Abstract
TorsinA is a member of the AAA+ ATPase family of proteins and, notably, is the only known ATPase localized to the ER lumen. It has been suggested to act as a molecular chaperone, while a mutant form associated with early-onset torsion dystonia, a dominantly inherited movement disorder, appears to result in a net loss of function in vivo. Thus far, no studies have examined the chaperone activity of torsinA in vitro. Here we expressed and purified both wild-type (WT) and mutant torsinA fusion proteins in bacteria and examined their ability to function as molecular chaperones by monitoring suppression of luciferase and citrate synthase (CS) aggregation. We also assessed their ability to hold proteins in an intermediate state for refolding. As measured by light scattering and SDS-PAGE, both WT and mutant torsinA effectively, and similarly, suppressed protein aggregation compared to controls. This function was not further enhanced by the presence of ATP. Further, we found that while neither form of torsinA could protect CS from heat-induced inactivation, they were both able to reactivate luciferase when ATP and rabbit reticulocyte lysate were added. This suggests that torsinA holds luciferase in an intermediate state, which can then be refolded in the presence of other chaperones. These data provide conclusive evidence that torsinA acts as a molecular chaperone in vitro and suggests that early-onset torsion dystonia is likely not a consequence of a loss in torsinA chaperone activity but might be an outcome of insufficient torsinA localization at the ER to manage protein folding or trafficking.
Keywords: AAA+ protein, Endoplasmic reticulum, Chaperone, Dystonia, TorsinA
Introduction
Early-onset torsion dystonia (or DYT1 dystonia) is a severe and heritable neurological disorder characterized by sustained muscle contractions and abnormal postures, typically beginning during a developmental window at an early age (Faun et al. 1998). Although there is no evidence of neurodegeneration (Rostasy et al. 2003), the disease may result from more subtle changes in neuronal function such as altered dopaminergic or cholinergic transmission, as has been observed in DYT1 mouse models (Dang et al. 2005; Shashidharan et al. 2005; Balcioglu et al. 2007; Zhao et al. 2008; Martella et al. 2009). The causative genetic mutation appearing in most cases of this disease is a loss of one of two adjacent glutamic acid residues (ΔE 302/303) resulting from deletion in the DYT1 gene encoding human torsinA. Heterozygous carriers with one WT copy and one mutant copy of torsinA (∆E) display the disorder in a dominant manner with only 30–40% penetrance (Risch et al. 1990; Bressman et al. 1994; Ozelius et al. 1997; Klein et al. 1999). Studies have shown the expression of torsinA to be ubiquitous and present in both neuronal and non-neuronal tissues (Augood et al. 1999; Ozelius et al. 1997; Rostasy et al. 2003). Importantly, investigations in Dyt1 mouse models have demonstrated that regulation of torsinA expression is critical as high levels of both WT torsinA and torsinA (∆E) result in comparable phenotypic defects (Grundmann et al. 2007). Furthermore, mice containing knockouts of torsinA do not survive long after birth (Dauer and Goodchild 2004).
TorsinA is a member of the AAA+ (ATPases associated with cellular activities) family of proteins (Ozelius et al. 1997). Many members of this family form hexameric oligomers and perform diverse cellular functions through chaperone-like activity with implications in membrane fusion, complex assembly and disassembly, and protein processing and folding (White and Lauring 2007). TorsinA appears to be no exception, with evidence demonstrating its involvement in protein processing in the secretory pathway, synaptic vesicle recycling, assembly/disassembly of the cytoskeletal network in the nuclear envelope, reduction of toxic misfolded protein aggregates, and serving in a neuroprotective capacity against α-synuclein-induced neurodegeneration (McLean et al. 2002; Cao et al. 2005; Hewett et al. 2007; Hamamichi et al. 2008; Granata et al. 2008; Nery et al. 2008). Interestingly, torsinA is the only AAA+ family member residing primarily in the ER lumen (Kustedjo et al. 2000; Hewett et al. 2003). An N-terminal hydrophobic domain is thought to anchor torsinA to the ER membrane; however, biochemical evidence suggests that torsinA remains peripherally associated (Callan et al. 2007; Liu et al. 2003). Considering that interaction partners of torsinA have been shown at the nuclear envelope (NE) (Goodchild and Dauer 2005; Nery et al. 2008), this peripheral association with the ER membrane suggests that torsinA may be involved in protein trafficking between the ER and NE. Indeed, a recent study has shown that LULL1, a known interacting partner of torsinA, directs it to the NE to displace certain LINC complex proteins, altering the structure of the NE (Heyden et al. 2009).
Current evidence suggests that mutant torsinA (ΔE) appears to result in a net cellular loss of torsinA function, as its presence may interfere with WT torsinA localization (Shashidharan et al. 2004; Torres et al. 2004). Firstly, evidence in both mice and cell cultures has shown torsinA (ΔE) to relocalize predominantly to the NE, similar to that of an ATP hydrolysis mutant E171Q in torsinA, which can bind, but not hydrolyze, ATP (Goodchild and Dauer 2004; Naismith et al. 2004). This suggests torsinA (ΔE) might act like an ATP hydrolysis mutant transferred to the NE by a permanently ATP bound substrate. Secondly, studies with human cell culture have shown that torsinA (ΔE) does not reduce misfolded protein aggregates as WT torsinA does (McLean et al. 2002). Finally, recent work has shown that the torsinA (ΔE) mutation results in a significantly reduced half-life compared to WT torsinA, indicating a high turnover rate (Giles et al. 2008; Gordon and Gonzalez-Alegre 2008). While these data suggest that expression of torsinA (ΔE) alone exerts reduced activity, the presence of both forms together, as in DYT1 patients, tells a different story. Importantly, studies have shown that cultured DYT1 patient fibroblasts display a significant reduction in secretory pathway efficiency compared to normal human cells (Hewett et al. 2008). However, knockdown of torsinA (ΔE) in these fibroblasts resulted in an increase in the secretory pathway efficiency, suggesting that torsinA (ΔE) is partially inhibiting the endogenous function of the WT form. Thus, it is speculated that the disease state may result from a loss of torsinA function due to association with the mutant form. Considering the implications for torsinA in the secretory pathway, a reduction in the activity of this suspected molecular chaperone may have substantial consequences on the proper processing of proteins and homeostasis in the ER, mechanisms that have significant consequences on neuronal transmission (Verkhratsky 2005).
Some, but not all members of the AAA+ family, display chaperone function through either ATP-dependent or ATP-independent mechanisms, thus supporting their role as classical molecular chaperones (Zolkiewski 1999; Zhou et al. 2001; Song et al. 2007). The ATP-independent chaperone activities include the ability to recognize misfolded proteins, as well as hold them in an intermediate state for refolding (Ben-Zvi and Goloubinoff 2001). While much cellular evidence suggests that torsinA may function as a molecular chaperone, studies to date on torsinA have been limited to indirect observations of co-localization and reduction of misfolded aggregates in vivo or in cell cultures only; they have not assessed whether torsinA has the ability to recognize misfolded proteins itself or if it participates in a complex to exhibit this chaperone-like activity. Here, for the first time, we report direct biochemical evidence of the chaperone function of torsinA in vitro using established and well characterized assays for chaperone activity involving luciferase and citrate synthase (Jakob et al. 1995; Herbst et al. 1997; Morrow et al. 2006). We assessed not only the capacity of both torsinA and mutant torsinA (ΔE) to function as chaperones in the recognition of non-native misfolded proteins but also their ability to hold them in an intermediate state for proper refolding.
