Abstract
Pregnant women are infected by specific variants of Plasmodium falciparum that adhere and accumulate in the placenta. Using serological and molecular approaches, we assessed the global antigenic diversity of surface antigens expressed by placenta-binding isolates to better understand immunity to malaria in pregnancy and evolution of polymorphisms and to inform vaccine development. We found that placenta-binding isolates originating from all major regions where malaria occurs were commonly recognized by antibodies in different populations of pregnant women. There was substantial antigenic overlap and sharing of epitopes between isolates, including isolates from distant geographic locations, suggesting that there are limitations to antigenic diversity; however, differences between populations and isolates were also seen. Many women had cross-reactive antibodies and/or a broad repertoire of antibodies to different isolates. Studying VAR2CSA as the major antigen expressed by placenta-binding isolates, we identified antibody epitopes encoded by variable sequence blocks in the DBL3 domain. Analysis of global var2csa DBL3 sequences demonstrated that there was extensive sharing of variable blocks between Africa, Asia, Papua New Guinea, and Latin America, which likely contributes to the high level of antigenic overlap between different isolates. However, there was also evidence of geographic clustering of sequences and differences in VAR2CSA sequences between populations. The results indicate that there is limited antigenic diversity in placenta-binding isolates and may explain why immunity to malaria in pregnancy can be achieved after exposure during one pregnancy. Inclusion of a limited number of variants in a candidate vaccine may be sufficient for broad population coverage, but geographic considerations may also have to be included in vaccine design.
Plasmodium falciparum malaria is more prevalent and severe in pregnancy, especially among primigravid (PG) women, despite immunity that may have been acquired prior to pregnancy. Infection is characterized by the accumulation of mature-stage parasite-infected erythrocytes (IEs) in the placenta (6, 59), which is mediated through adhesion to chondroitin sulfate A (CSA) (8, 28) and possibly other molecules in the placenta, such as hyaluronic acid (HA) and nonimmune immunoglobulins (7, 14, 26, 45). In the first pregnancy women generally lack antibodies to placenta-binding IEs as the parasites express novel variant surface antigens (VSA) to which women have not been exposed previously (8, 30, 41, 46). Reduced susceptibility to placental malaria is observed with increasing numbers of malaria-exposed pregnancies due to the acquisition of specific immunity (29). Antibodies to antigens expressed on the IE surface of placental isolates (8, 30) and isolates that adhere to CSA (11, 41, 44, 46) are acquired. Multigravid (MG) women generally have a higher prevalence of these antibodies than PG women (8, 30, 41, 46), which corresponds to a reduced risk of malaria. Antibody levels have been associated with improved pregnancy outcomes in some studies (23, 56), further suggesting that they play an important role in immunity to pregnancy malaria.
The major target of antibodies to the surface of IEs is thought to be P. falciparum erythrocyte membrane protein 1 (PfEMP1), a highly diverse protein encoded by the var multigene family (15, 37, 55). However, a number of other antigens have been proposed (27, 36, 60). Antigenic variation of PfEMP1 mediated through switching of expression of different var genes facilitates evasion of host immune responses. A specific var gene, var2csa, has been shown to contain CSA-binding domains (31), and this gene is upregulated in a range of CSA-binding and placental parasite isolates (21, 51, 52). Additionally, exposed pregnant women acquire antibodies to recombinant VAR2CSA proteins (51). Disruption of var2csa inhibits the ability of IEs to bind to CSA (22, 58), and antibody binding to the surface of CSA-binding parasites is greatly reduced when PfEMP1 expression is inhibited (39, 40). Together, these findings suggest that VAR2CSA PfEMP1 mediates adhesion of IEs in the placenta and is an important target of acquired and possibly protective antibodies.
VAR2CSA is comprised of six extracellular Duffy-binding-like (DBL) domains and an intracellular acidic terminal segment. Recombinant DBL2 and DBL3 domains have been shown to bind CSA (20, 31). Although the var2csa gene is relatively conserved, substantial sequence polymorphisms are present (52). This gene appears to be subject to diversifying selection, suggesting that it is a target of protective immune responses (17). Sequence polymorphisms are concentrated in flexible loops (17) and appear to result in antigenic and functional differences between variants (7, 10, 12, 31). However, the significance of the var2csa sequence diversity is difficult to determine because many epitopes are likely to be conformational, and there is limited information about the structure of PfEMP1 domains and the identity of target epitopes; recent studies have suggested that antibodies recognize both polymorphic and conserved regions of VAR2CSA (1, 2, 4, 20). Furthermore, sequence analysis has been limited largely to African populations, and there is limited data for other populations where var2csa diversity may have evolved differently.
The degree of diversity or conservation of antigens expressed by placenta-type parasites is unclear. Sera from women in different geographic regions inhibited adhesion to CSA of the same placental isolates, suggesting that there is some restriction in the global diversity of placenta-binding IEs (30). Moreover, antibodies generated by immunization with CSA-binding IEs exhibited cross-reactivity with different CSA-binding isolates expressing var2csa-type genes (24), and recent studies have shown that pregnant women can acquire antibodies that cross-react with different isolates (10). However, other findings suggest that a large component of the antibody response acquired by pregnant women targets diverse or variant-specific epitopes (4, 8). Antigenic differences between isolates have clearly been demonstrated (8, 10, 57), although only limited comparisons have been performed, and there is significant diversity in the adhesion phenotypes of parasites isolated from pregnant women (14, 47). Two key questions are whether immunity is mediated by a repertoire of antibodies with different specificities or by acquisition of cross-reactive antibodies and whether antibody cross-reactivity is related to the expression of strictly conserved epitopes or polymorphic epitopes that are shared by isolates.
Further studies are needed to define the antigenic diversity of placenta-binding P. falciparum, the global distribution of antigenic determinants, and the breadth of reactivity of acquired antibodies against placenta-binding parasites. Such knowledge is important for understanding acquired immunity and the evolution of polymorphisms and possible constraints on diversity and for potential development of pregnancy-specific malaria vaccines. To investigate the global antigenic diversity of antibody epitopes in VSA expressed by placenta-binding P. falciparum isolates, we developed a panel of defined placenta-binding parasites from each of the major regions where malaria occurs, Africa, Asia, Oceania (Papua New Guinea [PNG]), and Latin America, in which var2csa was shown to be the dominant var gene transcribed. We examined antibody recognition of these isolates in geographically separated populations in Malawi and PNG, where any evolution of antigen diversity would be expected to have occurred independently. Furthermore, we characterized the isolate specificity and cross-reactivity of acquired antibodies to different placenta-binding isolates in pregnant women. In order to further understand the molecular basis of antigenic diversity and the forces of diversifying selection, we then focused on the DBL3 domain of VAR2CSA, which is responsible for CSA-binding activity (33, 54). We determined whether the most polymorphic regions of DBL3 are important antibody epitopes and examined the global sequence diversity of this domain.
MATERIALS AND METHODS
Study populations and ethics statement.
Serum or plasma samples were separated from the venous blood of pregnant women and men living in regions of Malawi and PNG where malaria is endemic. Both populations experience year-round malaria transmission with seasonal variations (18, 50). The great majority of malaria in Malawi is caused by P. falciparum, whereas both P. falciparum and P. vivax are prevalent in PNG.
In Malawi, samples were collected from January 1998 to November 2000 from pregnant women in the delivery suite of Queen Elizabeth Central Hospital in Blantyre and from fathers of children admitted to the same hospital with malaria (11). For women, placental infection was determined by histology, and the women were classified as infected individuals (P. falciparum IEs present), as uninfected individuals (no parasite or pigment present and placental, peripheral, and cord blood smears negative), or as individuals with a previous or cleared infection (malaria pigment present, but no parasites found and negative placental, peripheral, and cord blood smears) (49). Women with an active placental infection were matched on the basis of age and gravidity with women who had no evidence of placental infection and also were peripheral blood smear negative for malaria (11). The cohort comprised 35 PG women, 35 MG women, and 18 men. The numbers of subjects used in specific experiments are indicated below.
In PNG, serum samples were collected from pregnant women in the second trimester who visited the antenatal clinic for routine care at the Yagaum Health Centre and the Modilon Hospital in Madang in 2001 and 2002. Samples were also collected from men accompanying children visiting clinics at Modilon Hospital, the Madang town clinic, and the Yagaum immunization service. Written informed consent was obtained from all human subjects. The cohort comprised 29 PG women, 27 MG women, and 28 men. The numbers of subjects used in specific experiments are indicated below.
Samples were also collected from nonexposed donors living in Melbourne, Australia. Ethical approval for this study was obtained from the College of Medicine Research Committee, University of Malawi, Blantyre, Malawi; the Medial Research Advisory Committee, Department of Health, Port Moresby, PNG; the Human Research Ethics Committee, Melbourne Health Research Directorate, Victoria, Australia; and the Human Research and Ethics Committee of the Walter and Eliza Hall Institute of Medical Research, Australia.
P. falciparum isolates and culture.
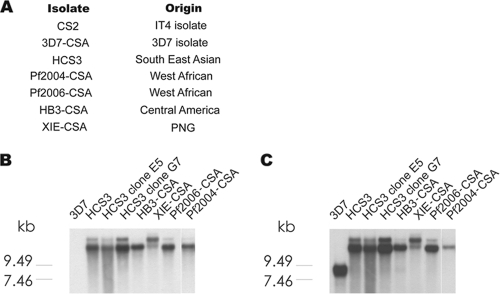
P. falciparum isolates were maintained in continuous culture as described previously (8). Cultures were regularly synchronized by resuspending culture pellets in d-sorbitol (5% in distilled H2O), and knob-expressing parasites were enriched by gelatin flotation. Cultures were free of Mycoplasma spp., as determined by PCR. P. falciparum isolates CS2, HCS3, 3D7-CSA, and HB3-CSA were obtained by selection for adhesion to CSA as described previously (2, 7) (Fig. 1A shows the origins of isolates); CS2, HB3, and 3D7 are long-established reference isolates. Pf2004 and Pf2006 were isolated from peripheral blood of travelers who were thought to have contracted malaria in Ghana, West Africa (25), and were adapted in in vitro culture over several weeks and then selected for adhesion to immobilized CSA (24) to generate isolates Pf2004-CSA and Pf2006-CSA. Isolate XIE was collected from the peripheral blood of a pregnant woman in PNG (Madang Province), adapted to in vitro culture over several weeks, and then selected for adhesion to immobilized CSA, which yielded isolate XIE-CSA. HCS3 was isolated from a traveler who contracted malaria in Southeast Asia and was selected for adhesion to CSA, as described previously (2, 7); it was then cloned by limiting dilution, and two clones, E5 and G7, were used in this study. All isolates were regularly reselected for CSA binding, and the identities of isolates were confirmed by sequencing the var2csa and ama1 genes.
