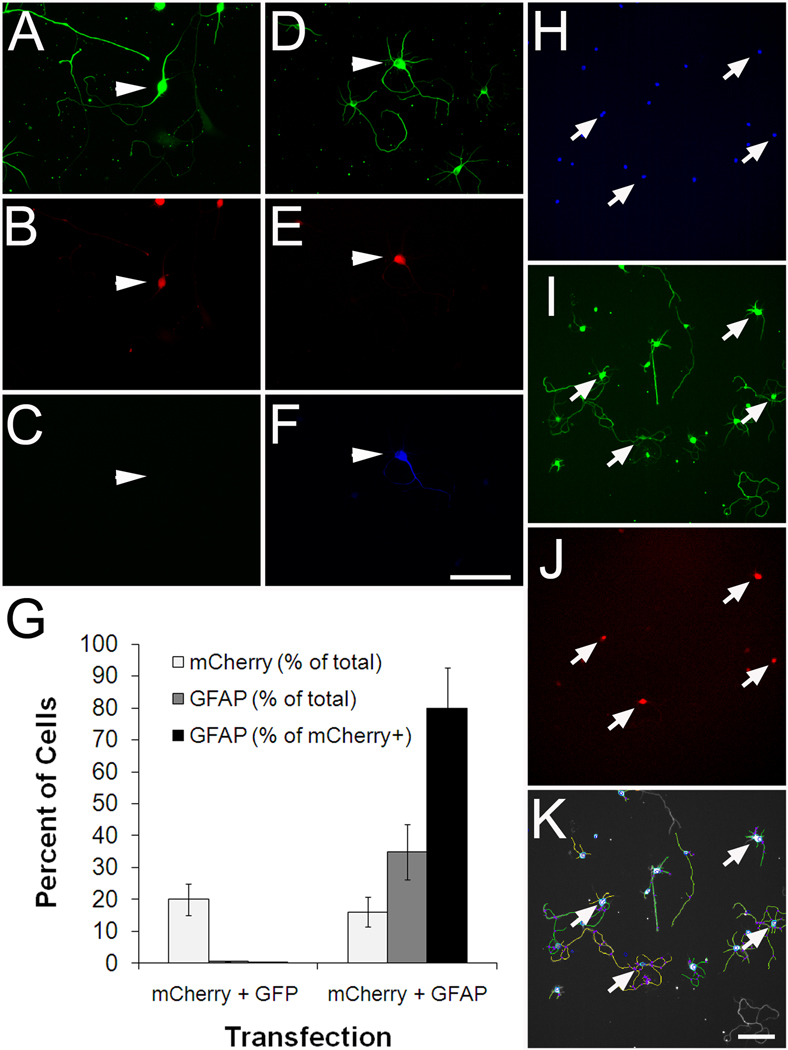
In principle, gene function can be queried either by overexpression or knockdown. Here we focused on gene overexpression. We obtained a library of nearly 16,000 mammalian expression vectors (OpenBiosystems), which contained plasmid DNA corresponding to 445 of our candidate genes represented redundantly by 743 clones. To screen these hundreds of candidate genes we adopted the strategy of electroporation in 96-well format, based on (Buchser et al., 2006), which allows many genes to be tested in a single experiment. We established conditions for transfecting cortical neurons with about 50% efficiency, as assessed by expression of EGFP and mCherry fluorescent reporters (Supplemental Figure 1). Although relatively high, 50% transfection efficiency still requires some means to specifically identify transfected neurons for downstream analysis. Thus we optimized a strategy of co-transfection with a fluorescent reporter plasmid. To test rates of co-transfection we used an OpenBiosystems clone encoding GFAP, a glial-specific intermediate filament protein chosen mainly because it is not expressed endogenously in neurons. As expected, we did not detect GFAP by immunohistochemistry in untransfected neurons (Figure 1). Three days after transfection with pCMV-SPORT6 GFAP, we detected GFAP expression in 34% of neurons. When GFAP was co-transfected with mCherry plasmid at a ratio of 6:1, more than 80% of mCherry-positive neurons also expressed GFAP. We obtained similar rates of co-transfection with other clones from the library, including one of our “hit” genes, doublecortin (data not shown and Figure 3). These results validate the effectiveness of transfection by electroporation in 96-well format, verify that plasmids from the expression library drive protein expression in cortical neurons, and establish that in our system, detection of a co-transfected fluorescent reporter is a high-probability indicator of expression of a candidate gene.
Figure 1. Cortical neurons transfected in 96-well format are identified by co-transfected mCherry reporter and efficiently traced by Cellomics instrumentation.
(A–G) P1 cortical neurons were co-transfected with mCherry and either EGFP or GFAP. (A–C) Control neurons transfected with mCherry and EGFP express βIII tubulin (arrowhead, A) and mCherry (arrowhead, B), but do not express GFAP (arrowhead, C). (D–E) Neurons transfected with mCherry and GFAP express both mCherry (arrow, E) and GFAP (arrow, F). (G) Quantification shows that 80% of mCherry-positive neurons express GFAP after co-transfection. More than 300 neurons were quantified per condition. (H–K) P1 cortical neurons transfected with mCherry were cultured for 3 days prior to fixation, staining, and automated scanning (Kineticscan, Cellomics). Only cells identified as neurons by βIII tubulin staining (I), and successfully transfected with mCherry reporter (arrows, J) were included in subsequent analysis. (D) Digitized overlay shows effective tracing of cell processes. Scale bars, 100µm.
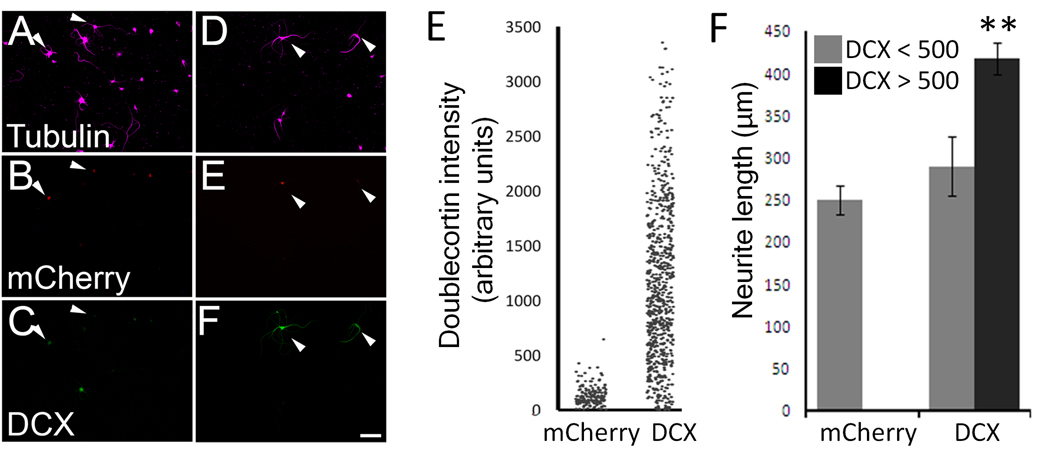
Figure 3. DCX overexpression correlates with longer neurites.
P5 cortical neurons were transfected with mCherry ± doublecortin. (A–F). Control neurons express mCherry (arrows, B) and are faintly immunoreactive for DCX. (C). Neurons co-transfected with mCherry and doublecortin (D–F) express mCherry reporter (E) and are brightly immunoreactive for DCX (F). (G) Each dot represents the average DCX immunofluorescence intensity of a single transfected (mCherry+) neuron. Nearly 100% of control neurons had an average intensity below 500 arbitrary units, while 87% of DCX-transfected neurons had an average intensity above 500. (H) Average neurite length of transfected neurons (βIII tubulin+, mCherry+). Neurons that were brightly immunofluorescent for DCX extended longer neurites that those with dimmer immunofluorescence (P<0.01, paired t-test, n>300 in each treatment). Scale bar is 50µm.