Materials and methods
Plasmid constructs
The DYT1 WT (wild type) and DYT1 (∆E 302/303) cDNAs, encoding torsinA and mutant torsinA (∆E), respectively, were obtained from Ben Cravatt (Scripps Research Institute, La Jolla, CA, USA) (Kustedjo et al. 2000). The DYT1 E171Q mutant was obtained from Phyllis Hanson (Naismith et al. 2004). All torsinA constructs were truncated 50 amino acids from the N-terminus and PCR-amplified using torsinA Fwd and Rev primers from Table 1 containing BamHI and HindIII sites. PCR products were blunt-end-ligated to pGEMT easy vector (Promega). The PCR fragments were then cut out of the pGEMT easy vector with BamHI and HindIII and gel-purified followed by ligation into a maltose binding protein (MBP) vector (a gift from Steve Pascal, Palmerston North Campus, New Zealand) (Alexandrov et al. 2001) precut with BamHI and HindIII. MBP was PCR-amplified from the MBP vector using MBP GTWY5 and MBP GTWY3 in Table 1 and cloned into pDONR221 to create an entry vector through the Gateway Cloning System (Invitrogen). MBP was then cloned into pDEST17, which contains an N-terminal histidine tag. All final products were verified by sequencing (MWG Sequencing).
Table 1.
Primers used for cloning MBP and torsinA variants
| Name | Primer sequence |
|---|---|
| MBP GTWY 5 | ggggacaagtttgtacaaaaaagcaggctggaaaatcgaagaaggtaaactgg |
| MBP GTWY 3 | ggggaccactttgtacaagaaagctgggtgtcaagtctgcgcgtctttcagg |
| TorsinA Fwd | gttggtttcggatccggaggtggaggtgggcagaagcggagccttag |
| TorsinA Rev | gaaaccaacaagctttcaatcatcgtagtaataatc |
Primers are listed in the 5′→3′ direction
Protein expression/purification
Triton X-114 (Sigma) was precondensed by adding 2 ml of Triton X-114 to 98 ml of 20 mM Tris–HCl 150 mM NaCl (pH 8.0) (buffer A). The solution was mixed at 4°C for 30 min and incubated overnight at 37°C. The detergent condensed and separated into two phases, and the upper aqueous phase was removed and replaced by the same buffer and brought back to 100 ml. The detergent was condensed twice more as above, and the concentration was determined at 268 nm.
Expression of recombinant WT torsinA, torsinA (ΔE), torsinA (E171Q), and MBP was carried out in Escherichia coli strain BL21-AI (Invitrogen). Transformed cells were induced with 10 μM IPTG at 20°C for 12 h. Cells were spun down and resuspended in buffer A containing 1 mM DTT, 0.1 mM PMSF, 1 mM EDTA, and 0.1 mM meta-bisulfite and frozen at −80°C. They were then lysed by sonication followed by centrifugation at 4°C (25,000×g) in a Sorvall centrifuge using a SS-34 rotor to eliminate insoluble proteins and cellular debris. Soluble extracts were then ultracentrifuged (222,000×g) at 8°C in a Beckman Ultracentrifuge using a 55.2Ti rotor. E. coli membranes containing torsinA fusions were then resuspended in buffer A, and precondensed Triton X-114 was added to 1.5% along with 10% glycerol. The membranes were then stirred at 4°C for 1 h then incubated at 25°C for 10 min until turbidity was observed. The phases were separated by centrifugation (1,800×g) at room temperature for 10 min. The upper aqueous phase containing torsinA fusions was removed and re-extracted with Triton X-114 and glycerol as described above. The upper aqueous phase from the second extraction was loaded on an amylose resin column and washed with buffer A until the absorbance of detergent at 268 nm was negligible. The column was washed with buffer A containing 1 M NaCl and eluted with buffer A containing 10 mM maltose according to manufacturer’s instructions (New England Biolabs).
Soluble extracts of MBP were loaded directly on an amylose resin column and purified as described above. After purification, the proteins were dialyzed against 40 mM HEPES-KOH (pH 7.5) and 150 mM NaCl and visualized by SDS-PAGE followed by Coomassie staining. Protein concentrations were determined by BCA assays using BSA as a standard.
Materials
Citrate synthase (CS) from porcine heart mitochondria was obtained from Roche and prepared as described previously (Buchner et al. 1998). CS concentration was determined using the published extinction coefficient of 1.78 for a 1-mg/ml solution (Buchner et al. 1998). Luciferase and the luciferase assay system were purchased from Promega. Oxaloacetate, acetyl-CoA, and dithio-1,4-nitrobenzoic acid were purchased from Sigma.
CS inactivation
Thermal inactivation of CS occurred by incubating the native protein at 43°C. CS (15 μM) was first diluted 1:100 into 40 mM HEPES-KOH (pH 7.5) and 0.1 mM EDTA in the presence or absence of additional proteins at 25°C. Inactivation was initiated by shifting the sample to 43°C. Aliquots were taken at specific time points, and CS activity was measured (Srere et al. 1963).
Aggregation assays
To measure CS aggregation, CS (15 μM) was diluted 1:100 in 40 mM HEPES-KOH (pH 7.5) and equilibrated at 44°C in the presence and absence of chaperones or controls. Luciferase (10 μM) was diluted as above and equilibrated at 42°C with and without chaperones or controls. To monitor kinetics of thermal aggregation, light scattering was measured in a PerkinElmer LS50-B luminescence spectrophotometer at 370 nm with a spectral bandwidth of 2.5 nm. For aggregation assays involving ATP, MBP or torsinA, constructs were incubated with luciferase in the presence and absence of 3 mM ATP, and aggregation was measured as described above.
For monitoring aggregation of luciferase and CS by SDS-PAGE and Coomassie staining, luciferase (0.8 μM) or CS (1.6 μM) was incubated at 42°C and 44°C, respectively, for 30 min in the presence and absence of MBP, WT torsinA, or torsinA (∆E). Reactions were then spun down at 13,000 rpm in a microcentrifuge to separate the soluble fraction from the insoluble pellet. The pellet was resuspended in an equal volume to the supernatant, and bands were visualized by SDS-PAGE and Coomassie staining.
Luciferase refolding assay
Luciferase refolding assays were performed as described by Lee et al. (1997) and as modified by Abdulle et al. (2002). Luciferase (0.17 μM) was incubated with various concentrations of torsinA, torsinA (ΔE), and MBP in refolding buffer [5 mM MgCl2, 10 mM KCl, 50 mM HEPES-KOH (pH 7.5), and 2 mM DTT] at 22°C or 42°C for 15 min, then cooled to room temperature. To prevent luciferase adsorption to walls, tubes were treated with 1 mg/ml BSA for 15 min then rinsed with water before use. Ten microliters of the mixtures was added to a solution pre-incubated at 30°C containing 18 μl of rabbit reticulocyte lysate (RRL) (Promega) and 2 μl of 0.1 M ATP. At various times, 2 μl of the sample was removed, added to 48 μl of 50 mM HEPES-KOH (pH 7.5), vortexed, and diluted 10-fold into luciferase assay reagent (Promega). Luciferase activity was measured with a Promega GloMAX 20/20 luminometer.
ATPase assay
WT torsinA, torsinA (∆E), torsinA (E171Q), and MBP, all at 11 μM, were incubated in 20 mM Tris, 150 mM NaCl pH 8.0 along with 2 mM ATP (Sigma) and 5 μCi of α32P-ATP (Perkin Elmer) for 1 h at 37°C in a total reaction volume of 10 μl. Soluble lysate from E. coli was used as a positive control. At the end of the reaction period, PEI-cellulose thin-layer chromatographic plates (Sorbent Technologies) were spotted with 1 μl of the reaction mixture and developed with 1 M formic acid (Sigma) and 0.5 M LiCl (Sigma) as previously performed (Liberek et al. 1991; McClellan et al. 1998). After drying, radiolabeled ATP and ADP spots were located by autoradiography and cut out, and radioactivity was determined by scintillation counting.
Statistics
Statistical analysis for chaperone activity was performed using the Student’s t test to compare torsinA and torsinA (ΔE) endpoint results in various assays. Significance was determined as P < 0.05.