FIG. 1.
var2csa transcription by various CSA-binding parasite isolates. (A) Origins of isolates used in this study. (B and C) RNA from ring-stage parasites was detected with a var2csa-specific probe (B) or var exon 2, which binds to most var genes (C). The dominant transcript in all isolates is var2csa. Note the presence of the double band with the var2csa-specific probe, indicating expression of two variants of var2csa. In both blots Pf2004-CSA was exposed slightly longer due to its lower RNA content. In addition to the parental HCS3 isolate, two clones of HCS3 parasites, E5 and G7, were included.
Antibodies to recombinant VAR2CSA domains.
Rabbits were immunized with recombinant protein or var2csa plasmid DNA. Recombinant VAR2CSA domains were expressed in Pichia pastoris, and expression was performed as previously described (2). In brief, rabbits were immunized using Freund's complete adjuvant for the first immunization and were boosted four times using Freund's incomplete adjuvant. For immunization with var2csa plasmid DNA, rabbits were injected six or seven times intramuscularly with DNA in Vaxfectin adjuvant (2). Preimmune and immune rabbit sera were heat inactivated for 45 min at 57°C and stored at −20°C.
Antibodies to surface antigens of P. falciparum IEs.
Testing for IgG binding to the surface of IEs in serum samples by flow cytometry was performed using previously described methods (11). Briefly, cells were sequentially incubated with test serum or plasma (1/20), rabbit anti-human IgG (1/100; Dako), and Alexa Fluor 488-conjugated anti-rabbit IgG (1/500; Molecular Probes) with ethidium bromide (10 μg/ml; Bio-Rad). All incubations were performed in phosphate-buffered saline (PBS) with 0.1% casein (Sigma) for 30 min at room temperature (RT). Samples were analyzed with a Becton-Dickinson FACSCalibur flow cytometer. Using FlowJo (TriStar), the IgG binding for each sample was expressed as the geometric mean fluorescence intensity (MFI) for IEs (ethidium bromide positive) after subtraction of the MFI for uninfected erythrocytes. Sera from nonexposed donors and positive-control samples were included in each assay. Samples were tested in two separate assays.
P. falciparum adhesion assays.
Adhesion of P. falciparum IEs was examined using pigmented trophozoite IEs at 3 to 8% parasitemia and 2 to 5% hematocrit, as previously described (8, 9, 11). The receptor concentrations were as follows: CSA (bovine trachea; Sigma), 10 μg/ml; CD36 [rhCD36(SR-B3)/Fc chimera; R&D], 10 μg/ml; ICAM-1 (Bender MedSystems), 5 to 8 μg/ml; and CD44 (rhCD44/Fc chimera, R&D), 10 μg/ml.
Mixed-agglutination assays.
Mixed-agglutination assays were performed using highly synchronized pigmented trophozoite-stage IEs (5 to 8% parasitemia) as described previously (10). Two different parasite isolates at similar parasitemias were stained separately with either ethidium bromide (20 μg/ml; Bio-Rad) or 4′,6′-diamidino-2-phenylindole (DAPI) (10 μg/ml). They were then mixed together and incubated with serum (1/10 dilution) with rolling at RT for 1 h. Agglutination was assessed by fluorescence microscopy. Agglutinates were defined as a minimum of three IEs for single-color agglutinates or a minimum of two IEs of each color for mixed-color agglutinates.
Peptides and peptide ELISAs.
VAR2CSA DBL3 variable block 3 (vb3), vb4, and vb5 (linear, cyclized, or biotin or bovine serum albumin [BSA] conjugated) of the IT4 isolate (equivalent to CS2), as well Dd2 DBL3 vb5, were used in peptide enzyme-linked immunosorbent assays (ELISAs). IT4 DBL3 vb3 (IIEKGTPQQKDKIGGVGS), vb4 (biotin-SGSGAITKINKKNNNSIFNGD), vb5 (TINGKNEKKCINSKSGQGDKIQGA, linear or biotin or BSA conjugated), and Dd2 DBL3 vb5 (INKDMEQNKCINSKNNNEKEIQGD) were synthesized by Mimotopes (Clayton, Victoria, Australia). All peptides had a free amine at the N terminus and a free acid at the OH terminus. Cyclized IT4 DBL3 vb5 (REACTINGNEKKSINSKSGQGDKIQGAC with a disulfide bridge between amino acids 4 and 28 [indicated by bold type]) was synthesized by Genscript (Piscataway, NJ).
Synthetic peptides in PBS (1 μg/ml) were coated on flat-bottom MaxiSorp plates (catalog no. 40422; Nunc) at 4°C overnight. The plates were blocked with PBS with 1% casein for 2 h at 37°C and then sequentially incubated with test sera diluted 1/250 in PBS with 0.1% casein and 0.05% Tween 20 and with sheep anti-human IgG-horseradish peroxidase (HRP) (1/1,000; Chemicon). All incubations were performed at RT for 1 h with three washes in between. IgG was detected by using 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) (Sigma). All samples were run in duplicate. We tested linear peptides and peptides conjugated to either biotin (which was then bound to commercially obtained or self-coated streptavidin plates) or BSA, as well as cyclized vb5, which resembled the natural folding of the flexible loop as presented on DBL3. All methods gave similar results (data not shown). For competition ELISA, antisera were preincubated with 5 μg/ml DBL3 recombinant protein produced in P. pastoris (2) before a standard ELISA was performed as described above. Unless stated otherwise, a sample was considered positive when the value for it was equal to or greater than the mean plus 2 standard deviations (SD) of the control values.
Northern blots.
Northern blots of mRNA from highly synchronous ring-stage parasite cultures were probed with var exon 2- and var2csa-specific sequences as previously described (22). Blots were washed at 55°C with 2× SSC and 0.1% SDS prior to autoradiography (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Sequencing of var2csa and analysis.
The DBL3 domain portion of var2csa was sequenced for isolates HCS3, XIE-CSA, Pf2004-CSA, and Pf2006-CSA, as well as DNA extracted from peripheral blood samples collected from P. falciparum-infected children and pregnant women visiting health services in Madang Province, PNG. Sequences for CS2, 3D7, and HB3 were already available. The DBL3 domain portion of var2csa was amplified using primers VAR2CSA DBL3F (5′ ATATAGGATCTGATACGTTTG) and DBL3R (5′ ACATATACTGCTATAATCTCC), which flank the DBL3 domain portion. PCR products were cloned into Escherichia coli and sequenced using standard methods. Analysis of HCS3, XIE-CSA, Pf2004-CSA, and Pf2006-CSA yielded 4 (from 15 clones), 6 (from 10 clones), 1 (from 10 clones), and 3 (from 10 clones) different sequences, respectively. Sixteen different samples from infected individuals in PNG (XHA or XJA) were sequenced, which yielded 48 unique sequences. Sequence alignment was performed using BioEdit (and DNAStar for Fig. 5), and segmentation and phylogenetic analyses were performed as previously described (16, 17). In brief, a segmentation analysis investigates correlations among nearby polymorphic segments. Starting with a multiple alignment, the segmentation algorithm was performed using a maximum segment length of 25 amino acids and a maximum of five types per segment. For phylogenetic analysis we used the dnam1 program of the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html). Trees were constructed using type-expanded alignment, which takes the output from the segmentation analysis and specifically compares the nucleotide variation that occurs within each segment type as a means to account for gene recombination. To examine whether sequences clustered by geography, a permutation test was performed with 2,000 iterations. Given the input tree (T) with tips labeled by geography, we computed the average distance (D) between the members of all pairs of sequences from the same geographic region. The permutation step fixes the topology of the T constant and randomly rearranges the geographical labels on the tips. For each permutation, the average within-geographic-region distance was computed, resulting in 2,000 values (V1 to V2000). The P value was determined by calculating the fraction of these values that were less than or equal to D.
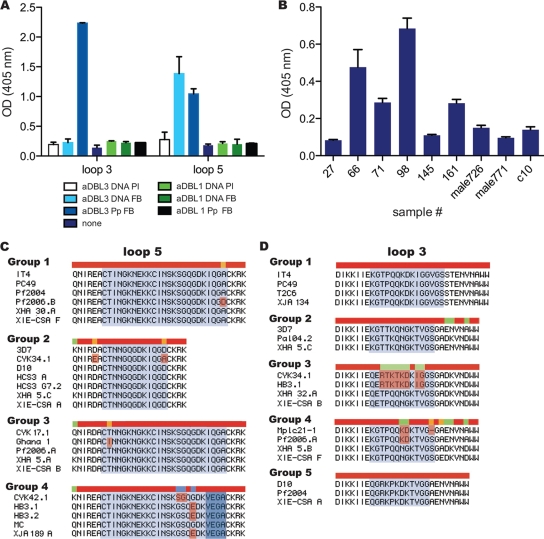
FIG. 5.
Polymorphic loops formed by variable sequence blocks of DBL3 were recognized by vaccine-induced and acquired antibodies. (A) Rabbit antiserum raised against the DBL1 or DBL3 domain of IT4 VAR2CSA was tested by ELISA for reactivity against synthetic peptides, which represent polymorphic loops of the DBL3 domain (IT4). Antibodies generated by DNA vaccination (aDBL3 DNA) recognized only loop 5, whereas antibodies generated by immunization with recombinant protein (aDBL3 Pp) reacted with IT4 DBL3 loops 5 and 3. Negative control sera against the VAR2CSA DBL1 domain were generated by DNA vaccination (aDBL1 DNA) or with recombinant protein (aDBL1 Pp) and did not react with either DBL3 loop. Subsequently, human sera from malaria-exposed Malawian (n = 72) and PNG (n = 56) women were tested for antibodies to IT4 DBL3 loop 5 peptide by an ELISA. The peptide was recognized by PG women, MG women, and several sympatric men from both countries, but not by Melbourne controls. Results for a representative selection of sera from Malawi (B) are shown. All women included were in the top quartile of responders to CS2 in FACS-based assays (see Table 1). In panel B samples 27, 71, 98, and 161 were samples from MG women, samples 66 and 145 were samples from PG women, and sample c10 was a sample from a Melbourne control donor. All samples were run in duplicate; means and standard deviations are shown. FB, final bleed; PI, preimmunization serum; OD (405), optical density at 405 nm. (C and D) Identical or nearly identical variable block 5 (C) and variable block 3 (D) sequences could be identified for isolates from diverse geographic origins. Each group contains sequences from isolates from different geographic regions, and the sequences were grouped using sequences obtained from one of the placenta-binding isolates used in antibody studies (3D7-CSA, CS2/IT4, HCS3, XIE-CSA, HB3-CSA, Pf2004-CSA, and Pf2006-CSA). Sequence alignments were constructed on the basis of identity in the region corresponding to synthetic peptides used in immunoassays (A and B). Blue shading indicates the predicted loop (additional residues are also shown), and red shading indicates an amino acid change compared to the majority of the sequences. The overall sequence alignment is indicated by bars; red indicates a 100% match, while orange, green, and blue indicate increasing numbers of sequence differences. The origins of the parasites are as follows: CYK34.1, CYK37, and CYK42.1, Senegal; D10, PNG; MC, Malaysia; Mplc21-1, Malawi; PC49, Peru; T2C6, Thailand; XHA clones, PNG; XIE par (non-CSA-selected XIE clones), PNG; XJA clones, PNG.