To quantify neurite outgrowth we used an automated microscope (Cellomics KSR, and subsequently Cellomics ArrayScan VTI) and Cellomics Neuronal Profiling software. Transfected neurons were cultured in 96-well culture plates in a stereotyped pattern, such that neurons transfected with each plasmid were located in six replicate wells on three separate plates. After three days neurons were fixed and visualized using immunohistochemistry for neuron-specific βIII tubulin. The automated microscope acquired images of neurons, automatically traced βIII-tubulin positive neurites (Figure 1), and reported morphological parameters for each cell, including the total length of neurites and fluorescence intensities of mCherry and βIII tubulin. Data were exported to Spotfire DecisionSite software for further analysis (see Methods and below). Throughput in the screen was limited by immunohistochemistry, which was done by hand using multichannel pipettes, and, to a lesser extent, the preparation of the cortical neurons. Nevertheless, by combining electroporation in 96-well format with automated image analysis and streamlined data analysis, we could comfortably test around 50 plasmids and acquire detailed morphological data from around 50,000 neurons per week.
Hit genes include both positive and negative regulators of neurite growth
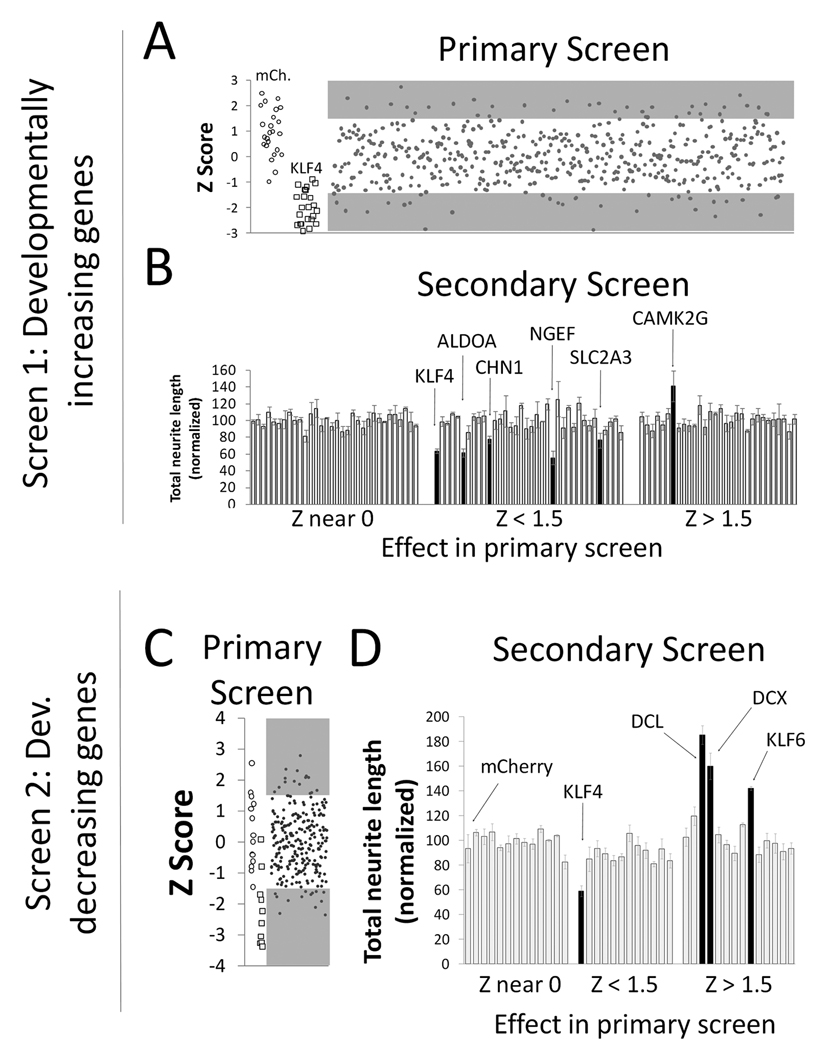
We first screened developmentally increasing genes, predicting that genes expressed highly in mature neurons, which regenerate poorly, might act to suppress growth when overexpressed in immature neurons. We obtained 499 plasmids corresponding to 310 different genes whose mRNA expression increased at least 3-fold in CSMNs between E18 and P14 (Arlotta et al., 2005). P1 cortical neurons were co-transfected with test plasmids and mCherry reporter, cultured for three days, and subjected to automated image analysis to quantify neurite length of transfected (mCherry-positive) neurons. Plasmids were prepared and screened in groups of 24, including three controls. The first control received no plasmid, and was used to set a threshold for background fluorescence to identify mCherry-positive cells in other wells. The second control received only mCherry plasmid, which is not expected to affect neurite length. The third control received plasmid encoding KLF4, a transcription factor that we recently identified as a reliable suppressor of neurite length ((Moore et al., 2009); Figure 2). Compared to the average of other plasmids, KLF4 decreased neurite length by a minimum of 20% in all experiments, with an average decrease of 30.1% (+/−6.4% SEM). Thus KLF4 serves as an essential positive control, and its presence in each set of experiments confers high confidence in the ability of our screen to detect changes in neurite outgrowth.
Figure 2. An overexpression screen of developmentally regulated genes.
Cortical neurons (P1, A, B; P5, C,D) were co-transfected with test genes and mCherry, and cultured for 3 days. (A, C) In primary screens of both up-regulated (A) and down-regulated (C) genes, plasmids were tested in sets of 24 that included three controls (no plasmid, mCherry (open circles), and growth-suppressive KLF4 (open squares)). Average neurite length was calculated from at least 100 transfected neurons, and a Z-scores was calculated for each gene based on the average and standard deviation across all test genes in the set (see Methods). (A, C) Each point represents the Z score from a plasmid in one experiment. , and shaded areas indicate plasmids selected for re-screening (Z scores > or < 1.5). (B,D) In secondary screens, genes were tested in sets of 24 that included mCherry (leftmost bar) and eight genes with no effect in the primary screen (Z near 0; see Methods). Neurite length was normalized to the average of the control genes. Bars represent the average normalized length across three replicate experiments. Black bars represent genes that differed significantly from controls (P < 0.05, ANOVA with post-hoc Dunnett’s).
Despite intense effort to control experimental variability, average neurite length differed considerably among experiments, even in control wells that received no plasmid. Therefore, in order to compare plasmids across the entire screen, we normalized the data using Z-scores based on neurite lengths within each group of 24 genes (see Methods). Using a cutoff of Z-score +/− 1.5, our primary screen of 310 genes identified 30 genes with increased neurite lengths and 40 genes with decreased neurite length (Figure 2A and Supplementary Table 1). Because of unavoidable noise in the data, we anticipated that only some of these represented true “hits.” To reduce the number of false positives and identify genes with large and statistically significant effects we re-tested these preliminary hits in a secondary screen. Plasmids were again tested in groups of 24 containing “no plasmid”, mCherry, and KLF4 controls. In addition, we also included eight genes that had produced Z-scores near 0 in the primary screen; as predicted by the primary screen, none of these genes significantly affected neurite length compared to mCherry control (p>.05, ANOVA). Therefore, for the purpose of the secondary screen, we refer to this set of genes collectively as control genes. Including these multiple control genes in triplicate experiments enabled statistical tests (ANOVA with post-hoc Dunnett’s) to determine whether the effects of the preliminary hit genes rose above the level of variability observed in controls. As shown in Figure 2B, we identified four genes that reliably decreased neurite length, and one that increased neurite length. Importantly, a viability assay detected no change in cell survival after transfection with these hit genes (Table 1). In sum, high content analysis of developmentally increasing genes identified five genes with strong and reproducible effects on neurite length. Of these, four significantly decreased neurite growth, as predicted by our hypothesis.
Table 1. Hit genes do not affect cell survival.
P5 cortical neurons were untransfected (no plasmid), or transfected with EGFP (control) or with hit genes, prior to culture for 3 days and measurement of cell death using Sytox orange.
| Gene |
% survival |
|---|---|
| No transfection | 51.4 (+/−5.6) |
| No supplements | 5.6 (+/−1.8)*** |
| EGFP | 46.9 (+/−5.6) |
| ALDOA | 47.3 (+/−5.8) |
| CAMK2G | 46.8 (+/−8.2) |
| CHN1 | 46.4 (+/−7.6) |
| DCL | 46.2 (+/−7.9) |
| DCX | 47.1 (+/−6.1) |
| KLF6 | 43.7 (+/−5.7) |
| NGEF | 45.6 (+/−5.8) |
| SLC2A3 | 44.7 (+/−7.2) |
95% of neurons cultured without supplements died (***p<.001, ANOVA with post-hoc Dunnett’s). Transfection efficiencies were above 50% in all experiments. No hit genes significantly altered cell survival compared to controls (n > 1000, N=3, ANOVA with post-hoc Dunnett’s).
