Abstract
Localized protein synthesis is increasingly recognized as a means for polarized cells to modulate protein levels in subcellular regions and the distal reaches of their cytoplasm. The axonal and dendritic processes of neurons represent functional domains of cytoplasm that can be separated from their cell body by vast distances. This separation provides a biological setting where the cell uses locally synthesized proteins to both autonomously respond to stimuli and to retrogradely signal the cell body of events occurring is this distal environment. Other cell types undoubtedly take advantage of this localized mechanism, but these have not proven as amenable for isolation of functional subcellular domains. Consequently, neurons have provided an appealing experimental platform for study of mRNA transport and localized protein synthesis. Molecular biology approaches have shown both the population of mRNAs that can localize into axons and dendrites and an unexpectedly complex regulation of their transport into these processes. Several lines of evidence point to similar complexities and specificity for regulation of mRNA translation at subcellular sites. Proteomics studies are beginning to provide a comprehensive view of the protein constituents of subcellular domains in neurons and other cell types. However, these have currently fallen short of dissecting temporal regulation of new protein synthesis in subcellular sites and mechanisms used to ferry mRNAs to these sites.
In eukaryotes, polarized cells have evolved mechanisms for targeting macromolecules to the correct subcellular locales, allowing these cells to establish and maintain the unique domains needed for their specialized functions. Temporally altering the composition of these cellular domains allows dynamic responses to environmental stimuli, such as growth factors for proliferation and differentiation, guidance cues for migration, and peptides and neurotransmitters for cell-cell communication. Many research efforts have focused on how proteins are delivered to subcellular domains because the proteins provide much of the enzymatic and structural components needed for cellular function. Over the past three decades, transport of mRNAs to subcellular domains with subsequent localized translation has been recognized as a means to regulate local protein composition (1).
Since the first identification of localized mRNAs in ascidian eggs (2), mRNA localization has been suggested as an effective means to target proteins where and when they are needed in the cell. Given that a single mRNA can give rise to multiple copies of a protein, localized protein synthesis effectively provides a means for rapid responses to extracellular stimuli, particularly for cells with large distances separating domains. Newer functional genomics-based studies using high throughput technologies are being applied to address the extent and diversity of mRNA localization. These approaches are establishing a comprehensive view of the overall prevalence and complexity of subcellular protein synthesis. These approaches also emphasize that subcellular targeting of mRNAs is a widespread phenomenon that has far reaching implications for cellular development and function. Transport of mRNAs to subcellular sites may be the norm for most mRNAs rather than an exception seen for a surprising few.
NEURONS AS EXPERIMENTAL PLATFORM FOR ANALYSES OF SUBCELLULAR PROTEIN SYNTHESIS
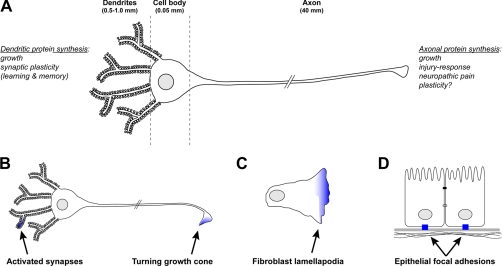
For neuronal cells, tremendous distances separate subcellular domains from the cell body where most protein synthesis was thought to occur (Fig. 1A). Dendrites, the highly branched efferent processes that receive synaptic input in the nervous system, extend for hundreds of micrometers from the neuronal perikaryon. Early ultrastructural studies in rodent hippocampus pointed to protein synthetic capacity for dendrites because ribosomes were seen concentrated at the base of dendritic spines near the synapse (3). Subsequent studies have shown that protein synthesis in dendrites contributes to the synaptic plasticity that is thought to underlie learning and memory. Indeed, neurons transport mRNAs into and locally translate mRNAs within their dendritic processes in response to transsynaptic stimuli, including various neurotransmitters and trophic factors (4). Although there were a few early examples of neuronal mRNAs in axons (5–7), the presynaptic processes that can extend for thousands to tens of thousands of micrometers from the perikaryon even in the mouse, initial ultrastructural studies did not show any ribosomes in these processes in the central nervous system (3).
Fig. 1.
Local protein translation in subcellular domains. A, the neuron is comprised of highly specialized subdomains that extend great distances from the cell body. Local protein synthesis within the dendrites plays a critical role in growth and synaptic plasticity. Within the axon, local protein synthesis is involved in growth, mediating the injury response, regeneration, and neuropathic pain and may play a role in plasticity. B, local protein synthesis allows the growth cone to respond autonomously to environmental cues. Attractive turning of the growth cone leads to local translation of mRNAs (blue). In dendrites, translation is activated by transsynaptic stimuli. C, in migrating fibroblasts, mRNAs encoding the microfilament regulating complex actin-related protein 2/3 localize to protrusions at the leading migration edge. Similarly, translation of β-actin in this region contributes to cell polarity and subsequent mobility. D, in epithelial cells, β-actin mRNA is localized to focal adhesions upon integrin binding with the plasma membrane.
Lack of ribosomes was considered a hallmark of axons, but several lines of evidence now indicate that axons of the peripheral nervous system (PNS)1 contain ribosomes and mRNAs (8). Ultrastructural evidence for ribosomes in myelinated PNS axons has been demonstrated in a few vertebrate organisms (9–13). Early studies with squid giant axon provided a relatively pure preparation of purified axonal contents to show the presence of rRNA, polyadenylated RNAs (likely mRNA), and tRNAs in these invertebrate axons (14–16). With classic biochemical techniques for fractionation of translationally active mRNAs, actively translating mRNAs were identified in purified axoplasm and in presynaptic terminals from the optic lobe of the squid (17–19). Although these findings were suggested to be a feature of incomplete polarity of these invertebrate neurons, subsequent works in embryonic and adult vertebrate neurons have shown that protein synthesis in axons contributes to both developmental and regenerative axonal growth (20).
As more has been learned of protein synthesis in these neuronal compartments, it has become clear that the neuron provides an excellent experimental platform for analyses of mRNA localization and local protein synthetic mechanisms. The geographic separation of neuronal processes from the cell body provides an experimental model where subcellular domains can be isolated to purity for molecular and biochemical studies. Here, we will summarize some of the recent advances in understanding the complexities in regulation of localized neuronal protein synthesis. We will focus on the axonal compartment here as the link of axonal protein synthesis to growth and injury responses bears similarities with other cellular systems and likely has broad applications to biology and health. However, work on protein synthesis in synapses, which has classically been attributed to the postsynaptic side of the synapse (i.e. dendritic), may have more direct implications for disease pathogenesis, and synaptic preparations have been better scrutinized by modern protein chemistry approaches.
LOCALIZED PROTEIN SYNTHESIS CONTRIBUTES TO DIRECTIONAL GROWTH OF AXONS
Studies using general protein synthesis inhibitors have pointed to a requirement for localized protein synthesis in response to some guidance cues. Developing axons are guided to their innervation targets by interaction with a variety of attractive and repulsive guidance cues (for a review, see Ref. 21). These soluble and membrane-bound guidance cues drive the turning response of the terminal axon or growth cone. This behavior of the growth cone, which can be separated from its cell body by the large distances outlined above, provides a compelling argument for the utility of local protein synthesis in axons (Fig. 1B). That is, the growth cone must respond to its environment well before any additional supply of proteins could be secured from the cell body through the known rates of anterograde transport (≤5 μm/s for vesicular transport and ≤8 mm/day for transport of most cytoskeletal elements). This theory was supported by observations that even after severing from the cell body the distal axon still responds to guidance cues by turning its growth cone toward gradients of Netrin-1 or away from gradients of Semaphorin; this turning response of severed axons is blocked by protein synthesis inhibitors (22). Other guidance cues, such as brain-derived neurotrophic factor (BDNF), Slit2, and engrailed-2, have also been shown to require local protein synthesis to elicit a turning response (22–25). Thus, localized protein synthesis can be required for the distal axon to respond to guidance cues and is autonomous from cell body-derived proteins.