Results
Production of recombinant WT and mutant torsinA fusions
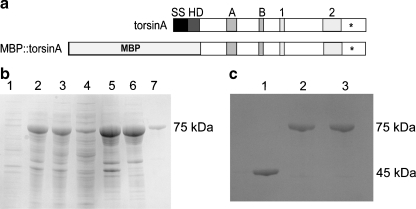
To examine the potential molecular chaperone activity of torsinA, we used a previously published torsinA fusion protein construct whereby the first 50 amino acids were truncated from the N-terminus to eliminate the hydrophobic domain (Hewett et al. 2003) (Fig. 1a). Both WT torsinA and torsinA (∆E) were prepared in this manner and fused to MBP. Ultracentrifugation was used to clean up the soluble protein extracts from E. coli. Surprisingly, however, we found that both WT torsinA and torsinA (ΔE) fusion proteins remained associated with E. coli membranes despite the removal of the hydrophobic domain. TorsinA fusions were pulled away from the membranes using Triton X-114 and found to be in the aqueous fraction after phase separation, suggesting a peripheral association (Fig. 1b). The purified fusion proteins were resolved on SDS-PAGE and migrated as 75-kDa bands as expected (Fig. 1b, c). All chaperone assays were based on the monomeric 75-kDa form.
Fig. 1.
Purification of WT torsinA and torsinA (ΔE) fusions. a Structural organization of torsinA before and after cloning modifications. Domains are indicated by abbreviations as follows: SS (signal sequence), HD (hydrophobic domain), A (Walker A domain), B (Walker B domain—also the location of the E171Q hydrolysis mutant), 1 (sensor 1 domain), 2 (sensor 2 domain); asterisks indicate location of ΔE 302/303 mutation. The SS and HD domains were removed from torsinA and replaced with the MBP for purification purposes. b Uninduced E. coli (lane 1), induced E. coli (lane 2), supernatant from 25,000×g spin (lane 3), supernatant from 222,000×g spin (lane 4), membrane fraction from 222,000×g spin (lane 5), aqueous fraction from Triton X-114 separation (lane 6), purified torsinA (lane 7) (75 kDa). c MBP (45 kDa) from the pDEST17 vector purified by amylose resin column (lane 1). WT torsinA (lane 2) and torsinA (ΔE) fusions (lane 3) (75 kDa) from MBP vector were purified by amylose resin column chromatography and analyzed by gel electrophoresis with Coomassie staining
WT and torsinA (ΔE) prevent aggregation of luciferase and CS
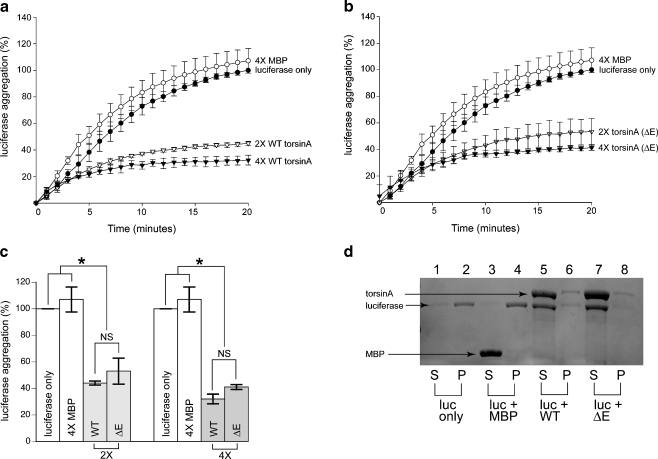
To determine if torsinA displays molecular chaperone activity, we investigated whether or not it could recognize and bind misfolded proteins directly. We made use of luciferase and CS as substrates for our chaperone assays. Luciferase and CS are two proteins that have the tendency to aggregate at temperatures greater than 40°C. Using light scattering, their aggregation can be measured over time. Figure 2a illustrates the suppression of luciferase aggregation by WT torsinA at 42°C through a time course of 20 min. WT torsinA effectively suppressed 55.1 ± 1.5% aggregation of luciferase at a 2× molar excess concentration; this suppression increased to 67.8 ± 3.6% at higher concentrations (4×). Surprisingly, torsinA (ΔE) also suppressed luciferase aggregation with 46.7 ± 9.8% suppression at 2× molar excess and 58.8 ± 1.9% suppression at 4× molar excess. There was no significant difference in luciferase aggregation suppression between the WT and ΔE forms of torsinA at either concentration tested (2×, P = 0.24, 4×, P = 0.10) (Fig. 2a, b). The negative control MBP revealed aggregation levels similar to that of luciferase alone demonstrating that MBP does not suppress aggregation in any way (Fig. 2a, b). Figure 2c depicts a comparative analysis of final time points, demonstrating that there was no significant difference between WT and mutant torsinA (∆E). To verify the results of the light scattering data, we heated luciferase at 42°C in the presence and absence of MBP, WT torsinA, and torsinA (∆E). The soluble and insoluble fractions were separated by centrifugation and analyzed by SDS-PAGE and Coomassie staining. Consistent with the light scattering assay, we found that both WT and mutant (∆E) torsinA significantly suppressed luciferase aggregation, while MBP had no effect (Fig. 2d).
Fig. 2.
WT torsinA and torsinA (ΔE) inhibit heat-induced aggregation of luciferase. Luciferase (0.1 μM) was incubated at 42°C for 20 min either alone (filled circles), in the presence of MBP (0.4 μM, open circles), WT torsinA (0.2 μM, open triangles; 0.4 μM, filled triangles) in a, or torsinA (ΔE) (0.2 μM, open triangles; 0.4 μM, filled triangles) in b. Aggregation was determined by light scattering at 370 nm. c There was no significant difference in the aggregation of luciferase between WT torsinA and torsinA (ΔE). Comparative analysis among torsinA constructs, MBP, and luciferase at 20-min time points. For all time points, data are representative of three trials and were calculated as a percentage of the maximum aggregation of luciferase after 20 min for each trial and are expressed as the mean ± the standard deviation. d Suppression of luciferase aggregation monitored by SDS-PAGE and Coomassie staining. Lanes 1, 3, 5, and 7 represent the soluble fractions. Lanes 2, 4, 6, and 8 represent the insoluble fractions. Lanes 1 and 2 are 0.8 μM of luciferase only. Lanes 3 and 4 are 4.8 μM of MBP and 0.8 μM of luciferase. Lanes 5 and 6 are 4.8 μM of WT torsinA and 0.8 μM of luciferase. Lanes 7 and 8 are 0.8 μM luciferase and 4.8 μM torsinA (∆E)
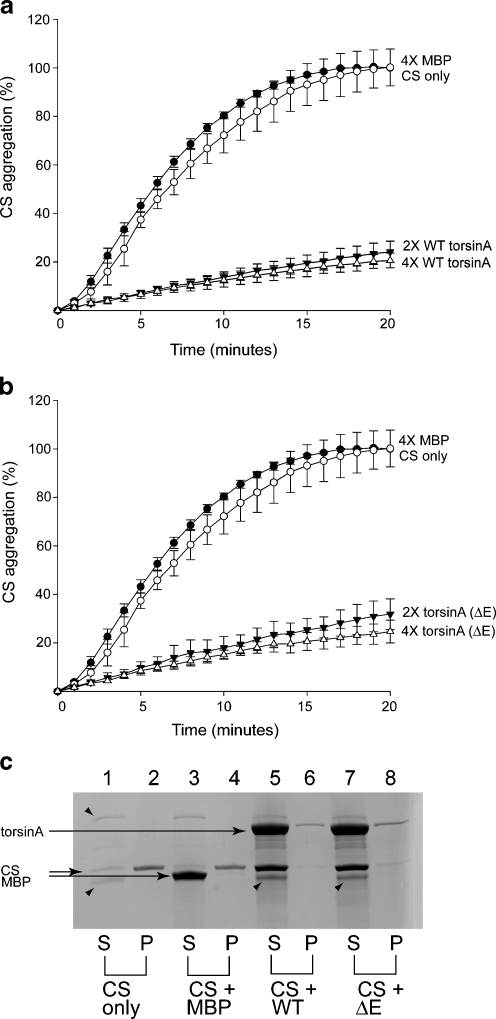
To confirm that torsinA was not specific for just one misfolded substrate, we also tested whether or not WT torsinA and torsinA (ΔE) could suppress the aggregation of CS. Figure 3 demonstrates that both WT and torsinA (ΔE) could also suppress CS aggregated at 44°C at equimolar concentrations. WT torsinA (Fig. 3a) decreased aggregation by 76.7 ± 4.4% at 2× molar excess and 79.5 ± 3.0% at 4× molar excess. Likewise, torsinA (ΔE) exhibited a similar protective effect of 69.1 ± 6.3% at 2× molar excess and 75.4 ± 4.6% at 4× molar excess (Fig. 3b). Thus, both WT torsinA and torsinA (ΔE) significantly suppressed CS aggregation (P < 0.01). There was no difference in CS aggregation prevention between WT torsinA and torsinA (ΔE) at 2× molar excess (P = 0.17) or at higher concentrations of protein (P = 0.28) at the 20-min time point. The suppression of CS aggregation as monitored by light scattering was confirmed by SDS-PAGE and Coomassie staining of CS incubated with and without MBP, WT torsinA, or torsinA (∆E) as described above (Fig. 3c). Taken together, these data revealed that both WT torsinA and torsinA (ΔE) can recognize and bind directly to misfolded protein substrates with general specificity.
