Abstract
The orphan receptor LRH-1 and the oxysterol receptors LXRα and LXRβ are established transcriptional regulators of lipid metabolism that appear to control inflammatory processes. Here, we investigate the anti-inflammatory actions of these nuclear receptors in the hepatic acute phase response (APR). We report that selective synthetic agonists induce SUMOylation-dependent recruitment of either LRH-1 or LXR to hepatic APR promoters and prevent the clearance of the N-CoR corepressor complex upon cytokine stimulation. Investigations of the APR in vivo, using LXR knockout mice, indicate that the anti-inflammatory actions of LXR agonists are triggered selectively by the LXRβ subtype. We further find that hepatic APR responses in small ubiquitin-like modifier-1 (SUMO-1) knockout mice are increased, which is due in part to diminished LRH-1 action at APR promoters. Finally, we provide evidence that the metabolically important coregulator GPS2 functions as a hitherto unrecognized transrepression mediator of interactions between SUMOylated nuclear receptors and the N-CoR corepressor complex. Our study extends the knowledge of anti-inflammatory mechanisms and pathways directed by metabolic nuclear receptor–corepressor networks to the control of the hepatic APR, and implies alternative pharmacological strategies for the treatment of human metabolic diseases associated with inflammation.
Keywords: LRH-1, LXR, GPS2, acute phase response, liver inflammation
Members of the nuclear receptor (NR) family of transcription factors are regulators of metabolic gene expression that also appear to play important roles in the control of inflammatory responses. Various ligand-activated NRs exert potent anti-inflammatory activities in macrophages and other tissues by antagonizing cytokine-mediated gene expression via transcriptional cross-talk with proinflammatory transcription factors, a process referred to as transrepression (Glass and Ogawa 2006).
The nuclear oxysterol receptors LXRα and LXRβ (NR1H3 and NR1H2) are key regulators of lipid metabolism and transport, and have been demonstrated more recently to regulate inflammatory signaling in macrophages (Joseph et al. 2003; Ghisletti et al. 2007, 2009). Efforts to elucidate molecular mechanisms revealed that naturally occurring oxysterols and synthetic LXR agonists repress lipopolysaccharide (LPS)-induced activation of proinflammatory genes in a small ubiquitin-like modifier (SUMO)-dependent transrepression pathway. In this pathway, ligand activation of LXR promotes specific conjugation of SUMO-2/3 to the LXR ligand-binding domain (LBD). This modification is necessary to recruit LXRs to proinflammatory genes and to prevent the dissociation of the N-CoR/HDAC3 corepressor complex upon cytokine stimulation. These findings, together with related observations made with respect to mechanisms of PPARγ transrepression (Pascual et al. 2005; Ghisletti et al. 2007), raised a variety of intriguing issues that need to be addressed to better understand this pathway at the molecular level (Treuter and Gustafsson 2007).
The first issue deals with the enigmatic molecular details and consequences of NR SUMOylation. Notably, it has not been clarified which components specifically recognize the SUMO status of the respective modified NRs and thus are necessary to bridge these receptors to the N-CoR corepressor complex. Also, there might be molecular features that enable SUMOylated NRs to enter the transrepression pathway, but perhaps exclude them from conventional activation.
The second issue deals with the generality of the SUMO/corepressor-dependent mechanism in anti-inflammatory pathways governed by NRs other than LXRs and PPARγ. Various NRs can be modified by SUMO, which is thought to be linked to repression but has not yet been investigated with regard to its relevance for anti-inflammatory transrepression. However, SUMO modification is not a prerequisite for all NRs to inhibit inflammatory gene expression, as, for example, the glucocorticoid receptor can interfere directly with NF-κB activation in a SUMO/corerepressor-independent manner (Glass and Ogawa 2006).
The third issue deals with the generality of the SUMO-dependent mechanism in cell types other than macrophages. While the initial studies on PPARγ (Pascual et al. 2005) and LXRs (Ghisletti et al. 2007) have characterized the parallel SUMO pathways using macrophages, work by others provided some indications that related yet distinct mechanisms may occur in other cell types (Lee et al. 2009; Saijo et al. 2009). Intriguingly, activation of LXRs by synthetic agonists has been demonstrated to inhibit the expression of C-reactive protein (CRP) gene, a key inflammatory marker of human liver (Blaschke et al. 2006). The obvious link between LXR activation and corepressor complex stabilization at the CRP promoter possibly suggests that LXR SUMOylation plays conserved roles in the control of inflammatory pathways in liver.
A recently identified negative modulator of hepatic inflammatory processes is the orphan receptor LRH-1 (NR5A2), a key metabolic sensor that has overlapping physiological roles with LXRs by regulating the expression of genes involved in bile acid synthesis, cholesterol homeostasis, and triglyceride synthesis (Fayard et al. 2004). While initially thought to function ligand-independently, LRH-1 became adopted with the identification of phospholipid ligands (Krylova et al. 2005; Ortlund et al. 2005). The description of cis-bicyclo[3.3.0]-oct-2-ene (GR8470) as the first synthetic agonist of LRH-1 suggests that it is possible to pharmacologically modulate LRH-1 activity (Whitby et al. 2006). How such modulation occurs at the molecular level and in which biological LRH-1 pathways it is relevant remain to be elucidated. One possible pathway seems to be the negative regulation of the hepatic acute phase response (APR), in which LRH-1 antagonizes the expression of cytokine-induced gene expression (Venteclef et al. 2006; Venteclef and Delerive 2007).
In response to infection and inflammation, a wide range of metabolic alterations occur that commonly is referred to as APR (Gabay and Kushner 1999; Khovidhunkit et al. 2004; Chait et al. 2005). APR characteristically induces changes in the concentration of specific plasma proteins called acute phase proteins (APPs), protecting the host from further injury and facilitating tissue repair. APPs are produced exclusively in hepatocytes in response to inflammatory signals, a process that is directly regulated at the transcriptional level by proinflammatory transcription factors NF-κB, STAT3, and C/EBPs. The APR can perturb lipoprotein metabolism and produces a variety of changes, which can be proatherogenic if the cause of infection or inflammation is prolonged. Thus, anti-inflammatory activities of candidate receptors such as LRH-1 and LXRs, and their respective ligands, are likely to have a considerable impact on the APR-dependent progression of human diseases.
Here, we aim to address the above issues by investigating the molecular mechanisms by which LRH-1 and LXR inhibit the inflammatory APR in human hepatocyte cultures and in mice. Our key findings indicate that (1) selective synthetic agonists induce SUMOylation-dependent recruitment of either LRH-1 or LXR onto hepatic APP promoters, thereby preventing corepressor complex clearance; (2) in vivo anti-inflammatory actions of LXR agonists in liver are selectively mediated by the LXRβ subtype; and (3) the corepressor complex subunit GPS2 (Zhang et al. 2002; Sanyal et al. 2007; Jakobsson et al. 2009) acts as a hitherto unrecognized mediator of transrepression by SUMOylated NRs.
Results
Ligand-activated LRH-1 or LXR inhibit hepatic APR by preventing dissociation of the N-CoR corepressor complex
To investigate the consequences of LRH-1 ligand activation, as well as the involvement of LXRs in the control of the hepatic APR, human primary hepatocytes (Fig. 1A–C) or the hepatoma Huh7 cell line (Supplemental Fig. S1A–C) were treated with the respective synthetic agonists for each receptor and subsequently stimulated by the cytokines IL1β and IL6. As expected, cytokine treatment induced the expression of key proinflammatory APR genes such as haptoglobin and Serum Amyloid A (SAA) (Fig. 1A,B; Supplemental Fig. S1A,B), fibrinogen β, PLAT, and α-glycoprotein (data not shown). However, pretreatment with LRH-1 agonist (GR8470) or LXR agonist (GW3965) significantly inhibited inflammatory gene expression. Interestingly, not all proinflammatory APR genes appear to be inhibited by LRH-1/LXR, as Plasminogen Activator Inhibitor 1 (PAI-1) (Venteclef et al. 2006) was “resistant” to ligand-induced inhibition (Fig. 1C; Supplemental Fig. S1C).
Figure 1.
LRH-1 and LXRs inhibit hepatic APR gene expression. (A–C) The potential of LRH-1 (GR8470) and LXR (GW3965) agonists to repress cytokine-induced APR gene expression was analyzed in human primary hepatocytes. Hepatocytes were pretreated with vehicle (DMSO), GR8470 (10 μM), or GW3965 (2 μM) for 24 h and stimulated with 10 nM IL1β + IL6 for 16 h. Haptoglobin, SAA, and PAI-1 mRNA levels were quantified by qPCR. Data are presented as mean ± SD of three independent experiments. (D) IL1β + IL6 induce the dissociation of N-CoR or SMRT corepressor complexes from APR promoters. Huh7 cells were stimulated with 10 nM IL1β + IL6 for 1 h. Protein recruitment to APR promoters was analyzed by ChIP. (E,F) Activation of LRH-1 (E) or LXR (F) prevents the dissociation of N-CoR complexes from the haptoglobin promoter. Huh7 cells were pretreated by GR8470 (10 μM) or GW3965 (2 μM) and treated with 10 nM IL1β + IL6 for 1 h. Protein recruitment was analyzed by ChIP and re-ChIP.