Consistent with the different morphological responses of attraction versus repulsion of the growth cone, such localized protein synthetic responses can require translational activation of specific axonal mRNAs. Similar to the motile front of migrating cells, the growth cone is an actin-rich structure, and the actin cytoskeleton appears to be a point of convergence for regulating growth cone turning through localized protein synthesis. Local translation of RhoA mRNA in axons is required for response to Semaphorins (26). RhoA is a small GTPase that can trigger depolymerization of actin microfilaments. Attractive turning of growth cones in response to Netrin-1 and BDNF leads to an asymmetrical translation of β-actin mRNA in the growth cone with a focal concentration of β-actin mRNA in the same region where filamentous actin increases (27, 28). Neurotrophin 3 also increases translation and transport of β-actin mRNA in axons of cortical neurons, and blocking the axonal localization of β-actin mRNA causes growth cone retraction (29, 30). Other effectors of the actin cytoskeleton have also been shown to be locally translated, although it is not known whether they exhibit the same type of asymmetric translation. The actin-depolymerizing protein (also termed cofilin) is increased in a translation-dependent manner in the presence of the guidance cue Slit-2, which leads to growth cone collapse (24). These studies point to a common role for locally synthesized axonal proteins in modifying the actin cytoskeleton for axonal guidance with attractive cues favoring polymerization and repulsive cues favoring depolymerization. Axonal levels of actin-modifying proteins under basal conditions may also be regulated through localized protein synthesis because axonal translation of Lymnaea β-thymosin mRNA, which encodes a small protein that sequesters actin monomers, can modulate basal neurite growth (31).
Regulation of the cytoskeleton is also a feature of localized protein synthesis in non-neuronal cells. Translation of localized β-actin mRNA in migrating fibroblasts contributes to polarity of the motility of these cells (32). mRNAs encoding for the multiprotein actin-related protein 2/3 complex, a complex that regulates microfilament nucleation, similarly localize to protrusions of migrating fibroblasts (Fig. 1C) (33). Subcellular localization of several mRNAs encoding different components of the cytoskeleton and proteins that directly modify cytoskeleton has been detected in numerous cellular systems (34–38). Interestingly, β-actin mRNA is also recruited to focal adhesions upon integrin binding in epithelial cells (Fig. 1D) (39), so localized synthesis of cytoskeletal modifying proteins may be a common means to manipulate subcellular domains.
The above studies point to a level of specificity for localized translational regulation because individual stimuli can trigger local synthesis of proteins that favor depolymerization or polymerization of filamentous actin. Extracellular stimuli can also regulate the local synthesis of non-structural proteins. Cultured neurons that are exposed to constant levels of attractant (Netrin-1 and BDNF) or repellant guidance cues (Semaphorins) undergo adaptation, becoming unresponsive to these stimuli. Resensitization of the axons requires localized mRNA translation, so locally generated proteins allow the axon to regain their initial sensitivity to these stimuli (25, 40). Exactly what protein(s) mediates this process remains unknown, but it is appealing to speculate that the local synthesis of cell surface receptors could resensitize the responsiveness of the axon. There is precedence for localized synthesis of receptor proteins in axons because localized translation of EphA2 mRNA, which encodes a receptor for the ephrin guidance cues, is seen in commissural axons, making a pathfinding decision in spinal cord (41). Although the classic ultrastructurally defined rough endoplasmic reticulum and Golgi apparatus needed for co-translational secretion of proteins have not been seen in axons, recent work indicates that functional equivalents of these organelles exist in the axonal compartment (42). Thus, locally synthesized proteins can be targeted into secretory pathways. Similar outposts of rough endoplasmic reticulum and Golgi have been documented in other cellular systems, suggesting that localized synthesis of secreted and membrane proteins can contribute to responses in other biological systems (for a review, see Ref. 43). Consistent with this notion, localized synthesis of the extracellular matrix receptor component integrin α3 facilitates migration of neoplastic cells (44).
LOCALLY SYNTHESIZED PROTEINS CAN GENERATE LONG DISTANCE SIGNALING
In addition to growth cone guidance, mRNA translation in axons is known to facilitate regenerative responses, both locally in the injured axon and distally in the cell body. Intra-axonal translation and proteolysis are needed to locally initiate growth cone formation after axotomy (45). In mature PNS axons, axonally synthesized proteins generate a retrograde signaling complex that is critical for an injury response in the cell body (46). Translation products of importin β1 and RanBP1 mRNAs allow formation of an importin α/β heterodimer that retrogradely transports signaling proteins to the cell body through interaction with dynein (47, 48). A proteolytic fragment of locally translated vimentin mRNA further provides a scaffold to couple activated mitogen-activated protein kinase to importin α/β complex (49). The sum of these distally placed protein synthetic events signals the cell body that its axon has been injured and initiates a new program of gene expression to support regeneration.
Neurotransmitters have also been shown to trigger retrograde transport of importins in dendrites of cultured cortical neurons, but it is not entirely clear whether this is through localized translation of importin mRNAs (50). Synaptic plasticity-invoking stimuli were recently shown to trigger retrograde transport of CREB2, a transcriptionally repressing member of the CREB family, from distal processes through importins, but this is independent of translation (51). However, translation of another member of the CREB family (i.e.“CREB” or CREB1) was detected in axons of neonatal sympathetic neurons (52). In this system, nerve growth factor stimulates retrograde transport of the axonally synthesized CREB, which seems to be required for nerve growth factor-dependent survival in these neurons. It is quite possible that distinct proteins are needed for retrograde signaling, formation of a growth cone, and turning of a growth cone; this would imply that localized translation of different mRNAs is specifically coupled to the metabolic and functional needs of the axon.
NEURONAL PROCESSES CONTAIN UP TO HUNDREDS OF DIFFERENT mRNAs
The studies outlined above point to the importance of localized protein synthesis in axonal responses to extracellular stimuli and injury, but these analyses have largely been limited to individual mRNAs and proteins. With the ability to isolate neuronal processes to purity combined with sensitivity provided by amplifying nucleic acids, we now know that hundreds of individual mRNAs are transported into distal neuronal processes (Table I). Early studies in the squid giant axon estimated that up to 200 different polypeptides could be encoded by the localized mRNAs based on a differential RNA-DNA hybridization approach (53). Analyses of sequences in a cDNA library generated from neurites of Aplysia sensory neurons also supported this level of complexity for localized mRNAs (54). We have used a modified Boyden chamber for isolating axonal processes from cultured sensory neurons (55). By hybridization to DNA arrays, we identified a similar number of mRNAs in the axons of vertebrate sensory neurons using very conservative measures for presence calling (56). These sensory neurons are unique in that they only extend axonal processes in culture, making their isolation from cell body and non-neuronal cells much easier than in neurons with both dendrites and axons. Taylor et al. (57) developed a microfluidic culture device to effectively separate distal axons from dendrites of mammalian central nervous system neurons. Using this approach, they recently provided a sum of several hundred mRNAs in hippocampal axons (58).