Fig. 3.
WT torsinA and torsinA (ΔE) inhibit heat-induced aggregation of citrate synthase (CS). CS (0.15 μM) was incubated at 44°C for 20 min either alone (open circles), in the presence of MBP (0.6 μM, filled circles), WT torsinA (0.3 μM, filled triangles; 0.6 μM, open triangles) in a, or torsinA (ΔE) (0.3 mM, filled triangles; 0.6 mM, open triangles) in b. Aggregation was determined by light scattering at 370 nm. Data are representative of three trials and were calculated as a percentage of the maximum aggregation of CS after 20 min for each trial and are expressed as the mean ± standard deviation. c Suppression of CS aggregation monitored by SDS-PAGE and Coomassie staining. Lanes 1, 3, 5, and 7 represent the soluble fractions. Lanes 2, 4, 6, and 8 represent the insoluble fractions. Lanes 1 and 2 are 1.6 μM of CS only. Lanes 3 and 4 are 6.4 μM of MBP and 1.6 μM of CS. Lanes 5 and 6 are 6.4 μM of WT torsinA and 1.6 μM of CS. Lanes 7 and 8 are 1.6 μM CS and 6.4 μM torsinA (∆E). Arrowheads represent slight contaminants and/or degradation products present in CS preparation from Roche
ATP does not affect the chaperone function of torsinA
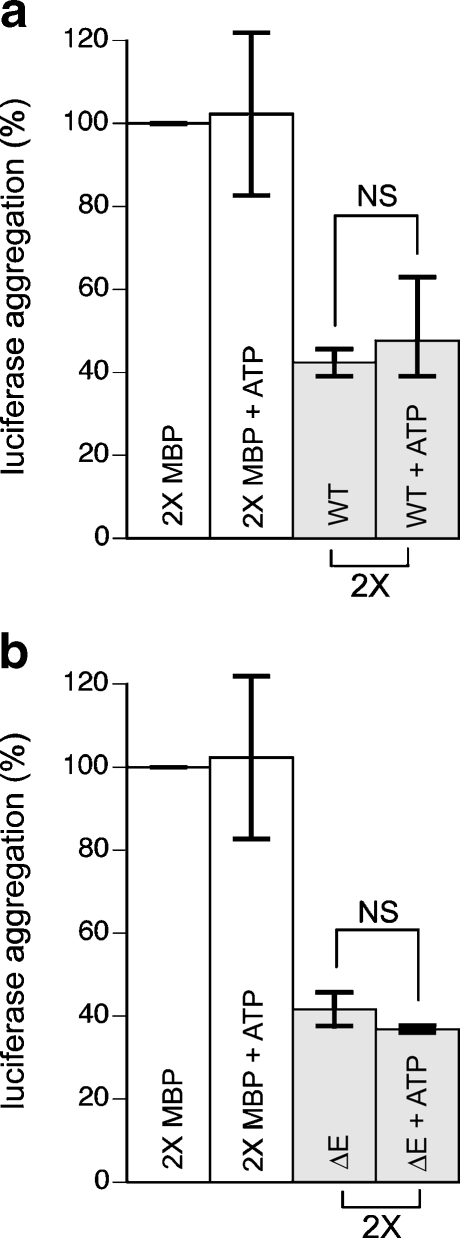
Since torsinA belongs to the AAA+ family of ATPases, we investigated whether or not the presence of ATP affected the ability of torsinA to suppress aggregation. We incubated luciferase with both WT torsinA and torsinA (∆E) in the presence and absence of 3 mM ATP. However, we found no significant difference in the chaperone function of torsinA in suppressing luciferase aggregation with or without ATP (Fig. 4a,b). Furthermore, we did not obtain any detectable level of ATPase activity with WT torsinA or torsinA (∆E); both of these were compared to the negative control, the torsinA E171Q hydrolysis mutant (data not shown). This suggests that ATP does not affect the chaperone function of torsinA alone, but may require the addition of a cofactor to activate its ATPase activity and influence its chaperone abilities as others have suggested (Zhu et al. 2008; Giles et al. 2009).
Fig. 4.
ATP does not influence the prevention of luciferase aggregation by WT torsinA or torsinA (∆E). Luciferase (0.1 μM) was heated at 42°C for 20 min in the presence and absence of 3 mM ATP with 0.2 μM MBP or 0.2 μM WT torsinA in a or 0.2 μM torsinA (∆E) in b. Aggregation was determined by light scattering at 370 nm. Data are representative of two trials and were calculated as a percentage of the maximum aggregation of luciferase after 20 min for each trial and are expressed as the mean ± standard deviation
WT torsinA and torsinA (ΔE) maintain denatured luciferase in a folding competent state
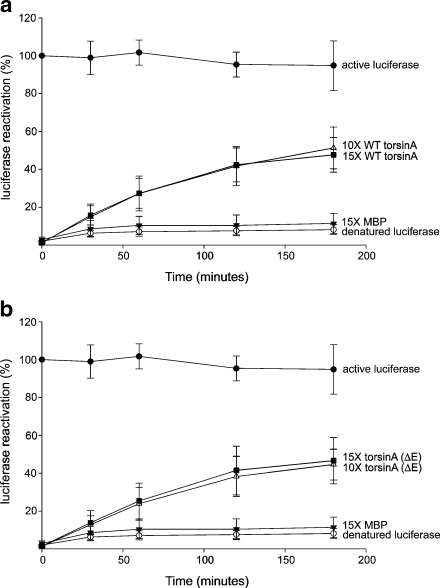
To determine whether or not the partially unfolded proteins captured by these chaperones were in a refoldable state, an in vitro luciferase refolding assay was employed. In this assay, luciferase is heat-denatured in the presence or absence of chaperones, and then allowed to refold at a lower temperature in the presence of ATP and other chaperone machinery present in RRL. Luciferase maintained at 25°C remained fairly stable throughout the refolding period (active luciferase—Fig. 5). However, when luciferase was incubated at 42°C for 15 min in the absence of any chaperones, or with 15× molar excess MBP, the activity decreased to around 10% with no recovery throughout the time course of luciferase refolding (Fig. 5a, b). In the presence of a 10× and 15× molar excess concentration of WT torsinA, luciferase regained 51.2 ± 11.0% and 47.6 ± 9.1% activity, resulting in significantly more reactivation than heated luciferase alone or in the presence of MBP (P < 0.05).