Statistical analysis.
The chi-square test (or Fisher's exact test when appropriate) was used to compare proportions, and t tests and Mann-Whitney U tests were used to compare means and medians, respectively. The correlation of antibody levels for two isolates was determined by using Spearman's rho. Principal component analysis (PCA) was used to identify relationships for antibody recognition of multiple isolates. The components identified linear combinations of the antibody data that explained the greatest proportion of the combined variation. Components with an eigenvalue of >1 were extracted. Rotation (Varimax with Kaiser normalization) was performed to maximize the differences between components and facilitate interpretation. Factor loading that was >0.4 was considered significant. The statistical analyses were performed using GraphPad Prism (GraphPad Software Inc.), except for PCA, which was done using SPSS for Windows (release 16.0, 2007; SPSS Inc., Chicago, IL).
Nucleotide sequence accession numbers.
The nucleotide sequences for the DBL3 domains for P. falciparum PNG field isolates XHA and XJA and for isolates XIE-CSA (PNG), Pf2004-CSA (Ghana), Pf2006-CSA (Ghana), and HCS3 (Southeast Asia) have been deposited in GenBank under accession no. GQ465324 to GQ465427 and GQ903562 to GQ903572.
RESULTS
Generation of isolates and determination of phenotypes and genotypes.
To study the antigenic properties of placenta-binding parasites and the acquisition of antibodies that protect against placental malaria, we developed a panel of seven different placenta-type P. falciparum isolates with clearly defined phenotypes; four of these isolates (3D7-CSA, CS2, HCS3, and HB3-CSA) had been described previously (7, 9, 17, 19, 35, 43, 48), while Pf2004-CSA and Pf2006-CSA (from West Africa) and XIE-CSA (from PNG) were generated for this study. This panel provided a representative selection of CSA-binding isolates originating from different regions where malaria is endemic, including Africa, Southeast Asia, Oceania, and Central America (Fig. 1A). All isolates adhered to CSA but did not adhere or adhered only weakly to CD36 or ICAM-1. The adhesion phenotypes for 3D7-CSA, CS2, and HCS3 have been reported previously (7). Levels of adhesion to CSA for isolates were as follows (at 10% parasitemia and 7% hematocrit): Pf2004-CSA, 2,826.1 bound IEs/mm2; Pf2006-CSA, 4,100 bound IEs/mm2; HB3-CSA, 3,400 bound IEs/mm2; and XIE-CSA, 7,907.6 bound IEs/mm2. Additionally, all isolates expressed var2csa as the dominant var gene transcript; this has been reported previously for 3D7-CSA, CS2, and HCS3 (21, 24). We established that var2csa was the dominant transcript expressed by XIE-CSA, HB3-CSA, Pf2004-CSA, and Pf2006-CSA by using Northern blots (Fig. 1). Probes specific for var2csa labeled bands in RNA extracts from all of the isolates except non-CSA-binding 3D7, which was used as a control (Fig. 1B). The same bands were labeled with a probe for the conserved acidic terminal sequence (ATS) second exon of var genes, which detected many transcribed var genes (Fig. 1C). Two var2csa bands were observed for some isolates. Even after cloning this was evident for HCS3, suggesting that some isolates possess more than one var2csa gene, as previously found for HB3 (35) and other lines (53).
Common recognition of geographically diverse isolates in pregnant women from Malawi and PNG.
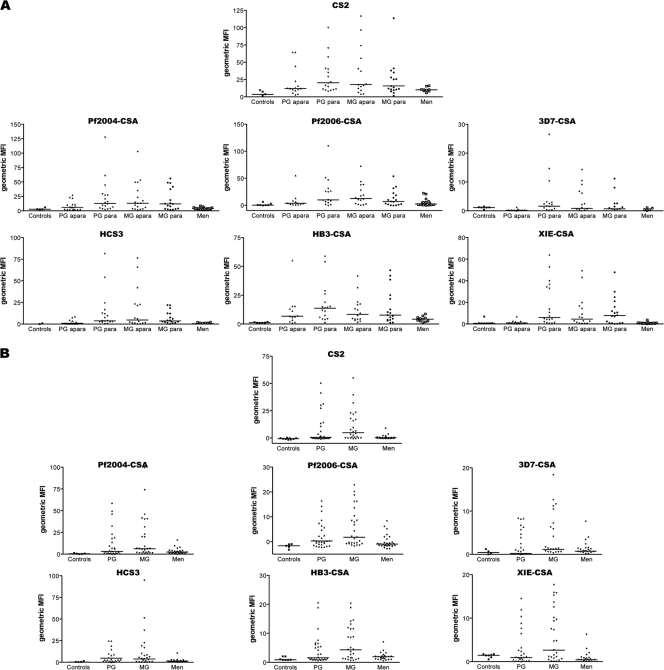
To assess the recognition of geographically diverse isolates in different populations, we tested the reactivities of serum antibodies from pregnant women from PNG (n = 56) and Malawi (n = 62), as well as from men (11 to 18 men for Malawi and 20 to 26 men for PNG) and from nonexposed Melbourne donors as controls (n = 7), with seven P. falciparum isolates originating from the major regions where malaria occurs by using flow cytometry. The geometric MFI of IgG binding to each isolate for women in the two study populations was stratified for gravity (Fig. 2). Malawi women were also stratified by placental parasitemia status (Fig. 2A), but this type of stratification was not possible for pregnant PNG women (Fig. 2B) as samples were not collected at term.
FIG. 2.
Antibodies to a global selection of placenta-type CSA-binding isolates in malaria-exposed pregnant women in Malawi and PNG. (A) Parasite isolates from different parts of the world selected for CSA adhesion were tested with sera from malaria-exposed women in Malawi by using FACS. The results of one of two independent experiments for each isolate are shown. The geometric mean fluorescence intensities (MFI) for IgG to VSA in sera from aparasitemic (apara) and parasitemic (para) PG and MG women, men, and Melbourne controls are shown. Isolates were recognized in a gender- and gravidity-dependent fashion. Many samples from pregnant women had antibodies to the isolates, whereas there was no or little reactivity for men (with the exception of Pf2006-CSA) and there was no reactivity for Melbourne controls. Aparasitemic MG women showed higher median MFI (horizontal lines) than aparasitamic PG women and men. (B) A global panel of P. falciparum isolates was selected for CSA adhesion and tested with sera from pregnant women and men from PNG and Melbourne controls by using FACS. The results of one of two independent experiments for each isolate are shown. Isolates were recognized in a gender- and gravidity-dependent fashion. For each isolate, a subset of pregnant women had antibodies, whereas the sera of men exhibited little or no recognition and the sera of Melbourne controls uniformly lacked antibodies. The horizontal lines indicate the median MFI. Note that in some cases the y axis starts below zero so that all data points can be shown.
All isolates tested were recognized by women in both populations, regardless of the origin of the isolate. Pregnant women had higher levels of IgG to VSA than men in the same population and nonexposed Australian adults, and the antibody levels for MG women were higher than those for PG women for most isolates. Recognition of isolates from distant regions was clearly demonstrated; for example, recent PNG isolate XIE-CSA was recognized by Malawian women (Fig. 2A), and recent West African isolates Pf2004-CSA and Pf2006-CSA were recognized by PNG women (Fig. 2B).
In Malawi, MG and PG women had significantly higher levels of IgG to all isolates than men (P < 0.05 for all comparisons, Mann-Whitney U test) (Fig. 2A). We observed that some men reacted to Pf2006-CSA, but men did not react to the other isolates. When individuals with and without placental parasitemia were compared, PG women with placental parasitemia (PG para) had consistently higher levels of antibody binding than PG women without parasitemia (PG apara), and the levels of significance (P values) ranged from 0.008 to 0.21. While MG apara generally exhibited higher median MFI than PG apara (P < 0.05 for most isolates), a difference was not seen for parasitemic individuals (P = 0.32).
In the PNG cohort (Fig. 2B), the IgG value was significantly higher for pregnant women than for men for all isolates except 3D7-CSA. The levels of IgG to CS2, Pf2004-CSA, Pf2006-CSA, HB3-CSA, and XIE-CSA were significantly higher for MG women than for men (P = 0.003, P = 0.009, P = 0.019, P = 0.018, and P = 0.0001, respectively). For the two remaining isolates (HCS3 and 3D7-CSA), the IgG levels also tended to be higher for MG women than for men, but the levels of statistical significance were lower (P = 0.069 and P = 0.124, respectively). The recognition of PNG isolate XIE-CSA by MG women was significantly greater than that by men (P = 0.0001) or PG women (P = 0.05), and the recognition by PG women was greater than that by men (P = 0.044). Unlike the findings for the Malawi cohort, sera from a number of PNG men recognized several of the isolates tested, although the antibody levels were generally low.
Repertoire of antibodies against different isolates.
The common recognition of placenta-type isolates by sera from distinct geographical regions raised important questions, including what is the repertoire or breadth of antibody reactivity to different placenta-type VSA among pregnant women? To investigate this, we focused on women with high levels of IgG by identifying samples with IgG levels in the top quartile for each cohort and classifying these women as high responders for each isolate (Table 1). In both study populations the recognition of isolates was diverse, and there was variation in the breadth of antibody reactivity among samples. While few women were high responders to all isolates (5/62 Malawi women [8.1%] and 4/56 PNG women [7.1%]), a substantial number of pregnant women had high levels of antibodies to multiple (≥2) isolates (21/62 Malawi women [33.9%] and 20/56 PNG women [35.7%]; note that these proportions are higher than the proportions of samples in the top quartile for each isolate as different women were high responders to different combinations of isolates). Many individuals were high responders to only a single isolate (16/62 Malawi women [25.8%] and 10/56 PNG women [17.9%]), and these individuals included both PG and MG women; this suggests that a substantial component of the acquired antibody response is variant specific. Previously, we reported that some sera have substantial strain-specific activity when they are tested for inhibition of adhesion to CSA, and sera that are cross-reactive against different isolates are not necessarily cross-inhibitory (10). Many samples had high levels of antibodies to isolates from different geographic regions (e.g., Africa and PNG), further indicating the limited global diversity of isolates.
TABLE 1.