Table I. Profiling of neuronal subdomain mRNA populations.
| Experimental system | Subcellular domain | Method for RNA analyses | mRNA profile |
|---|---|---|---|
| Squid giant axon | Extruded axoplasm (14) | Kinetic analyses of DNA-RNA hybrids | ∼200 different mRNAs, identity unknown |
| Sensory neurons (Aplysia) | Dissected neurites (54) | cDNA library sequencing | 198 different mRNA species |
| Sensory neurons (rodent) | Isolated axons (modified Boyden chamber) (56) | DNA array (absolute hybridization) | >200 different mRNA species (with transcripts identified) |
| Hippocampal neurons (rodent) | Isolated axonal processes (microfluidic chamber) (58) | DNA array (hybridization with advanced presence-calling algorithms) | ≥800 different mRNA species (with transcripts identified) |
| Hippocampal neurons (rodent) | Isolated dendrites (modified Boyden chamber) (110) | DNA arrays (differential hybridization, dendrite vs. cell body) | >100 mRNAs enriched in dendritic processes |
| Rodent brain | Synaptosome fractionation plus polysome sedimentations (79) | DNA arrays (differential hybridization) | ≥50 mRNAs enriched in heavy polysome fraction (likely within RNA granules) |
| Human brain (control and incipient AD) | Synaptosome fractionation from post-mortem prefrontal cortex (80) | DNA arrays (differential hybridization) | >400 mRNAs with different levels in control vs. AD at p < 0.01 |
| Fibroblasts (human) | Isolated pseudopodia (59) | DNA array (differential hybridization) | ≥50 mRNAs with enrichment upon lysophosphatidic acid vs. fibronectin treatment |
Taken together, these studies paint a picture of the neuronal process as an elaborate cellular subdomain with a complex population of mRNAs available for localized translation. Dendritic mRNA populations are likely to be similarly complex. Subdomains of other cellular types have not been as easily studied as neurons. However, a recent differential analysis of protrusions from mammalian fibroblasts showed at least 50 distinct mRNAs (59). Importantly, the local transcriptomes in both neurons and other cell types encode proteins with diverse cellular functions including metabolism and protein synthesis. This could provide these distal cellular regions with a degree of functional autonomy from the more proximal regions of the cell cytoplasm and organelles.
REGULATING SPECIFICITY OF SUBCELLULAR PROTEIN SYNTHESIS
mRNA profiles of subcellular sites have proven informative to show what proteins can be produced locally, but these studies do not address how and when translation of these mRNAs occurs. Transport of mRNAs to subcellular sites, with extracellular stimuli modifying the population of localized mRNAs, is one means to regulate specificity of translation (60). However, once localized, individual mRNAs are also subjected to translational regulation in response to specific stimuli. An example of this is seen with translation of axonal importin β1, vimentin, and RanBP1 mRNAs; the rapid appearance of these proteins in the axons with injury argues that their mRNAs are locally stored in a translationally dormant state until needed. Despite evidence for local translational regulation, the vast majority of analyses of axons and dendrites have been limited to individual proteins rather than the needed systematic approach that proteomics studies would provide.
Nonetheless, analyzing the proteome of neuronal processes has given insight into protein composition of these subdomains. Recent proteomics analysis of neurites fractionated from differentiated NIE-115 neuroblastoma cells mapped the spatial relationship of nearly 5000 proteins for neurites versus cell body (61). This study revealed networks of signaling proteins involved in neuritogenesis, cytoskeletal organization, axonal guidance, and cell polarity. Subsequent analysis of growth cones and membrane fractions from growth cones isolated from developing rat forebrain further established the proteomes of these regions of growing axons (62). Synaptosomes, an enriched subcellular fraction isolated from mature brain that contains both presynaptic and postsynaptic membrane-bound compartments, have given new insight into the synaptic proteome (63, 64). Even further interrogation of the synapse has been provided by purification of the postsynaptic density (PSD), an electron-dense postsynaptic region involved in afferent signaling (65). Several presynaptic proteins are also included in these PSD isolates, and correlation profiling has recently been used to show that scaffolding proteins and cytoskeletal proteins are highly enriched in the PSD compared with mitochondrial proteins and transporters (66). Mass spectrometry of co-immunoprecipitating synaptosome proteins has provided clues into temporal and spatial patterns of protein-protein interactions in the synapse and may also help to distinguish pre- and postsynaptic components (67). Because isolated axons, growth cones, and synaptosomes all have protein synthetic activity, at least some of the proteins identified in these preparations are generated locally. For the PSD, ribosomes are concentrated further beneath the synapse, but some of the proteins within the PSD are locally synthesized in the dendrite.
Although the studies outlined above do not differentiate between locally synthesized and cell body-synthesized proteins, they have been informative by identifying proteins that are highly enriched in these compartments. Several locally synthesized proteins have been identified in synaptosomes from squid optic lobe and in isolated axons from adult rodents through isotopic metabolic labeling, but the requirement for two-dimensional electrophoretic separation limited these studies to the most abundant locally synthesized proteins (68, 69). As noted above, both pre- and postsynaptic processes contain hundreds of different mRNAs (see Table I), so localized translation likely contributes to enrichment of many proteins in presynaptic and postsynaptic compartments, and this would not be detected by standard isotopic labeling approaches. Future integration of proteomics data with transcriptomics work from subcellular compartments will further point to the potential for localized synthesis modulating protein levels, but better methodologies are needed to enrich for locally synthesized proteins prior to mass spectrometry.
Both the synapse and growth cones are dynamic structures where protein levels and activities are rapidly altered by extracellular stimuli. Protein activity is often regulated by post-translational modifications, and phosphorylation of PSD proteins has been implicated in their function, localization, and interactions with other proteins (70). Phosphorylation can also directly modulate activity of protein synthesis machinery, including localized machinery, by activating and inactivating signaling networks that converge on translation factors and ribosomal proteins (71). Identification of phosphorylated peptides has been technically difficult given the relative low abundance of these peptides within the larger PSD proteome. Affinity chromatography has been used to enrich phosphopeptides from PSD fractions prior to mass spectrometry (72, 73), and analyzing different regions of the mouse brain has provided a unique view of protein and phosphoprotein levels in brain structures with distinct functions (74).
Together, the above and other studies are beginning to establish a comprehensive view of protein constituents and basal post-translational modifications in functional subdomains of the neuron. Quantitative mass spectrometry approaches have shown changes in synaptosome and PSD proteins and phosphoproteins in biological models of disease and injury processes including Alzheimer disease, Down syndrome, fragile X mental retardation, and Wallerian degeneration (75–78). These studies point to potential pathophysiology for neuronal subdomain dysfunction. However, they fall short of determining the dynamic regulation of proteins that may occur through changes in protein synthesis. As noted above, alterations in axonal mRNA levels have been shown to occur in response to extracellular stimulation (56), and electroconvulsive shock of rodents was also shown to change mRNA composition of synaptosomes (79). These dynamic transitions in axonal mRNA levels would have direct impact on what proteins can be produced locally.
