Abstract
When Ebola virus nucleoprotein (NP) is expressed in mammalian cells, it assembles into helical structures. Here, the recombinant NP helix purified from cells expressing NP was characterized biochemically and morphologically. We found that the recombinant NP helix is associated with non-viral RNA, which is not protected from RNase digestion and that the morphology of the helix changes depending on the environmental salt concentration. The N-terminal 450 aa residues of NP are sufficient for these properties. However, digestion of the NP-associated RNA eliminates the plasticity of the helix, suggesting that this RNA is an essential structural component of the helix, binding to individual NP molecules via the N-terminal 450 aa. These findings enhance our knowledge of Ebola virus assembly and understanding of the Ebola virus life cycle.
Viruses belonging to the order Mononegavirales have a non-segmented negative-sense single-strand RNA for their genome. Each viral genomic RNA is always associated with multiple copies of a viral nucleoprotein (NP), forming a helical NP–RNA complex, the morphology of which differs among viruses (Lamb & Parks, 2007). The NP–RNA complex, rather than the naked genomic RNA, serves as a template for viral RNA synthesis, which is carried out by the RNA-dependent RNA polymerase (Lamb & Parks, 2007). When NP of mononegaviuses is expressed alone in bacteria or eukaryotic cells, it binds non-specifically to cellular RNAs and forms recombinant helical NP–RNA complexes (Bhella et al., 2002; Errington & Emmerson, 1997; Heggeness et al., 1980; Iseni et al., 1998; Mavrakis et al., 2002; Moyer et al., 1991; Préhaud et al., 1990; Spehner et al., 1991), which have the same morphology and stoichiometry as the authentic NP–viral RNA complexes (Fooks et al., 1995; Iseni et al., 1998; Spehner et al., 1991). Therefore, the recombinant NP–RNA complex of mononegaviruses could be a useful model to use to examine the properties of an authentic NP–viral RNA complex.
Ebola virus belongs to the family Filoviridae of the order Mononegavirales (Sanchez et al., 2007). Its NP consists of 739 aa that can be divided into a hydrophobic N-terminal half (approx. 350 aa) and a hydrophilic C-terminal half (approx. 400 aa) (Sanchez et al., 2007). The C-terminal region is responsible for the interaction with the matrix protein VP40 and the incorporation of NP into virions (Licata et al., 2004; Noda et al., 2007). The N-terminal region is thought to be important for RNA binding via its interaction with the phosphodiester backbone that is independent of the nucleotide sequences, similar to paramyxo- and rhabdoviruses (Albertini et al., 2006; Green et al., 2006; Iseni et al., 1998; Tawar et al., 2009). We recently found that when NP is expressed in mammalian cells in the absence of viral RNA and the other viral proteins, it assembles into helices of approximately 20 nm in diameter that are morphologically distinct from authentic nucleocapsids and that the N-terminal 450 aa residues are necessary for the NP–NP interaction to form those helices (Noda et al., 2006; Watanabe et al., 2006). Co-expression of NP with the minor matrix protein VP24 and polymerase cofactor VP35 leads to the formation of nucleocapsid-like structures of approximately 50 nm in diameter that are morphologically indistinguishable from the authentic nucleocapsids inside the filamentous virions (Huang et al., 2002; Noda et al., 2006). We also showed that if the NP helices do not form, the nucleocapsid-like structures are also not formed, suggesting that formation of the NP helix is likely to be the first step in nucleocapsid assembly and that the NP helix may be the core structure of the nucleocapsid (Watanabe et al., 2006). Yet, despite the significance of the NP helix in nucleocapsid morphogenesis, little more is known about its properties.