In our second screen we predicted that genes more abundant in embryonic neurons, which regenerate robustly, might enhance neurite outgrowth when transfected into older neurons. Ideally, this screen would be performed in mature neurons that display reduced baseline outgrowth compared to P1. However, low viability of older neurons rendered this impractical. We therefore used P5 cortical neurons, which represent the oldest age that, in our hands, allowed sufficient cell survival for high content analysis (data not shown). In these cells we screened developmentally decreasing genes, using 244 plasmids corresponding to 135 unique genes. As in the previous screen, plasmids were tested in groups of 24, each containing no-plasmid, mCherry, and KLF4 controls. KLF4 again reliably suppressed neurite growth, demonstrating the sensitivity of our assay (Figure 2C). The primary screen identified 13 plasmids that suppressed growth (Z-score <−1.5), and 15 that increased growth (Z-score >1.5) (Supplementary Table 1). Re-testing of these preliminary hits identified three genes with robust effects, all of which increased neurite length (Figure 2D) without affecting cell survival (Table 1). One of these genes, KLF6, is a transcription factor from the same family as our growth-suppressive control gene KLF4. Subsequent experiments on this family of transcription factors identified an important role for KLFs in controlling neurite growth of CNS neurons during development; these results are reported elsewhere (Moore et al., 2009). The other two genes are two closely related microtubule associated factors, doublecortin (DCX) and doublecortin-like (DCL; see below).
DCX and DCL enhance axonal outgrowth in postnatal CNS neurons
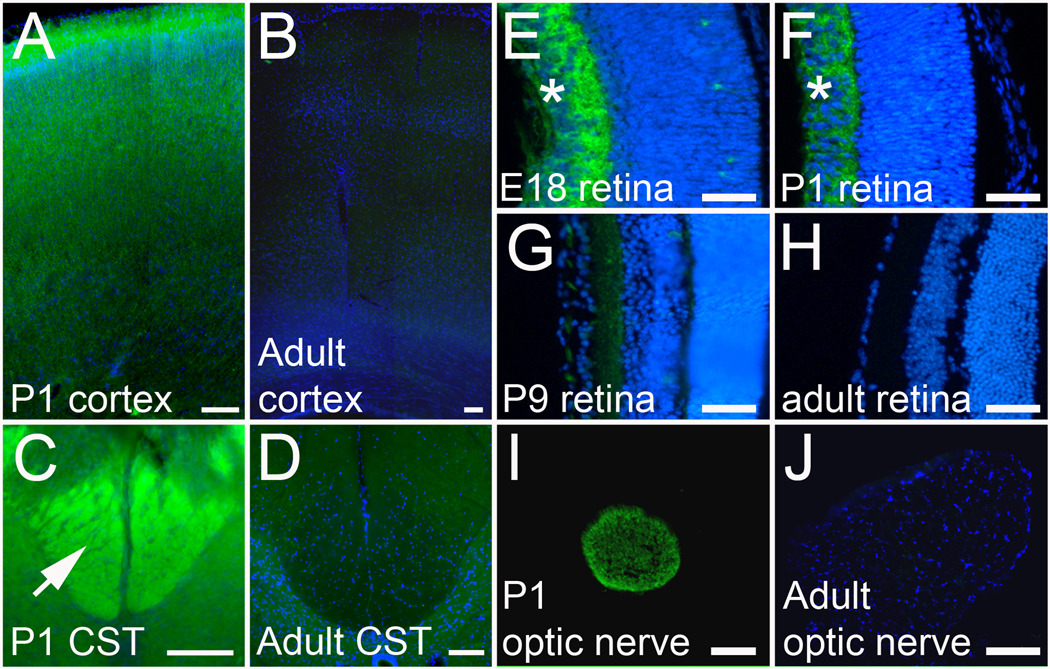
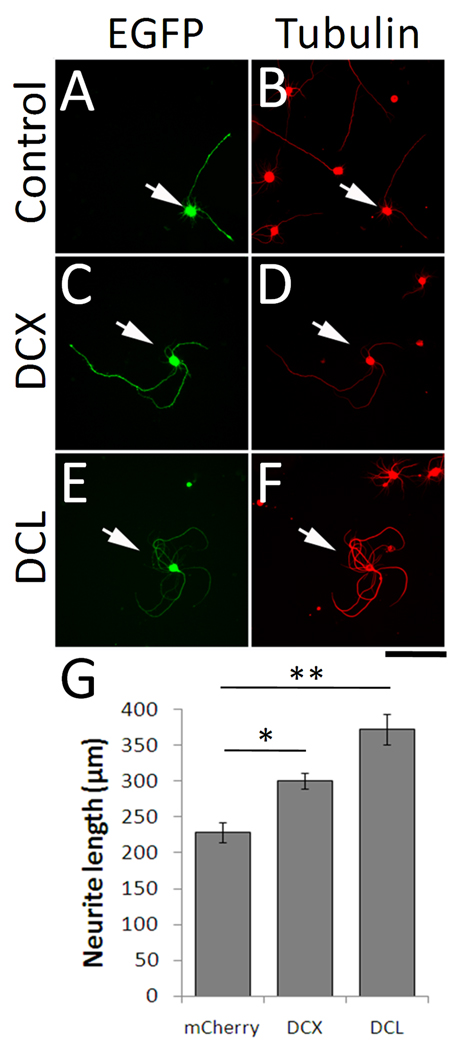
To validate and further characterize the effects of our “hit” genes on neurite growth, we selected a subset for further analysis. To take advantage of existing reagents and to facilitate validation, we prioritized genes previously described in the neurite outgrowth literature. DCX is a microtubule binding protein with a well established role in neuronal migration and an emerging role in neurite outgrowth (Kawauchi and Hoshino, 2008). Doublecortin-like kinase (DCLK) contains an N-terminal domain that is highly homologous to DCX, and a C-terminal kinase domain. However, sequencing of the doublecortin-like kinase clone that emerged as a “hit” gene showed it to be isoform 4, a truncated isoform that lacks the kinase domain entirely. This isoform is 79% homologous to DCX, and is thus referred to as doublecortin-like (DCL) (Vreugdenhil et al., 2007). DCX expression is known to decrease postnatally throughout the brain (Francis et al., 1999; Gleeson et al., 1999); our immunohistochemistry confirmed the developmental downregulation of DCX in the cortex and in the corticospinal tract in the spinal cord (Figure 6). In transfected cortical neurons in culture we used an antibody to directly detect the presence of over-expressed DCX. Importantly, neurons that expressed high levels of exogenous DCX (i.e. with average DCX signal that was higher than untransfected control neurons) showed an increase in average neurite length (Figure 3). These data directly correlate increased DCX expression with enhanced neurite length in postnatal cortical neurons. Furthermore, because the effects on neurite outgrowth were similar regardless of whether transfected cells were identified based on DCX or mCherry expression, these results further validate co-transfection with mCherry as an effective screening strategy.
Figure 6. DCX is regulated in developing cortex and in RGCs.
(A–D) Horizontal cortical sections and transverse spinal cord sections from P1 and adult animals were immunostained for DCX (green). DCX Immunoreactivity is bright throughout P1 cortex (A) and in the CST in P1 spinal cord (arrow, C), but is very dim in adult cortex (B) and spinal cord (D). (E–J) Retinas (E–H) and optic nerves (I,J) were cryosectioned and stained for DCX. DCX immunoreactivity (green) is abundant in the retinal ganglion cell layer at E15 and P1 (asterisk, E,F) but is downregulated by P9 (G) and undetectable in the adult (H). In the optic nerve, DCX is readily detectable at P1 (I), but undetectable in the adult (J). Scale bars are 100µm (A–D, I,J) and 50 µm (E–H).