Recent studies have suggested that LXRs inhibit inflammatory responses in macrophages by preventing the dissociation of N-CoR corepressor complexes from proinflammatory gene promoters (Ghisletti et al. 2007, 2009). To investigate whether LRH-1 and LXRs inhibit the APR in hepatocytes via related mechanisms, we analyzed corepressor complex recruitment by chromatin immunoprecipitation (ChIP) assays under the above experimental conditions (Fig. 1D–F). The experiments revealed first that the promoters of SAA and haptoglobin are under the control of the conventional NR corepressor complex containing N-CoR and HDAC3, which rapidly dissociates upon cytokine stimulation in order to induce gene transcription (Fig. 1D). An interesting observation was that the N-CoR-related corepressor SMRT, implicated in repressing a subset of inflammatory genes in macrophages (Ghisletti et al. 2009), was not recruited onto the hepatic SAA or haptoglobin promoters. However, SMRT was recruited together with HDAC3 on the PAI-1 promoter and released upon cytokine treatment (Fig. 1D). N-CoR and SMRT were not the only corepressor complex subunits that displayed selective recruitment profiles, since we found that GPS2, a stoichiometric corepressor complex subunit (Zhang et al. 2002), co-occupied the SAA and haptoglobin promoters but not the PAI-1 promoter, and was released upon cytokine stimulation (Fig. 1D).
Second, these experiments revealed that pretreatment with GR8470 or GW3965 prevented dissociation of the N-CoR complex core subunits (i.e., N-CoR, GPS2, TBLR1, and HDAC3) and triggered recruitment of LRH-1 or LXRs to the complex, as demonstrated by sequential (re-)ChIP assays on the haptoglobin promoter (Fig. 1E,F). Similar results were observed at the SAA promoter and verified by quantitative PCR (qPCR)-ChIP analysis (data not shown). Additionally, treatment kinetics in the absence of agonists indicated that cytokines do not induce recruitment of LRH-1 or LXRs to APR promoters (Supplemental Fig. S1D).
Third, ChIP profiling revealed that RXR, the obligatory heterodimer partner of LXRs in classical activation pathways (for example, see Jakobsson et al. 2009), was not present on APR promoters (Fig. 1F). This suggests that RXRs do not participate in transrepression by LXRs, and that both LXRs and LRH-1 act as monomeric receptors in this pathway.
To provide further evidence for the specificity of the synthetic LRH-1 agonist, we depleted endogenous LRH-1 using RNAi (Supplemental Fig. S2A). This resulted in a lack of inhibition, proving that LRH-1 was required to specially mediate the anti-inflammatory effect of GR8470. As another control for ligand specificity, GR8470 treatment up-regulated LRH-1 target genes such as APOA1 and SHP (Supplemental Fig. S2B). Additionally, transrepression of haptoglobin expression by GR8470 was dose-dependent in the range of 1–10 μM (Supplemental Fig. S2C,D). Consistent with observations made during the initial characterization of the LRH-1 (Whitby et al. 2006), GR8470 did not enhance the expression of endogenous LRH-1 target genes in mouse hepatoma cells (Supplemental Fig. S2E), and only poorly activates overexpressed mouse LRH-1 (Supplemental Fig. S2F).
Taken together, these data demonstrate that selective ligand activation of LRH-1 or LXR attenuates the inflammatory response in human hepatocytes by antagonizing dissociation of the N-CoR corepressor complex from the promoters of proinflammatory APR genes.
APR transrepression by LRH-1 and LXR is linked to distinct SUMOylation pathways
Recent work has revealed that ligand-dependent modification by the small ubiquitin-like modifiers SUMO-1 or SUMO-2/3 is a prerequisite allowing activated PPARγ and LXRs, respectively, to enter transrepression pathways in macrophages (Pascual et al. 2005; Ghisletti et al. 2007). To assess whether the SUMOylation pathway is also required for hepatic APR transrepression, hepatocytes were transfected with specific siRNAs targeting SUMO-1 or SUMO-2/3 and treated with receptor agonists under inflammatory conditions (Fig. 2A,B). The data indicate that knockdown of SUMO-1 specifically affected the inhibition by LRH-1 of cytokine-induced haptoglobin expression, and that knockdown of SUMO-2/3 specifically affected the anti-inflammatory activity of LXRs (Fig. 2A). These results were confirmed in transrepression assays using a haptoglobin promoter–luciferase reporter, demonstrating that conjugation of LRH-1 by SUMO-1 and LXR by SUMO-2/3 is required for transcriptional repression (Fig. 2B).
Figure 2.
APR transrepression by LRH-1 and LXR is dependent on SUMOylation. (A,B) siRNA depletion of SUMO-1 or SUMO-2/3 reverse transrepression of haptoglobin expression (A) and haptoglobin reporter activity (B) in Huh7 cells. Huh7 cells were pretreated with DMSO, GR8470 (10 μM), or GW3965 (2 μM) for 24 h and 10 nM IL1β + IL6 for 3 h. (C,D) siRNA depletion of SUMO-1 or SUMO-2/3 prevents recruitment of LRH-1 or LXR to the haptoglobin promoter. Huh7 cells were transfected with siRNA according to the figure, and were treated with LRH-1 or LXR agonists for 24 h and 10 nM IL1β + IL6 for 1 h. Protein recruitment was analyzed by ChIP. (E,F) SUMOylation of LRH-1 and LXRβ is required for transrepression. Huh7 cells were transfected by Myc-LRH-1 wild type or K224R mutant (E) or Flag-LXRβ wild type or K410R/K448R double mutant (F). After transfection, cells were treated with DMSO or agonists for 24 h and 10 nM IL1β + IL6 for 1 h. Receptor recruitment at the haptoglobin promoter was measured by ChIP using Myc or Flag antibodies.
ChIP assays revealed that ligand activation of LRH-1 or LXRs was correlated with the recruitment of known components of the SUMOylation pathways (Fig. 2C,D). Of interest was the mutually exclusive association of LRH-1 with SUMO-1, and of LXRs with HDAC4, the putative E3 ligase for SUMO-2/3 conjugation (Ghisletti et al. 2007). As expected, Ubc9, the SUMO-conjugating E2 enzyme, was detected under both conditions (data not shown). Importantly, the ChIP assays indicated that recruitment of LRH-1 or LXR onto the haptoglobin promoter was SUMOylation-dependent (Fig. 2C,D). Specifically, knockdown of SUMO-1 but not SUMO-2/3 (Fig. 2C for LRH-1), or of SUMO-2/3 but not SUMO-1 (Fig. 2D for LXRs), or of Ubc9 (data not shown for both LRH-1 and LXRs) abolished recruitment of the ligand-activated receptors, and thereby their capacity to prevent the dissociation of the corepressor complex upon agonist treatment.
SUMOylation of LRH-1 and LXR is required for APR inhibition
We next wanted to see that the above transcriptional effects were directly dependent on site-specific SUMO modification of LRH-1 or LXRs, respectively. This was particularly crucial in the case of LRH-1, which has not yet been implicated in SUMO-dependent transrepression pathways. A previous study has suggested that SUMOylation of LRH-1 at lysine residue 224 translocated LRH-1 to transcriptionally inactive subnuclear domains (Chalkiadaki and Talianidis 2005). Whereas such a mechanism is unlikely to explain transrepression, which should involve recruitment of SUMOylated LRH-1 to chromatin sites prone to rapid activation, we considered LRH-1 SUMOylation to be a conserved feature and investigated its involvement in APR transrepression.
First, LRH-1 wild type and SUMO site mutants were cotransfected with the haptoglobin reporter under inflammatory conditions (Supplemental Fig. S3A). As expected from previous work (Venteclef et al. 2006), overexpression of LRH-1 wild type inhibited cytokine-mediated haptoglobin reporter activity; a similar finding was observed with the K146R and K264R mutants. In contrast, LRH-1 K224R was not capable of inhibiting cytokine-induced activity, suggesting that SUMOylation of Lys 224 is required for transrepression. Control experiments revealed that the transactivation ability of LRH-1 K224R is not different from that of LRH-1 wild type (data not shown). In addition, we found that overexpression of a SUMO protease 1 (SuPr-1) significantly inhibits the anti-inflammatory activity of LRH-1.