Fig. 5.
Luciferase can be refolded in the presence of either WT torsinA or torsinA (ΔE). Active luciferase (0.17 μM, filled circles) was measured after incubation at 25°C for the indicated times. All other measurements were made after incubation at 42°C for 15 min. Luciferase (0.17 μM) was incubated either alone (open circles), or in the presence of 15× MBP (2.5 μM; filled triangles). In a, luciferase was incubated with WT torsinA (1.7 and 2.5 μM), and in b, luciferase was incubated with torsinA (ΔE) (1.7 and 2.5 μM). The refolding step was performed at 30°C for 120 min in RRL supplemented with ATP, and luciferase activity was determined at the time points indicated. Data are representative of three trials and are presented as percentage of luciferase activity after 15 min of incubation at 42°C. Data are expressed as mean ± standard deviation
However, no enhanced effect was observed with increasing torsinA concentrations (P = 0.68) (Fig. 5a).
When luciferase was denatured in the presence of a 10× and 15× molar excess of torsinA (ΔE), 44.5 ± 8.1% and 46.5 ± 12.2% activity was recovered, demonstrating considerably more reactivation than heated luciferase alone or in the presence of MBP (P < 0.05). Further increasing concentrations of torsinA (ΔE) had no effect (P = 0.82) (Fig. 5b). Similar to the aggregation assays, there was no significant difference between WT torsinA and torsinA (ΔE) in restoring luciferase activity at either concentration tested (10×, P = 0.44; 15×, P = 0.91) (Fig. 5a, b). From these data, it is clear that both WT torsinA and torsinA (ΔE) can maintain heat-denatured luciferase in a refoldable state that allows it to be refolded by other chaperones into a fully active enzyme.
WT and ΔE torsinA cannot protect CS from thermal inactivation
We next sought to investigate whether torsinA can protect CS from inactivation at increased temperatures. If a chaperone is capable of binding and efficiently releasing early intermediate states of CS during heating, the result will be an extended activity of CS during longer heating periods. We thus tested whether or not torsinA could also hold CS in an intermediate folding state as well as release it without the aid of other chaperones, as in the luciferase refolding assay.
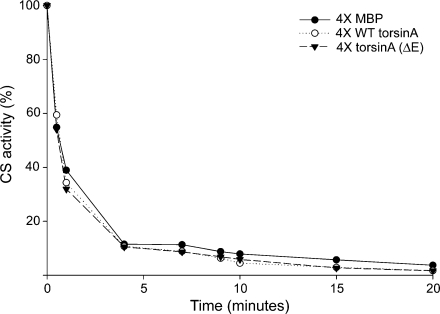
We found that neither WT torsinA nor torsinA (ΔE) could protect CS from inactivation when compared to the negative control MBP (Fig. 6). Further, we observed no effect at higher concentrations tested (data not shown). The loss of CS activity followed first-order enzyme kinetics with an inactivation rate of 16.1 × 10−3 s−1 for MBP. Both WT torsinA and torsinA (ΔE) had slightly higher rates (Table 2). These data, combined with the aggregation results, suggest that torsinA must be holding CS in a very stable complex and may require other chaperones to remove the misfolded protein from torsinA for proper refolding.
Fig. 6.
WT torsinA and torsinA (ΔE) do not affect the rate of heat-induced inactivation of CS. CS (0.15 μM) was incubated for 30 min at 43°C in the presence of 0.6 μM MBP (filled circles), 0.6 μM WT torsinA (open circles), or 0.6 µM torsinA (ΔE) (filled triangles), and CS activity was measured at various time points. The enzymatic activity of CS was expressed as a percentage of initial CS activity
Table 2.
Influence of various proteins on the rate of thermal inactivation of CS at 43°C
| Addition to inactivation buffer | Rate constant (×10−3 s−1) of CS thermal inactivationa |
|---|---|
| 0.6 µM MBP | 16.1 |
| 0.6 μM WT torsinA | 17.0 |
| 0.6 μM torsinA (ΔE) | 19.1 |
aRate constants were obtained from the inactivation kinetics shown in Fig. 5
Discussion
The large and diverse AAA+ family of proteins to which torsinA belongs contains a variety of proteins that function with chaperone-like activity (White and Lauring 2007). However, not all members of this family have been demonstrated to behave as classical molecular chaperones in recognizing misfolded proteins and holding them in an intermediate state for refolding. For example, the AAA+ protein, valosin/p97, can recognize and suppress aggregation of non-native misfolded proteins, while another, NSF, lacks this capability (Zhou et al. 2001; Song et al. 2007). While the precise cellular function of torsinA remains to be elucidated, it has been speculated that this essential protein can function as a molecular chaperone based on cellular assays (McLean et al. 2002; Caldwell et al. 2003).
We have shown that torsinA does indeed possess the ability to recognize non-native misfolded proteins and bind them directly using two different substrates. The data presented from the aggregation assays involving luciferase and CS demonstrate that torsinA prevents aggregation considerably and with general specificity for misfolded proteins, thus fulfilling one of the basic criteria of a classical molecular chaperone (Ben-Zvi and Goloubinoff 2001). The comparative activity of torsinA in suppressing aggregation is similar to that observed for hsp90, as well as some small heat shock proteins, further demonstrating its effectiveness as a molecular chaperone (Jakob et al. 1993; Ehrnsperger et al. 1997; Buchner et al. 1998). Strikingly, we found that the DYT1 dystonia-associated form of this protein, torsinA (ΔE), also exhibited chaperone activity equal to that of the WT form. Since both WT and torsinA (ΔE) are fully capable of acting as molecular chaperones, this suggests the cellular dysfunction associated with dystonia may be more related to mislocalization of the protein to the NE and/or absence of sufficient torsinA function at the ER.
Although torsinA is a member of the AAA+ family, we did not detect any ATPase activity of the torsinA constructs or influence of chaperone function by the presence of ATP. This is not surprising though, for a number of reasons. Firstly, studies of the torsinA worm homolog ooc-5, revealed no detectable ATPase activity when compared to its ATP hydrolysis mutant (Zhu et al. 2008). Secondly, as Zhu and others have pointed out, previous studies that have investigated the ATPase activity of WT torsinA and torsinA (∆E) have been contradictory and did not have an ATP hydrolysis mutant as a negative control. These studies have also shown much lower rates of activity than that of other AAA+ proteins as well as conflicting results between the ATPase activity of WT torsinA and torsinA (∆E) (Kustedjo et al. 2000; Konakova and Pulst 2005; Pham et al. 2006).
Finally, studies have shown that the torsinA family lacks critical residues needed for ATP binding and hydrolysis (Zhu et al. 2008; Nagy et al. 2008). An elegant study by Nagy and others found that while most AAA+ family members such as ClpB contain a threonine or serine directly flanking the critical lysine residue in the Walker A binding motif, the torsinA family contains an asparagine residue instead. Their data showed that in ClpB, replacing the threonine with asparagine compromised its ATPase activity as well as its ATP-dependent chaperone function. Thus, the threonine residue may be involved in nucleotide binding (Nagy et al. 2008). Further, DnaC also contains an asparagine residue in the same position as torsinA in its Walker A motif and, in addition, lacks ATPase activity without the presence of other factors such as ssDNA or DnaB (Davy et al. 2002; Nagy et al. 2008). Finally, many AAA+ members contain a highly conserved arginine finger located near the sensor II motif, which is needed for ATP hydrolysis (Ogura et al. 2003). As others have found, the torsinA family also lacks this critical residue. It remains possible, however, that torsinA may require some unknown cofactor to activate its ATPase activity, as previously suggested (Zhu et al. 2008; Giles et al. 2009). This in turn could influence its chaperone abilities, although further studies will be needed to test this hypothesis.