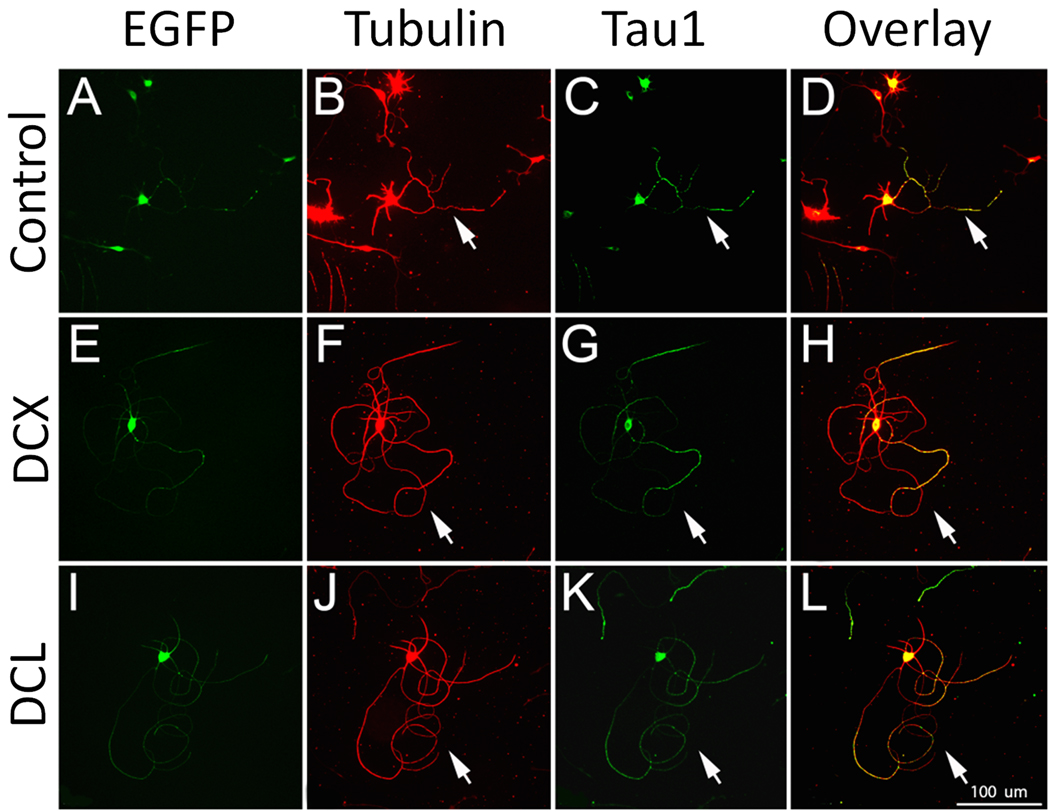
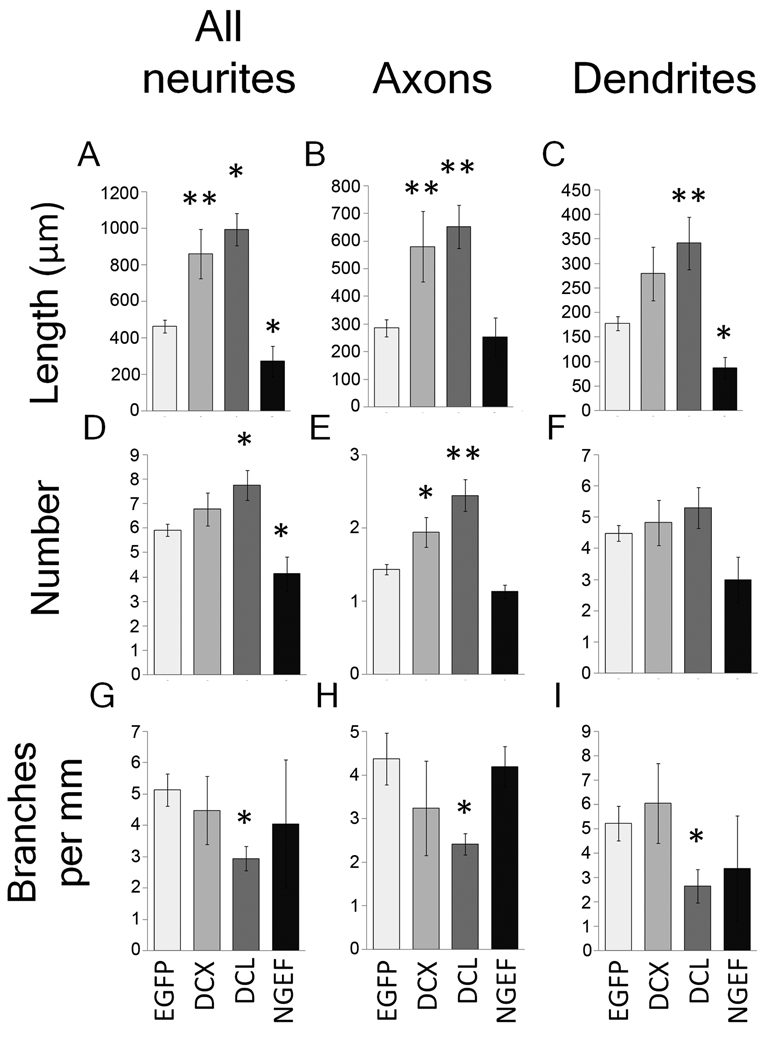
We next hand-traced neurons transfected with DCX or DCL, using an anti-Tau1 antibody to differentiate effects on axons and dendrites. DCX and DCL caused similar phenotypes characterized by long neurites with looped trajectories (Figure 4). Hand tracing confirmed that total neurite length was significantly increased (Figure 5), as was the length and number of Tau1-positive axons. Branching of axons was significantly suppressed by DCL, with a non-significant trend in the same direction by DCX. Thus, increased expression of DCX and DCL in postnatal cortical neurons increased the number and length of axons while suppressing axon branching.
Figure 4. Overexpression of DCX and DCL increase axon length in postnatal cortical neurons.
P5 cortical neurons were transfected with EGFP (A–D), or EGFP plus DCX (E–H) or DCL (I–L). After three days, neurons were immunostained for βIII tubulin (B,F,J) and Tau1 (C,G,K). Arrows identify axons from transfected (EGFP+) neurons. Compared to controls, neurons transfected with DCX or DCL extend long axons with looped trajectories (arrows, H, L). Scale bar is 100µm.
Figure 5. DCX, DCL, and NGEF affect length, number, and branching of axons and dendrites.
P5 cortical neurons were transfected with EGFP (control), or co-transfected with EGFP and DCX, DCL, or NGEF. Images of EGFP+ neurons were acquired and traced using Neurolucida. All EGFP+ neurons with neurites were included in A, D and G. For other graphs, only polarized neurons with clear tau1-positive processes were included. (A–C) NGEF decreased and DCX and DCL increased average neurite length. DCX and DCL significantly increased the length of axons, and DCL significantly increased dendrite length. (D–F) NGEF decreased and DCL increased the average number of neurites, whereas DCX and DCL significantly increased the average number of axons (E), but not the number of dendrites (F). (G–H) DCL significantly decreased the amount of branching for both axon and dendrites. N ≥ 40. *p<0.05, **p<0.01 ANOVA with post-hoc Dunnett’s.
We next asked whether the effects of overexpression of DCX and DCL might be generalized to other postnatal CNS neuronal populations, using retinal ganglion cells (RGCs) as a test case. DCX expression decreases strongly throughout the postnatal brain, but developmental regulation specifically in RGCs is less clear (Francis et al., 1999; Gleeson et al., 1999; Lee et al., 2003). Immunohistochemistry for DCX in the embryonic retina produced strong labeling in the ganglion cell layer and in the optic nerve (Figure 6). Because only RGCs project axons to the optic nerve, we conclude that embryonic RGCs express DCX. Immunoreactivity for DCX declined after birth, and was undetectable in the adult ganglion cell layer or optic nerve, demonstrating downregulation of DCX protein by maturing RGCs. To test the effects of overexpression of DCX and DCL we purified postnatal RGCs using established immunopanning techniques previously shown to generate >99% pure cultures (Barres et al., 1988; Goldberg et al., 2002; Moore et al., 2009). In postnatal RGCs in culture, overexpression of DCX and DCL produced phenotypes similar to those observed in cortical neurons, with an increase in total neurite length and looped trajectories of neurites (Figure 7). Thus, overexpression of DCX in at least two different types of postnatal CNS neurons strongly enhances neurite growth.