To investigate the molecular reasons for the loss of transrepression capacity of the LRH-1 K224R mutant, we analyzed LRH-1 recruitment onto the haptoglobin promoter by ChIP assays (Fig. 2E). We found that, under inflammatory conditions, LRH-1 wild type, but not the SUMOylation-deficient mutant, was recruited onto the haptoglobin promoter. Control ChIPs revealed that the recruitment of both variants onto the promoters of target genes up-regulated by LRH-1 was similar (Supplemental Fig. S4A). Re-ChIP assays with antibodies against suspected core components of the hepatic “LRH-1 transrepression complex” revealed co-occupancy of LRH-1 with GPS2, N-CoR (but not SMRT), TBLR1, HDAC3 (but not HDAC1 or HDAC4), and TAB2 (Supplemental Fig. S4B). Moreover, the specific association of LRH-1 with SUMO-1 was verified by ChIP assays from hepatocytes treated with control versus LRH-1 siRNAs (Supplemental Fig. S3B).
An interesting observation was that LRH-1 recruitment was enhanced by the synthetic agonist GR8470 (Fig. 2E). In contrast to LXRs, agonist binding to LRH-1 is not known to induce SUMOylation, consistent with the apparent constitutive LRH-1 modification by SUMO-1 (Chalkiadaki and Talianidis 2005), or to change the receptor conformation (Whitby et al. 2006). To shed light into the mechanisms of LRH-1 activation by GR8470, we investigated the fate of endogenous LRH-1 upon agonist treatment in hepatocytes. We found by immunological analysis that GR8470 treatment increases the levels of LRH-1 species corresponding to unmodified (wild-type) and SUMO-modified receptors (Supplemental Fig. S5A). Immunoprecipitation of endogenous LRH-1 followed by immunoblotting to detect SUMO-1 confirmed the time-dependent increase of SUMOylated LRH-1 by GR8470 exposure (Supplemental Fig. S5B). These results are consistent with the documented compound effects on LRH-1 recruitment profiles and APR transcription. They suggest that LRH-1 agonists may act anti-inflammatorily, at least in part, by stabilizing both free and SUMOylated receptor levels, thereby increasing the cellular pools of transrepression-competent LRH-1.
In the case of LXRs, previous work has identified two putative lysine residues within the LBD that become modified by SUMO-2 upon treatment with agonists, and that are necessary to inhibit LPS-induced iNos gene expression in macrophages (Ghisletti et al. 2007). In confirmation of the general requirement of these sites for the hepatic APR, we demonstrate that mutation of lysine residues 410 and 448 in LXRβ was sufficient to generate a mutant receptor that could no longer be recruited to the haptoglobin promoter in vivo (Fig. 2F). Expression levels and activation capacity of wild-type and mutated LXRβ were similar (data not shown).
Taken together, these data implicate that direct receptor SUMOylation, with distinct conjugation sites and mechanisms of ligand action, controls related transrepression pathways of LRH-1 and LXR during the hepatic APR.
Alterations of the hepatic APR and of LRH-1 transrepression in SUMO-1 knockout (KO) mice
Our present data and a previous study (Venteclef et al. 2006) demonstrate that LRH-1 attenuates the hepatic APR, and that action of LRH-1 is linked to its SUMO-1 conjugation. To investigate the role of the SUMO-1 pathway in the control of hepatic inflammatory response in vivo, we treated SUMO-1 KO mice (Zhang et al. 2008) with LPS and analyzed the expression levels of key acute phase genes by qPCR (Fig. 3A–C). The inflammatory response was significantly more pronounced in SUMO-1 KO mice than in wild-type mice, as documented by increased levels of haptoglobin, SAA (Fig. 3A,B), and CRP (Supplemental Fig. S6) transcripts. However, the LPS-mediated expression of PAI-1, which is not repressed by LRH-1 or LXR, was identical in SUMO-1 KO and wild-type mice (Fig. 3C).
Figure 3.
SUMO-1 KO mice have increased APR. (A–C) Wild-type and SUMO-1 KO mice were treated with 10 mg/kg LPS for 6 h. APR gene expression was analyzed by qPCR. (D,E) Protein recruitment onto the mouse haptoglobin promoter was analyzed by ChIP from liver extracts. (F,G) Gene expression of LRH-1 target genes SHP and CYP7A1 (left panel) and LRH-1 recruitment to the promoters (right panel) were analyzed in wild-type and SUMO-1 KO mice. Data are presented as mean ± SD from five individual mice per group.
Because the absence of SUMOylated LRH-1 in SUMO-1 KO mice could explain the exacerbated inflammatory response, we assessed the recruitment of N-CoR, GPS2, SUMO-1, SUMO-2/3, LRH-1, LXRs, and PPARα onto the haptoglobin promoter by ChIP assays and quantified the results by using qPCR (Fig. 3D,E). First, in wild-type mice, LRH-1 (fivefold) and SUMO-1 (fourfold) were recruited onto the haptoglobin promoter, suggesting that SUMO-1-conjugated LRH-1 interacts with the N-CoR complex already in absence of inflammatory stimuli. Second, LPS treatment induced partial dissociation of N-CoR and GPS2, but not of SUMO-1 and LRH-1. In contrast, LRH-1 was not recruited onto the haptoglobin promoter in SUMO-1 KO mice, demonstrating the importance of SUMO-1 conjugation for the recruitment of LRH-1 in vivo. LPS challenge induced a more complete dissociation of the N-CoR complex from the promoter in SUMO-1 KO mice in comparison with wild-type mice. Notably, no significant differences were detected regarding the recruitment of LXRs, PPARα, and SUMO-2/3, providing support for the specific link of LRH-1 to SUMO-1. A final interesting observation was that the expression levels of endogenous LRH-1 target genes (e.g., SHP and CYP7A), as well as LRH-1 recruitment to their promoters, were increased in SUMO-1 KO mice, indicating SUMO-1 modification also influence LRH-1 repression of direct target genes (Fig. 3 F,G).
Collectively, these data indicate that SUMOylated LRH-1 interacts with the N-CoR/GPS2 complex, and blocks its dismissal from the haptoglobin promoter also under in vivo conditions.
The LXR–SUMO–corepressor pathway negatively regulates the hepatic APR in vivo
While the study of LRH-1 KO mice has supported the specific involvement of LRH-1 for the hepatic APR in vivo (Venteclef et al. 2006; Venteclef and Delerive 2007), the biological relevance of the anti-inflammatory properties of LXRs, and of synthetic LXR agonists, in liver remained to be established.
To address these issues, we investigated the APR in wild-type and LXRα/β double-KO mice and upon ligand activation of LXRs. Whereas LPS treatment in wild-type mice resulted in an induction of APR genes such as haptoglobin, SAA, and CRP (Fig. 4A; Supplemental Fig. S7A,D), this inflammatory response was strongly reduced in wild-type mice treated with GW3965. Quantification of plasma SAA protein levels agreed with the gene expression data; i.e., a significant reduction of inflammatory response in mice treated with the LXR agonist (Supplemental Fig. S7C). Hepatic LXR expression was monitored to demonstrate that LXRα and LXRβ expression were not modified by the LPS challenge (Supplemental Fig. S9D). Moreover, LXR was required, as GW3965 treatment did not inhibit the LPS-mediated haptoglobin, SAA, and CRP gene expression, and protein levels in LXRα/β KO mice. Notably, the inflammatory response was more pronounced in these mice, demonstrating that LXRs play a crucial role in the control of the hepatic APR in vivo even in the absence of exogenously added synthetic agonists (Fig. 4A; Supplemental Fig. S7A,D).
Figure 4.
LXRβ mediates APR transrepression upon activation of the LXRs. (A,B) Wild-type, LXRαβ double-KO, or LXRα and LXRβ single-KO mice were pretreated with 30 mg/kg per day GW3965 for 4 d and then treated with 2 mg/kg LPS for 2.5 h. Gene expression was analyzed by qPCR. (C–F) Protein recruitment to the mouse haptoglobin promoter was analyzed by ChIP from liver extracts. Data are presented as mean ± SD from five individual mice per group. (G) LXRβ but not LXRα is recruited to the haptoglobin promoter. Huh7 cells were transfected with Flag-LXRα or Flag-LXRβ, and were treated with GW3965 (2 μM) for 24 h and 10 nM IL1β + IL6 for 1 h. Recruitment of LXRs to haptoglobin or ABCA1 promoters was analyzed by ChIP using Flag or LXR antibody. (H) LXRβ but not LXRα is modified by SUMO-2. Huh7 cells were cotransfected with Flag-LXRα or Flag-LXRβ and Myc-SUMO-2, and were treated with GW3965 (2 μM) for 6 h. Whole-cell lysates were immunoblotted using Flag antibody to detect modified LXR, or immunoprecipitated using Flag antibody followed by immunoblotting using Myc antibody to detect SUMO-2-conjugated LXRs.