Alterations in localized mRNA populations may have a role in disease pathogenesis. Interestingly, mRNA profiles in synaptosomes were very recently shown to be altered in the incipient stages of Alzheimer disease (AD) (80). By in vivo imaging techniques using localized reporter mRNAs, translation in dendrites was shown to be altered in AD model mice directly adjacent to amyloid plaques that are seen in AD (81). Decreased axonal transport of β-actin mRNA has also been demonstrated in spinal muscular atrophy (82). Although one can generate a compelling argument for analyses of dendritic and axonal mRNA profiles and translation in other neurological diseases, our knowledge of how and when translation is regulated is limited and cries out for more sensitive means to systematically and temporally analyze new protein synthesis. Although the stable isotope labeling by amino acids in cell culture (SILAC) technique was used to quantify synaptosome proteins in the fragile X neurons mentioned above, this required steady-state labeling so that changes in protein synthesis elicited by the absence of dendritic fragile X mental retardation protein were not addressed (77). Therefore, new labeling and enrichment methods are needed to analyze localized protein synthesis. Currently available labeling and tagging methods for mass spectrometry are summarized in Table II. Although RNA-based methods have the advantage of amplification of protein analysis, it is important to note that eukaryotes use organelle-like structures, stress granules, and processing bodies to segregate mRNAs for storage and degradation (83). Axonal importin β1, RanBP1, and vimentin are prime examples of localized mRNAs that are sequestered until needed. Thus, an increase in mRNA localization does not always equate to temporal regulation of translation (Table II).
Table II. Non-biased approaches to analyze local protein synthesis.
2D, two-dimensional; iTRAQ, isotope tags for relative and absolute quantification; BONCAT, bioorthogonal noncanonical amino acid tagging.
| Advantages | Disadvantages | |
|---|---|---|
| RNA-based analyses | ||
| Microarray hybridization (56, 58) | Widely available; can be non-biased (at least for genome-wide analyses) | Only addresses mRNA levels not translation; background hybridization signals complicate presence calling; limited to expression of specific target genes |
| Deep sequencing (“454 sequencing”) (90) | Non-biased; can allow identification of small, non-coding RNAs; eases presence calling (RNA is either there or not there) | Only addresses mRNA levels, not translation; sensitivity for subcellular domains remains undetermined; expense |
| Polysomal RNA fractionation (18, 55, 89) | Identifies translationally active mRNAs; can be applied to small samples by using microultracentrifugation | Fractionation can be difficult to normalize; cannot distinguish mRNAs with stalled ribosomes; must confirm with microarrays or 454 sequencing |
| TRAP (91, 92) | Identifies translationally active mRNAs; cell-specific mRNA profiling; potential to isolate translationally active mRNAs from in vivo preparations | Sensitivity for subcellular domains remains undetermined; cannot distinguish mRNAs with stalled ribosomes; need to generate/obtain transgenic mouse for in vivo study |
| Protein-based approaches | ||
| Classic isotopic labeling (e.g. [35S]methionine) (68, 69) | Identifies translation products from small quantities of starting materials; potential to scale up by adding more isotope | Limited to most abundant proteins; requires 2D gel-based fractionations to separate labeled proteins; poor sensitivity for small quantities |
| SILAC (77) | Quantitation of synthesized proteins; steady-state labeling useful for studying post-translational modifications (e.g. phosphopeptides) | Requires steady-state labeling (labeling period of days); limited number of samples can be compared at one time |
| Non-isotopic labeling (AHA; BONCAT) (87, 88) | Reported high level of incorporation with short labeling times (2 h); allows for affinity enrichment of labeled proteins | Sensitivity is unproven for subcellular samples; no quantitation possible unless coupled with additional methods (e.g. iTRAQ) |
| ICAT (65, 66) | Allows for relative quantitation; useful for less complex samples (i.e. subdomains) and small amount of material | Cannot distinguish between locally and non-locally synthesized proteins; targets only cysteine-containing peptides leading to under-representation of some proteins |
| iTRAQ (74, 111) | Relative quantitation of protein levels between samples; multiplex analyses (8-plex); affinity enrichment of tagged proteins | Cannot distinguish between locally and non-locally synthesized proteins; every sample must be screened by tandem MS, increasing analysis time and amount of sample needed |
METHODS FOR UNBIASED ANALYSES OF LOCALIZED TRANSLATIONAL REGULATION
Local protein synthesis in intact and severed axons has largely been detected through incorporation of radioactively labeled amino acids. Isotopic labeling with [35S]methionine has been used to demonstrate stimulus-dependent changes in protein synthesis in axons (84). However, systematic proteomics analysis of locally synthesized axonal proteins has been an unmet challenge considering the excessively limited amounts of proteins synthesized locally. Metabolic labeling of axons indicated that ∼5% of total cellular protein synthesis occurs in these distal processes (85). Thus, at least 95% of protein isolated from neuronal processes and other subcellular compartments has been transported into these processes rather than synthesized de novo. In our hands, isolated axons only remain visibly intact for about 6 h after severing from the cell body (55). Thus, this relatively meager fraction of protein is further limited by the survival of isolated subcellular domains. Considering that newly synthesized proteins can only be labeled over a time period of a few hours and they represent only a small fraction of the proteins present in the processes, proteomics approaches for identifying and quantifying locally synthesized proteins will require significant enrichment similar to the approaches used for phosphoproteomes. A newly developed methionine analog, azido-homoalanine (AHA), may prove amenable to this because AHA-labeled proteins can be covalently tagged with epitopes for purification through a copper-catalyzed azide-alkyne ligation (86). Dieterich et al. (87) found that a 1–2-h labeling period generates sufficient protein for affinity purification and mass spectrometry using non-fractionated cells (i.e. whole cell lysates rather than subcellular domains). AHA has recently been used with similar labeling coupled with fluorescent tagging to visualize protein synthesis in axonal growth cones (88). Nonetheless, RNA-based approaches remain appealing with the ability to exponentially amplify signals through RT-PCR.
Translational activity of individual mRNAs can be addressed through testing for their association with ribosomes. This classic ultracentrifugation technique, or polysome fractionation, has been used to show translation of mRNAs in squid axoplasm, synaptosome preparations, and regenerating axons by showing that the transcripts are associated with ribosomes (19, 55, 89). Zheng et al. (55) further used an RNA co-immunoprecipitation (RIP) by using the Y10B monoclonal antibody to 28 S rRNA and axonal lysates prepared under conditions that maintain ribosome subunit interactions. Dissociation of 80 S ribosome particles by application of EDTA or puromycin prior to fractionation by centrifugation of RIP provides a control for these analyses. Combining polysome fractionation techniques with RT-PCR and DNA microarray or deep sequencing technologies could be used to quantitatively address the relative translation state of localized mRNAs in cell culture preparations. The deep sequencing approach was recently used to quantify translation of mRNAs in Saccharomyces cerevisiae by comparing ribosome bound to total mRNA isolates (90).
A limitation for much of the mRNA profiling work outlined above is that it is limited to preparations of cultured neurons. A recently developed method, termed “targeted translating ribosome affinity purification” (TRAP) (91, 92), holds promise for quantifying mRNA-polysome association in subcellular domains from complex in vivo preparations. Heintz and co-workers (91, 92) developed transgenic mice that express enhanced GFP-tagged ribosomal protein L10a (GFP-L10a) using bacterial artificial chromosomes containing regulatory elements for gene expression to provide transgene expression that is remarkably restricted to cell types and subtypes. For example, neuronal subtype-specific expression was achieved for pyramidal neuron subtypes in different layers of the cerebral cortex, regions that could never be completely separated by manual dissection or culture. Taking advantage of the GFP-tagged ribosomes, cell type-specific ribosome-mRNA complexes could be immunoprecipitated from whole brain lysates using anti-GFP antibodies. With amplification of the mRNAs followed by DNA microarray hybridization, a translational profile of individual cell populations can be determined (91, 92).