Figure 7. Overexpression of DCX or DCL increases neurite length in postnatal RGCs.
Purified P4 RGCs were transfected with EGFP (control) or EGFP plus DCX or DCL, cultured for 2 days, and stained for tubulin. Transfected RGCs were identified by EGFP (arrows). Compared to control (A,B), RGCs transfected with DCX (C,D) or DCL (E,F) showed increased neurite outgrowth and curved/looped neurite trajectories. (G) Total neurite length was significantly increased in neurons transfected with DCX or DCL (*p<0.05, **p<0.01, ANOVA with post-hoc Dunnet’s). N= 3 experiments, with > 100 transfected neurons/treatment. Error bars show SEM. Scale bar, 100µm.
NGEF overexpression leads to defects in morphological differentiation
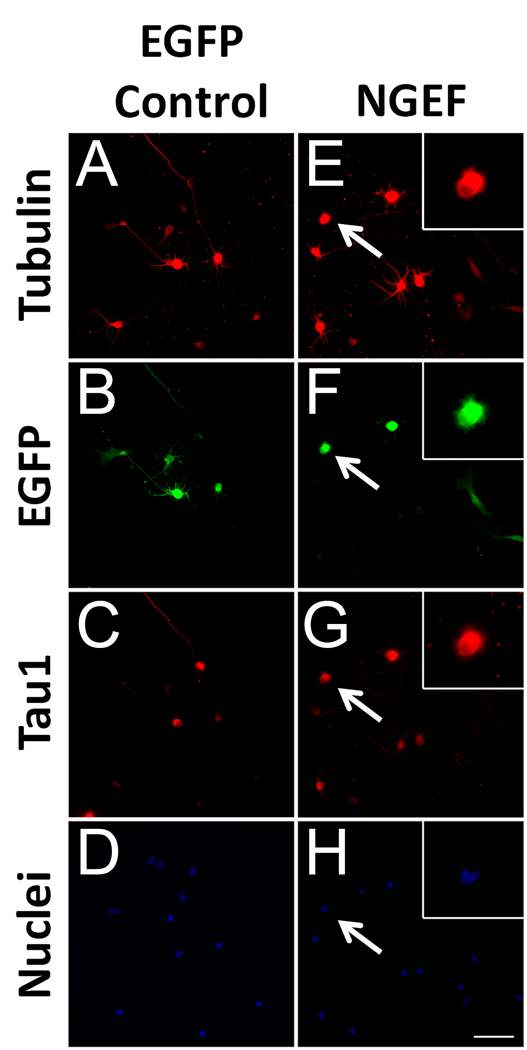
NGEF (also called ephexin) emerged as a strong suppressor of neurite length in our screen. NGEF is a guanylnucleotide exchange factor that acts downstream of ephrin/EphA4 signaling to activate the small GTPases Cdc42, Rac1, and RhoA (Shamah et al., 2001). In accordance with the microarray data from purified CSMNs (Arlotta et al., 2005), northern blotting has also demonstrated strong upregulation of NGEF in postnatal brain (Shamah et al., 2001). To examine the phenotype produced by NGEF overexpression in more detail, we manually traced transfected P1 cortical neurons using anti-βIII tubulin to label all neurites and anti-Tau1 to label axons. As shown in Figure 8, the most striking effect of overexpression was a failure to extend neurites; although 92.7% of control neurons extended neurites >10µm after three days in culture, only 47.6% of NGEF-transfected neurons extended neurites (p<.001, Chi-square, n>30 per experiment, N=3). NGEF-transfected neurons appeared healthy, with large uncondensed nuclei, and often extended lamellaepodial protrusions (arrows, Figure 10). Furthermore, when NGEF-transfected neurons did extend neurites, these neurites were shorter on average than control neurons (Figure 5), and also displayed defects in polarization. Whereas 79.6% of control neurons displayed asymmetric Tau1 localization in neurites (a hallmark of neuronal polarization), only 48.3% of neurite-bearing neurons transfected with NGEF had a clear Tau1-positive axon (p<.001, Chi-square, n>30 per experiment, N=3). We conclude that overexpression of NGEF in early postnatal cortical neurons produces a variety of defects in neurite initiation, polarization, and extension.
Figure 8. NGEF suppresses neurite growth in postnatal cortical neurons.
P1 cortical neurons were co-transfected with EGFP (control, A–D) or NGEF plus EGFP (NGEF, E–H) and cultured for 3 days. Control neurons (A–D) extend long processes, some of which are identified as axons by Tau1 immunoreactivity (C). Neurons transfected with NGEF (E–H, arrows) often extended no neurites but appeared viable with large, uncondensed nuclei and abundant lamellar protrusions (insets, E–H). Scale bar is 50µm.
Discussion
We have developed a screening method using high content analysis to test large numbers of genes in overexpression assays in postnatal cortical neurons. By screening developmentally regulated genes we identified eight genes that robustly affect neurite growth. Interestingly, these genes follow a pattern in which most of those whose expression increases during this postnatal period reduced neurite growth, and all of those whose expression decreases during development increased neurite growth. Follow-up experiments demonstrated novel effects of NGEF in suppressing neuritogenesis, and of DCX and DCL in enhancing axon growth in postnatal neurons.
High content analysis identifies neurite growth genes
Advances in automated microscopy and image analysis, robotic liquid handling, and data analysis have combined to make high content analysis (HCA) a powerful strategy to test the functions of large numbers of genes and bioactive compounds. We have harnessed these technologies to study gene regulation of neurite outgrowth. We previously used HCA to screen genes in embryonic hippocampal neurons, and identified the transcription factor KLF4 as a potent suppressor of neurite outgrowth (Moore et al., 2009). The current study is among the first examples of a large-scale overexpression screen in postnatal CNS neurons. Primary neurons, particularly postnatal CNS neurons, have stringent survival requirements, are relatively difficult to transfect, and display high intra-experiment variability of neurite outgrowth. Previous solutions to these problems have included the use of cell lines as substitutes for neurons (Yamada et al., 2007; Loh et al., 2008), the use of non-vertebrate neurons more amenable to RNAi treatments (Sepp et al., 2008), or screens of chemical libraries (Sivasankaran et al., 2004). These screens have yielded important insights into the regulation of neurite outgrowth, but it is likely that some important information can be obtained only by specific study of gene perturbation in primary mammalian neurons. Indeed, as we discuss below, DCX overexpression appears to have cell-specific effects that differ even between embryonic hippocampal neurons and postnatal cortical neurons, highlighting the importance of studying neurite outgrowth in a specific neuronal population of interest.
Potential limitations of our screening approach
We queried gene function only by overexpression, and not by knockdown. Overexpressed proteins can display non-physiological functions, while knockdown, which confers higher confidence in biological relevance, risks both false positive and false negative results due to slow protein turnover, protein redundancy, and off-target effects. The most powerful approach would be complementary knockdown and overexpression screens, but despite considerable effort, we have not yet developed a knockdown method that is reliable enough in postnatal cortical neurons to support a screen. Thus complementary knockdown screens await technique development.
A second issue concerns the use of P5 neurons, rather than fully mature neurons, in our screen for growth enhancers. In our hands, cortical neurons older than P5 exhibit poor survival in culture, which complicates interpretation of reductions in neurite outgrowth and also makes HCA impractical. Prior to initiating these experiments, a significant concern was that in the absence of a control gene that enhanced neurite outgrowth, our use of P5 neurons, which retain a relatively high capacity for neurite outgrowth, might make the assay insensitive to growth-promoting genes. Importantly, however, our results indicate otherwise. Although it is possible that our screen missed genes with activities that are specific to more fully mature neurons, the results of this screen and our previous work (Moore et al., 2009) clearly demonstrate the ability of our assay to detect growth promotion.