To confirm that LXRs were required to prevent dissociation of the corepressor complex from acute phase gene promoters in vivo, we analyzed the recruitment of LXRs, N-CoR, GPS2, HDAC3, and HDAC4 onto the haptoglobin, SAA, and CRP promoters in wild-type and LXRα/β KO mice by ChIP assays from liver tissue extracts (Fig. 4C,D; Supplemental Fig. S8). In agreement with the results derived from human hepatocytes (Fig. 1), GW3965 treatment prevented the dissociation of N-CoR and GPS2 from the haptoglobin promoter in LXR wild-type but not LXRα/β KO mice upon LPS challenge. Importantly, GW3965 treatment induced the recruitment of LXRs onto the promoter, again only in LXR wild-type but not LXRα/β KO mice. This was associated with corecruitment of HDAC4 and SUMO-2/3 (Fig. 4C), suggesting that SUMOylation-dependent LXR transrepression occurs in both mouse and human hepatocytes. Finally, recruitment profiles in the LXRα/β KO mice treated with LXR agonist GW3965 confirm the strict LXR dependence of transrepression complex formation on APR promoters (Fig. 4D).
The LXRβ subtype selectively inhibits hepatic APR in a SUMOylation-dependent manner
Although initial in vitro studies in macrophages have suggested that both LXRs are SUMOylated and thereby are capable of transrepression (Ghisletti et al. 2007), the important question of LXR subtype selectivity needed to be reinvestigated for in vivo relevance in light of hepatic LXR pathways presented here.
We addressed this issue by treating groups of LXRα or LXRβ single-KO mice as described above (Fig. 4B,E,F; Supplemental Fig. S9). We found that GW3965 inhibited LPS-mediated haptoglobin gene expression in LXRα KO mice, whereas no effect was seen in LXRβ KO mice (Fig. 4B). Analysis of additional APR genes confirmed their general LXR requirement, as well as the lack of PAI-1 inhibition in both human and mouse hepatocytes (Supplemental Fig. S9C). Also, LXR levels were unchanged in wild-type mice under different treatments (Supplemental Fig. S9D).
ChIP analysis at the mouse haptoglobin promoter (Fig. 4E,F) and additional APR promoters (data not shown) essentially confirmed the expression data by demonstrating that LXR recruitment was observed in LXRα KO mice, but not in LXRβ KO mice. The notion that LXRβ, HDAC4, and SUMO-2/3 co-occupied the haptoglobin promoter in mice lacking LXRα suggests that SUMOylated LXRβ was responsible for APR inhibition in vivo.
siRNA-mediated depletion of each LXR in human hepatocytes (Supplemental Fig. S10A–D) revealed that LXRβ subtype selectivity is a conserved feature of human and mouse LXR transrepression but not of activation, as demonstrated for ABCA1. The individual recruitment of LXRα or LXRβ onto haptoglobin or ABCA1 promoters (Jakobsson et al. 2009) was analyzed in hepatocytes transfected with either Flag-LXRα or Flag-LXRβ and then treated with GW3965 under inflammatory conditions (Fig. 4G). We found that agonist treatment induced the specific recruitment of Flag-LXRβ, but not that of Flag-LXRα, to the haptoglobin promoter, as demonstrated by ChIP assay. Overexpressed Flag-LXRs were both functional, as they were recruited onto the ABCA1 promoter upon ligand treatment in the same experiments (Fig. 4G). These results suggested that LXR-dependent transrepression is LXRβ-specific, while activation is not. To understand these differences at the molecular level, we analyzed the SUMOylation status of both receptors. Huh7 cells were cotransfected with Flag-LXRα or Flag-LXRβ and Myc-SUMO-2, and then treated with GW3965. Two approaches—direct immunoblotting of lysates using Flag (LXR) antibody or Flag (LXR) immunoprecipitation followed by detection of SUMO-2 using Myc antibody (Fig. 4H)—suggest that SUMO-2 modification occurs only on LXRβ upon ligand activation. As similar results were observed in HeLa cells (Supplemental Fig. S10E), it can be assumed that subtype selectivity is not limited to the hepatocyte cellular environment. Collectively, these results link LXRβ selectivity in the hepatic APR to the preferential SUMO-2 conjugation of this particular subtype.
GPS2 is required to prevent agonist-dependent corepressor complex dissociation
Recent studies have highlighted the importance of the N-CoR complex subunit GPS2 in regulating transcriptional activation of LRH-1 and LXR in pathways linked to cholesterol metabolism and transport (Sanyal et al. 2007; Jakobsson et al. 2009). Since our initial results (Fig. 1D–F) suggested an additional involvement for GPS2 in the hepatic APR, we investigated the importance of GPS2 in relation to N-CoR in the APR pathway.
In the initial set of experiments, human hepatoma cells were transfected with siRNAs targeting N-CoR or GPS2 and then treated with agonists under inflammatory conditions (Fig. 5). We found that, in N-CoR-deficient cells, neither LRH-1 nor LXR activation could inhibit cytokine-mediated expression of endogenous haptoglobin (Fig. 5A) or exogenous haptoglobin promoter–luciferase reporter activity (Fig. 5B). These results were consistent, in the case of LXRs, with N-CoR depletion studies in macrophages (Ghisletti et al. 2007). Surprisingly, similar results were observed in GPS2-depleted cells (Fig. 5A,B), suggesting an additional requirement of GPS2 in either recruiting the corepressor complex to inflammatory promoters (repression), or preventing corepressor complex dissociation by recruiting LRH-1 or LXRs (transrepression).
Figure 5.
APR transrepression by LRH-1 and LXR requires GPS2. (A,B) siRNA depletion of N-CoR or GPS2 abolishes transrepression of haptoglobin expression (A) and haptoglobin reporter gene activity (B) in Huh7 cells. Cells were transfected with siRNA according to figure, and were treated with DMSO or agonists for 24 h and 10 nM IL1β + IL6 for 3 h. Data are represented as mean ± SD of triplicate experiments. (C) GPS2 is required for transrepression of the haptoglobin promoter by LRH-1 (top panel) and LXR (bottom panel). (D) Mapping of the GPS2 “transrepression” domain. Huh7 cells expressing siRNA targeting GPS2 were transfected with different HA-GPS2 derivatives (wild type, 1–105, and 100–327). Recruitment of LRH-1 and LXR to the haptoglobin promoter was analyzed by ChIP.
These possibilities were investigated further by ChIP assays from cells treated under the same experimental conditions (Fig. 5C). While knockdown of GPS2 did not affect the recruitment of N-CoR and HDAC3 onto the haptoglobin promoter in the absence of cytokine stimulation, interleukin treatment induced clearance of N-CoR and HDAC3 in GPS2-deficient cells. In contrast, N-CoR depletion resulted in removal of the corepressor complex already in the absence of cytokine stimulation. These combined results suggest that repression of proinflammatory gene expression and the mechanisms that trigger proteasomal corepressor complex removal upon stimulation do not depend on GPS2, but require N-CoR. It was, however, intriguing to observe that, in GPS2-deficient hepatocytes, LRH-1 (Fig. 5C, top panel) or LXRs (Fig. 5C, bottom panel) were no longer recruited onto the haptoglobin promoter, and thus are not capable of inhibiting corepressor complex dissociation. Lack of receptor recruitment was also demonstrated in the N-CoR-deficient cells, consistent with the absence of the corepressor complex under all conditions.
To further investigate the potential role of GPS2 in recruiting SUMO-NRs to the promoter-bound N-CoR complex, we performed rescue experiments by expressing HA-tagged GPS2 derivatives in hepatocytes upon siRNA-mediated depletion of endogenous GPS2 (Fig. 5D). The assays revealed that HA-GPS2 wild type (amino acids 1–327) and, surprisingly, HA-GPS2 N (amino acids 1–105) could restore the recruitment of LRH-1 or LXR onto the haptoglobin promoter under inflammatory conditions. In contrast, expression of a HA-GPS2 derivative that lacks the N-terminal repressor/N-CoR-binding domain (Zhang et al. 2002), but contains the C-terminal interaction domain (amino acids 100–327) with LRH-1 (Sanyal et al. 2007) and LXRs (Jakobsson et al. 2009), failed to restore recruitment of these receptors. This indicates that GPS2 interactions with N-CoR are necessary to recruit NRs to the complex, and that the previously identified NR interaction domain is insufficient for targeting these NRs under inflammatory conditions.
In conclusion, these results suggest that GPS2 and N-CoR are required for distinct steps during derepression of inflammatory genes, and that GPS2 could be the link between ligand-activated NRs and the N-CoR complex to mediate transrepression.