Pattern of antibody reactivity to different placenta-binding isolates for high respondersa
| Samplea | Gravidity | Placental parasitemia | High responder tob: |
Total no. of isolates recognized | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CS2 | Pf2004-CSA | Pf2006-CSA | HCS3 | 3D7-CSA | HB3-CSA | XIE-CSA | ||||
| Malawi women | ||||||||||
| 39 | PG | − | x | 1 | ||||||
| 73 | PG | − | x | 1 | ||||||
| 107 | PG | − | x | 1 | ||||||
| 145 | PG | − | x | 1 | ||||||
| 153 | PG | − | x | x | x | 3 | ||||
| 30 | PG | + | x | 1 | ||||||
| 81 | PG | + | x | 1 | ||||||
| 110 | PG | + | x | 1 | ||||||
| 114 | PG | + | x | 1 | ||||||
| 122 | PG | + | x | 1 | ||||||
| 126 | PG | + | 1 | |||||||
| 104 | PG | + | x | x | 2 | |||||
| 116 | PG | + | x | x | 2 | |||||
| 38 | PG | + | x | x | x | x | x | 5 | ||
| 121 | PG | + | x | x | x | x | x | 5 | ||
| 98 | PG | + | x | x | x | x | x | x | 6 | |
| 66 | PG | + | x | x | x | x | x | x | x | 7 |
| 69 | PG | + | x | x | x | x | x | x | x | 7 |
| 103 | PG | + | x | x | x | x | x | x | x | 7 |
| 25 | MG | − | x | 1 | ||||||
| 34 | MG | − | x | 1 | ||||||
| 151 | MG | − | x | x | 1 | |||||
| 35 | MG | − | x | x | 2 | |||||
| 161 | MG | − | x | x | x | x | 4 | |||
| 86 | MG | − | x | x | x | x | x | 5 | ||
| 77 | MG | − | x | x | x | x | x | x | 6 | |
| 71 | MG | − | x | x | x | x | x | x | x | 7 |
| 80 | MG | − | x | x | x | x | x | x | x | 7 |
| 112 | MG | + | x | 1 | ||||||
| 162 | MG | + | x | 1 | ||||||
| 164 | MG | + | x | 1 | ||||||
| 115 | MG | + | x | x | 2 | |||||
| 146 | MG | + | x | x | 2 | |||||
| 128 | MG | + | x | x | x | 3 | ||||
| 173 | MG | + | x | x | x | 3 | ||||
| 27 | MG | + | x | x | x | x | x | 5 | ||
| 105 | MG | + | x | x | x | x | x | x | 6 | |
| Papua New Guinea women | ||||||||||
| 59 | PG | x | 1 | |||||||
| 75 | PG | x | 1 | |||||||
| 82 | PG | x | 1 | |||||||
| 92 | PG | x | 1 | |||||||
| 104 | PG | x | 1 | |||||||
| 16 | PG | x | x | 2 | ||||||
| 83 | PG | x | x | 2 | ||||||
| 7 | PG | x | x | x | 3 | |||||
| 78 | PG | x | x | x | 3 | |||||
| 17 | PG | x | x | x | x | x | x | 6 | ||
| 22 | PG | x | x | x | x | x | x | 6 | ||
| 90 | PG | x | x | x | x | x | x | 6 | ||
| 27 | PG | x | x | x | x | x | x | x | 7 | |
| 29 | MG | x | 1 | |||||||
| 69 | MG | x | 1 | |||||||
| 70 | MG | x | x | 1 | ||||||
| 118 | MG | x | 1 | |||||||
| 124 | MG | x | 1 | |||||||
| 21 | MG | x | x | 2 | ||||||
| 102 | MG | x | x | 2 | ||||||
| 112 | MG | x | x | 2 | ||||||
| 30 | MG | x | x | x | 3 | |||||
| 103 | MG | x | x | x | 3 | |||||
| 15 | MG | x | x | x | x | 4 | ||||
| 87 | MG | x | x | x | x | 4 | ||||
| 119 | MG | x | x | x | x | x | 4 | |||
| 2 | MG | x | x | x | x | x | x | 6 | ||
| 4 | MG | x | x | x | x | x | x | x | 7 | |
| 40 | MG | x | x | x | x | x | x | x | 7 | |
| 93 | MG | x | x | x | x | x | x | x | 7 | |
Samples from Malawi women were collected at the time of delivery; placental parasitemia was diagnosed by histology. Samples from PNG women were collected during the second trimester.
For each isolate high responders were defined as samples with IgG binding measured by flow cytometry in the top quartile (based on normalized average data for two assays). x, sample classified as a high responder with the isolate.
For the Malawi samples, there were fewer PG apara samples that were highly reactive to any one isolate (n = 5; 8.1%) than PG para samples (n = 14; 22.6%) (P < 0.05). Among the PG para 42.9% were high responders to only one isolate, while an equal proportion had high antibody reactivities to ≥4 isolates. In contrast, none of the PG apara recognized four or more isolates. For MG women, equal proportions of infected and uninfected women recognized only one isolate (33.3%). On average, PG apara and PG para recognized 1.6 and 3.4 isolates, respectively, whereas MG apara and MG para recognized 3.8 and 2.7 isolates, respectively.
In the PNG group a slightly lower percentage of PG women than of MG women (23.2% versus 30.4%) were highly reactive against one or more of the seven isolates. Of the PG women, 38.5% were high responders to only one isolate, and 30.7% were high responders to ≥4 isolates. Of the MG women, 29.4% had high levels of antibody to only one isolate, and 23.5% had high levels of antibodies to ≥4 isolates. On average, PG and MG women recognized 2.9 and 3.3 isolates, respectively.
Relationships between findings for antibody recognition of different isolates.
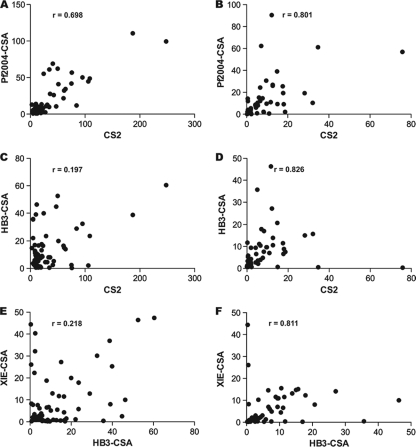
To gain insight into antigenic overlap between isolates and into the relatedness of responses of women to different isolates, we examined the correlation between antibodies to different isolates based on the flow cytometric data (Table 2 and Fig. 3). The majority of antibody binding data were strongly positively correlated for parasite isolates, suggesting that there is substantial antigenic overlap between isolates. Interestingly, for both cohorts the best correlations were the correlations between isolates having very different geographic origins (Pf2004-CSA and XIE-CSA for Malawi [rs = 0.899; P < 0.001; Pf2006-CSA and XIE-CSA for PNG [rs = 0.833; P < 0.001]) (Table 2). This suggests that the antigenic overlap is not restricted to isolates from similar regions. Also, the correlation between two recent West African isolates, Pf2004-CSA and Pf2006-CSA, was high for both cohorts (for Malawi, rs = 0.777 and P < 0.001; for PNG, rs = 0.788 and P < 0.001). However, while the correlations for many combinations of isolates were quite similar for the different cohorts, the correlations were different for other combinations. For example, the correlation between CS2 and Pf2004-CSA was high when it was determined for the Malawi (rs = 0.698, P < 0.001) and PNG (rs = 0.801, P < 0.001) cohorts. On the other hand, HB3-CSA and XIE-CSA showed poor correlation when they were tested with the Malawi samples (rs = 0.218, P = 0.0921), but the correlation was high for the PNG samples (rs = 0.811, P < 0.001). The same was true for CS2 and HB3-CSA (for Malawi samples, rs = 0.197 and P < 0.142; for PNG samples, rs = 0.826 and P < 0.001). For the Malawi cohort HB3-CSA generally correlated poorly with all of the other isolates (Table 2). The differences between the Malawi and PNG cohorts in correlations between isolates suggest that there were differences in the level or nature of exposure to different variants in the populations.
TABLE 2.
Correlation between IgG responses to combinations of P. falciparum isolates tested against sera from Malawi and PNGa
| Isolate | Correlation coefficient with isolate: |
|||||
|---|---|---|---|---|---|---|
| Pf2004-CSA | Pf2006-CSA | HCS3 | 3D7-CSA | HB3-CSA | XIE-CSA | |
| Tested against sera from Malawi | ||||||
| CS2 | 0.698 | 0.702 | 0.576 | 0.551 | 0.197d | 0.717 |
| Pf2004-CSA | 0.777 | 0.537 | 0.443 | 0.25b | 0.899 | |
| Pf2006-CSA | 0.581 | 0.512 | 0.265b | 0.826 | ||
| HCS3-CSA | 0.741 | 0.409 | 0.537 | |||
| 3D7-CSA | 0.418 | 0.404c | ||||
| HB3-CSA | 0.218d | |||||
| Tested against sera from PNG | ||||||
| CS2 | 0.801 | 0.776 | 0.630 | 0.758 | 0.826 | 0.822 |
| Pf2004-CSA | 0.788 | 0.599 | 0.570 | 0.785 | 0.745 | |
| Pf2006-CSA | 0.651 | 0.597 | 0.717 | 0.833 | ||
| HCS3-CSA | 0.482 | 0.596 | 0.622 | |||
| 3D7-CSA | 0.654 | 0.736 | ||||
| HB3-CSA | 0.811 | |||||
The values are correlation coefficients calculated by using Spearman's rho; n ranged from 57 to 63 for different comparisons for the sera from Malawi and was 54 or 55 for different comparisons for the sera from PNG. Bold type indicates isolates combinations for which corresponding graphs are shown in Fig. 3. All P values are ≤0.001 unless indicated otherwise.
P ≤0.05 (two-tailed).
P ≤0.01 (two-tailed).
Not statistically significantly different.
FIG. 3.
Antigenic overlap and differences between isolates of different geographic origins. The correlations between antibody reactivities to different isolates are shown for selected isolate combinations. Antibodies to isolates were measured by FACS for samples from Malawi (A, C, and E) and PNG (B, D, and F). Data points represent the mean values resulting from testing samples with each isolate in two independent experiments. All correlations were calculated by using Spearman's rho and are statistically significant (P < 0.0001), except for the correlations shown in panels C (P = 0.142) and E (P = 0.092).
To investigate the relationship between antibody responses to multiple isolates, the multivariate statistical method principal component analysis (PCA) was used. PCA was performed with the Malawi cohort using results obtained by measuring the IgG responses to the different isolates. PCA identified two significant components that accounted for 80.8% of the variance in the original set of variables (Table 3). Component 1 (44.7%) loaded strongly on Pf2004-CSA, Pf2006-CSA, and XIE-CSA, indicating that antibody recognition of these isolates is related, and component 2 (36.1%) loaded strongly on HCS3, 3D7-CSA, and HB3-CSA. These findings suggest that the isolates can be grouped on the basis of their antigenic properties and that 6 of the 7 isolates can be broadly classified into two groups that are not related to their geographic origins. For CS2 loading was significant for components 1 and 2, suggesting that antigenic characteristics of CS2 are related to antigenic characteristics of both groups of isolates.
TABLE 3.
Principal component analysis of levels of antibody binding to multiple isolates for Malawia
| Isolate | Factor loadingb |
|
|---|---|---|
| Component 1 | Component 2 | |
| CS2 | 0.710 | 0.618 |
| Pf2004-CSA | 0.892 | 0.310 |
| Pf2006-CSA | 0.845 | 0.288 |
| HCS3 | 0.362 | 0.856 |
| 3D7-CSA | 0.328 | 0.865 |
| HB3-CSA | 0.087 | 0.683 |
| XIE-CSA | 0.933 | 0.143 |
The total percentage of the variance explained by component 1 was 44.7%, and the total percentage of the variance explained by component 2 was 36.1%. Rotation converged in three iterations.