Our use of a heterogeneous population of cortical neurons affects interpretation of our findings. One potential concern is the presence of GABAergic interneurons, but we find that nearly 80% of transfected neurons in our assay are non-GABAergic (Supplemental Figure 1). Nevertheless, cortical projection neurons are highly heterogeneous, and it is unlikely that our assay would detect genes that affected any one population (such as CST neurons) specifically. Our screen is therefore expected to detect genes that act generally across diverse populations of CNS neurons. In this regard it is interesting to note that “hit” genes identified in these mixed cortical assays, including KLF transcription factors and now DCX, exert similar effects in RGCs (Moore et al., 2009). Thus, our mixed cortical system appears effective in identification of general regulators of CNS neurite outgrowth.
DCX and DCL kinase enhance neurite growth in vitro
DCX and DCL are microtubule associated proteins best known for their roles in supporting neuronal migration (Kawauchi and Hoshino, 2008). However, it is increasingly clear that DCX and DCL also play essential roles in neurite outgrowth, possibly by acting to bundle and stabilize microtubules in the “wrist” region at the base of growth cones, or alternatively by mediating microtubule/actin interactions in the actinrich periphery of growth cones (Tsukada et al., 2005; Shmueli et al., 2006; Bielas et al., 2007; Tint et al., 2009). In vivo, double knockout of DCX and DCLK (although not single knockout of either) dramatically disrupts the growth of efferent cortical projections, establishing a necessary role for these MAPs in axon growth in vivo (Deuel et al., 2006; Koizumi et al., 2006). Here we have shown for the first time that overexpression of DCX or DCL is sufficient to enhance neurite outgrowth from postnatal neurons, which normally express low levels of DCX. In contrast to the striking phenotype we observe in postnatal neurons, previous studies and our own unpublished observations show that overexpression of DCX has no effect on neurite length in embryonic hippocampal neurons (Cohen et al., 2008; Tint et al., 2009). Because the expression of DCX declines in CNS neurons postnatally, it may be that overexpression of DCX and DCL in postnatal neurons reveals functions that are masked in embryonic neurons with higher endogenous levels of DCX. This point illustrates the importance of screening candidate growth-promoting genes in postnatal neurons; a screen performed in embryonic neurons may have missed the growth-promoting effects of DCX and DCL.
Because DCX and DCL are required for axon growth during embryogenesis and are absent in most mature CNS neurons, they are in principle candidates for therapeutic overexpression after axonal injury. Studies exploring this therapeutic potential will need to address a number of questions. First, although DCX expression is low in mature neurons, we don’t know how it responds to injury. Second, because the activities of DCX and DCL are highly regulated, simple overexpression may not be sufficient to restore function. The affinities of DCX and DCL for microtubules and actin are regulated by phosphorylation, involving multiple kinases, phosphatases, and scaffolding proteins (Gdalyahu et al., 2004; Schaar et al., 2004; Tanaka et al., 2004; Shmueli et al., 2006; Bielas et al., 2007). Furthermore, components of this regulatory network are themselves developmentally regulated (Tsukada et al., 2003; Shmueli et al., 2006; Bielas et al., 2007). Manipulating this regulatory network after injury to maximize DCX/DCL’s growth-enhancing activities will likely be essential to developing any DCX/DCL-based therapy. Finally, it will be important to explore how DCX/DCL, which target axon growth regulation at the level of axonal cytoskeletal dynamics, (Zhou and Snider, 2006; Erturk et al., 2007; Pak et al., 2008), might synergize with strategies that target the cell body response at the level of gene transcription or metabolism (Park et al., 2008; Moore et al., 2009).
NGEF suppresses neurite growth in vitro
We have shown, for the first time, that forced overexpression of NGEF leads to substantial suppression of neurite outgrowth. NGEF (originally named Ephexin) is a guanylnucleotide exchange factor of the Dbl family, which acts as a downstream effector of ephrin/EphA4 signaling. By regulating the activities of small GTPases cdc42, rac1, and RhoA, NGEF is well positioned to regulate actin dynamics and axonal outgrowth (Shamah et al., 2001). In agreement with the microarray data used here, Northern blotting has shown strong upregulation of NGEF in the postnatal brain (Shamah et al., 2001). One model of NGEF activity is that unphosphorylated NGEF activates Cdc42, Rac1, and RhoA indiscriminately, but that upon ephrin binding to EphA4 receptors, NGEF is phosphorylated and preferentially stimulates RhoA activity, causing growth cone collapse and a blockade of axon growth (Sahin et al., 2005). Ephrins are expressed in the spinal cord after injury and act to suppress axon regeneration (Benson et al., 2005; Niclou et al., 2006), so the increased expression of NGEF in mature neurons may confer a higher sensitivity to growth-suppressive ephrin signaling.
Our finding that overexpression of NGEF suppresses neurite growth in the absence of exogenous ephrins is somewhat unexpected. It has been proposed previously that in the absence of ephrin signaling NGEF’s basal activation of small GTPases acts to support neurite growth. Indeed, knockdown of NGEF in retinal ganglion cells that were not treated with ephrin caused a decrease in axon growth (Sahin et al., 2005). It is possible that ephrin A’s may be present in the culture media. Alternatively, our results may be due to a difference in cell types. NGEF has not been previously studied in postnatal cortical neurons, and we have not characterized the relative abundance or activation of rho-family GTPases in control or NGEF-overexpressing neurons in culture. It is possible that in this signaling context, high levels of NGEF expression act to suppress neurite growth even in the absence of ephrin stimulation. Final conclusions regarding NGEF function in postnatal cortical neurons must await loss of function experiments. Nevertheless, given the striking increase in NGEF expression during postnatal development, we speculate that abundant NGEF in mature neurons might act to suppress regeneration after injury. It would be particularly interesting to test whether axonal regeneration is enhanced in mature neurons that lack NGEF expression.
Other hits
Although we have yet to perform follow-up experiments on all the hit genes identified in our screens, previous reports suggest potential roles in regulating neurite outgrowth during development. For instance, our finding that overexpression of CaMKIIG enhances neurite outgrowth is consistent with findings regarding other CaMK family members (reviewed in (Wayman et al., 2008). CaMKI family members appear to regulate axonal outgrowth (Wayman et al., 2004), whereas CaMKII family members are prominent in regulating dendritic morphogenesis. For instance, overexpression of CaMKIIB has been shown previously to increase dendritic arborization (Fink et al., 2003). Thus, our finding that a member of CaMK family affects neurite outgrowth might be predicted from the literature, although this particular family member (CaMKIIG) has not previously been shown to regulate neurite growth. Similarly, CHNI (alpha-chimaerin) negative regulates the small GTPase Rac1, and is a mediator of growth cone collapse in response to sempahorins and ephrins (Brown et al., 2004; Shi et al., 2007). Another hit gene, ALDOA (aldolase A) belongs to a family of glycolytic enzymes and was initially a surprising hit. Intriguingly, however, aldolases have recently been shown to have an unexpected ability to bind and stabilize mRNA in neurons, thereby increasing the expression of genes including neurofilaments (Canete-Soler et al., 2005; Stefanizzi and Canete-Soler, 2007). Future experiments can determine whether the ability of ALDOA to suppress neurite outgrowth might be related to this mRNA-binding activity.
In summary, our screening of developmentally regulated genes has identified a number of genes that regulate neurite outgrowth. Follow-up experiments validated the ability of the screen to identify both positive and negative growth regulators. By using primary postnatal neurons, we have identified previously unappreciated effects on neurite outgrowth by relatively well-studied genes such as DCX/DCL and NGEF. Notably (with a single exception), developmentally upregulated genes suppressed neurite growth, and developmentally downregulated genes enhanced growth. These intriguing results support the concept that a coordinated transcriptional program underlies the transition between a high growth state and a low growth state (see (Moore et al., 2009)). By expanding our screening efforts to cover more of the genome we will be able to test this idea in detail.