GPS2 functions as the SUMO-NR sensor of the transrepression complex
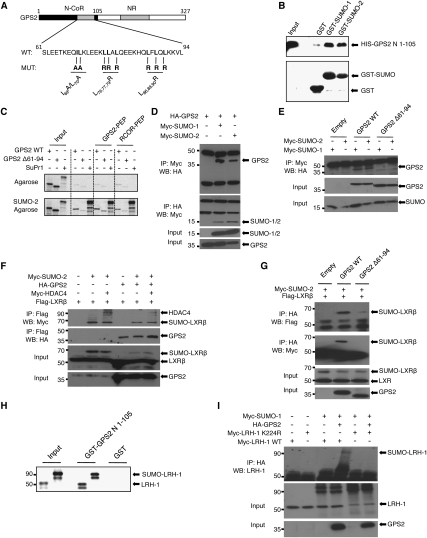
Our demonstration that GPS2, in addition to N-CoR, was required for both LRH-1 and LXRβ to inhibit hepatic APR gene expression, together with the obvious requirement of receptor SUMOylation, raised the question of whether these two observations are mechanistically linked to each other. Considering that transrepression targeting might involve as yet unrecognized interaction features of GPS2 with SUMOylated NRs, we noticed that the predicted N-terminal coiled-coil domain (Zhang et al. 2002) revealed some similarities; i.e., patterns of hydrophobic residues with acidic regions adjacent (Fig. 6A) to recently identified SUMO-interacting motifs (SIMs) (Kerscher 2007; Ouyang et al. 2009).
Figure 6.
GPS2 interacts with SUMO and SUMOylated LRH-1 or LXRβ. (A) Schematic structure of human GPS2 and mutations within the N-terminal transrepression domain. (B) Purified recombinant HIS-GPS2 (amino acids 1–105) was incubated with purified GST, GST-SUMO-1, or GST-SUMO-2 protein. Precipitated GPS2 was visualized using HIS antibody. (C) Agarose-SUMO-2 was incubated with 35S-labeled GPS2 or SUMO protease 1 (SuPr1) and GPS2 or RCOR peptides as indicated. Proteins were separated by SDS-PAGE and detected using autoradiography. (D) Lysates from HeLa cells transfected with Myc-SUMO-1 or Myc-SUMO-2 and HA-GPS2 were immunoprecipitated using HA or Myc antibodies. Precipitates were subjected to Western blot as indicated. (E) Lysates from Cos-7 cells transfected with Myc-SUMO-1 or Myc-SUMO-2 and HA-GPS2 derivatives (wild type and Δ61–94) were immunoprecipitated using Myc antibody. Precipitates were subjected to Western blot using HA antibody. (F) Lysates from Cos-7 cells transfected with Myc-SUMO-2, Myc-HDAC4, Flag-LXRβ, and HA-GPS2 were immunoprecipitated using Flag antibody. Precipitates were subjected to Western blot using HA and Myc antibodies. (G) Lysates from Cos-7 cells transfected with Myc-SUMO-2, Flag-LXRβ, and HA-GPS2 derivatives (wild type and Δ61–94) were immunoprecipitated using HA antibody. Precipitates were subjected to Western blot using Flag and Myc antibodies. (H) GST or GST-GPS2-N (amino acids 1–105) were bound to gluthathione sepharose and incubated with 35S-labeled LRH-1 and SUMO-1–LRH-1 for 2 h. Proteins were separated by SDS-PAGE and detected using autoradiography. (I) Lysates from Cos-7 cells transfected with Myc-SUMO-1, HA-GPS2, and Myc-LRH-1 (wild type or K224R) were immunoprecipitated using HA antibody. Precipitates were subjected to Western blot using LRH-1 antibody.
Various in vitro assays were carried out to confirm the functional relevance of these implications. First, we demonstrate using purified proteins that the GPS2 N terminus (amino acids 1–105) could indeed bind directly to GST-SUMO-1 or GST-SUMO-2 fusion proteins (Fig. 6B). Second, specific binding was verified using SUMO-2 agarose, with in vitro translated 35S-labeled GPS2 and SUMO protease 1 (SuPr1) for comparison (Fig. 6C; Supplemental Fig. S11A). In these experiments, peptides corresponding to the characterized SIM of the corepressor CoREST1 (Ouyang et al. 2009) or to shorter parts of the GPS2 SUMO-binding domain (amino acids 61–94) were inefficient in competing for binding, suggesting the SUMO-binding surface of GPS2 to be distinct from and more complex than those of classic short SIM peptide motifs. Third, specific interactions could also be detected in mammalian cells by coimmunoprecipitation of HA-tagged GPS2 and Myc-tagged SUMO-1 or SUMO-2, respectively (Fig. 6D). Fourth, while deletion of the GPS2 SUMO-binding domain (amino acids 61–94) reduced or abolished interactions in coimmunoprecipitation or pull-down assays (Fig. 6E; Supplemental Fig. S11A,C,D), mutations of key hydrophobic residues (Fig. 6A) that distantly conform to the SIM consensus had little effect on the in vitro interactions (Supplemental Fig. S11B,C,E). This suggests that the entire domain, rather than individual SIMs, determines SUMO-dependent interactions of GPS2. Fifth, coimmunoprecipitations from cell extracts expressing LXRβ or LRH-1 together with Myc-tagged SUMO revealed specific precipitation of LXRβ–SUMO-2 or LRH-1–SUMO-1 by GPS2 (Fig. 6G,I). Deletion of GPS2 SUMO binding (amino acids 61–94) reduced but did not abolish NR interactions of GPS2, consistent with the contribution of a separate C-terminal NR-binding domain to these in vitro interactions (Fig. 6G). Inclusion of HDAC4 in the interaction assays revealed the existence of complexes consisting of LXRβ–SUMO-2, HDAC4, and GPS2 in solution (Fig. 6F), consistent with our above-described ChIP data and with the suspected role of HDAC4 in triggering SUMO-2 modification of LXRs (Ghisletti et al. 2007). Sixth, purified GST-GPS2-N (amino acids 1–105) interacted with an in vitro translated SUMO-1–LRH-1 fusion protein as well as with LRH-1 wild type, suggesting an additional binding site for both modified and free LRH-1 within the GPS2 N terminus (Fig. 6H).
Complications for the analysis and interpretation of GPS2-dependent SUMO (and SUMO-NR) targeting arise from the fact that the same GPS2 N-terminal region mediates interactions with N-CoR (Zhang et al. 2002). Thus, we asked whether it is possible to separate N-CoR binding from NR-SUMO recruitment. We performed ChIP rescue experiments using GPS2 wild type and mutants, and this time analyzed GW3965-dependent recruitment of LXR, GPS2, and N-CoR in both the presence or absence of cytokine stimulation on the haptoglobin promoter. The key results for LXR (Fig. 7A) were as follows: (1) Co-occupancy of all three proteins was observed only in the presence of cytokines and GW3965 in cells expressing HA-GPS2 wild type, while none of the GPS2 mutants could rescue endogenous GPS2 depletion. This suggests that all mutants were deficient in either N-CoR binding and/or NR-SUMO targeting. (2) Analysis in the absence of cytokine stimulation demonstrated that one GPS2 mutant (L76,77,79R) could be recruited to the N-CoR complex. Thus, in this case, the lack of LXR recruitment upon cytokine stimulation was likely to be a direct consequence of abolished interactions with SUMOylated LXR, and not due to defective N-CoR binding. To further support this assumption, we analyzed the corecruitment of GPS2 wild type and derivatives with N-CoR at the LXR-regulated ABCA1 promoter (Jakobsson et al. 2009). Unlike at APR promoters, ABCA1 transcription is repressed by GPS2–N-CoR complexes in the absence of LXR ligand activation, likely to be independent of SUMOylation. Intriguingly, GPS2 (L76,77,79R) was capable of rescuing GPS2 deficiency, and was corecruited together with N-CoR to the LXRE of ABCA1 (Fig. 7B).
Figure 7.
In vivo characterization of transrepression domains in GPS2 and LRH-1. (A,B) Mapping of the GPS2 transrepression domain. Huh7 cells, depleted for GPS2 by siRNA, were transfected with different HA-GPS2 derivatives and treated with DMSO, GR8470 (10 μM), or GW3965 (2 μM) for 24 h and 10 nM IL1β + IL6 for 1 h. Recruitment of LXR, N-CoR, and GPS2 (HA) to the haptoglobin (A) and ABCA1 (B) promoters were analyzed by ChIP. (C) Mapping of the LRH-1 transrepression domain. The LRH-1 LBD is required to prevent the degradation of the N-CoR complex from the APR promoter. Huh7 cells were transfected with Myc-LRH-1 wild type, Myc-LRH-1 ΔLBD, or Myc-SUMO–1-LRH-1 and treated with DMSO or GR8470 (10 μM) (top panel), or cotransfected with GFP-SUMO-1 (bottom panel) for 24 h. Cells were treated with 10 nM IL1β + IL6 for 1 h. Recruitment of Myc-tagged LRH-1 derivatives, N-CoR, SUMO-1, and GPS2 to the haptoglobin promoter was analyzed by ChIP. (D) LRH-1 transrepression at APR promoters can be dissected into two separate steps. (Step 1) Ligand- and SUMO-dependent “docking” to the corepressor complex via GPS2. (Step 2) LRH-1-dependent inhibition of corepressor complex degradation.