Since both torsinA forms were capable of suppressing aggregation, we were curious to know whether or not the torsinA–luciferase complex was a “dead-end complex” that rendered the misfolded substrate incapable of being refolded, or whether torsinA could in fact hold the luciferase in a state for proper refolding by other chaperones. To this end, a luciferase reactivation assay was employed. We found that both WT and torsinA (ΔE) could maintain luciferase in a state for refolding, resulting in 40–50% reactivation of luciferase. This evidence indicates that the torsinA–luciferase complex was not in a dead-end state, and the misfolded protein could be salvaged.
The results of the luciferase reactivation assay prompted us to further examine the intermediate folding states of the misfolded protein, as well as the efficiency of torsinA in binding and releasing it. The CS inactivation assay differs from the luciferase reactivation because the refolding of the early intermediate states of CS depends upon their equilibrium with native CS. Thus, this assay depends on the ability of torsinA to bind and release these intermediates efficiently, thereby creating a reservoir of early CS intermediates, rather than requiring additional chaperones to remove it from torsinA and refold it themselves (Buchner et al. 1998). If torsinA can bind and release CS efficiently during heating, the activity of CS should be significantly extended compared to heating of CS with the negative control MBP. However, neither WT nor torsinA (ΔE) could protect CS from inactivation, suggesting that torsinA holds CS in a very stable complex that is not readily dissociated. This is not too surprising, as a number of small heat shock proteins can suppress CS aggregation but are unable to release it; these data often are suggestive of a requirement for other chaperones (Lee et al. 1995; Bose et al. 1996; Chang et al. 1996; Ehrnsperger et al. 1997).
Members of the AAA+ family share a highly conserved AAA domain responsible for their ATPase activity. The number of AAA domains in the protein classifies them as class I (containing two AAA domains) or class II (containing a single AAA domain) members, with a few proteins such as dynein containing six AAA domains (White and Lauring 2007). TorsinA is classified as a class II AAA+ protein containing a single AAA domain. Previous studies have found that the AAA domain is involved in the prevention of protein aggregation in both class I and II members that are capable of this function (Wawrzynow et al. 1995; Weibezahn et al. 2003; Song et al. 2007).
Past and present structural model predictions suggest that the structure of torsinA is most similar to that of the second AAA domain of ClpA and ClpB (Kock et al. 2006; Zhu et al. 2008). ClpB is an ATP-dependent chaperone present in E. coli, which is capable of preventing the aggregation of misfolded proteins as well as disaggregating them (Zolkiewski 1999). Although studies have shown that substrate interaction with ClpB occurs within the N-terminal AAA domain, its full chaperone capabilities require communication between both AAA domains in a cycle of ATP binding and hydrolysis, which is linked to substrate binding and release (Schlieker et al. 2004; Doyle et al. 2007).
ClpA is another bacterial protein with ATP-dependent chaperone abilities similar to ClpB (Doyle et al. 2007). However, in contrast to ClpB, ClpA can also prevent aggregation in the absence of ATP (Wickner et al. 1994). Our study demonstrates that torsinA, which contains a single AAA domain, may function similar to ClpA in its ability to prevent aggregation in the absence of ATP. This is also similar to that of another AAA+ protein, valosin p97 (Song et al. 2007). Furthermore, both torsinA and ClpA can protect luciferase from thermal inactivation, although they require other chaperones for luciferase reactivation (Wickner et al. 1994). Despite the lack of altered chaperone function in the presence of ATP, it remains possible that torsinA, with the addition of activating cofactors of ATPase activity, might still act as an ATP-dependent chaperone similar to ClpA and ClpB.
The realization that torsinA (ΔE) possesses chaperone activity equal to that of the WT torsinA protein is interesting considering that previous studies have suggested that torsinA (ΔE) appears to result in loss of function (Hewett et al. 2000; McLean et al. 2002; Caldwell et al. 2003; Misbahuddin et al. 2004). Prior reports demonstrated that overexpressed torsinA (ΔE) was not only unable to recognize and prevent the aggregation of misfolded proteins but also formed membranous inclusions itself in cell cultures. In contrast, our in vitro data conclusively show that mutant torsinA (ΔE) was fully capable of recognizing and suppressing the aggregation of both luciferase and CS, as well as able to hold luciferase in an intermediate state for refolding.
It is possible that the overall reduction of chaperone activity seen in previous studies is due to sequestration of torsinA (ΔE) into membranous inclusions when overexpressed in cell culture and in vivo, thus rendering it incapable of activity. Notably, membranous inclusions are not observed when experiments are performed using DYT1 dystonia patient cells that have endogenous levels of torsinA (ΔE) (Walker et al. 2002; Rostasy et al. 2003; Bragg et al. 2004). Thus, the absence of torsinA (ΔE) chaperone function associated with overexpression in cell cultures could possibly be an artifact of the expression levels employed, as has been suggested by others (Gordon and Gonzalez-Alegre 2008).
Studies have shown that WT torsinA is a relatively long-lived protein that is degraded by autophagy while torsinA (∆E) is short lived, and is prematurely degraded through the proteasome and autophagy (Giles et al. 2008; Gordon and Gonzalez-Alegre 2008). Furthermore, human genetic studies have found that torsinA contains a single nucleotide polymorphism (SNP) that changes aspartic acid (D) to histidine (H) at residue 216. Interestingly, this SNP is found more often in torsinA (∆E) carriers lacking dystonia symptoms but less often in those displaying symptoms (Kock et al. 2006; Risch et al. 2007). Likewise, studies have shown that co-expression of ∆216H and torsinA (∆E) in trans, but not in cis, leads to an overall reduction in inclusion formation (Kock et al. 2006). In the context of our work, where we show that both the WT and torsinA (∆E) proteins retain chaperone function, it is possible that the multimeric complex of these proteins together in vivo leads to an instability of the oligomer resulting in its early degradation.
To date, much of the accumulated evidence on torsinA suggests potentially distinct functions or differential interactions at both the NE and the ER. On one hand, the interaction of torsinA with LULL1 regulates its trafficking to, and subsequent restructuring of, the NE through interacting LINC complex proteins (Goodchild and Dauer 2005; Heyden et al. 2009). On the other hand, evidence has clearly shown torsinA to have implications on secretory pathway function, with the WT protein serving a necessary role in maintaining secretion efficiency in the ER (Hewett et al. 2007). The biochemical outcomes of our study lend solid evidence to the hypothesis that torsinA does in fact function as a molecular chaperone, irrespective of its intracellular locale or mutated state in dystonia. In this regard, torsinA chaperone activity need not presuppose a mutual exclusivity of function at either the ER or NE, as the diversity of chaperone functions in protein folding and trafficking are well established. Nevertheless, it remains important to further discern the consequences of torsinA activity in the context of its cellular localization, especially as it may pertain to the disease state.
Acknowledgments
We wish to acknowledge the cooperative spirit of all Caldwell Lab members. Special thanks go to Lindsay Faircloth for technical advice, as well as Carol Duffy and Janis O’Donnell for use of their luminometer and spectrophotometer, respectively. This research was supported by the Dystonia Medical Research Foundation Cure Dystonia Initiative (PFC, GAC, KAC) and a CAREER Award from the National Science Foundation (KAC).