To further characterize the NP helix, we analysed it biochemically and morphologically. Human embryonic kidney 293T cells were transfected with 5 μg of a plasmid expressing Zaire Ebolavirus NP protein (Watanabe et al., 2006) by use of Trans IT 293 reagent (Mirus Corporation). Three days post-transfection, the cells were lysed in lysis buffer [10 mM Tris/HCl (pH 7.8), 0.15 M NaCl, 1 mM EDTA, 1 % NP-40 and Protease inhibitor cocktail (Roche)], incubated at 4 °C for 30 min, and centrifuged at 20 000 g at 4 °C for 10 min to remove insoluble components. The clear supernatant was loaded onto a discontinuous 25–40 % (w/w) CsCl gradient and centrifuged at 250 000 g at 20 °C for 1 h. A visible band was collected, subjected to centrifugation at 200 000 g at 4 °C for 30 min, and the resultant pellet was then resuspended in PBS.
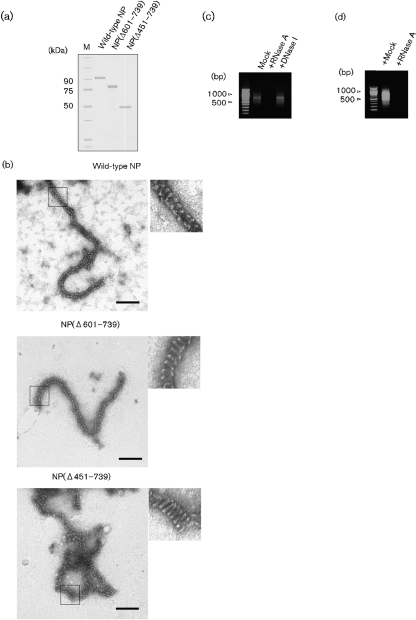
SDS-PAGE followed by Coomassie blue staining showed a single protein component with a molecular mass corresponding to that of Ebola virus NP (approx. 110 kDa; Fig. 1a), which was confirmed as NP by Western blot analysis by using rabbit anti-NP serum (data not shown). Electron microscopy (EM) by negative staining with uranyl acetate revealed serpentine-curved, helical and filamentous structures of approximately 20 nm in diameter (Fig. 1b), which were similar to those observed previously in the cytoplasm of cells expressing NP (Noda et al., 2006). Previous studies showed that removal of the C-terminal domain of paramyxovirus NP by trypsin treatment results in tightening of the helical pitch of the NP–RNA complex (Heggeness et al., 1980, 1981; Mountcastle et al., 1974). To investigate the morphological effects of the C-terminal region of Ebola virus NP on the NP helix, two C-terminal deletion mutants, NP(Δ601–739) that lacks the C-terminal aa 601–739 of NP and NP(Δ451–739) that lacks the C-terminal aa 451–739 of NP (Watanabe et al., 2006), were expressed in 293T cells and were purified as described above (Fig. 1a). NP(Δ601–739), which supports the replication of minigenomic RNA (Watanabe et al., 2006) and, therefore, presumably binds to viral RNA, formed curved helical structures of approximately 20 nm in diameter, which were similar to the wild-type NP helices, whereas NP(Δ451–739) formed more tightly coiled helices with shorter pitch (Fig. 1b). These results suggest that the hydrophilic C-terminal region (aa 451–739) of Ebola virus NP influences the conformation of the recombinant NP helix. Such morphological changes would be a common property among the NPs of non-segmented negative-sense single-strand RNA viruses (Heggeness et al., 1980, 1981; Mountcastle et al., 1974).
Fig. 1.
Characterization of the purified NP helix. (a) SDS-PAGE of the visible band isolated from the CsCl gradient. NP(Δ601–739) lacks C-terminal aa 601–739 of NP. NP(Δ451–739) lacks C-terminal aa 451–739 of NP. M, Molecular mass marker. (b) EM of negatively stained NP helices composed of wild-type NP, NP(Δ601–739) and NP(Δ451–739). Bars, 100 nm. A magnified area of the black rectangle is shown in each micrograph (b, insets). (c) Agarose gel electrophoresis of nucleic acids extracted from wild-type NP helices. The extracted nucleic acids were treated with mock, RNase A or DNase I. (d) Agarose gel electrophoresis of the RNA fraction extracted from wild-type NP helices after RNase A treatment.