Similar results were observed with LRH-1 on the haptoglobin promoter (Supplemental Fig. S12A,B), supporting the relevance of the above findings for the in vivo situation. However, attempts to reconstitute the differences between GPS2 wild type and (L76,77,79R) in vitro were not satisfactory. For example, mutation of (L76,77,79R) in the context of GST-GPS2 N (amino acids 1–105) did not abolish the interactions with in vitro translated LRH-1 wild type or SUMO-1–LRH-1 (Supplemental Fig. S11E; data not shown). This points at the existence of additional specificity determinants—such as the GPS2 C terminus, N-CoR, or TBLs—that may allosterically modulate interactions within the GPS2 N terminus. Collectively, these data suggest that the GPS2 coiled-coil domain encompassing amino acids 61–94 is responsible for mediating interactions between LRH-1/LXR and N-CoR, and that it is possible to separate N-CoR binding from NR-SUMO recruitment by mutation of distinct hydrophobic GPS2 surface residues.
Our data suggested that transrepression by LRH-1, similar to LXR (Ghisletti et al. 2007; this study), follows a two-step mechanism: (1) ligand- and SUMO-dependent NR docking to the corepressor complex via GPS2, and (2) inhibition of complex dismissal upon cytokine stimulation. We therefore finally asked whether these two steps can be separated, and wanted, in particular, to address the specificity of SUMO-dependent NR docking in step 1. ChIP analysis of LRH-1 and SUMO derivatives—including SUMO fusion proteins to LRH-1 and GFP, and LRH-1 (amino acids 1–267) lacking the LBD but retaining the SUMO acceptor Lys K224—revealed the following (Fig. 7C): (1) SUMO fusion could overcome the ligand requirement for LRH-1 SUMOylation and did mimic activated SUMO–LRH-1 in both steps; i.e., docking occurred without cytokine treatment and corepressor release was inhibited upon stimulation (Fig. 7C, top panel; Supplemental Fig. S12E,F). (2) Deletion of the LBD abolished the functionality of LRH-1 in both steps, but docking of this derivative could be rescued by coexpression of SUMO-1, presumably by overcoming the ligand dependence of LRH-1 SUMOylation and restoring corepressor docking via GPS2 (Fig. 7C, bottom panel). (3) SUMO fusions to GFP could not substitute for LRH-1 because they were not capable of docking (Supplemental Fig. S12C,D). These results indicate that efficient docking of SUMOylated LRH-1 to the GPS2 complex requires both SUMO and the receptor, thereby highlighting the NR selectivity of the SUMO-dependent transrepression mechanism.
Discussion
LRH-1 and LXRβ: key regulators of the hepatic APR, drug targets for anti-atherogenic strategies?
In the present study, we demonstrate that selective activation of LRH-1 and LXRβ by synthetic ligands represses cytokine-mediated APR gene expression in human hepatocytes and in mice. Our results imply that appropriate activation of LRH-1 or LXRβ might be pivotal to the body's systemic response by suppressing multiple inflammatory APR pathways. They suggest the APR as a hitherto unrecognized transrepression pathway of metabolic NR action in liver that should encourage receptor-selective pharmacological strategies.
In the case of LXRs, recent development has put forward drug strategies that selectively activate, or inhibit, one of the two subtypes. This was necessary since LXRα activation by synthetic ligands increases plasma trigylceride levels, an independent risk factor of cardiovascular disease, due to hepatic lipogenesis (Schultz et al. 2000). Consequently, LXRβ-selective agonists are believed to offer promising pharmacological intervention to improve reverse cholesterol transport without affecting hepatic lipogenesis. That such strategies are feasible is supported by findings in LXRα/apoE-deficient mice, where selective ligand activation of LXRβ reversed atherosclerosis development, probably due to the repression of proinflammatory gene expression in macrophages (Bradley et al. 2007). Furthermore, N-acylthiadiazolines have been described as a new class of LXRβ-selective agonists, which induce ABCA1-dependent cholesterol efflux in macrophages without activating lipogenesis in liver (Molteni et al. 2007). Notable is the recent identification of transrepression-selective LXRβ compounds that act anti-inflammatorily in macrophages (Chao et al. 2008). These examples, together with our demonstration of LXRβ selectivity in the hepatic APR, suggest that it might be possible to develop compounds with improved subtype and pathway selectivity.
In the case of LRH-1, it has been unclear whether or not this receptor can be used for pharmacological development. Recently, synthetic ligands have been identified that bind with high affinity to human (not mouse) LRH-1, and work agonistically by stimulating LRH-1-mediated expression of its target gene, SHP, in human hepatoma cells (Whitby et al. 2006). Our demonstration that activation of LRH-1 by this compound can potently inhibit inflammatory APR gene expression therefore substantially extends the repertoire of LRH-1 ligand action in liver.
APR modulation via selective activation of LRH-1 and/or LXRβ transrepression pathways, linked to the N-CoR/GPS2 corepressor complex, may be complementary or even add to previously identified areas of LXR action. It is important to consider consequences and benefits of pharmacological APR inhibition with regard to human diseases. During infection and inflammation, biosynthesis of APPs alters the composition of HDL particles (e.g., depletion of ApoA1 and enrichment of haptoglobin and SAA) to generate acute-phase HDL, which is believed to be proatherogenic (Navab et al. 2005). Acute-phase HDL may not protect against LDL oxidation, and thereby cannot reduce macrophage foam cell formation. In addition, epidemiologic studies have demonstrated a greater risk of coronary artery disease in subjects with high levels of APPs such as CRP and SAA (Chait et al. 2005). Thus, the selective activation of LRH-1/LXRβ transrepression pathways to reduce the production of hepatic APPs could lead to the discovery of new classes of synthetic ligands to treat inflammatory diseases.
SUMOylation appears to be a common feature of diverse anti-inflammatory NR cross-talk pathways
SUMOylation of NRs such as PPARγ, LXRs, and NURR1 has only recently been linked to unique mechanisms of anti-inflammatory transrepression in macrophages, microglia, and astrocytes (Pascual et al. 2005; Ghisletti et al. 2007, 2009; Saijo et al. 2009). Here, we show that the repressive action of LXRβ in liver, as in macrophages, is associated with the SUMO-2/3 pathway, and we demonstrate that the LRH-1 transrepression pathway in liver, similar to the PPARγ pathway in macrophages, is dependent on SUMO-1. We note that PPARγ and LRH-1 are modified by SUMO-1 at acceptor lysine(s) within the consensus sequence ψKxE, while LXRβ and Nurr1 are modified by SUMO-2/3 at nonconsensus sites. Thus, it is possible that different NRs use alternative strategies to ensure signal or ligand-dependent SUMO modification despite divergent primary sequences.
The availability of SUMO-1 KO mice (Zhang et al. 2008) enabled us to investigate the importance of SUMO-1 pathways in the regulation of the hepatic inflammatory APR response. We demonstrate here for the first time a crucial and nonredundant role of SUMO-1 in the control of inflammatory processes in vivo. The observed exacerbated inflammatory response in the SUMO-1-deficient mice is likely a result, at least in part, of reduced anti-inflammatory activity of LRH-1, similar to that seen in LRH-1 heterozygote mice (Venteclef et al. 2006). An obvious question for future research is to determine the extent to which loss of SUMO-1-dependent LRH-1 transrepression would modulate SUMO-2/3-dependent LXR transrepression in vivo. Interestingly, at least in interferon-stimulated brain astrocytes, both LXR subypes can also be differentially modified (Lee et al. 2009).
Another key finding of our study is that RXR appears to be excluded from the LXR transrepression complex. Interference with RXR heterodimerization by SUMOylation is conceivable, since putative conjugation sites, including K410, which is unique to LXRβ, are located near the dimerization surface. An intriguing consequence would be that SUMOylated LXRs cannot be recruited anymore to classic metabolic target genes. Interestingly, LRH-1 does not form RXR heterodimers, and thus presumably uses another molecular strategy to increase pools of transrepression-competent SUMOylated receptors. Perhaps, the exclusion of SUMOylated NRs from classic activation pathways could be necessary to ensure that limiting pools of modified NRs can be effective in triggering transrepression pathways.