Combining TRAP with established and developing techniques for isolating the axonal subdomain would be a powerful tool for profiling the population of translationally active axonal mRNAs. Recently developed methods for isolation of axoplasm from peripheral nerve, where axons are widely separated from cell body and other neurons, offer a particularly appealing approach for quantifying in vivo axonal mRNA translation (93). A drawback for both TRAP and the Y10B RIP approaches is that a fraction of the purified mRNA could represent transcripts with stalled ribosomes. However, this should be a relatively minor fraction that may be separated by their relative levels compared with other mRNAs. Additionally, the deep sequencing approach brings the potential for “footprinting” mRNA sequences by identifying sites that are protected from nuclease digestion through binding by protein or other nuclease-resistant macromolecules. Ingolia et al. (90) successfully used this approach to gauge translational efficiency for yeast mRNAs, and translational efficiency can be extrapolated to give an indication of true polysome association of the mRNAs.
MECHANISTIC INSIGHTS INTO mRNA TRANSPORT
mRNAs are actively transported to subcellular sites as RNA-protein complexes, or RNPs, that interact with motor proteins (94). For example, subcellular localization of β-actin mRNA requires binding of zip code-binding protein 1 (ZBP1) to its 3′-untranslated region localization element (zip code) (95). Studies in Xenopus oocytes argue that initial mRNA-protein interactions for localizing mRNAs occur in the nucleus with remodeling of the transport RNP as it enters the cytoplasm and then engages with motor proteins (96). Addition and exchange of protein constituents are the basis of this remodeling. Such remodeling has also been seen for mammalian β-actin mRNA where the transcript is initially bound by the nuclear protein KH-type splicing regulatory protein (termed ZBP2 in chick); this initial binding by KH-type splicing regulatory protein provides cooperative binding for ZBP1 that is needed for subcellular localization of β-actin mRNA (97). Interestingly, ZBP1 serves as a translational suppressor while bound to mRNA; affinity of ZBP1 for mRNA is decreased by Src kinase-dependent phosphorylation, resulting in translational derepression of β-actin mRNA (98). Translational repression within transport RNPs is a now a common theme in RNA localization with post-translational modifications frequently serving as the basis to alter RNA-binding protein activity (99).
Despite remarkably increased knowledge of the subcellular transcriptome, a huge gap in knowledge remains when it comes to proteins responsible for their transport. Attempts to isolate transport RNPs from whole cell lysates have generated new insight into composition of these complexes through proteomics and ribonomics studies (100–102). Cell biology-based approaches show that these isolates likely contain multiple different RNP populations when individual neuronal processes are queried (103). Thus, how specificity is achieved for mRNA transport still remains to be solved. Applying proteomics to RNPs from subcellular regions, such as axons or dendrites, will help to enrich for transported RNPs and potentially provide a more pure population for study. The yield of proteins from isolated axonal and dendritic RNPs will be even more challenging than the RNA profiling studies, but the increasing sensitivity of proteomics instrumentation offers hope for meeting this challenge in the near future.
Studies of single RNA-binding proteins have shown that most can bind to a surprisingly large number of different mRNAs. Immunoprecipitates of the human ortholog of ZBP1, termed IMP1 (insulin-like protein 2 mRNA-binding protein), were shown to contain >350 different mRNAs in HEK293 cells (104). Similarly, for the neuronal RNA binding protein HuD, that recognizes AU-rich elements and stabilizes bound mRNAs, several hundred mRNA targets were identified by array profiling (105). It is not always clear whether such interactions are due to direct protein recognition of RNA motifs, protein-protein interactions, or nonspecific binding from in vitro preparations. Addition of in vivo cross-linking to RIP followed by nuclease digestion (i.e. cross-linking with co-immunoprecipitation (106)) has advantages of avoiding both protein-protein interactions and nonspecific RNA-protein interactions that can occur in cell lysates (107). Deep sequencing of RNAs isolated through cross-linking and immunoprecipitation brings the potential to identify RNA motifs that are directly targeted by RNA-binding proteins because the protected regions are indicative of protein binding sites (108, 109). Considering that many RNA motifs for localization are determined by secondary structure that have proven difficult to analyze through bioinformatics approaches (for a review, see Ref. 60), large scale analyses of RNA-protein interaction sites through cross-linking and immunoprecipitation will likely uncover functional RNA motifs that have been unrecognized to date. It is not clear whether these RNA co-immunoprecipitation approaches are of sufficient sensitivity to distinguish where the mRNA-protein interactions occur within the cell; i.e. will they be of sufficient sensitivity for the low amounts of protein and RNA isolated from subcellular compartments like axons and dendrites? Additionally, proteins indirectly binding to mRNAs through protein-protein interactions likely contribute to specificity for when individual mRNAs are transported. So for dissecting the transported RNP composition, both the target mRNAs and the protein constituents will need to be considered; this will require seeking new means to integrate high sensitivity ribonomics and proteomics data sets.
CONCLUSIONS
Polarized cells spatially regulate mRNA translation by delivering transcripts and protein synthesis machinery to subcellular regions. Molecular biology approaches have been used to dissect this spatial regulation by providing profiles of localized mRNAs, and cell biology approaches have been used to both detect localized synthetic machinery and show its functionality. These same approaches indicate that localized protein synthesis is temporally regulated both in neurons and other cell types. A key question for understanding the biology of localized protein synthesis is how specificity is achieved. Several lines of evidence point to specific translation of different mRNAs in response different stimuli. Cell biology and biochemical approaches thus far have been limited to the study of individual proteins or mRNAs. Enhanced sensitivity of mass spectrometry, new approaches to enrich metabolically labeled proteins, and continued effort by cell biologists to isolate subcellular domains should provide means to systematically profile localized protein synthesis and mRNA transport (see Table II). This will be key in any future approaches to therapeutically modulate axonal and dendritic protein synthesis. The roles of axonal protein synthesis in regeneration and dendritic (and possibly axonal) protein synthesis in neuronal function are appealing targets for intervention. Understanding how specificity is achieved in neuronal processes may also lead to new developments for general cell migration where translational control can have biomedical implications for metastasis of cancer cells (44).
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants K00-NR010797 (to D. E. W.), R01-NS041596 (to J. L. T.), and R01-NS049041 (to J. L. T.).
1 The abbreviations used are:
- PNS
- peripheral nervous system
- AD
- Alzheimer disease
- AHA
- azido-homoalanine
- BDNF
- brain-derived neurotrophic factor
- PSD
- postsynaptic density
- RIP
- RNA co-immunoprecipitation
- RNP
- ribonucleoprotein particle
- SILAC
- stable isotope labeling by amino acids in cell culture
- TRAP
- targeted translating ribosome affinity purification
- ZBP1
- zip code-binding protein 1
- CREB
- cAMP-response element-binding protein
- GFP
- green fluorescent protein.