Factor loading values of >0.4 were considered significant and are indicated by bold type.
Cross-reactive antibodies and sharing of epitopes between isolates.
Recognition of isolates that had diverse origins by the same individual could be explained either by the presence of antibodies specific for shared epitopes expressed on different isolates or by the presence of antibodies with different specificities for multiple nonshared epitopes. To examine these possibilities, we tested samples using mixed-agglutination assays which measured the presence of antibodies to shared or conserved epitopes (10). The samples tested included samples that had significant IgG reactivities to two or more isolates as determined by fluorescence-activated cell sorting (FACS). Analysis by fluorescence microscopy revealed that some women did indeed have antibodies that recognized shared epitopes, which resulted in the formation of mixed-color agglutinates, whereas other women did not have such antibodies (Fig. 4).
FIG. 4.
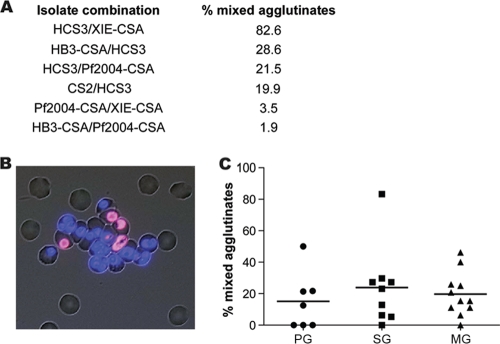
Cross-reactive and variant-specific antibodies in pregnant women detected using mixed-agglutination assays. Serum samples from Malawian women were tested for the ability to agglutinate isolates in mixed-agglutination assays. (A) Percentages of agglutinates that were mixed agglutinates for a selection of isolate combinations. The values are the means resulting from testing serum samples from different women. Mixed agglutinates were defined as agglutinates containing ≥2 IEs of each parasite line and indicate the presence of cross-reactive antibodies to the two isolates. (B) Typical mixed-color agglutinate of two isolates, resulting from the presence of cross-reactive antibodies. The two isolates were CS2 (DAPI, blue) and HCS3 (ethidium bromide, red). (C) Samples from primigravid (PG), secundigravid (SG), and multigravid (MG) women from Malawi were tested for the ability to agglutinate CS2 and HCS3. Shown are the proportions of agglutinates that were mixed agglutinates (the horizontal lines indicate the medians).
When a number of individual samples from Malawi were tested with CS2 and HCS3, both of which originated in Southeast Asia, 19.9% of the agglutinates were mixed agglutinates, and when Southeast Asia isolate HCS3 and PNG isolate XIE-CSA were tested, 82.6% of the agglutinates were mixed agglutinates (Fig. 4A). Substantial proportions of mixed agglutinates were also obtained with combinations of isolates from distant regions (HB3-CSA and HCS3 or HCS3 and Pf2004-CSA). In contrast, when Pf2004-CSA and XIE-CSA or HB3-CSA and Pf2004-CSA were tested, there were few mixed agglutinates. Consistent with these observations, the correlation coefficient for antibodies to Pf2004-CSA and XIE-CSA (rs = 0.49) as determined by FACS was much lower than that for antibodies to CS2 and HCS3 (rs = 0.75) or HCS3 and XIE-CSA (rs = 0.75). We confirmed that cross-reactive antibodies were also present in PNG samples; single-color and mixed-color agglutinates were observed for HCS3 and XIE-CSA and for Pf2004-CSA and XIE-CSA. These data indicate that epitopes are shared by isolates having diverse origins but that there is variation in the extent of epitope sharing by isolates; the results suggest that mixed agglutination was greater when isolates from nearby regions were used than when isolates from distant regions were used.
Isolate-specific antibodies, as well as cross-reactive antibodies, were observed for PG and MG women when different isolate combinations were used. To further evaluate the acquisition of cross-reactive antibodies by women of different gravidities, a more extensive assessment of one combination of isolates, isolates CS2 and HCS3, was performed. Samples from PG women, secundigravid (SG) women, and MG women were first separately tested for agglutination of CS2 or HCS3; 54 of 96 samples were positive for CS2 and/or HCS3. Of the positive samples, 32 (59%) agglutinated both CS2 and HCS3, and these samples were then tested using mixed-agglutination assays. Mixed agglutinates were observed for the majority of samples tested, but the levels of mixed agglutination varied widely (Fig. 4; see Table S1 in the supplemental material). The presence of cross-reactive antibodies was not strongly associated with gravidity; however, mixed agglutinates were observed for a higher proportion of SG and MG women than of PG women (90% versus 57.1%; P = 0.0501).
Antibody epitopes are encoded by highly polymorphic segments of VAR2CSA DBL3.
Having characterized antigenic diversity and antibody specificity using whole IEs, we next aimed to understand the molecular basis for our observations and the forces driving diversifying selection. We focused on the VAR2CSA DBL3 domain because it exhibits CSA-binding activity (20, 31) and shows evidence of diversifying selection, suggesting that it is a target of protective immunity (17, 20). This domain is also the only domain for which a substantial amount of sequence data is available and is one of only two domains encoded by var genes for which the crystal structure has been solved (32, 54). Structural studies have revealed that the sequence diversity of DBL3 is concentrated in 5 variable blocks, all of which are surface exposed (1, 17, 33, 54). Several of these variable blocks include flexible loop regions in the tertiary structure of DBL3 (33, 54). They can therefore be used as linear synthetic peptides in immunoassays.
We first tested variable blocks (expressed as synthetic peptides) for reactivity with antibodies raised in rabbits against recombinant DBL3 (IT4 sequence, which is identical to the CS2 sequence). Anti-DBL3 antibodies reacted with vb3 and vb5 of IT4 DBL3 (Fig. 5A), whereas they did not bind to DBL3 vb5 from another parasite isolate, Dd2 (from Southeast Asia), and IT4 DBL3 vb4 (data not shown). Antiserum generated by vaccination with var2csa DBL3 plasmid DNA reacted only with IT4 DBL3 vb5 and not with vb3. Neither prevaccination serum nor antiserum raised against IT4 DBL1 tested positive for either IT4 vb3 or vb5. We did not study vb1 or vb2 because they are very short in IT4 VAR2CSA, whereas both vb5 and vb3 form flexible loops in the tertiary structure of DBL3 (32, 54).
Sera from the Malawian study group were then tested for antibodies to the peptides (Fig. 5B). We found that 27.8% (15/54) of the MG women recognized vb5, whereas 20.8% (10/48) of the PG women reacted. Among the PG women 33.3% (9/27) of the parasitemic individuals but only 4.8% (1/21) of the aparasitemic individuals were positive (P = 0.021). The specificity of IgG to IT4 vb5 was confirmed by performing a competition ELISA; preincubation of positive sera with recombinant IT4 DBL3 (5 μg/ml) reduced IgG binding to vb5 by 30 to 100% (data not shown). Some samples from men (4/24 [16.7%]) were also reactive; antibodies to recombinant full-length DBL3 have also been reported for men (5). However, the antibody levels for positive men were significantly lower than the antibody levels for positive MG women (P < 0.01). Furthermore, in competition ELISAs, preincubation of samples from MG women with recombinant DBL3 substantially inhibited IgG reactivity to vb5, whereas the inhibition of IgG reactivity of samples from men was minimal (data not shown). When tested against IT4 DBL3 vb3, several samples from Malawian women were positive (data not shown).
Isolates having diverse origins share common sequence motifs in VAR2CSA.
To further understand the extent and evolution of antigenic diversity of VAR2CSA, we analyzed DBL3 sequences from different geographic regions. Many sequences from Africa were already available in the public database. However, the sequence data for regions outside Africa were limited; a small number of sequences were available for isolates from Asia and Latin America, but only one sequence was available for PNG. Therefore, we sequenced DBL3 domains for P. falciparum infections in PNG donors, as well DBL3 domains for isolates XIE-CSA, Pf2004-CSA, Pf2006-CSA, and HCS3; sequences for other isolates that we used in our antibody assays were already available.
A total of 124 DBL3 sequences were compared (15 sequences from Southeast Asia, 54 sequences from PNG, 43 sequences from Senegal, 8 sequences from other African countries, 3 sequences from Central and South America, and 1 unknown sequence [3D7]) (see Fig. S1 and Table S2 in the supplemental material). Sequences of isolates from geographically distant regions were found to share many features, and the same sequences in variable blocks 1 to 5 were observed for isolates from Africa, PNG, Asia, and South America (Fig. 6; see Fig. S1 in the supplemental material). Many identical and closely related sequences corresponding to vb5 and vb3, which are antibody epitopes, were found in isolates from different regions, indicating that some polymorphic epitopes are shared by isolates (Fig. 5C and D show representative examples grouped according to identity with the different placenta-binding isolates used in antibody assays). These findings support serological findings indicating that there is limited global antigenic diversity that originates from sharing of common sequences, as even the most polymorphic epitopes appear to be shared by isolates having different origins and wide geographic distributions.
FIG. 6.
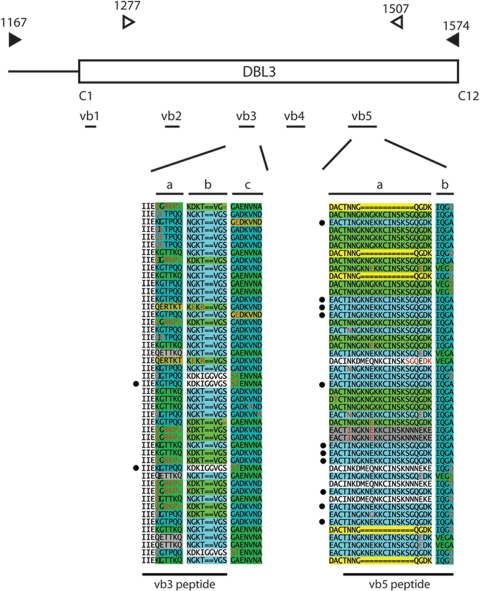
Globally diverse isolates share DBL3 variable block sequences. Analysis of VAR2CSA DBL3 variable block 3 and 5 amino acid sequences demonstrated that isolates having diverse geographic origins shared the same polymorphic loop sequences. A diagram of the DBL3 domain is shown at the top. The positions and lengths of variable blocks 1 to 5 in the DBL3 sequences are indicated. The locations of primers used for PCR amplification are indicated by arrowheads. The filled arrowheads indicate primers used to amplify PNG sequences in this study, and the open arrowheads indicate the locations of primers used previously to amplify sequences from Senegal isolates (1). The amino acid position of the primers is counted from the first amino acid in the IT4 VAR2CSA sequence (accession no. AAQ73926). The segmentation structures for variable blocks 3 and 5 in representative PNG sequences are shown below the diagram of the DBL3 domain (a full alignment of all sequences is shown in Fig. S1 in the supplemental material). Variable block 3 is composed of three segments, segments a, b, and c, and variable block 5 is composed of two segments, segments a and b. The locations of variable block peptides used for serological analysis are indicated under the alignments. PNG sequences that were identical to the IT4 VAR2CSA peptide sequences are indicated by filled circles.