To examine whether the recombinant NP helix is associated with cellular nucleic acid, as occurs with the recombinant NP helices of paramyxo- and rhabdoviruses (Bhella et al., 2002; Errington & Emmerson, 1997; Iseni et al., 1998; Moyer et al., 1991; Préhaud et al., 1990; Spehner et al., 1991), a UV absorption spectrum was taken. The absorption spectrum had a peak at 260 nm (not shown), suggesting association with cellular nucleic acid. To identify the nucleic acid in the recombinant NP helices, it was phenol/chloroform extracted from the recombinant NP helix, treated with 10 ng RNaseA μl−1, 0.2 Kunitz U DNase I μl−1 or mock treated at 37 °C for 30 min, and then analysed on a 1 % agarose gel. The nucleic acid with mock treatment ran as a smear with bands around 500–1000 nt (Fig. 1c) and was unaffected by DNase I treatment (Fig. 1c), but was digested by RNase A treatment (Fig. 1c), demonstrating that the recombinant NP helix binds to non-viral RNAs, similar to recombinant paramyxo- and rhabdovirus NPs.
To examine whether the NP-associated RNA in the NP helix is protected from RNase digestion, as occurs with paramyxoviruses (Lamb & Parks, 2007), the recombinant NP–RNA complex was treated with a high concentration of RNase A (10 ng μl−1) in PBS at 37 °C for 30 min. After excess RNase inhibitor (160 U; Promega) was added, the RNA was phenol/chloroform extracted from the complex as described above. Unlike the recombinant NP–RNA complexes of paramyxoviruses (Heggeness et al., 1980), but as has been reported for some rhabdoviruses (Iseni et al., 1998; Masters & Banerjee, 1988), the NP-associated RNA was sensitive to RNase A and was digested (Fig. 1d). The same result was obtained using micrococcal nuclease (100 U; Takara Bio) in the presence of 2 mM CaCl2 followed by chelation of the calcium ions with 5 mM EGTA prior to RNA extraction, confirming that the RNA in recombinant Ebola virus NP–RNA complexes is sensitive to environmental nucleases.
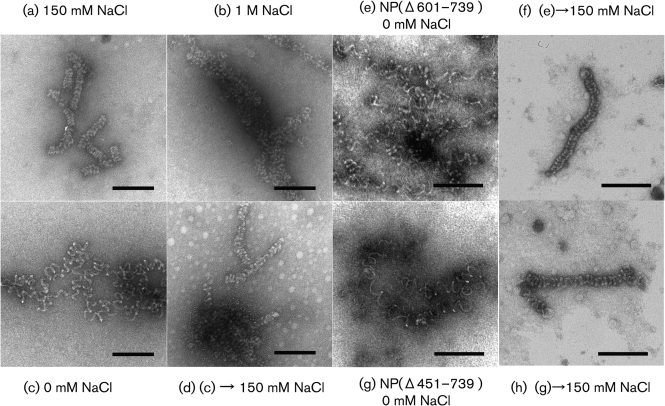
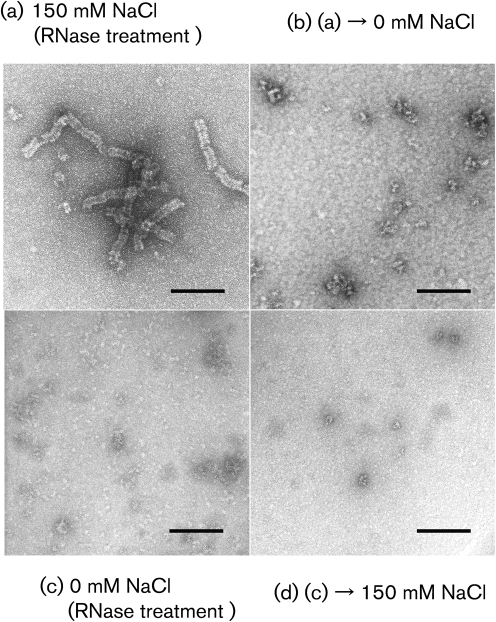
To determine whether the recombinant NP–RNA complex possesses the kind of conformational flexibility that has been observed with paramyxo- and rhabdoviruses (Heggeness et al., 1980), we next examined the effect of salt concentration on the conformation of the helix. Aliquots of purified sample were dialysed overnight at room temperature against 10 mM phosphate buffer (PB) with different NaCl concentrations (0–1000 mM), and observed by EM. In the presence of 150 mM NaCl, which most closely resembles intracellular ionic conditions, the recombinant NP–RNA complex retained its tight helical conformation (Fig. 2a). With increasing salt concentrations [500 mM (data not shown) and 1 M NaCl], the helix became mildly deformed, but it did not disassemble (Fig. 2b). By contrast, in low salt conditions (0 mM NaCl), the recombinant helical structure was extended and the helical turns became loose (Fig. 2c). Such a conformational change under low salt concentration was previously reported for authentic NP–RNA complexes of paramyxo- and rhabdoviruses (Heggeness et al., 1980), suggesting that the response to low salt is a