REFERENCES
- 1.Martin K. C., Ephrussi A. (2009) mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeffery W. R., Tomlinson C. R., Brodeur R. D. (1983) Localization of actin messenger RNA during early ascidian development. Dev. Biol 99, 408–417 [DOI] [PubMed] [Google Scholar]
- 3.Steward O., Levy W. B. (1982) Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci 2, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bramham C. R., Wells D. G. (2007) Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci 8, 776–789 [DOI] [PubMed] [Google Scholar]
- 5.Skutella T., Probst J. C., Blanco E., Jirikowski G. F. (1994) Localization of tyrosine hydroxylase mRNA in the axons of the hypothalamo-neurohypophysial system. Brain Res. Mol. Brain Res 23, 179–184 [DOI] [PubMed] [Google Scholar]
- 6.Svane P. C., Thorn N. A., Richter D., Mohr E. (1995) Effect of hypoosmolality on the abundance, poly(A) tail length and axonal targeting of arginine vasopressin and oxytocin mRNAs in rat hypothalamic magnocellular neurons. FEBS Lett 373, 35–38 [DOI] [PubMed] [Google Scholar]
- 7.Wensley C. H., Stone D. M., Baker H., Kauer J. S., Margolis F. L., Chikaraishi D. M. (1995) Olfactory marker protein mRNA is found in axons of olfactory receptor neurons. J. Neurosci 15, 4827–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Twiss J. L., Fainzilber M. (2009) Ribosomes in axons—scrounging from the neighbors? Trends Cell Biol 19, 236–243 [DOI] [PubMed] [Google Scholar]
- 9.Koenig E., Martin R., Titmus M., Sotelo-Silveira J. R. (2000) Cryptic peripheral ribosomal domains distributed intermittently along mammalian myelinated axons. J. Neurosci 20, 8390–8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koenig E., Martin R. (1996) Cortical plaque-like structures identify ribosome-containing domains in the Mauthner cell axon. J. Neurosci 16, 1400–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pannese E., Ledda M. (1991) Ribosomes in myelinated axons of the rabbit spinal ganglion neurons. J. Submicrosc. Cytol. Pathol 23, 33–38 [PubMed] [Google Scholar]
- 12.Zelená J. (1972) Ribosomes in myelinated axons of dorsal root ganglia. Z. Zellforsch. Mikrosk. Anat 124, 217–229 [DOI] [PubMed] [Google Scholar]
- 13.Zelená J. (1970) Ribosome-like particles in myelinated axons of the rat. Brain Res 24, 359–363 [DOI] [PubMed] [Google Scholar]
- 14.Capano C. P., Giuditta A., Castigli E., Kaplan B. B. (1987) Occurrence and sequence complexity of polyadenylated RNA in squid axoplasm. J. Neurochem 49, 698–704 [DOI] [PubMed] [Google Scholar]
- 15.Giuditta A., Cupello A., Lazzarini G. (1980) Ribosomal RNA in axoplasm of squid giant axon. J. Neurochem 34, 1757–1760 [DOI] [PubMed] [Google Scholar]
- 16.Black M. M., Lasek R. J. (1977) The presence of transfer RNA in the axoplasm of the squid giant axon. J. Neurobiol 8, 229–237 [DOI] [PubMed] [Google Scholar]
- 17.Crispino M., Capano C. P., Kaplan B. B., Giuditta A. (1993) Neurofilament proteins are synthesized in nerve endings in squid brain. J. Neurochem 61, 1144–1146 [DOI] [PubMed] [Google Scholar]
- 18.Crispino M., Castigli E., Perrone Capano C., Martin R., Menichini E., Kaplan B. B., Giuditta A. (1993) Protein synthesis in a synaptosomal fraction from squid brain. Mol. Cell. Neurosci 4, 366–374 [DOI] [PubMed] [Google Scholar]
- 19.Crispino M., Kaplan B. B., Martin R., Alvarez J., Chun J. T., Benech J. C., Giuditta A. (1997) Active polysomes are present in the large presynaptic endings of the synaptosomal fraction from squid brain. J. Neurosci 17, 7694–7702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piper M., Holt C. (2004) RNA translation in axons. Annu. Rev. Cell Dev. Biol 20, 505–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson B. J., Senti K. A. (2002) Axon guidance: growth cones make an unexpected turn. Curr. Biol 12, R218–220 [DOI] [PubMed] [Google Scholar]
- 22.Campbell D. S., Holt C. E. (2001) Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, 1013–1026 [DOI] [PubMed] [Google Scholar]
- 23.Brunet I., Weinl C., Piper M., Trembleau A., Volovitch M., Harris W., Prochiantz A., Holt C. (2005) The transcription factor Engrailed-2 guides retinal axons. Nature 438, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piper M., Anderson R., Dwivedy A., Weinl C., van Horck F., Leung K. M., Cogill E., Holt C. (2006) Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron 49, 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ming G. L., Wong S. T., Henley J., Yuan X. B., Song H. J., Spitzer N. C., Poo M. M. (2002) Adaptation in the chemotactic guidance of nerve growth cones. Nature 417, 411–418 [DOI] [PubMed] [Google Scholar]
- 26.Wu K. Y., Hengst U., Cox L. J., Macosko E. Z., Jeromin A., Urquhart E. R., Jaffrey S. R. (2005) Local translation of RhoA regulates growth cone collapse. Nature 436, 1020–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao J., Sasaki Y., Wen Z., Bassell G. J., Zheng J. Q. (2006) An essential role for beta-actin mRNA localization and translation in Ca(2+)-dependent growth cone guidance. Nat. Neurosci 9, 1265–1273 [DOI] [PubMed] [Google Scholar]
- 28.Leung K. M., van Horck F. P., Lin A. C., Allison R., Standart N., Holt C. E. (2006) Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci 9, 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H. L., Eom T., Oleynikov Y., Shenoy S. M., Liebelt D. A., Dictenberg J. B., Singer R. H., Bassell G. J. (2001) Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron 31, 261–275 [DOI] [PubMed] [Google Scholar]
- 30.Zhang H. L., Singer R. H., Bassell G. J. (1999) Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J. Cell Biol 147, 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Kesteren R. E., Carter C., Dissel H. M., van Minnen J., Gouwenberg Y., Syed N. I., Spencer G. E., Smit A. B. (2006) Local synthesis of actin-binding protein beta-thymosin regulates neurite outgrowth. J. Neurosci 26, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shestakova E. A., Singer R. H., Condeelis J. (2001) The physiological significance of beta-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. U.S.A 98, 7045–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mingle L. A., Okuhama N. N., Shi J., Singer R. H., Condeelis J., Liu G. (2005) Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci 118, 2425–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang L., Shav-Tal Y., Trcek T., Singer R. H., Goldman R. D. (2006) Assembling an intermediate filament network by dynamic cotranslation. J. Cell Biol 172, 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris E. J., Fulton A. B. (1994) Rearrangement of mRNAs for costamere proteins during costamere development in cultured skeletal muscle from chicken. J. Cell Sci 107, 377–386 [DOI] [PubMed] [Google Scholar]
- 36.Fulton A. B. (1993) Spatial organization of the synthesis of cytoskeletal proteins. J. Cell. Biochem 52, 148–152 [DOI] [PubMed] [Google Scholar]
- 37.Cripe L., Morris E., Fulton A. B. (1993) Vimentin mRNA location changes during muscle development. Proc. Natl. Acad. Sci. U.S.A 90, 2724–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hannan A. J., Schevzov G., Gunning P., Jeffrey P. L., Weinberger R. P. (1995) Intracellular localization of tropomyosin mRNA and protein is associated with development of neuronal polarity. Mol. Cell. Neurosci 6, 397–412 [DOI] [PubMed] [Google Scholar]
- 39.Chicurel M. E., Singer R. H., Meyer C. J., Ingber D. E. (1998) Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature 392, 730–733 [DOI] [PubMed] [Google Scholar]