GPS2 acts as an anchor between SUMOylated NRs and the N-CoR complex and is critical for transrepression
We demonstrate here that GPS2 acts as a necessary bridging and SUMO-sensing component of anti-inflammatory NR transrepression in the liver APR. GPS2 was identified originally as a stoichiometric subunit of the conserved N-CoR corepressor complex, along with additional exchangeable factors such as TBLR1/TBL1 (Zhang et al. 2002). Subsequent work has described GPS2 as a direct NR-binding protein capable of executing context-specific roles in regulating gene expression governing hepatic cholesterol metabolism (Sanyal et al. 2007) and ABCG1-dependent cholesterol efflux from macrophages (Jakobsson et al. 2009). Thus, our study extends the actions of GPS2 to hepatic transrepression and clarifies the molecular mechanism by which SUMOylated receptors interact with the N-CoR complex.
We propose the following molecular model (Fig. 8): Ligand activation increases cellular levels of SUMOylated NRs via either receptor stabilization (LRH-1) or conformational changes facilitating SUMOylation (LXRβ). Subsequently, SUMOylated NRs interact with GPS2, a process that may involve additional modifying components (e.g., protein modifiers or post-translational modifications). Intriguingly, so far, GPS2 is the only known subunit of the N-CoR complex that is capable of recognizing the ligand-activated NR conformation. Therefore, for the in vivo situation, it is reasonable to propose that it is this unique capability of GPS2 to recognize “free” (i.e., non-SUMOylated, liganded) NRs, that may predispose GPS2-NR complexes to rapidly enter the SUMO-dependent transrepression pathway in response to reversible SUMOylation cycles.
Figure 8.
Suggested roles of GPS2, LRH-1, and LXRβ in the transrepression of hepatic APR. Treatment with agonists either induces specific SUMOylation of LXRβ or increases the levels of SUMOylated LRH-1. SUMOylated LXRβ or LRH-1 interacts with GPS2, thereby associating with the N-CoR complex and preventing its dissociation in an inflammatory state. Considered is the involvement of additional modulators (MOD) of GPS2 interactions. Furthermore, as a result of the anti-inflammatory transrepression, LXR and LRH-1 inhibit the production of APPs during inflammation and infection, thereby potentially reducing the biosynthesis of proatherogenic acute-phase HDL. (APO) Apolipoproteins (e.g., APOAI).
We provide further evidence that GPS2-dependent NR-SUMO “docking” (the first cytokine-independent step in transrepression) is not sufficient to inhibit corepressor complex degradation (the second step in transrepression) in response to inflammatory stimuli. This second step appears to be a specific feature of NRs, but not of any SUMOylated protein, to interfere with the ubiquitin-mediated protesomal degradation. Since TBL factors, the F-box components of involved ubiquitin ligase complexes, also bind to GPS2 (Zhang et al. 2002), it remains an open issue to what extent GPS2 interactions influence TBL-dependent degradation processes.
Considering the molecular features of GPS2 action, we suspect broader roles of GPS2 in anti-inflammatory pathways in additional tissues and cell types. Indeed, we found that GPS2 is equally required for SUMO-dependent LXR transrepression in human macrophages (N Venteclef and E Treuter, unpubl.). As these pathways connect metabolism and inflammation, their molecular mechanisms are of immense relevance for the understanding and pharmacological management of metabolic syndrome diseases such as obesity, diabetes, and atherosclerosis. Future research, including the generation of tissue-specific mouse models, will be necessary to fully establish the role of GPS2 in modulating metabolic transactivation by LRH-1 (Sanyal et al. 2007) and LXRs (Jakobsson et al. 2009), as well as anti-inflammatory transrepression by these NRs (this study).
Materials and methods
Plasmid constructs
The human haptoglobin promoter construct, Flag-LXR constructs, and HA-GPS2 derivatives have been described previously (Venteclef et al. 2006; Sanyal et al. 2007; Jakobsson et al. 2009). Derivatives of human GPS2 and LXRs were generated using standard cloning techniques and the QuickChange site-directed mutagenesis kit (Stratagene). GST-SUMO constructs were provided by J. Palvimo, GFP-SUMO constructs were provided by H. Shih, and LRH-1 constructs and Flag-SuPr1 were provided by I. Talianidis. A detailed description of all constructs is available on request.
Antibodies
Antibodies against LXRs (Jakobsson et al. 2009) and LRH-1 (Chalkiadaki and Talianidis 2005) were produced as described previously. SUMO-1 (sc-9060), HDAC4 (sc6298), TAB2 (sc-20756), HDAC1 (sc-11418), and PPARα (sc-9000) antibodies were from Santa Cruz Biotechnologies; SMRT (#06-891) and N-CoR (#23750) antibodies were from Upstate Biotechnologies; and the SUMO-2/3 (M114-3) antibody was from MBL. The sources of all other antibodies used in this study were described previously (Jakobsson et al. 2009).
Animal models
Experimental protocols were approved by Karolinska Institutet Animal Care and by the University of Helsinki Review Board for animal experiments. The LXRαβ KO mouse model has been described previously (Alberti et al. 2001; Jakobsson et al. 2009). C57Bl/6J and KO mice (females, 10 wk old) were fed a chow diet and treated with GW3965 (30 mg/kg per day) for 4 d. Then the mice received an intraperitoneal injection of LPS (2 mg/kg) for 2.5 h. The generation of SUMO-1 KO mice has been described previously (Zhang et al. 2008). Wild-type and SUMO-1 KO mice (males, 10 wk old) were fed with chow diet and received an intraperitoneal injection of LPS (10 mg/kg) for 6 h. The liver was quickly removed, frozen in liquid nitrogen, and used for RNA extraction and ChIP assays.
Mammalian cell cultures
Huh7, Cos-7, and HeLa cells were cultured in Dulbecco's modified Eagle's medium (high glucose; Invitrogen) containing 10% (v/v) fetal bovine serum (Saveen Werner AB), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). Primary hepatocytes were isolated and cultured as described previously (Sanyal et al. 2007).
DNA transfections and luciferase reporter assays
For reporter assays, Huh7 cells were seeded in 24-well plates and transfected with indicated constructs and luciferase reporter the following day. Luciferase and β-galactosidase activities were measured using luciferine and ATP reagents (BioThema) and a Galacto-Star kit (Tropix), respectively, in a microplate luminometer (Thermo Electron Corp.). All transfections were performed using FuGENE 6 transfection reagent (Roche) according to the manufacturer's instructions.
siRNA transfections
Huh7 cells were transfected using 20 ng of control siRNA (siCTL), siRNA against hGPS2, hN-CoR, hLXRα, and hLXRβ (SMARTpool and single siRNAs, Dharmacon); and SUMO-1 (sc-29498) and SUMO-2/3 (sc-37167) (Santa Cruz Biotechnologies) according to the manufacturer's instructions.
qRT–PCR
RNA was isolated using EZNA RNA kit (Omega Biotek) according to the manufacturer's protocol. cDNA synthesis was performed using SuperScript II RT (Invitrogen) with random hexamers. mRNA expression was quantified on an Applied Biosystems 7500 instrument by using the SYBR green technology. Data were analyzed with the comparative CT method, using 18S as internal control.
Specific primer sequences are available on request.
ChIP assays
ChIP assays were performed essentially as described previously (Jakobsson et al. 2009). Quantification of the precipitated DNA regions was performed by PCR and qPCR. PCR amplification was performed using human and mouse haptoglobin and SAA, human PAI-1, mouse CRP, mouse SHP, and mouse CYP7A1 primer pairs as follows: human haptoglobin (5′-GCCTGGGCAACAGGAGTGAAA-3′ and 5′-CTTGGTTGGTCTTGCCTCTGG-3′), mouse haptoglobin (5′-GAGCCAGTACAAGAGTCCCA-3′ and 5′-CTGTGTTGCACAACGTCTC), human SAA (5′-GCACAACTGGGATAA-3′ and 5′-ATCTGTGCTGTAGCT-3′), mouse SAA (5′-ACTGCTACAGCACAGAAAAC-3′ and 5′-CTATGGGTGTAGGTGAGAGGA-3′), human PAI-1 (5′-ACACATGCCTCAGCAAGTCC-3′ and 5′-TCTTCTTGACAGCGCTCTTG-3′), mouse CRP (5′-GAGCTAGGCCAAGTGGAGCC-3′ and 5′-AGTCCTGGAACGCTTGCCTC-3′), mouse SHP (5′-CAGCCTGGGTTAATGACCCT-3′ and 5′-ATGCATACACGCTGACCCTG-3′), mouse CYP7A1 (5′-AGGGACAGACCTTCGGCTTA-3′ and 3′-TGGGTGACCAGAGCAAACAC-5′).