general property of the NP–RNA complex of mononegaviruses and that hydrophobic NP–NP interactions between neighbouring helical turns are important for NP helix morphology. Interestingly, the loose coil observed under low salt conditions (Fig. 2c) reverted to a tight helix when the sample was redialysed against PBS containing 150 mM NaCl (Fig. 2d). Helical structures composed of NP(Δ601–739) and NP(Δ451–739) still possessed this conformational flexibility and reversibility (Fig. 2e–h). These results indicate that the helical NP–RNA complex inherently possesses plasticity and that the hydrophobic N-terminal 450 aa of NP are sufficient for the plasticity.
Fig. 2.
EM of purified NP–RNA complex in different salt concentrations. Wild-type NP–RNA complex was dialysed against (a) 150 mM (b) 1 M (c) 0 mM NaCl in PB. Wild-type NP–RNA complex dialysed against 0 mM NaCl was redialysed against 150 mM NaCl (d). NP(Δ601–739)–RNA complex was dialysed against 0 mM NaCl in PB (e) and was redialysed against 150 mM NaCl (f). NP(Δ451–739)–RNA complex was dialysed against 0 mM NaCl in PB (g) and was redialysed against 150 mM NaCl (h). Bars, 100 nm.
To examine the effect of RNA digestion on the conformation of the recombinant NP–RNA complex, the complex was treated with 10 ng RNase A μl−1 at 37 °C for 30 min, excess RNase inhibitor (160 U) was then added, and the sample was observed by EM. Even though the RNA was completely digested (Fig. 1d), the tight helical structure was maintained in 150 mM NaCl and showed no appreciable difference from the sample that was not RNase A treated (compare Figs 2a and 3a), indicating that the NP–NP interactions were sufficient to maintain the tight helical conformation. To examine whether the recombinant NP–RNA complex treated with RNase possessed conformational flexibility under low salt conditions, as was shown for the intact recombinant NP–RNA complex without RNase treatment (Fig. 2c), the RNA-digested helix was dialysed against 10 mM PB with no salt and then observed by EM. The results showed that the helical structure did not stretch loosely but disassembled into oligomers (Fig. 3b), suggesting that the NP-associated RNA provides a backbone and stabilizes the helix. Finally, to reveal whether the NP-associated RNA is also responsible for the reversibility, as shown in Fig. 2(d), we treated the NP–RNA complex in 10 mM PB with 10 ng RNase A μl−1. We found that the loosely extended coil in 10 mM PB (Fig. 2c) totally disassembled into oligomers and could not maintain the helical conformation (Fig. 3c), unlike the NP–RNA complex treated with RNase in the presence of 150 mM NaCl (Fig. 3a). Although this sample was subsequently subjected to dialysis against PBS containing 150 mM NaCl, the NP oligomers remained disassembled (Fig. 3d), unlike the NP–RNA complex without RNase treatment (Fig. 2d). Taken together, these results suggest that the NP-associated RNA likely confers the plasticity to the helix and is important for the helical formation, although the NP–NP interaction is sufficient to maintain the tight helical structure under normal physiological salt conditions, after the tight helix structure is formed.