- 40.Piper M., Salih S., Weinl C., Holt C. E., Harris W. A. (2005) Endocytosis dependent desensitization and protein synthesis-dependent resensitization in retinal growth cone adaptation. Nat. Neurosci 8, 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brittis P. A., Lu Q., Flanagan J. G. (2002) Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell 110, 223–235 [DOI] [PubMed] [Google Scholar]
- 42.Merianda T. T., Lin A. C., Lam J. S., Vuppalanchi D., Willis D. E., Karin N., Holt C. E., Twiss J. L. (2009) A functional equivalent of endoplasmic reticulum and Golgi in axons for secretion of locally synthesized proteins. Mol. Cell. Neurosci 40, 128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerst J. E. (2008) Message on the web: mRNA and ER co-trafficking. Trends Cell Biol 18, 68–76 [DOI] [PubMed] [Google Scholar]
- 44.Adereth Y., Dammai V., Kose N., Li R., Hsu T. (2005) RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat. Cell Biol 7, 1240–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma P., Chierzi S., Codd A. M., Campbell D. S., Meyer R. L., Holt C. E., Fawcett J. W. (2005) Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J. Neurosci 25, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rishal I., Fainzilber M. (2010) Retrograde signaling in axonal regeneration. Exp. Neurol, in press [DOI] [PubMed] [Google Scholar]
- 47.Hanz S., Perlson E., Willis D., Zheng J. Q., Massarwa R., Huerta J. J., Koltzenburg M., Kohler M., van-Minnen J., Twiss J. L., Fainzilber M. (2003) Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron 40, 1095–1104 [DOI] [PubMed] [Google Scholar]
- 48.Yudin D., Hanz S., Yoo S., Iavnilovitch E., Willis D., Gradus T., Vuppalanchi D., Segal-Ruder Y., Ben-Yaakov K., Hieda M., Yoneda Y., Twiss J. L., Fainzilber M. (2008) Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron 59, 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perlson E., Hanz S., Ben-Yaakov K., Segal-Ruder Y., Seger R., Fainzilber M. (2005) Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron 45, 715–726 [DOI] [PubMed] [Google Scholar]
- 50.Thompson K. R., Otis K. O., Chen D. Y., Zhao Y., O'Dell T. J., Martin K. C. (2004) Synapse to nucleus signaling during long-term synaptic plasticity; a role for the classical active nuclear import pathway. Neuron 44, 997–1009 [DOI] [PubMed] [Google Scholar]
- 51.Lai K. O., Zhao Y., Ch'ng T. H., Martin K. C. (2008) Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc. Natl. Acad. Sci. U.S.A 105, 17175–17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox L. J., Hengst U., Gurskaya N. G., Lukyanov K. A., Jaffrey S. R. (2008) Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat. Cell Biol 10, 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capano C. P., Gioio A. E., Giuditta A., Kaplan B. B. (1986) Complexity of nuclear and polysomal RNA from squid optic lobe and gill. J. Neurochem 46, 1517–1521 [DOI] [PubMed] [Google Scholar]
- 54.Moccia R., Chen D., Lyles V., Kapuya E., E Y., Kalachikov S., Spahn C. M., Frank J., Kandel E. R., Barad M., Martin K. C. (2003) An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J. Neurosci 23, 9409–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng J. Q., Kelly T. K., Chang B., Ryazantsev S., Rajasekaran A. K., Martin K. C., Twiss J. L. (2001) A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J. Neurosci 21, 9291–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willis D. E., van Niekerk E. A., Sasaki Y., Mesngon M., Merianda T. T., Williams G. G., Kendall M., Smith D. S., Bassell G. J., Twiss J. L. (2007) Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J. Cell Biol 178, 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor A. M., Blurton-Jones M., Rhee S. W., Cribbs D. H., Cotman C. W., Jeon N. L. (2005) A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods 2, 599–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor A. M., Berchtold N. C., Perreau V. M., Tu C. H., Li Jeon N., Cotman C. W. (2009) Axonal mRNA in uninjured and regenerating cortical mammalian axons. J. Neurosci 29, 4697–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mili S., Moissoglu K., Macara I. G. (2008) Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453, 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vuppalanchi D., Willis D. E., Twiss J. L. (2009) Regulation of mRNA transport and translation in axons. Results Probl. Cell Differ 48, 193–224 [DOI] [PubMed] [Google Scholar]
- 61.Pertz O. C., Wang Y., Yang F., Wang W., Gay L. J., Gristenko M. A., Clauss T. R., Anderson D. J., Liu T., Auberry K. J., Camp D. G., 2nd, Smith R. D., Klemke R. L. (2008) Spatial mapping of the neurite and soma proteomes reveals a functional Cdc42/Rac regulatory network. Proc. Natl. Acad. Sci. U.S.A 105, 1931–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nozumi M., Togano T., Takahashi-Niki K., Lu J., Honda A., Taoka M., Shinkawa T., Koga H., Takeuchi K., Isobe T., Igarashi M. (2009) Identification of functional marker proteins in the mammalian growth cone. Proc. Natl. Acad. Sci. U.S.A 106, 17211–17216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schrimpf S. P., Meskenaite V., Brunner E., Rutishauser D., Walther P., Eng J., Aebersold R., Sonderegger P. (2005) Proteomic analysis of synaptosomes using isotope-coded affinity tags and mass spectrometry. Proteomics 5, 2531–2541 [DOI] [PubMed] [Google Scholar]
- 64.Witzmann F. A., Arnold R. J., Bai F., Hrncirova P., Kimpel M. W., Mechref Y. S., McBride W. J., Novotny M. V., Pedrick N. M., Ringham H. N., Simon J. R. (2005) A proteomic survey of rat cerebral cortical synaptosomes. Proteomics 5, 2177–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li K. W., Hornshaw M. P., Van Der Schors R. C., Watson R., Tate S., Casetta B., Jimenez C. R., Gouwenberg Y., Gundelfinger E. D., Smalla K. H., Smit A. B. (2004) Proteomics analysis of rat brain postsynaptic density: implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem 279, 987–1002 [DOI] [PubMed] [Google Scholar]
- 66.Li K., Hornshaw M. P., van Minnen J., Smalla K. H., Gundelfinger E. D., Smit A. B. (2005) Organelle proteomics of rat synaptic proteins: correlation-profiling by isotope-coded affinity tagging in conjunction with liquid chromatography-tandem mass spectrometry to reveal post-synaptic density specific proteins. J. Proteome Res 4, 725–733 [DOI] [PubMed] [Google Scholar]
- 67.Klemmer P., Smit A. B., Li K. W. (2009) Proteomics analysis of immuno-precipitated synaptic protein complexes. J. Proteomics 72, 82–90 [DOI] [PubMed] [Google Scholar]
- 68.Jiménez C. R., Eyman M., Lavina Z. S., Gioio A., Li K. W., van der Schors R. C., Geraerts W. P., Giuditta A., Kaplan B. B., van Minnen J. (2002) Protein synthesis in synaptosomes: a proteomics analysis. J. Neurochem 81, 735–744 [DOI] [PubMed] [Google Scholar]
- 69.Willis D., Li K. W., Zheng J. Q., Chang J. H., Smit A., Kelly T., Merianda T. T., Sylvester J., van Minnen J., Twiss J. L. (2005) Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J. Neurosci 25, 778–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamauchi T. (2002) Molecular constituents and phosphorylation-dependent regulation of the post-synaptic density. Mass Spectrom. Rev 21, 266–286 [DOI] [PubMed] [Google Scholar]
- 71.Proud C. G. (2007) Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem. J 403, 217–234 [DOI] [PubMed] [Google Scholar]
- 72.Trinidad J. C., Thalhammer A., Specht C. G., Schoepfer R., Burlingame A. L. (2005) Phosphorylation state of postsynaptic density proteins. J. Neurochem 92, 1306–1316 [DOI] [PubMed] [Google Scholar]
- 73.Trinidad J. C., Specht C. G., Thalhammer A., Schoepfer R., Burlingame A. L. (2006) Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics 5, 914–922 [DOI] [PubMed] [Google Scholar]