Coimmunoprecipitations
HeLa, Huh7, or Cos-7 cells were transfected with indicated constructs. Cells were lysed in NP-40 lysis buffer containing 50 mM Tris (pH 7.7), 150 mM NaCl, 1 mM EDTA, 1% NP-40, protease inhibitors, and 10 mM NEM (in the case of SUMO immunoprecipitations), subjected to immunoprecipitation (0.2% NP-40 Buffer) using indicated antibodies and Protein A Sepharose (GE Healthcare) for 2–3 h at 4°C, washed three times for 5 min, and subjected to Western blot analysis as indicated.
GST pull-down assays
GST, GST-SUMO-1, GST-SUMO-2, GST-GPS2-N (amino acids 1–105 wild type, L76,77,79R), and His-GPS2-N (amino acids 1–105 wild type) were bacterially expressed and purified using Glutathione Sepharose 4B (GE Healthcare). GST proteins were mixed, as indicated, with His-GPS2-N (amino acids 1–105) or [35S]-methionine-labeled proteins (GPS2 wild type, GPS2Δ61–94, SuPr1, LRH-1, SUMO-1–LRH-1) in pull-down buffer containing PBS, 0.1% Tween 20, and 1% BSA (Sanyal et al. 2007), and rotated for 2 h at 4°C. Glutathione Sepahrose 4B beads were added and rotated for 1 h at 4°C. Beads were washed three times for 10 min, and bound proteins were eluted with SDS sample buffer and subjected to Western blot analysis or autoradiography, respectively. For competition assays, peptides were prebound for 1 h before adding in vitro translated proteins. SIM peptides (Genscript) were as follows: GPS2-63–82, EETKEQILKLEEKLLALQEE; GPS2-81–100, EEKHQLFLQLKKVLHEEEKR; RCOR1-258–277, EDELEEANGNNPIDIEVDQN (Ouyang et al. 2009).
Acknowledgments
We are grateful to Drs. Iannis Talianidis for providing LRH-1 mutants and antibodies, Jorma J. Palvimo and Hsiu-Ming Shih for SUMO constructs, John R. Whitby and Timothy M. Willson for synthetic agonists, and John W. Schwabe for sharing unpublished structure information. We thank colleagues at the Department of Biosciences and Nutrition for sharing reagents, in particular Dr. Gudrun Toresson for purified proteins. This work was supported by grants from the Center for Biosciences (to E.T. and J.-Å.G.), the Swedish Research Council (to E.T., K.R.S., J.-Å.G., and P.P.), the Novo Nordisk Foundation (to E.T.), the Swedish Diabetes Society (to E.T.), the Academy of Finland (to O.A.J.), the Sigrid Jusélius Foundation (to O.A.J.), the Finnish Cancer Foundations (to O.A.J.), the European Union Integrated Project CRESCENDO (to O.A.J.), and the Swedish Heart-Lung Foundation (to P.P.), and a post-doctoral fellowship from the Swedish Research Council (to N.V.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.545110.
Supplemental material is available at http://www.genesdev.org.
References
- Alberti S, Schuster G, Parini P, Feltkamp D, Diczfalusy U, Rudling M, Angelin B, Bjorkhem I, Pettersson S, Gustafsson J-Å. Hepatic cholesterol metabolism and resistance to dietary cholesterol in LXRβ-deficient mice. J Clin Invest. 2001;107:565–573. doi: 10.1172/JCI9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke F, Takata Y, Caglayan E, Collins A, Tontonoz P, Hsueh WA, Tangirala RK. A nuclear receptor corepressor-dependent pathway mediates suppression of cytokine-induced C-reactive protein gene expression by liver X receptor. Circ Res. 2006;99:e88–e99. doi: 10.1161/01.RES.0000252878.34269.06. [DOI] [PubMed] [Google Scholar]
- Bradley MN, Hong C, Chen M, Joseph SB, Wilpitz DC, Wang X, Lusis AJ, Collins A, Hseuh WA, Collins JL, et al. Ligand activation of LXRβ reverses atherosclerosis and cellular cholesterol overload in mice lacking LXRα and apoE. J Clin Invest. 2007;117:2337–2346. doi: 10.1172/JCI31909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chait A, Han CY, Oram JF, Heinecke JW. Thematic review series: The immune system and atherogenesis. Lipoprotein-associated inflammatory proteins: Markers or mediators of cardiovascular disease? J Lipid Res. 2005;46:389–403. doi: 10.1194/jlr.R400017-JLR200. [DOI] [PubMed] [Google Scholar]
- Chalkiadaki A, Talianidis I. SUMO-dependent compartmentalization in promyelocytic leukemia protein nuclear bodies prevents the access of LRH-1 to chromatin. Mol Cell Biol. 2005;25:5095–5105. doi: 10.1128/MCB.25.12.5095-5105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao EY, Caravella JA, Watson MA, Campobasso N, Ghisletti S, Billin AN, Galardi C, Wang P, Laffitte BA, Iannone MA, et al. Structure-guided design of N-phenyl tertiary amines as transrepression-selective liver X receptor modulators with anti-inflammatory activity. J Med Chem. 2008;51:5758–5765. doi: 10.1021/jm800612u. [DOI] [PubMed] [Google Scholar]
- Fayard E, Auwerx J, Schoonjans K. LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARγ. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes & Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol. 2006;6:44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- Jakobsson T, Venteclef N, Toresson G, Damdimopoulos AE, Ehrlund A, Lou X, Sanyal S, Steffensen KR, Gustafsson J-Å, Treuter E. GPS2 is required for cholesterol efflux by triggering histone demethylation, LXR recruitment, and coregulator assembly at the ABCG1 locus. Mol Cell. 2009;34:510–518. doi: 10.1016/j.molcel.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- Kerscher O. SUMO junction—What's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khovidhunkit W, Kim MS, Memon RA, Shigenaga JK, Moser AH, Feingold KR, Grunfeld C. Effects of infection and inflammation on lipid and lipoprotein metabolism: Mechanisms and consequences to the host. J Lipid Res. 2004;45:1169–1196. doi: 10.1194/jlr.R300019-JLR200. [DOI] [PubMed] [Google Scholar]
- Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Lee JH, Park SM, Kim OS, Lee CS, Woo JH, Park SJ, Joe EH, Jou I. Differential SUMOylation of LXRα and LXRβ mediates transrepression of STAT1 inflammatory signaling in IFN-γ-stimulated brain astrocytes. Mol Cell. 2009;35:806–817. doi: 10.1016/j.molcel.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Molteni V, Li X, Nabakka J, Liang F, Wityak J, Koder A, Vargas L, Romeo R, Mitro N, Mak PA, et al. N-Acylthiadiazolines, a new class of liver X receptor agonists with selectivity for LXRβ. J Med Chem. 2007;50:4255–4259. doi: 10.1021/jm070453f. [DOI] [PubMed] [Google Scholar]
- Navab M, Anantharamaiah GM, Fogelman AM. The role of high-density lipoprotein in inflammation. Trends Cardiovasc Med. 2005;15:158–161. doi: 10.1016/j.tcm.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Lee Y, Solomon IH, Hager JM, Safi R, Choi Y, Guan Z, Tripathy A, Raetz CR, McDonnell DP, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12:357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S, Bavner A, Haroniti A, Nilsson LM, Lundasen T, Rehnmark S, Witt MR, Einarsson C, Talianidis I, Gustafsson J-Å, et al. Involvement of corepressor complex subunit GPS2 in transcriptional pathways governing human bile acid biosynthesis. Proc Natl Acad Sci. 2007;104:15665–15670. doi: 10.1073/pnas.0706736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, et al. Role of LXRs in control of lipogenesis. Genes & Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuter E, Gustafsson J-Å. Wrestling rules in transrepression: As easy as SUMO-1, -2, -3? Mol Cell. 2007;25:178–180. doi: 10.1016/j.molcel.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Venteclef N, Delerive P. Interleukin-1 receptor antagonist induction as an additional mechanism for liver receptor homolog-1 to negatively regulate the hepatic acute phase response. J Biol Chem. 2007;282:4393–4399. doi: 10.1074/jbc.M608993200. [DOI] [PubMed] [Google Scholar]
- Venteclef N, Smith JC, Goodwin B, Delerive P. Liver receptor homolog 1 is a negative regulator of the hepatic acute-phase response. Mol Cell Biol. 2006;26:6799–6807. doi: 10.1128/MCB.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby RJ, Dixon S, Maloney PR, Delerive P, Goodwin BJ, Parks DJ, Willson TM. Identification of small molecule agonists of the orphan nuclear receptors liver receptor homolog-1 and steroidogenic factor-1. J Med Chem. 2006;49:6652–6655. doi: 10.1021/jm060990k. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Jänne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]