Fig. 3.
EM of NP–RNA complex treated with RNase A. (a) NP–RNA complex in 150 mM NaCl in PB was treated with RNase A. (b) Sample (a) was then dialysed against 0 mM NaCl in PB. (c) NP–RNA complex in 0 mM NaCl in PB was treated with RNase A. (d) Sample (c) was then dialysed against 150 mM NaCl in PB. Bars, 100 nm.
Here, we demonstrated that recombinant Ebola virus NP forms a helical structure associated with non-viral RNAs, which are not protected from RNase digestion. The NP helix changes its conformation depending on the environmental salt concentration, if it is associated with the non-viral RNAs. As previously reported for paramyxo- and rhabdoviruses (Heggeness et al., 1980; Iseni et al., 1998), the tight helical NP–RNA structure of Ebola virus is maintained primarily by its NP–NP interaction, given that complete digestion of the residential RNA at 150 mM salt concentration does not affect the tight helical conformation (Fig. 3a). With RNase treatment in low salt, however, the coil cannot maintain its conformation and disassembles into oligomers (Fig. 3c), suggesting that the hydrophobic NP–NP interaction is essential for helix formation, while the RNA holds the NP molecules. RNA digestion experiments (Fig. 3) indicated that residential RNA, together with NP, forms the structural components of the helix and that the RNA provides plasticity for the helical complex. It is noteworthy that disassembled NPs after RNA digestion could not restore the helical structure even when the environmental salt concentration became physiological (Fig. 3d), suggesting that the RNA is likely indispensable for the formation of the helical structure, acting as a backbone to form the architecture upon which the helical structure is built up with concomitant replication of viral RNA.
In terms of conformational plasticity, both C-terminal deletion mutants tested here, NP(Δ601–739) and NP(Δ451–739), showed plasticity dependent on the environmental salt concentration (Fig. 2e–h), suggesting that the interaction of NP with RNA through its N-terminal 450 aa residues provides plasticity to the NP–RNA complex. Such plasticity represents the potential functional importance of conformational changes of authentic NP–viral RNA complex in the transcription and replication processes, as well as in the assembly process. Further analysis of the recombinant NP–RNA complex in the presence or absence of other viral components would contribute to our knowledge of the Ebola virus life cycle.
Acknowledgments
We thank Susan Watson for editing the manuscript. This work was supported by grants-in-aid from the Ministries of Education, Culture, Sports, Science, Japan, by National Institute of Allergy and Infectious Diseases Public Health Service research grants. We acknowledge membership within and support from the Region V ‘Great Lakes'.
References
- Albertini, A. A., Wernimont, A. K., Muziol, T., Ravelli, R. B., Clapier, C. R., Schoehn, G., Weissenhorn, W. & Ruigrok, R. W. (2006). Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313, 360–363. [DOI] [PubMed] [Google Scholar]
- Bhella, D., Ralph, A., Murphy, L. B. & Yeo, R. P. (2002). Significant differences in nucleocapsid morphology within the Paramyxoviridae. J Gen Virol 83, 1831–1839. [DOI] [PubMed] [Google Scholar]
- Errington, W. & Emmerson, P. T. (1997). Assembly of recombinant Newcastle disease virus nucleocapsid protein into nucleocapsid-like structures is inhibited by the phosphoprotein. J Gen Virol 78, 2335–2339. [DOI] [PubMed] [Google Scholar]
- Fooks, A. R., Schadeck, E., Liebert, U. G., Dowsett, A. B., Rima, B. K., Steward, M., Stephenson, J. R. & Wilkinson, G. W. (1995). High-level expression of the measles virus nucleocapsid protein by using a replication-deficient adenovirus vector: induction of an MHC-1-restricted CTL response and protection in a murine model. Virology 210, 456–465. [DOI] [PubMed] [Google Scholar]