Abbreviations
- CS
Citrate synthase
- RRL
Rabbit reticulocyte lysate
- ER
Endoplasmic reticulum
- DYT1
Early-onset torsion dystonia
- NE
Nuclear envelope
- MBP
Maltose binding protein
- WT
Wild type
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
Footnotes
Sentence summary
TorsinA is a member of the diverse AAA+ family of proteins that includes molecular chaperones and is an evolutionarily conserved, ER-resident protein for which a precise function remains to be elucidated. Importantly, a 3bp deletion in the DYT1 gene encoding human torsinA results in a movement disorder termed early-onset torsion dystonia, wherein this mutation leads to protein mislocalization and is thought to result in a net loss of torsinA function in vivo. Here, for the first time, we biochemically investigate the chaperone function of torsinA, as well as the dystonia-associated mutant form, and report that both proteins display comparable chaperone-like activity in vitro, thereby implying that the neuronal dysfunction linked to torsinA may not be a direct consequence of a deficit in chaperone capacity but is, perhaps, an outcome of insufficient torsinA localization at the ER to manage protein folding or trafficking.
References
- Abdulle R, Mohindra A, Fernando P, Heikkila JJ. Xenopus small heat shock proteins, Hsp30C and Hsp30D, maintain heat- and chemically denatured luciferase in a folding-competent state. Cell Stress Chaperones. 2002;7(1):6–16. doi: 10.1379/1466-1268(2002)007<0006:XSHSPH>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Dutta K, Pascal SM. MBP fusion protein with a viral protease cleavage site: one-step cleavage/purification of insoluble proteins. Biotechniques. 2001;30(6):1194–1198. doi: 10.2144/01306bm01. [DOI] [PubMed] [Google Scholar]
- Augood SJ, Martin DM, Ozelius LJ, Breakefield XO, Penney JB, Jr, Standaert DG. Distribution of the mRNAs encoding TorsinA and TorsinB in the normal adult human brain. Ann Neurol. 1999;46(5):761–769. doi: 10.1002/1531-8249(199911)46:5<761::AID-ANA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Balcioglu A, Kim MO, Sharma N, Cha JH, Breakefield XO, Standaert DG. Dopamine release is impaired in a mouse model of DYT1 dystonia. J Neurochem. 2007;102(3):783–788. doi: 10.1111/j.1471-4159.2007.04590.x. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi AP, Goloubinoff P. Review: mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J Struct Biol. 2001;135(2):84–93. doi: 10.1006/jsbi.2001.4352. [DOI] [PubMed] [Google Scholar]
- Bose S, Weikl T, Hans B, Buchner J. Chaperone function of Hsp90-associated proteins. Sci Rep. 1996;274(5293):1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- Bragg DC, Camp SM, Kaufman CA, Wilbur JD, Boston H, Schuback DE, Hanson PI, Sena-esteves M, Breakefield XO. Perinuclear biogenesis of mutant torsinA inclusions in cultured cells infected with tetracycline-regulated herpes simplex virus type 1 amplicon vectors. Neuroscience. 2004;125(3):651–661. doi: 10.1016/j.neuroscience.2004.01.053. [DOI] [PubMed] [Google Scholar]
- Bressman SB, Deleon MS, Kramer PL, Ozelius LJ, Brin MF, Greene PE, Fahn S, Breakefield XO, Risch NJ. Dystonia in Ashkenazi Jews: clinical characterization of a founder mutation. Ann Neurol. 1994;36(5):771–777. doi: 10.1002/ana.410360514. [DOI] [PubMed] [Google Scholar]
- Buchner J, Grallert H, Jakob U. Analysis of chaperone function using citrate synthase as nonnative substrate protein. In: Abelson JN, Simon MI, Lorimer GH, Baldwin TO, editors. Methods in enzymology. San Diego: Academic; 1998. pp. 323–338. [DOI] [PubMed] [Google Scholar]
- Caldwell GA, Cao S, Sexton EG, Gelwix CC, Bevel JP, Caldwell KA. Suppression of polyglutamine-induced protein aggregation in Caenorhabditis elegans by torsin proteins. Hum Mol Genet. 2003;12(3):307–319. doi: 10.1093/hmg/ddg027. [DOI] [PubMed] [Google Scholar]
- Callan AC, Bunning S, Jones OT, High S, Swanton E. Biosynthesis of the dystonia-associated AAA+ ATPase torsinA at the endoplasmic reticulum. Biochem J. 2007;401(2):607–612. doi: 10.1042/BJ20061313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Gelwix CC, Caldwell KA, Caldwell GA. Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J Neurosci. 2005;25(15):3801–3812. doi: 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Primm TP, Jakana J, Lee IH, Serysheva I, Chiu W, Gilbert HF, Quiocho FA. Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J Biol Chem. 1996;271(12):7218–7223. doi: 10.1074/jbc.271.12.6658. [DOI] [PubMed] [Google Scholar]
- Dang MT, Yokoi F, McNaught KS, Jengelley TA, Jackson T, Li J, Li Y. Generation and characterization of Dyt1 DeltaGAG knock-in mouse as a model for early-onset torsion dystonia. Exp Neurol. 2005;196(2):452–463. doi: 10.1016/j.expneurol.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Dauer W, Goodchild R. Mouse models of torsinA dysfunction. Adv Neurol. 2004;94:67–72. [PubMed] [Google Scholar]
- Davy MJ, Fang L, McInerney P, Georgescu RE, O’ Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;21(12):3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14(2):114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16(2):221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faun S, Bressman SB, Marsden CD. Classification of dystonia. In: Fahn S, Marsden CD, DeLong M, editors. Dystonia 3: advances in neurology. Philadelphia: Lippincott-Raven; 1998. [PubMed] [Google Scholar]
- Giles LM, Chen J, Li L, Chin LS. Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type-specific mislocalization to the nuclear envelope. Hum Mol Genet. 2008;17(17):2712–2722. doi: 10.1093/hmg/ddn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles LM, Lian L, Chin LS. Printor, a novel torsina-interacting protein implicated in dystonia pathogenesis. J Biol Chem. 2009;284(32):21765–21775. doi: 10.1074/jbc.M109.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci USA. 2004;101(3):847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol. 2005;168(6):855–862. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon KL, Gonzalez-Alegre P. Consequences of the DYT1 mutation on torsinA oligomerization and degradation. Neuroscience. 2008;157(3):588–595. doi: 10.1016/j.neuroscience.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granata A, Watson R, Collinson LM, Schiavo G, Warner TT. The dystonia-associated protein torsina modulates synaptic vesicle recycling. J Biol Chem. 2008;238(12):7568–7579. doi: 10.1074/jbc.M704097200. [DOI] [PubMed] [Google Scholar]
- Grundmann K, Reischmann B, Vanhoutte G, Hübener J, Teismann P, Hauser TK, Bonin M, Wilbertz J, Horn S, Nguyen HP, Kuhn M, Chanarat S, Wolburg H, Linden A, Riess O. Overexpression of human wildtype torsinA and human DeltaGAG torsinA in a transgenic mouse model causes phenotypic abnormalities. Neurobiol Dis. 2007;27(2):190–206. doi: 10.1016/j.nbd.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc Natl Acad Sci USA. 2008;105(2):728–733. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst R, Schafer U, Seckler R. Equilibrium intermediates in the reversible unfolding of firefly (Photinus pyralis) luciferase. J Biol Chem. 1997;272(11):7099–7105. doi: 10.1074/jbc.272.11.7099. [DOI] [PubMed] [Google Scholar]
- Hewett J, Gonzalez-Agosti C, Slater D, Ziefer P, Li S, Bergeron D, Jacoby DJ, Ozelius LJ, Ramesh V, Breakefield XO. Mutant torsinA, responsible for early-onset torsion dystonia, forms membranous inclusions in cultured neural cells. Hum Mol Genet. 2000;9(9):1403–1413. doi: 10.1093/hmg/9.9.1403. [DOI] [PubMed] [Google Scholar]