Supplementary Material
P1 cortical neurons were transfected with mCherry and cultured for 3 days. (A) Immunohistochemistry for βIII tubulin identifies neurons. (B) mCherry identifies transfected cells. (C–D) Immunohistochemistry for GAD67 identifies GABAergic interneurons (arrowhead) and non-GABAergic neurons (arrows). (E) Cellomics instrumentation and software quantified the percent of βIII-positive neurons that were also GAD67-positive and mCherry-positive. 80% of total neurons, and 78% of transfected neurons, were non-GABAergic. More than 1000 neurons were measured in a single experiment. Scale bar is 100µm.
Acknowledgements
This work was supported by the C.H. Neilsen Foundation, the R. Wilson Foundation, and The Buoniconti Fund to Cure Paralysis (VPL/JLB); and by NINDS (R01-NS061348, JLG), the Seigal Foundation (JLG), NEI (P30-EY014801), and an unrestricted grant from Research to Prevent Blindness to the University of Miami. V. Lemmon holds the Walter G. Ross Distinguished Chair in Developmental Neuroscience at the University of Miami. D.L.M. is a Lois Pope LIFE Fellow, with support from NINDS training grants T32 S07492 and T32 NS007459. We are indebted to Roger Tsien (UCSD) for the generous gift of mCherry plasmid. We are grateful to Guerline Lambert for her technical assistance, as well as to lab members Yan Shi, Eli Weaver, Eleut Hernandez, and Raul Corredor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Barres BA, Silverstein BE, Corey DP, Chun LL. Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron. 1988;1:791–803. doi: 10.1016/0896-6273(88)90127-4. [DOI] [PubMed] [Google Scholar]
- Benson MD, Romero MI, Lush ME, Lu QR, Henkemeyer M, Parada LF. Ephrin-B3 is a myelin-based inhibitor of neurite outgrowth. Proc Natl Acad Sci U S A. 2005;102:10694–10699. doi: 10.1073/pnas.0504021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, Serneo FF, Chechlacz M, Deerinck TJ, Perkins GA, Allen PB, Ellisman MH, Gleeson JG. Spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist. Cell. 2007;129:579–591. doi: 10.1016/j.cell.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore M, Letourneau PC. Changes within maturing neurons limit axonal regeneration in the developing spinal cord. J Neurobiol. 2006;66:348–360. doi: 10.1002/neu.20224. [DOI] [PubMed] [Google Scholar]
- Bregman BS, Kunkel-Bagden E, McAtee M, O'Neill A. Extension of the critical period for developmental plasticity of the corticospinal pathway. J Comp Neurol. 1989;282:355–370. doi: 10.1002/cne.902820304. [DOI] [PubMed] [Google Scholar]
- Brown M, Jacobs T, Eickholt B, Ferrari G, Teo M, Monfries C, Qi RZ, Leung T, Lim L, Hall C. Alpha2-chimaerin, cyclin-dependent Kinase 5/p35, and its target collapsin response mediator protein-2 are essential components in semaphorin 3A-induced growth-cone collapse. J Neurosci. 2004;24:8994–9004. doi: 10.1523/JNEUROSCI.3184-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchser WJ, Pardinas JR, Shi Y, Bixby JL, Lemmon VP. 96-well electroporation method for transfection of mammalian central neurons. Biotechniques. 2006;41:619–624. doi: 10.2144/000112279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canete-Soler R, Reddy KS, Tolan DR, Zhai J. Aldolases a and C are ribonucleolytic components of a neuronal complex that regulates the stability of the light-neurofilament mRNA. J Neurosci. 2005;25:4353–4364. doi: 10.1523/JNEUROSCI.0885-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LC, Tessier-Lavigne M. Regeneration of the adult central nervous system. Curr Biol. 2005;15:R749–R753. doi: 10.1016/j.cub.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Chen DF, Jhaveri S, Schneider GE. Intrinsic changes in developing retinal neurons result in regenerative failure of their axons. Proc Natl Acad Sci U S A. 1995;92:7287–7291. doi: 10.1073/pnas.92.16.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Segal M, Reiner O. Doublecortin supports the development of dendritic arbors in primary hippocampal neurons. Dev Neurosci. 2008;30:187–199. doi: 10.1159/000109862. [DOI] [PubMed] [Google Scholar]
- De Hoop MJ, Meyn L, Dotti CG. Culturing hippocampal neurons and astrocytes from fetal rodent brain. San Diego: Academic; 1998. [Google Scholar]
- Deuel TA, Liu JS, Corbo JC, Yoo SY, Rorke-Adams LB, Walsh CA. Genetic interactions between doublecortin and doublecortin-like kinase in neuronal migration and axon outgrowth. Neuron. 2006;49:41–53. doi: 10.1016/j.neuron.2005.10.038. [DOI] [PubMed] [Google Scholar]
- Dusart I, Airaksinen MS, Sotelo C. Purkinje cell survival and axonal regeneration are age dependent: an in vitro study. J Neurosci. 1997;17:3710–3726. doi: 10.1523/JNEUROSCI.17-10-03710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk A, Hellal F, Enes J, Bradke F. Disorganized microtubules underlie the formation of retraction bulbs and the failure of axonal regeneration. J Neurosci. 2007;27:9169–9180. doi: 10.1523/JNEUROSCI.0612-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink CC, Bayer KU, Myers JW, Ferrell JE, Jr, Schulman H, Meyer T. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 2003;39:283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Francis F, Koulakoff A, Boucher D, Chafey P, Schaar B, Vinet MC, Friocourt G, McDonnell N, Reiner O, Kahn A, McConnell SK, Berwald-Netter Y, Denoulet P, Chelly J. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- Gdalyahu A, Ghosh I, Levy T, Sapir T, Sapoznik S, Fishler Y, Azoulai D, Reiner O. DCX, a new mediator of the JNK pathway. Embo J. 2004;23:823–832. doi: 10.1038/sj.emboj.7600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, Guyer M, Peck AM, Derge JG, Lipman D, Collins FS, Jang W, Sherry S, Feolo M, Misquitta L, Lee E, Rotmistrovsky K, Greenhut SF, Schaefer CF, Buetow K, Bonner TI, Haussler D, Kent J, Kiekhaus M, Furey T, Brent M, Prange C, Schreiber K, Shapiro N, Bhat NK, Hopkins RF, Hsie F, Driscoll T, Soares MB, Casavant TL, Scheetz TE, Brown-stein MJ, Usdin TB, Toshiyuki S, Carninci P, Piao Y, Dudekula DB, Ko MS, Kawakami K, Suzuki Y, Sugano S, Gruber CE, Smith MR, Simmons B, Moore T, Waterman R, Johnson SL, Ruan Y, Wei CL, Mathavan S, Gunaratne PH, Wu J, Garcia AM, Hulyk SW, Fuh E, Yuan Y, Sneed A, Kowis C, Hodgson A, Muzny DM, McPherson J, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madari A, Young AC, Wetherby KD, Granite SJ, Kwong PN, Brinkley CP, Pearson RL, Bouffard GG, Blakesly RW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Griffith M, Griffith OL, Krzywinski MI, Liao N, Morin R, Palmquist D, et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Klassen MP, Hua Y, Barres BA. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science. 2002;296:1860–1864. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- Hafidi A, Grumet M, Sanes DH. In vitro analysis of mechanisms underlying age-dependent failure of axon regeneration. J Comp Neurol. 2004;470:80–92. doi: 10.1002/cne.20003. [DOI] [PubMed] [Google Scholar]
- Hasan SJ, Keirstead HS, Muir GD, Steeves JD. Axonal regeneration contributes to repair of injured brainstem-spinal neurons in embryonic chick. J Neurosci. 1993;13:492–507. doi: 10.1523/JNEUROSCI.13-02-00492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kawano H, Li HP, Sango K, Kawamura K, Raisman G. Inhibition of collagen synthesis overrides the age-related failure of regeneration of nigrostriatal dopaminergic axons. J Neurosci Res. 2005;80:191–202. doi: 10.1002/jnr.20441. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Hoshino M. Molecular pathways regulating cytoskeletal organization and morphological changes in migrating neurons. Dev Neurosci. 2008;30:36–46. doi: 10.1159/000109850. [DOI] [PubMed] [Google Scholar]