Distribution of VAR2CSA DBL3 sequences across different regions.
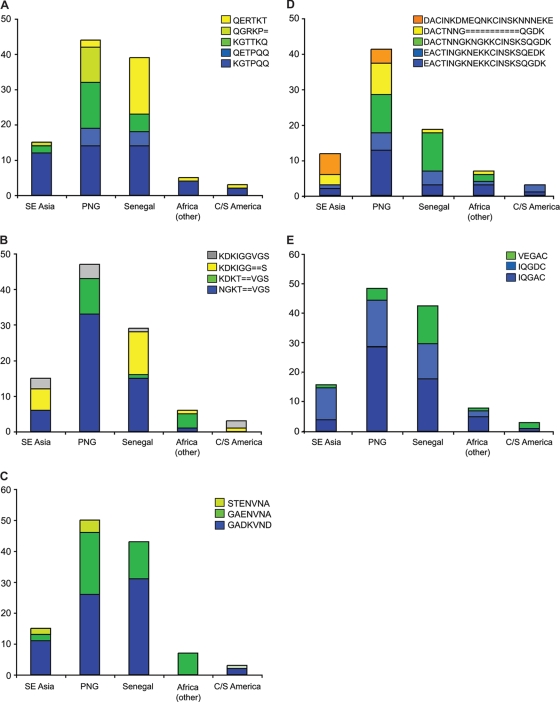
To better understand the distribution of polymorphism in variable blocks, we performed a segmentation analysis, which can identify linkages between nearby polymorphic residues (17). As determined by this analysis, vb3 was composed of three polymorphic segments, segments a, b, and c, and vb5 was composed of two polymorphic segments, segments a and b (Fig. 6). These regions correspond to the peptides used for serological analysis, and the findings demonstrate that it is not uncommon to identify isolates with identical or related variable blocks. In vb3, segment a starts at position 129 in the alignment (e.g., KIIEKGTTKQ), segment b starts at position 139 (e.g., NGKT--VGS), and segment c starts at position 148 (e.g., G-DKVND) (see Fig. S1 in the supplemental material). At each of these segment locations, there was limited diversity of major segment types that were shared across diverse geographic regions (Fig. 7). For instance, in segment a of vb3, five segment types accounted for 86% of the diversity in the sequence sets. Four of these segment types were shared by PNG and Senegal data sets, but the proportions in the two parasite populations were different (Fig. 7). The fifth type (QGRKP) was found in PNG sequences and one of the sequences from Southeast Asia, but it was not present in Senegal sequences. Overall, a few of the major segment types of vb3 and vb5 were unique to a geographic region (Fig. 7); however, they differed in frequency. When PNG (n = 54) and Senegal (n = 43) sequences were compared, there were statistically significant differences in the distribution of sequences in vb3 and vb5 (P < 0.0001 for differences in vb3 segments a and b; P = 0.045 for differences in vb3 segment c; P = 0.061 and P = 0.023 for differences in vb5 segments a and b, respectively). In addition to the most common segment types, there were also a few examples of minor segment types that were found in only a specific geographic region. Taken together, the results of this analysis demonstrate that there is extensive overlap of polymorphic segments between African and PNG isolates, which could contribute to the antibody cross-reactivity between CSA-binding lines, but they also suggest that there are differences between Senegal and PNG sequences due to new mutations in ancestral polymorphic segments and to the frequency distribution of polymorphic segments.
FIG. 7.
Overlap and differences in distribution of polymorphic segments in variable blocks 3 and 5. The major segment types found in variable blocks 3 and 5 in the VAR2CSA DBL3 domain were quantified. A total of 124 DBL3 sequences were compared (16 sequences from Southeast Asia, 54 sequences from PNG, 43 sequences from Senegal, 8 sequences from other African countries, and 3 sequences from Central and South America [C/S America], as well as one sequence of unknown origin (3D7). (A) Major segment types of vb3 segment a. (B) Major segment types of vb3 segment b. (C) Major segment types of vb3 segment c. (D) Major segment types of vb5 segment a. (E) Major segment types of vb5 segment b. When data for PNG (n = 54) and Senegal (n = 43) were compared, there were statistically significant differences in the distribution of sequences in variable block 3 and 5 (P < 0.0001 for differences in the proportions of vb3 segment a and vb3 segment b sequences; P = 0.045 for differences in the proportions of vb3 segment c sequences; P = 0.061 and P = 0.023 for differences in the proportions of vb5 segment a and vb5 segment b sequences, respectively [Fisher's exact test]).
To further investigate whether DBL3 sequences were diversifying between PNG and Senegal, a phylogenetic analysis was performed using a slightly truncated sequence of DBL3 (sequence without loop 1), which enabled inclusion of 43 sequences from Senegal. For tree building, a “population tree” approach based on the segmentation output was used (17). This approach was employed because standard phylogenetic approaches assume that sequences evolve through mutation and they are not appropriate for studying highly recombinogenic genes, such as var2csa (3). This analysis revealed that sequences from different geographic ȯrigins were spread throughout the phylogenetic tree, but there was also evidence of geographic clustering. Of the 124 DBL3 sequences, 18 were identical in two isolates, 4 were identical in three isolates, and 1 was identical in 4 isolates. Identical sequences appeared to be more common for parasites within a geographic region; only 3 of 18 sequences were identical in two different geographic regions, and it was as common to find identical sequences in the PNG data set as it was to find identical sequences in the Senegal data set. Furthermore, many of the nonidentical sequences from Senegal or PNG clustered together in the tree (see Fig. S2 in the supplemental material). To investigate whether clustering was statistically significant, we compared the average distances for all pairs of sequences for the same geographical region and performed a permutation test. When all sequences were compared, there was strong evidence for geographical clustering (P = 0.0015). Differences were not significant when the comparison was limited to one sequence per isolate due to the tendency of sequences from the same isolate to cluster together in the tree. Taken together, despite extensive gene mosaicism and overlap in polymorphic regions, there were differences in VAR2CSA sequences between parasite populations, suggesting that there is ongoing regional diversification. Therefore, population differences may need to be considered when vaccines are designed.
DISCUSSION
This study evaluated the global antigenic diversity of placenta-binding P. falciparum isolates. We studied placenta-binding isolates originating from the major regions where malaria occurs and performed a detailed examination of antibody specificity, the antibody repertoire, and cross-reactivity for pregnant women in two different regions. To gain insight into the molecular basis of antigenic diversity, we analyzed sequences of VAR2CSA DBL3 from different geographic regions and determined the extent of relatedness between them. Furthermore, we examined polymorphic sequences as antibody epitopes and examined their global distribution.
Our findings clearly demonstrate that there is limited global antigenic diversity in VSA expressed by placenta-binding IEs. Previous studies have shown that there is recognition of isolates in populations outside their geographic origins (10, 30, 46). We extended these findings by performing a detailed study of antibody specificity and cross-reactivity, testing a panel of defined placenta-binding isolates from the major regions where malaria occurs (Africa, Southeast Asia, PNG, and Latin America). There was recognition of all isolates by both Malawian and PNG populations, and the correlation for antibody reactivity was generally high between isolates. PNG pregnant women had antibodies that recognized recent isolates from Africa, and African women had antibodies that recognized recent isolates from PNG and Thailand. However, there were some differences in antibody recognition between women in Malawi and women in PNG (for example, recognition of 3D7-CSA was not strongly gravidity dependent in PNG). In other studies, we found that isolates CS2 and HCS3 are commonly recognized by antibodies in exposed pregnant women in Kenya and Thailand (M. Hommel and J. Beeson, unpublished observations). Additionally, cross-reactive antibodies to different isolates were present in many individuals, indicating that shared epitopes are expressed by isolates from different regions. Furthermore we found evidence that even the most polymorphic segments of var2csa are shared by isolates of distant geographic origins. Many antibody epitopes do not appear to be strictly conserved, as individuals exhibited strong isolate-specific responses rather than pan-reactive or cross-reactive responses. Antigenic differences between isolates have also been suggested by other studies (8, 10, 57). PCA data obtained by testing samples for antibodies to different isolates indicated that broad grouping of isolates may be possible based on antigenic similarities and differences. The results of the PCA suggested that six of the seven isolates tested could be classified into two groups based on antigenic properties, and the grouping did not reflect the geographic origins of the isolates. This approach may be useful for guiding vaccine development.
The broad recognition of different isolates by antibodies in pregnant women can be explained by antibodies to epitopes shared by isolates and by the presence of multiple antibodies with different specificities in serum. Mixed-agglutination assays and analysis of high responders by FACS suggested that a significant component of the antibody response is variant specific (i.e., a response to polymorphic epitopes). However, if global antigenic diversity is limited, exposure to one isolate may give rise to antibodies that are reactive with geographically diverse isolates. Since infections are typically polyclonal (34), exposure to multiple different placenta-binding variants may occur during a single episode and therefore result in a repertoire of antibodies to different isolates. Women of all gravidities appeared to acquire substantial cross-reactive antibodies, which were somewhat more prevalent in MG women than in PG women. A previous study suggested that MG women have a broader repertoire of antibodies to different isolates, which is consistent with greater exposure (10). Some evidence suggests that acquired antibodies may target conserved regions of VAR2CSA (1, 20), and some antibodies to whole IEs and VAR2CSA domains generated by immunization have broad reactivity with different isolates (24, 38). However, strictly conserved epitopes have not been defined yet. Here, we focused on defining antigenic properties using acquired human antibodies rather than antibodies to specific domains of VAR2CSA induced by immunization of animals. Vaccine-induced antibody responses may differ substantially from naturally acquired responses because recombinant proteins may not fully represent the native presentation of VAR2CSA domains and because immunization can induce responses with specificities different from those acquired through natural exposure.
In order to better understand the molecular basis of antigenic diversity and the forces driving diversifying selection in var2csa, we examined the most polymorphic regions of DBL3, which are concentrated in variable blocks, several of which include surface-exposed flexible loops (17, 20, 33). At present, VAR2CSA is the only clearly established antigen on the surface of placenta-binding IEs, and the DBL3 domain is important because it is responsible for CSA-binding activity (31, 51). Other antigens have been proposed but not yet confirmed (13, 27). Our findings provide evidence that variable blocks 3 and 5 of DBL3 are antibody epitopes, suggesting that acquired antibodies are likely driving the diversifying selection that is observed in these sequences. Remarkably, the same polymorphic epitopes could be identified in isolates from Africa, Asia, PNG, and Latin America, suggesting that the limited global diversity of placenta-binding isolates is partially explained by sharing of polymorphic epitopes that have a wide geographic distribution. This is an important proof of principle, but further studies of other polymorphic regions of VAR2CSA domains are needed.
