EMBO J (2010) 29 4, 795–805. doi:10.1038/emboj.2009.371 EMBO J (2009),29 4, 806–818. doi:10.1038/emboj.2009.385
DNA lesions or genomic regions that are difficult to traverse frequently hinder or block DNA replication. In response to replication fork stalling, the cell activates the replication stress response pathway, which acts to protect the fork from collapse, promotes the repair or bypass of the blockage and facilitates the resumption of DNA synthesis. In this issue of the EMBO Journal, two studies conducted by the Constantinou and Niedzwiedz laboratories shed light on how the DNA translocase FANCM acts to regulate the replication stress response (Luke-Glaser et al, 2009; Schwab et al, 2009). These studies help to explain how FANCM (mutated in the human cancer predisposition syndrome, Fanconi's anaemia (FA)) co-ordinately regulates checkpoint signalling and replication fork progression.
FA is a complex multigenic genomic instability disorder that can develop from a mutation in 1 of at least 13 different genes. Evidence suggests that the FA pathway acts in concert with nucleotide excision repair, translesion synthesis and homologous recombination to promote lesion repair/bypass, yet the mechanism by which this is achieved remains poorly understood. Cloning of the FANCM gene showed a putative DNA helicase/translocase that is related to Hef1, an archaebacteria protein that maintains replication fork integrity (Meetei et al, 2005; Mosedale et al, 2005). Several studies have since established that FANCM is a potent DNA translocase that can remodel replication fork structures to promote fork regression (Gari et al, 2008). Surprisingly, it was shown that DNA translocase activity of FANCM is dispensable for its role in the FA pathway (Singh et al, 2009), but is instead essential for maintaining the integrity of stalled replication forks as part of the replication stress response (Collis et al, 2008).
Studies published by Luke-Glaser et al (2009) and Schwab et al (2009) in this issue of the EMBO Journal provide insight into the mechanism through which FANCM controls replication fork progression and contributes to replication stress response. Using DNA fibre analysis to directly visualize replication forks in mammalian cells, Luke-Glaser et al (2009) established that the DNA translocase activity of FANCM is required to restrain replication elongation; replication forks progress more quickly in the absence of FANCM and this leads to increased genomic instability. Similarly, Schwab et al (2009) found that replication forks are unstable in FANCM-deficient chicken DT40 cells and this leads to the accumulation of single-stranded DNA. Both groups also show that the DNA translocase activity of FANCM is required for the efficient restart of stalled replication forks. Schwab et al found that an inability to restart stalled replication forks leads to inappropriate firing of nearby dormant origins, which is a hallmark of checkpoint deficiency.
FANCM was recently implicated in the ATR signalling pathway, which is central to the replication stress response (Collis et al, 2008). Studies by Luke-Glaser et al and Schwab et al corroborate this observation and provide further insight into the role of FANCM in this pathway. Both groups found that FANCM is important for eliciting efficient activation of the ATR checkpoint in response to various replication stress conditions: Schwab et al found that phosphorylation of Chk1 and Smc1 (ATR targets) is compromised in the absence of FANCM, and provide evidence that this defect is likely caused by a failure to efficiently recruit and/or retain TopBP1 on chromatin, which is essential for ATR activation (Kumagai et al, 2006). Luke-Glaser et al found that FANCM and Chk1 mutually stabilize each other. Thus, in the absence of FANCM, the ATR signalling pathway is not efficiently activated leading to replication fork instability (Figure 1; Collis et al, 2008; Luke-Glaser et al, 2009; Schwab et al, 2009).
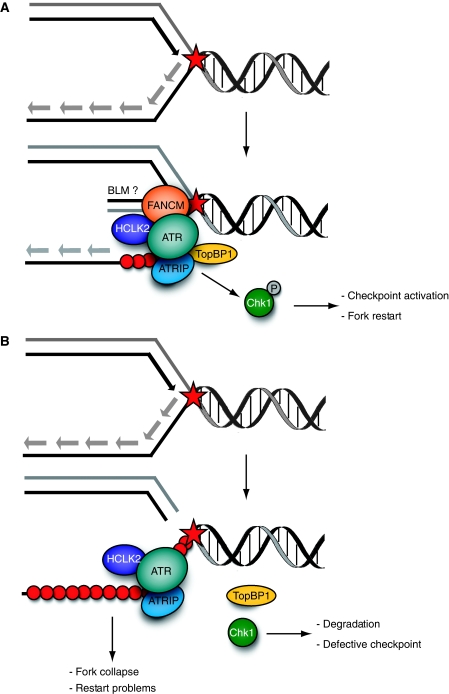
Figure 1.
(A) Replication forks can be stalled by secondary structures or DNA lesions (star) that must be removed or by-passed for replication to proceed. The DNA translocase activity of FANCM remodels the stalled fork to facilitate activation of the ATR–ATRIP kinase complex. FANCM also interacts with HCLK2 and TopBP1 to further facilitate efficient ATR activation. FANCM may also act with the BLM helicase to facilitate replication restart. (B) Stalled forks are unstable in the absence of FANCM and accumulate extensive regions of ssDNA-RPA (red circles). Although the ATR–ATRIP complex is recruited to these sites, the checkpoint is not efficiently activated because of a failure to retain TopBP1 on chromatin and/or loss of Chk1.
How does FANCM facilitate ATR activation and promote replication fork restart? It is possible that remodelling of stalled replication forks by FANCM may facilitate interactions between TopBP1 and ATR, to promote its activation (Figure 1). FANCM may also act as a scaffold to coordinately recruit at least three key complexes to stalled replication forks; the ATR kinase complex (through direct binding to HCLK2), the FA core complex (through direct binding to FANCF) and Bloom's syndrome complex (through direct binding to RMI1 and TopoIIIα; Deans and West (2009)). Indeed, loss of FANCM confers phenotypes consistent with defects in all three complexes (Collis et al, 2008; Deans and West, 2009).
In summary, the current studies provide further insight into the role of FANCM in controlling replisome progression and fork dynamics in response to replication stress. The challenge over the next few years will be to determine how these critical functions of FANCM are regulated during cell cycle to ensure genome integrity.
Footnotes
The authors declare that they have no conflict of interest.
References
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ (2008) FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 32: 313–324 [DOI] [PubMed] [Google Scholar]
- Deans AJ, West SC (2009) FANCM connects the genome instability disorders Bloom's syndrome and Fanconi anemia. Mol Cell 36: 1–11 [DOI] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A (2008) Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci USA 105: 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG (2006) TopBP1 activates the ATR-ATRIP complex. Cell 124: 943–955 [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B, Grossi S, Constantinou A (2009) FANCM regulates DNA chain elongation and is stabilized by S phase checkpoint signaling. EMBO J 29: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W (2005) A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 37: 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, Johnson M, Langevin F, Pace P, Patel KJ (2005) The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol 12: 763–771 [DOI] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W (2009) ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, Ali AM, Du CH, Rooimans MA, Fan Q, Wahengbam K, Steltenpool J, Andreassen PR, Williams DA, Joenje H, de Winter JP, Meetei AR (2009) Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood 114: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]