- Koizumi H, Tanaka T, Gleeson JG. Doublecortin-like kinase functions with doublecortin to mediate fiber tract decussation and neuronal migration. Neuron. 2006;49:55–66. doi: 10.1016/j.neuron.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Kim IB, Lee E, Kwon SO, Oh SJ, Chun MH. Differential expression and cellular localization of doublecortin in the developing rat retina. Eur J Neurosci. 2003;17:1542–1548. doi: 10.1046/j.1460-9568.2003.02583.x. [DOI] [PubMed] [Google Scholar]
- Loh SH, Francescut L, Lingor P, Bahr M, Nicotera P. Identification of new kinase clusters required for neurite outgrowth and retraction by a loss-of-function RNA interference screen. Cell Death Differ. 2008;15:283–298. doi: 10.1038/sj.cdd.4402258. [DOI] [PubMed] [Google Scholar]
- Meyer-Franke A, Kaplan MR, Pfrieger FW, Barres BA. Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron. 1995;15:805–819. doi: 10.1016/0896-6273(95)90172-8. [DOI] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niclou SP, Ehlert EM, Verhaagen J. Chemorepellent axon guidance molecules in spinal cord injury. J Neurotrauma. 2006;23:409–421. doi: 10.1089/neu.2006.23.409. [DOI] [PubMed] [Google Scholar]
- Nothias F, Boyne L, Murray M, Tessler A, Fischer I. The expression and distribution of tau proteins and messenger RNA in rat dorsal root ganglion neurons during development and regeneration. Neuroscience. 1995;66:707–719. doi: 10.1016/0306-4522(94)00598-y. [DOI] [PubMed] [Google Scholar]
- Pak CW, Flynn KC, Bamburg JR. Actin-binding proteins take the reins in growth cones. Nat Rev Neurosci. 2008;9:136–147. doi: 10.1038/nrn2236. [DOI] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin M, Greer PL, Lin MZ, Poucher H, Eberhart J, Schmidt S, Wright TM, Shamah SM, O'Connell S, Cowan CW, Hu L, Goldberg JL, Debant A, Corfas G, Krull CE, Greenberg ME. Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron. 2005;46:191–204. doi: 10.1016/j.neuron.2005.01.030. [DOI] [PubMed] [Google Scholar]
- Saunders NR, Kitchener P, Knott GW, Nicholls JG, Potter A, Smith TJ. Development of walking, swimming and neuronal connections after complete spinal cord transection in the neonatal opossum, Monodelphis domestica. J Neurosci. 1998;18:339–355. doi: 10.1523/JNEUROSCI.18-01-00339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar BT, Kinoshita K, McConnell SK. Doublecortin microtubule affinity is regulated by a balance of kinase and phosphatase activity at the leading edge of migrating neurons. Neuron. 2004;41:203–213. doi: 10.1016/s0896-6273(03)00843-2. [DOI] [PubMed] [Google Scholar]
- Senut MC, Tuszynski MH, Raymon HK, Suhr ST, Liou NH, Jones KR, Reichardt LF, Gage FH. Regional differences in responsiveness of adult CNS axons to grafts of cells expressing human neurotrophin 3. Exp Neurol. 1995;135:36–55. doi: 10.1006/exnr.1995.1064. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Hong P, Lizarraga SB, Liu JS, Mejia LA, Walsh CA, Perrimon N. Identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet. 2008;4:e1000111. doi: 10.1371/journal.pgen.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamah SM, Lin MZ, Goldberg JL, Estrach S, Sahin M, Hu L, Bazalakova M, Neve RL, Corfas G, Debant A, Greenberg ME. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell. 2001;105:233–244. doi: 10.1016/s0092-8674(01)00314-2. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shi L, Fu WY, Hung KW, Porchetta C, Hall C, Fu AK, Ip NY. Alpha2-chimaerin interacts with EphA4 and regulates EphA4-dependent growth cone collapse. Proc Natl Acad Sci U S A. 2007;104:16347–16352. doi: 10.1073/pnas.0706626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli A, Gdalyahu A, Sapoznik S, Sapir T, Tsukada M, Reiner O. Site-specific dephosphorylation of doublecortin (DCX) by protein phosphatase 1 (PP1) Mol Cell Neurosci. 2006;32:15–26. doi: 10.1016/j.mcn.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Sivasankaran R, Pei J, Wang KC, Zhang YP, Shields CB, Xu XM, He Z. PKC mediates inhibitory effects of myelin and chondroitin sulfate proteoglycans on axonal regeneration. Nat Neurosci. 2004;7:261–268. doi: 10.1038/nn1193. [DOI] [PubMed] [Google Scholar]
- So KF, Schneider GE, Ayres S. Lesions of the brachium of the superior colliculus in neonate hamsters: correlation of anatomy with behavior. Exp Neurol. 1981;72:379–400. doi: 10.1016/0014-4886(81)90231-4. [DOI] [PubMed] [Google Scholar]
- Stefanizzi I, Canete-Soler R. Coregulation of light neurofilament mRNA by poly(A)-binding protein and aldolase C: implications for neurodegeneration. Brain Res. 2007;1139:15–28. doi: 10.1016/j.brainres.2006.12.092. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Tseng HC, Kulkarni AB, Tsai LH, Gleeson JG. Cdk5 phosphorylation of oublecortin ser297 regulates its effect on neuronal migration. Neuron. 2004;41:215–227. doi: 10.1016/s0896-6273(03)00852-3. [DOI] [PubMed] [Google Scholar]
- Tint I, Jean D, Baas PW, Black MM. Doublecortin associates with microtubules preferentially in regions of the axon displaying actin-rich protrusive structures. J Neurosci. 2009;29:10995–11010. doi: 10.1523/JNEUROSCI.3399-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Prokscha A, Oldekamp J, Eichele G. Identification of neurabin II as a novel doublecortin interacting protein. Mech Dev. 2003;120:1033–1043. doi: 10.1016/s0925-4773(03)00177-1. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Prokscha A, Ungewickell E, Eichele G. Doublecortin association with actin filaments is regulated by neurabin II. J Biol Chem. 2005;280:11361–11368. doi: 10.1074/jbc.M405525200. [DOI] [PubMed] [Google Scholar]
- Vreugdenhil E, Kolk SM, Boekhoorn K, Fitzsimons CP, Schaaf M, Schouten T, Sarabdjitsingh A, Sibug R, Lucassen PJ. Doublecortin-like, a microtubule-associated protein expressed in radial glia, is crucial for neuronal precursor division and radial process stability. Eur J Neurosci. 2007;25:635–648. doi: 10.1111/j.1460-9568.2007.05318.x. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Lee YS, Tokumitsu H, Silva A, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Kaech S, Grant WF, Davare M, Impey S, Tokumitsu H, Nozaki N, Banker G, Soderling TR. Regulation of axonal extension and growth cone motility by calmodulin-dependent protein kinase I. J Neurosci. 2004;24:3786–3794. doi: 10.1523/JNEUROSCI.3294-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Uchimura E, Ueda T, Nomura T, Fujita S, Matsumoto K, Funeriu DP, Miyake M, Miyake J. Identification of twinfilin-2 as a factor involved in neurite outgrowth by RNAi-based screen. Biochem Biophys Res Commun. 2007;363:926–930. doi: 10.1016/j.bbrc.2007.09.069. [DOI] [PubMed] [Google Scholar]
- Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7:617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FQ, Snider WD. Intracellular control of developmental and regenerative axon growth. Philos Trans R Soc Lond B Biol Sci. 2006;361:1575–1592. doi: 10.1098/rstb.2006.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P1 cortical neurons were transfected with mCherry and cultured for 3 days. (A) Immunohistochemistry for βIII tubulin identifies neurons. (B) mCherry identifies transfected cells. (C–D) Immunohistochemistry for GAD67 identifies GABAergic interneurons (arrowhead) and non-GABAergic neurons (arrows). (E) Cellomics instrumentation and software quantified the percent of βIII-positive neurons that were also GAD67-positive and mCherry-positive. 80% of total neurons, and 78% of transfected neurons, were non-GABAergic. More than 1000 neurons were measured in a single experiment. Scale bar is 100µm.