- 74.Trinidad J. C., Thalhammer A., Specht C. G., Lynn A. J., Baker P. R., Schoepfer R., Burlingame A. L. (2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684–696 [DOI] [PubMed] [Google Scholar]
- 75.Gillardon F., Rist W., Kussmaul L., Vogel J., Berg M., Danzer K., Kraut N., Hengerer B. (2007) Proteomic and functional alterations in brain mitochondria from Tg2576 mice occur before amyloid plaque deposition. Proteomics 7, 605–616 [DOI] [PubMed] [Google Scholar]
- 76.Fernandez F., Trinidad J. C., Blank M., Feng D. D., Burlingame A. L., Garner C. C. (2009) Normal protein composition of synapses in Ts65Dn mice: a mouse model of Down syndrome. J. Neurochem 110, 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liao L., Park S. K., Xu T., Vanderklish P., Yates J. R., 3rd. (2008) Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice. Proc. Natl. Acad. Sci. U.S.A 105, 15281–15286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wishart T. M., Paterson J. M., Short D. M., Meredith S., Robertson K. A., Sutherland C., Cousin M. A., Dutia M. B., Gillingwater T. H. (2007) Differential proteomics analysis of synaptic proteins identifies potential cellular targets and protein mediators of synaptic neuroprotection conferred by the slow Wallerian degeneration (Wlds) gene. Mol. Cell. Proteomics 6, 1318–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsumoto M., Setou M., Inokuchi K. (2007) Transcriptome analysis reveals the population of dendritic RNAs and their redistribution by neural activity. Neurosci. Res 57, 411–423 [DOI] [PubMed] [Google Scholar]
- 80.Williams C., Mehrian Shai R., Wu Y., Hsu Y. H., Sitzer T., Spann B., McCleary C., Mo Y., Miller C. A. (2009) Transcriptome analysis of synaptoneurosomes identifies neuroplasticity genes overexpressed in incipient Alzheimer's disease. PLoS One 4, e4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyer-Luehmann M., Mielke M., Spires-Jones T. L., Stoothoff W., Jones P., Bacskai B. J., Hyman B. T. (2009) A reporter of local dendritic translocation shows plaque-related loss of neural system function in APP-transgenic mice. J. Neurosci 29, 12636–12640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rossoll W., Jablonka S., Andreassi C., Kröning A. K., Karle K., Monani U. R., Sendtner M. (2003) Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol 163, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balagopal V., Parker R. (2009) Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol 21, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang W., van Niekerk E., Willis D. E., Twiss J. L. (2007) RNA transport and localized protein synthesis in neurological disorders and neural repair. Dev. Neurobiol 67, 1166–1182 [DOI] [PubMed] [Google Scholar]
- 85.Lee S. K., Hollenbeck P. J. (2003) Organization and translation of mRNA in sympathetic axons. J. Cell Sci 116, 4467–4478 [DOI] [PubMed] [Google Scholar]
- 86.Rostovtsev V. V., Green L. G., Fokin V. V., Sharpless K. B. (2002) A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl 41, 2596–2599 [DOI] [PubMed] [Google Scholar]
- 87.Dieterich D. C., Lee J. J., Link A. J., Graumann J., Tirrell D. A., Schuman E. M. (2007) Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc 2, 532–540 [DOI] [PubMed] [Google Scholar]
- 88.Roche F. K., Marsick B. M., Letourneau P. C. (2009) Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J. Neurosci 29, 638–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weiler I. J., Greenough W. T. (1993) Metabotropic glutamate receptors trigger postsynaptic protein synthesis. Proc. Natl. Acad. Sci. U.S.A 90, 7168–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heiman M., Schaefer A., Gong S., Peterson J. D., Day M., Ramsey K. E., Suárez-Fariñas M., Schwarz C., Stephan D. A., Surmeier D. J., Greengard P., Heintz N. (2008) A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Doyle J. P., Dougherty J. D., Heiman M., Schmidt E. F., Stevens T. R., Ma G., Bupp S., Shrestha P., Shah R. D., Doughty M. L., Gong S., Greengard P., Heintz N. (2008) Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rishal I., Michaelevski I., Rozenbaum M., Shinder V., Medzihradszky K. F., Burlingame A. L., Fainzilber M. (2010) Axoplasm isolation from peripheral nerve. Dev. Neurobiol 70, 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kiebler M. A., Bassell G. J. (2006) Neuronal RNA granules: movers and makers. Neuron 51, 685–690 [DOI] [PubMed] [Google Scholar]
- 95.Farina K. L., Huttelmaier S., Musunuru K., Darnell R., Singer R. H. (2003) Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol 160, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lewis R. A., Mowry K. L. (2007) Ribonucleoprotein remodeling during RNA localization. Differentiation 75, 507–518 [DOI] [PubMed] [Google Scholar]
- 97.Pan F., Hüttelmaier S., Singer R. H., Gu W. (2007) ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol. Cell. Biol 27, 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hüttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G. J., Condeelis J., Singer R. H. (2005) Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–515 [DOI] [PubMed] [Google Scholar]
- 99.Wells D. G. (2006) RNA-binding proteins: a lesson in repression. J. Neurosci 26, 7135–7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Elvira G., Wasiak S., Blandford V., Tong X. K., Serrano A., Fan X., del Rayo Sánchez-Carbente M., Servant F., Bell A. W., Boismenu D., Lacaille J. C., McPherson P. S., DesGroseillers L., Sossin W. S. (2006) Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics 5, 635–651 [DOI] [PubMed] [Google Scholar]
- 101.Krichevsky A. M., Kosik K. S. (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32, 683–696 [DOI] [PubMed] [Google Scholar]
- 102.Kanai Y., Dohmae N., Hirokawa N. (2004) Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 [DOI] [PubMed] [Google Scholar]
- 103.Miller L. C., Blandford V., McAdam R., Sanchez-Carbente M. R., Badeaux F., DesGroseillers L., Sossin W. S. (2009) Combinations of DEAD box proteins distinguish distinct types of RNA:protein complexes in neurons. Mol. Cell. Neurosci 40, 485–495 [DOI] [PubMed] [Google Scholar]
- 104.Jønson L., Vikesaa J., Krogh A., Nielsen L. K., Hansen T., Borup R., Johnsen A. H., Christiansen J., Nielsen F. C. (2007) Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell. Proteomics 6, 798–811 [DOI] [PubMed] [Google Scholar]
- 105.Bolognani F., Contente-Cuomo T., Perrone-Bizzozero N. I. (2010) Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res 38, 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ho S., Clipstone N., Timmermann L., Northrop J., Graef I., Fiorentino D., Nourse J., Crabtree G. R. (1996) The mechanism of action of cyclosporin A and FK506. Clin. Immunol. Immunopathol 80, S40–S45 [DOI] [PubMed] [Google Scholar]
- 107.Jensen K. B., Darnell R. B. (2008) CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol. Biol 488, 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Licatalosi D. D., Mele A., Fak J. J., Ule J., Kayikci M., Chi S. W., Clark T. A., Schweitzer A. C., Blume J. E., Wang X., Darnell J. C., Darnell R. B. (2008) HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chi S. W., Zang J. B., Mele A., Darnell R. B. (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Poon M. M., Choi S. H., Jamieson C. A., Geschwind D. H., Martin K. C. (2006) Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J. Neurosci 26, 13390–13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Michaelevski I., Medzihradszky K. F., Lynn A., Burlingame A. L., Fainzilber M. (November14, 2009) Axonal transport proteomics reveals mobilization of translation machinery to the lesion site in injured sciatic nerve. Mol. Cell. Proteomics 10.1074/mcp.M900369-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]