Our results provide insights into the evolution of the antigenic diversity of VSA expressed by placenta-binding IEs and P. falciparum antigens more broadly. At present, there is little data on the global diversity and molecular epidemiology of important P. falciparum antigens, particularly VSA. The rationale for comparing Africa and PNG populations is that there is great separation between these populations in terms of time and distance, which provided an opportunity to examine evidence of regional diversification in antigenic determinants or differences in the distribution of specific variants (differences in the global distribution of serotypes have been observed for many bacterial and viral infections). There appears to have been little parasite gene flow between PNG and other regions (42, 61). We demonstrated that antibodies in the two populations recognize isolates with diverse origins and that there is extensive sharing of sequences corresponding to variable blocks 3 and 5, which we have shown to be antibody epitopes, as well as other polymorphic segments, between all regions where malaria occurs. Nevertheless, there also appeared to be differences between VAR2CSA sequences when an African population (Senegal) and a PNG population were compared. While the same major polymorphic segment types were generally found in both locations, the frequencies of these segments differed between populations, and there were also examples of rare new mutant types that were unique to a specific region. Using phylogenetic approaches that account for gene recombination, we also found that DBL3 sequences from a geographic region frequently clustered together, suggesting that var2csa sequences in the different populations are diverging. Our findings support the hypothesis that there was an ancient origin of polymorphisms that are shared across global isolates but are gradually evolving through gene recombination and new mutations. These features help account for the great range of maternal antibody responses to placenta-binding isolates and may explain why there were differences in the profiles of antibody reactivity between Malawian and PNG sera. Differences in antibody reactivity may also be influenced by the level of malaria transmission and the timing of sample collection, which were different for the two populations. The exposure to antigenic determinants expressed by placenta-binding parasites may have been different in the different populations due to dynamic changes or ongoing mutation of ancient polymorphic segments.
In conclusion, our findings demonstrated that there is limited antigenic diversity of placenta-binding P. falciparum isolates and defined the specificity, cross-reactivity, and repertoire of acquired antibodies to these isolates in pregnant women. The molecular basis for limited diversity is partially explained by the finding that the most polymorphic regions of VAR2CSA are antibody epitopes and the finding that these epitopes are commonly shared across the global P. falciparum population. These findings have important implications for understanding immunity to malaria in pregnancy, the evolution of antigenic diversity in VSA, and potential vaccine development. The limited diversity of placenta-binding isolates and the wide geographic distribution of epitopes may explain why a substantial level of immunity to malaria in pregnancy can be achieved by exposure during one pregnancy. Inclusion of a limited number of variants in a candidate vaccine may be sufficient for broad coverage, provided that geographic differences are considered in vaccine design.
Supplementary Material
Acknowledgments
We gratefully acknowledge the participants in the studies and the staffs of the Queen Elizabeth Central Hospital (Blantyre, Malawi), the Malawi-Liverpool-Wellcome Trust Clinical Research Programme (College of Medicine, University of Malawi), the PNG Institute of Medical Research (Yagaum Health Centre, PNG), and Modilon Hospital (Madang, PNG) for assistance with enrollment of participants and sample collection and processing. We also acknowledge Marianne Hathaway (SBRI) for excellent lab assistance and Alfred Cortes for helpful discussions and advice. Human erythrocytes and serum samples used for in vitro culture were kindly provided by the Australian Red Cross Blood Service (Melbourne, Australia).
Funding was provided by the National Health and Medical Research Council of Australia (career development award to J. Beeson; project grants to J. Beeson, S. Rogerson, and M. Hommel; IRIISS grant 361646), the Wellcome Trust, United Kingdom (career development fellowship to S. Rogerson), the Bill & Melinda Gates Foundation (J. Smith), the Miller Fellowship of the Walter and Eliza Hall Institute (to J. Beeson), and a Victorian State Government Operational Infrastructure Support grant.
Editor: J. H. Adams
Footnotes
Published ahead of print on 16 February 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Andersen, P., M. A. Nielsen, M. Resende, T. S. Rask, M. Dahlbäck, T. Theander, O. Lund, and A. Salanti. 2008. Structural insight into epitopes in the pregnancy-associated malaria protein VAR2CSA. PLoS Pathog. 15:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avril, M., B. R. Kulasekara, S. O. Gose, C. Rowe, M. Dahlbäck, P. E. Duffy, M. Fried, A. Salanti, L. Misher, D. L. Narum, and J. D. Smith. 2008. Evidence for globally shared, cross-reacting polymorphic epitopes in the pregnancy malaria vaccine candidate VAR2CSA. Infect. Immun. 74:1791-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awadalla, P. 2003. The evolutionary genomics of pathogen recombination. Nat. Rev. Genet. 4:50-60. [DOI] [PubMed] [Google Scholar]
- 4.Barfod, L., N. L. Bernasconi, M. Dahlback, D. Jarrossay, P. H. Andersen, A. Salanti, M. F. Ofori, L. Turner, M. Resende, M. A. Nielsen, T. G. Theander, F. Sallusto, A. Lanzavecchia, and L. Hviid. 2007. Human pregnancy-associated malaria-specific B cells target polymorphic, conformational epitopes in VAR2CSA. Mol. Microbiol. 63:335-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barfod, L., M. A. Nielsen, L. Turner, M. Dahlback, A. T. Jensen, L. Hviid, T. G. Theander, and A. Salanti. 2006. Baculovirus-expressed constructs induce immunoglobulin G that recognizes VAR2CSA on Plasmodium falciparum-infected erythrocytes. Infect. Immun. 74:4357-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beeson, J. G., N. Amin, M. Kanjala, and S. J. Rogerson. 2002. Selective accumulation of mature asexual stages of Plasmodium falciparum-infected erythrocytes in the placenta. Infect. Immun. 70:5412-5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beeson, J. G., and G. V. Brown. 2004. Plasmodium falciparum-infected erythrocytes demonstrate dual specificity for adhesion to hyaluronic acid and chondroitin sulfate A and have distinct adhesive properties. J. Infect. Dis. 189:169-179. [DOI] [PubMed] [Google Scholar]
- 8.Beeson, J. G., G. V. Brown, M. E. Molyneux, C. Mhango, F. Dzinjalamala, and S. J. Rogerson. 1999. Plasmodium falciparum isolates from infected pregnant women and children are associated with distinct adhesive and antigenic properties. J. Infect. Dis. 180:464-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beeson, J. G., W. Chai, S. J. Rogerson, A. M. Lawson, and G. V. Brown. 1998. Inhibition of binding of malaria-infected erythrocytes by a tetradecasaccharide fraction from chondroitin sulfate A. Infect. Immun. 66:3397-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beeson, J. G., E. J. Mann, T. J. Byrne, A. Caragounis, S. R. Elliott, G. V. Brown, and S. J. Rogerson. 2006. Antigenic differences and conservation among placental Plasmodium falciparum-infected erythrocytes and acquisition of variant-specific and cross-reactive antibodies. J. Infect. Dis. 193:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beeson, J. G., E. M. Mann, S. R. Elliott, V. M. Lema, E. Tadesse, M. E. Molyneux, G. V. Brown, and S. J. Rogerson. 2004. Antibodies to variant surface antigens of Plasmodium falciparum-infected erythrocytes and adhesion inhibitory antibodies are associated with placental malaria and have overlapping and distinct targets. J. Infect. Dis. 189:540-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beeson, J. G., F. Ndungu, K. E. M. Persson, G. Kelly, S. Uyoga, S. L. Hallamore, T. Williams, J. C. Reeder, G. V. Brown, and K. Marsh. 2007. Antibodies among men and children to placental-type Plasmodium falciparum-infected erythrocytes that adhere to chondroitin sulfate A and express var2csa. Am. J. Trop. Med. Hyg. 77:22-28. [PubMed] [Google Scholar]
- 13.Beeson, J. G., F. H. Osier, and C. R. Engwerda. 2008. Recent insights into humoral and cellular immune responses against malaria. Trends Parasitol. 24:578-584. [DOI] [PubMed] [Google Scholar]
- 14.Beeson, J. G., S. J. Rogerson, B. M. Cooke, J. C. Reeder, W. Chai, A. M. Lawson, M. E. Molyneux, and G. V. Brown. 2000. Adhesion of Plasmodium falciparum-infected erythrocytes to hyaluronic acid in placental malaria. Nat. Med. 6:86-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biggs, B. A., L. Goozé, K. Wycherley, W. Wollish, B. Southwell, J. H. Leech, and G. V. Brown. 1991. Antigenic variation in Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 88:9171-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bockhorst, J., and N. Joiic. 2007. Discovering patterns in biological sequences by optimal segmentation, p. 17-24. In Proceedings of the 23rd Annual Conference on Uncertainty in Artificial Intelligence (UAI-07). AUAI Press, Corvallis, OR.