- Green, T. J., Zhang, X., Wertz, G. W. & Luo, M. (2006). Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313, 357–360. [DOI] [PubMed] [Google Scholar]
- Heggeness, M. H., Scheid, A. & Choppin, P. W. (1980). Conformation of the helical nucleocapsids of paramyxoviruses and vesicular stomatitis virus: reversible coiling and uncoiling induced by changes in salt concentration. Proc Natl Acad Sci U S A 77, 2631–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heggeness, M. H., Scheid, A. & Choppin, P. W. (1981). The relationship of conformational changes in the Sendai virus nucleocapsid to proteolytic cleavage of the NP polypeptide. Virology 114, 555–562. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Xu, L. & Nabel, G. J. (2002). The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell 10, 307–316. [DOI] [PubMed] [Google Scholar]
- Iseni, F., Barge, A., Baudin, F., Blondel, D. & Ruigrok, R. W. (1998). Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J Gen Virol 79, 2909–2919. [DOI] [PubMed] [Google Scholar]
- Lamb, R. A. & Parks, G. D. (2007). Paramyxoviridae: the viruses and their replication. In Fields Virology, 5th edn, pp. 1449–1498. Edited by D. M. Knipe & P. M. Howley. Philadelphia, PA: Lippincott/Williams & Wilkins.
- Licata, J. M., Johnson, R. F., Han, Z. & Harty, R. N. (2004). Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J Virol 78, 7344–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, P. S. & Banerjee, A. K. (1988). Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J Virol 62, 2658–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis, M., Kolesnikova, L., Schoehn, G., Becker, S. & Ruigrok, R. W. (2002). Morphology of Marburg virus NP-RNA. Virology 296, 300–307. [DOI] [PubMed] [Google Scholar]
- Mountcastle, W. E., Compans, R. W., Lackland, H. & Choppin, P. W. (1974). Proteolytic cleavage of subunits of the nucleocapsid of the paramyxovirus simian virus 5. J Virol 14, 1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, S. A., Smallwood-Kentro, S., Haddad, A. & Prevec, L. (1991). Assembly and transcription of synthetic vesicular stomatitis virus nucleocapsids. J Virol 65, 2170–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Ebihara, H., Muramoto, Y., Fujii, K., Takada, A., Sagara, H., Kim, J. H., Kida, H., Feldmann, H. & Kawaoka, Y. (2006). Assembly and budding of Ebola virus. PLoS Pathog 2, e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Watanabe, S., Sagara, H. & Kawaoka, Y. (2007). Mapping of the VP40-binding regions of the nucleoprotein of Ebola virus. J Virol 81, 3554–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Préhaud, C., Harris, R. D., Fulop, V., Koh, C. L., Wong, J., Flamand, A. & Bishop, D. H. (1990). Expression, characterization, and purification of a phosphorylated rabies nucleoprotein synthesized in insect cells by baculovirus vectors. Virology 178, 486–497. [DOI] [PubMed] [Google Scholar]
- Sanchez, A., Geisbert, T. W. & Feldmann, H. (2007). Filoviridae: Marburg and Ebola viruses. In Fields Virology, 5th edn, pp. 1409–1448. Edited by D. M. Knipe & P. M. Howley. Philadelphia, PA: Lippincott/Williams & Wilkins.
- Spehner, D., Kim, A. & Drillien, R. (1991). Assembly of nucleocapsid-like structures in animal cells infected with a vaccinia virus recombinant encoding the measles virus nucleoprotein. J Virol 65, 6296–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawar, R. G., Duquerroy, S., Vonrhein, C., Varela, P. F., Damier-Piolle, L., Castagne, N., MacLellan, K., Bedouelle, H., Bricogne, G. & other authors (2009). Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326, 1279–1283. [DOI] [PubMed] [Google Scholar]
- Watanabe, S., Noda, T. & Kawaoka, Y. (2006). Functional mapping of the nucleoprotein of Ebola virus. J Virol 80, 3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]