- 17.Bockhorst, J., F. Luc, J. H. Janes, J. Keebler, B. Gamain, P. Awadalla, X. Suc, R. Samudrala, N. Jojic, and J. D. Smith. 2007. Structural polymorphism and diversifying selection on the pregnancy malaria vaccine candidate VAR2CSA. Mol. Biochem. Parasitol. 155:103-112. [DOI] [PubMed] [Google Scholar]
- 18.Cattani, J., J. Tulloch, H. Vrbova, D. Jolley, F. Gibson, J. Moir, P. Heywood, M. Alpers, A. Stevenson, and R. Clancy. 1986. The epidemiology of malaria in a population surrounding Madang, Papua New Guinea. Am. J. Trop. Med. Hyg. 35:3-15. [DOI] [PubMed] [Google Scholar]
- 19.Cooke, B. M., S. J. Rogerson, G. V. Brown, and R. L. Coppel. 1996. Adhesion of malaria-infected red blood cells to chondroitin sulfate A under flow conditions. Blood 88:4040-4044. [PubMed] [Google Scholar]
- 20.Dahlbäck, M., T. S. Rask, P. H. Andersen, M. A. Nielsen, N. T. Ndam, M. Resende, L. Turner, P. Deloron, L. Hviid, O. Lund, A. G. Pedersen, T. G. Theander, and A. Salanti. 2006. Epitope mapping and topographic analysis of VAR2CSA DBL3X involved in P. falciparum placental sequestration. PLoS Pathog. 2:e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duffy, M. F., T. J. Byrne, S. R. Elliott, D. W. Wilson, S. J. Rogerson, J. G. Beeson, R. Noviyanti, and G. V. Brown. 2005. Broad analysis reveals a consistent pattern of var gene transcription in Plasmodium falciparum repeatedly selected for a defined adhesion phenotype. Mol. Microbiol. 56:774-788. [DOI] [PubMed] [Google Scholar]
- 22.Duffy, M. F., A. G. Maier, T. J. Byrne, A. J. Marty, S. R. Elliott, M. T. O'Neill, P. D. Payne, S. J. Rogerson, A. F. Cowman, B. S. Crabb, and G. V. Brown. 2006. VAR2CSA is the principal ligand for chondroitin sulfate A in two allogeneic isolates of Plasmodium falciparum. Mol. Biochem. Parasitol. 148:117-124. [DOI] [PubMed] [Google Scholar]
- 23.Duffy, P. E., and M. Fried. 2003. Antibodies that inhibit Plasmodium falciparum adhesion to chondroitin sulfate A are associated with increased birth weight and the gestational age of newborns. Infect. Immun. 71:6620-6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott, S., M. F. Duffy, T. J. Byrne, J. G. Beeson, E. J. Mann, D. W. Wilson, S. J. Rogerson, and G. V. Brown. 2005. Cross-reactive surface epitopes on chondroitin sulfate A-adherent Plasmodium falciparum-infected erythrocytes are associated with transcription of var2csa. Infect. Immun. 73:2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott, S. R., P. D. Payne, M. F. Duffy, T. J. Byrne, W. H. Tham, S. J. Rogerson, G. V. Brown, and D. P. Eisen. 2007. Antibody recognition of heterologous variant surface antigens after a single Plasmodium falciparum infection in previously naive adults. Am. J. Trop. Med. Hyg. 76:860-864. [PubMed] [Google Scholar]
- 26.Flick, K., C. Scholander, Q. Chen, V. Fernandez, B. Pouvelle, J. Gysin, and M. Wahlgren. 2001. Role of nonimmune IgG bound to PfEMP1 in placental malaria. Science 293:2098-2100. [DOI] [PubMed] [Google Scholar]
- 27.Francis, S. E., V. A. Malkov, A. V. Oleinikov, E. Rossnagle, J. P. Wendler, T. K. Mutabingwa, M. Fried, and P. E. Duffy. 2007. Six genes are preferentially transcribed by the circulating and sequestered forms of Plasmodium falciparum parasites that infect pregnant women. Infect. Immun. 75:4838-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fried, M., and P. E. Duffy. 1996. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science 272:1502-1504. [DOI] [PubMed] [Google Scholar]
- 29.Fried, M., and P. E. Duffy. 1998. Maternal malaria and parasite adhesion. J. Mol. Med. 76:162-171. [DOI] [PubMed] [Google Scholar]
- 30.Fried, M., F. Nosten, A. Brockman, B. J. Brabin, and P. E. Duffy. 1998. Maternal antibodies block malaria. Nature 395:851-852. [DOI] [PubMed] [Google Scholar]
- 31.Gamain, B., A. R. Trimnell, C. Scheidig, A. Scherf, L. H. Miller, and J. D. Smith. 2005. Identification of multiple chondroitin sulfate A (CSA)-binding domains in the var2CSA gene transcribed in CSA-binding parasites. J. Infect. Dis. 191:1010-1013. [DOI] [PubMed] [Google Scholar]
- 32.Higgins, M. 2008. Overproduction, purification and crystallization of a chondroitin sulfate A-binding DBL domain from a Plasmodium falciparum var2csa-encoded PfEMP1 protein. Acta Crystallogr. Sect. F Struct. Biol. Crystallogr. Commun. 64:221-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higgins, M. 2008. The structure of a chondroitin sulfate-binding domain important in placental malaria. J. Biol. Chem. 283:21842-21846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamwendo, D. D., F. K. Dzinjalamala, G. Snounou, M. C. Kanjala, C. G. Mhango, M. E. Molyneux, and S. J. Rogerson. 2002. Plasmodium falciparum: PCR detection and genotyping of isolates from peripheral, placental, and cord blood of pregnant Malawian women and their infants. Trans. R. Soc. Hyg. Trop. Med. 96:145-149. [DOI] [PubMed] [Google Scholar]
- 35.Kraemer, S. M., S. A. Kyes, G. Aggarwal, A. L. Springer, S. O. Nelson, Z. Christodoulou, L. M. Smith, W. Wang, E. Levin, C. I. Newbold, P. J. Myler, and J. D. Smith. 2007. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics 8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyes, S. A., J. A. Rowe, N. Kriek, and C. I. Newbold. 1999. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 96:9333-9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leech, J. H., J. W. Barnwell, L. H. Miller, and R. J. Howard. 1984. Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes. J. Exp. Med. 159:1567-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magistrado, P., A. Salanti, N. G. Tuikue Ndam, S. B. Mwakalinga, M. Resende, M. Dahlback, L. Hviid, J. Lusingu, T. G. Theander, and M. A. Nielsen. 2008. VAR2CSA expression on the surface of placenta-derived Plasmodium falciparum-infected erythrocytes. J. Infect. Dis. 198:1071-1074. [DOI] [PubMed] [Google Scholar]
- 39.Maier, A. G., M. Rug, M. T. O'Neill, J. G. Beeson, M. Marti, J. C. Reeder, and A. F. Cowman. 2007. Skeleton-binding protein 1 functions at the parasitophorous vacuole membrane to traffic PfEMP1 to the Plasmodium falciparum-infected erythrocyte surface. Blood 109:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maier, A. G., M. Rug, M. T. O'Neill, M. Brown, S. Chakravorty, T. Szestak, J. Chesson, Y. Wu, K. Hughes, R. L. Coppel, C. Newbold, J. G. Beeson, A. Craig, B. S. Crabb, and A. F. Cowman. 2008. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 134:48-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maubert, B., N. Fievet, G. Tami, M. Cot, C. Boudin, and P. Deloron. 1999. Development of antibodies against chondroitin sulfate A-adherent Plasmodium falciparum in pregnant women. Infect. Immun. 67:5367-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mu, J., P. Awadalla, J. Duan, K. M. McGee, D. A. Joy, G. A. McVean, and X. Z. Su. 2005. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 3:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noviyanti, R., G. V. Brown, M. E. Wickham, M. F. Duffy, A. F. Cowman, and J. C. Reeder. 2001. Multiple var gene transcripts are expressed in Plasmodium falciparum infected erythrocytes selected for adhesion. Mol. Biochem. Parasitol. 114:227-237. [DOI] [PubMed] [Google Scholar]
- 44.O'Neil-Dunne, I., R. N. Achur, S. T. Agbor-Enoh, M. Valiyaveettil, R. S. Naik, C. F. Ockenhouse, A. Zhou, R. Megnekou, R. Leke, D. W. Taylor, and D. C. Gowda. 2001. Gravidity-dependent production of antibodies that inhibit binding of Plasmodium falciparum-infected erythrocytes to placental chondroitin sulfate proteoglycan during pregnancy. Infect. Immun. 69:7487-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pouvelle, B., P. A. Buffet, C. Lepolard, A. Scherf, and J. Gysin. 2000. Cytoadhesion of Plasmodium falciparum ring-stage-infected erythrocytes. Nat. Med. 6:1264-1268. [DOI] [PubMed] [Google Scholar]
- 46.Ricke, C. H., T. Staalsoe, K. Koram, B. D. Akanmori, E. M. Riley, T. G. Theander, and L. Hviid. 2000. Plasma antibodies from malaria-exposed pregnant women recognize variant surface antigens on Plasmodium falciparum-infected erythrocytes in a parity-dependent manner and block parasite adhesion to chondroitin sulfate A. J. Immunol. 165:3309-3316. [DOI] [PubMed] [Google Scholar]
- 47.Rogerson, S. J., J. G. Beeson, C. Mhango, F. Dzinjalamala, and M. E. Molyneux. 2000. Plasmodium falciparum rosette formation is uncommon in isolates from pregnant women. Infect. Immun. 68:391-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogerson, S. J., S. C. Chaiyaroj, K. Ng, J. C. Reeder, and G. V. Brown. 1995. Chondroitin sulfate A is a cell surface receptor for Plasmodium falciparum-infected erythrocytes. J. Exp. Med. 182:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogerson, S. J., E. Pollina, A. Getachew, E. Tadesse, V. M. Lema, and M. E. Molyneux. 2003. Placental monocyte infiltrates in response to Plasmodium falciparum malaria infection and their association with adverse pregnancy outcomes. Am. J. Trop. Med. Hyg. 68:115-119. [PubMed] [Google Scholar]
- 50.Rogerson, S. J., N. R. van den Broek, E. Chaluluka, C. Qonqwane, C. G. Mhango, and M. E. Molyneux. 2000. Malaria and anemia in antenatal women in Blantyre, Malawi: a twelve month survey. Am. J. Trop. Med. Hyg. 62:335-340. [DOI] [PubMed] [Google Scholar]
- 51.Salanti, A., M. Dahlbäck, L. Turner, M. A. Nielsen, L. Barfod, P. Magistrado, A. T. Jensen, T. Lavstsen, M. F. Ofori, K. Marsh, L. Hviid, and T. G. Theander. 2004. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med. 200:1197-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salanti, A., T. Staalsoe, T. Lavstsen, A. T. Jensen, M. P. Sowa, D. E. Arnot, L. Hviid, and T. G. Theander. 2003. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 49:179-191. [DOI] [PubMed] [Google Scholar]
- 53.Sander, A. F., A. Ali Salanti, T. Lavstsen, M. A. Nielsen, P. Magistrado, J. Lusingu, N. Tuikue Ndam, and D. E. Arnot. 2009. Multiple var2csa-type PfEMP1 genes located at different chromosomal loci occur in many Plasmodium falciparum isolates. PLoS One 4:e6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh, K., A. G. Gittis, P. Nguyen, D. C. Gowda, L. H. Miller, and D. N. Garboczi. 2008. Structure of the DBL3x domain of pregnancy-associated malaria protein VAR2CSA complexed with chondroitin sulfate A. Nat. Struct. Mol. Biol. [DOI] [PMC free article] [PubMed]
- 55.Smith, J. D., C. E. Chitnis, A. G. Craig, D. J. Roberts, D. E. Hudson-Taylor, D. S. Peterson, R. Pinches, C. I. Newbold, and L. H. Miller. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staalsoe, T., C. E. Shulman, J. N. Bulmer, K. Kawuondo, K. Marsh, and L. Hviid. 2004. Variant surface antigen-specific IgG and protection against clinical consequences of pregnancy-associated Plasmodium falciparum malaria. Lancet 263:283-289. [DOI] [PubMed] [Google Scholar]
- 57.Tuikue Ndam, N. G., N. Fievet, G. Bertin, G. Cottrell, A. Gaye, and P. Deloron. 2004. Variable adhesion abilities and overlapping antigenic properties in placental Plasmodium falciparum isolates. J. Infect. Dis. 190:2001-2009. [DOI] [PubMed] [Google Scholar]
- 58.Viebig, N. K., B. Gamain, C. Scheidig, C. Lepolard, J. Przyborski, M. Lanzer, J. Gysin, and A. Scherf. 2005. A single member of the Plasmodium falciparum var multigene family determines cytoadhesion to the placental receptor chondroitin sulphate A. EMBO Rep. 6:775-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter, P. R., Y. Garin, and P. Blot. 1982. Placental pathologic changes in malaria. A histologic and ultrastructural study. Am. J. Pathol. 109:330-342. [PMC free article] [PubMed] [Google Scholar]
- 60.Winter, G., S. Kawai, M. Haeggstrom, O. Kaneko, A. von Euler, S. Kawazu, D. Palm, V. Fernandez, and M. Wahlgren. 2005. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J. Exp. Med. 201:1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu, D. I. Baruch, A. J. Magill, and X. Z. Su. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320-323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.