- Hewett J, Ziefer P, Bergeron D, Naismith T, Boston H, Slater D, Wilbur J, Schuback D, Kamm C, Smith N, Camp S, Ozelius LJ, Ramesh V, Hanson PI, Breakefield XO. TorsinA in PC12 cells: localization in the endoplasmic reticulum and response to stress. J Neurosci Res. 2003;72(2):158–168. doi: 10.1002/jnr.10567. [DOI] [PubMed] [Google Scholar]
- Hewett JW, Tannous B, Niland BP, Nery FC, Zeng J, Li Y, Breakefield XO. Mutant torsinA interferes with protein processing through the secretory pathway in DYT1 dystonia cells. Proc Natl Acad Sci USA. 2007;104(17):7271–7276. doi: 10.1073/pnas.0701185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett JW, Nery FC, Niland B, Ge P, Tan P, Hadwiger P, Tannous BA, Sah DW, Breakefield XO. siRNA knock-down of mutant torsinA restores processing through secretory pathway in DYT1 dystonia cell. Hum Mol Genet. 2008;17(10):1436–1445. doi: 10.1093/hmg/ddn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyden ABV, Naismith TV, Snapp EL, Hodzic D, Hanson PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cell. 2009;20:2661–2672. doi: 10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem Comm. 1993;268(3):1517–1520. [PubMed] [Google Scholar]
- Jakob U, Lilie H, Meyer I, Buchner J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J Biol Chem. 1995;270(13):7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- Klein C, Friedman J, Bressman S, Vieregge P, Brin MF, Pramstaller PP, Deleon D, Hagenah J, Sieberer M, Fleet C, Kiely R, Xin W, Breakefield XO, Ozelius LJ, Sims KB. Genetic testing for early-onset torsion dystonia (DYT1): introduction of a simple screening method, experiences from testing of a large patient cohort, and ethical aspects. Genet Test. 1999;3(4):323–328. doi: 10.1089/gte.1999.3.323. [DOI] [PubMed] [Google Scholar]
- Kock N, Naismith TV, Boston HE, Ozelius LJ, Corey DP, Breakefield XO, Hanson PI. Effects of the genetic variations in the dystonia protein torsinA: identification of polymorphism at residue 216 as protein modifier. Hum Mol Genet. 2006;15(8):1355–1364. doi: 10.1093/hmg/ddl055. [DOI] [PubMed] [Google Scholar]
- Konakova M, Pulst SM. Dystonia-associated forms of torsinA are deficient in ATPase activity. J Mol Neurosci. 2005;25:105–117. doi: 10.1385/JMN:25:1:105. [DOI] [PubMed] [Google Scholar]
- Kustedjo K, Bracey MH, Cravatt BF. TorsinA and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem. 2000;275(36):27933–27939. doi: 10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995;270(18):10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. Embo J. 1997;16(3):659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88(7):2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zolkiewska A, Zolkiewski M. Characterization of human torsinA and its dystonia-associated mutant form. Biochem J. 2003;374(pt1):117–122. doi: 10.1042/BJ20030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella G, Tassone A, Sciamanna G, Platania P, Cuomo D, Viscomi MT, Bonsi P, Cacci E, Biagioni S, Usiello A, Sharma N, Standaert DG, PIsani A. Impairment of bidirectional synaptic plasticity in the striatum of a mouse model of DYT1 dystonia: role of endogenous acetylcholine. Brain. 2009;132:2336–2349. doi: 10.1093/brain/awp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Endres JB, Vogel JP, Palazzi D, Rose MD, Brodsky JL. Specific molecular chaperone interactions and an ATP-dependent conformational change are required during posttranslational protein translocation into the yeast ER. Mol Biol Cell. 1998;9(12):3533–3545. doi: 10.1091/mbc.9.12.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean PJ, Kawamata H, Shariff S, Hewett J, Sharma N, Ueda K, Breakefield XO, Hyman BT. TorsinA and heat shock proteins act as molecular chaperones: suppression of α-synuclein aggregation. J Neurochem. 2002;83(4):846–854. doi: 10.1046/j.1471-4159.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- Misbahuddin A, Placzek MR, Taanman JW, Gschmeissner S, Schiavo G, Cooper MJ, Warner TT. Mutant torsinA, which causes early-onset primary torsion dystonia, is redistributed to membranous structures enriched in vesicular monoamine transporter in cultured human SH-SY5Y cells. Mov Disord. 2004;20(4):432–440. doi: 10.1002/mds.20351. [DOI] [PubMed] [Google Scholar]
- Morrow G, Heikkila JJ, Tanguay RM. Differences in the chaperone-like activities of the four main small heat shock proteins of Drosophila melanogaster. Cell Stress Chaperones. 2006;11(1):51–60. doi: 10.1379/CSC-166.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy M, Wu HC, Liu Z, Mieszkowska SK, Zolkiewski M. Walker-A threonine couples nucleotide occupancy with the chaperone activity of the AAA+ ATPase ClpB. Protein Sci. 2008;18(2):287–293. doi: 10.1002/pro.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith TV, Heuser JE, Breakefield XO, Hanson PI. TorsinA in the nuclear envelope. Proc Natl Acad Sci USA. 2004;101(20):7612–7617. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Zeng J, Niland BP, Hewett J, Farley J, Irimia D, Li Y, Wiche G, Sonnenberg A, Breakefield XO. TorsinA binds the KASH domain of nespins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci. 2008;121(20):3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2003;146(1–2):106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, DeLeon D, Brin MF, Raymond D, Corey DP, Fahn S, Risch NJ, Buckler AJ, Gusella JF, Breakefield XO. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17(1):40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- Pham P, Frei KP, Woo W, Truong DD. Molecular defects of the dystonia-causing torsinA mutation. Mol Neurosci. 2006;17(16):1725–1728. doi: 10.1097/WNR.0b013e3280101220. [DOI] [PubMed] [Google Scholar]
- Risch NJ, Bressman SB, Deleon D, Brin MF, Burke RE, Greene PE, Shale H, Claus EB, Cupples LA, Fahn S. Segregation analysis of idiopathic torsion dystonia in Ashkenazi Jews suggests autosomal dominant inheritance. Am J Hum Genet. 1990;46(3):533–538. [PMC free article] [PubMed] [Google Scholar]
- Risch NJ, Bressman SB, Senthil G, Ozelius LJ. Intragenic cis and trans modification of genetic susceptibility in DYT1 torsion dystonia. Am J Hum Genet. 2007;80(6):1188–1193. doi: 10.1086/518427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostasy K, Augood SJ, Hewett JW, Leung JC, Sasaki H, Ozelius LJ, Ramesh V, Standaert DG, Breakefield XO, Hedreen JC. TorsinA protein and neuropathology in early onset generalized dystonia with GAG deletion. Neurobiol Dis. 2003;12(1):11–24. doi: 10.1016/S0969-9961(02)00010-4. [DOI] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, Bukau B, Mogk A (2004) Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol 11(7):607–615 [DOI] [PubMed]
- Shashidharan P, Paris N, Sandu D, Karthikeyan L, McNaught KS, Walker RH, Olanow CS. Overexpression of torsinA in PC12 cells protects against toxicity. J Neurochem. 2004;88(4):1019–1025. doi: 10.1046/j.1471-4159.2003.02233.x. [DOI] [PubMed] [Google Scholar]
- Shashidharan P, Sandu D, Potla U, Armata IA, Walker RH, McNaught KS, Weisz D, Sreenath T, Brin MF, Olanow CW. Transgenic mouse model of early-onset DYT1 dystonia. Hum Mol Genet. 2005;14(1):125–133. doi: 10.1093/hmg/ddi012. [DOI] [PubMed] [Google Scholar]
- Song C, Wang Q, Li CH. Characterization of the aggregation-prevention activity of p97/valosin-containing protein. Biochemistry. 2007;46(51):14889–14898. doi: 10.1021/bi700499j. [DOI] [PubMed] [Google Scholar]
- Srere PA, Brazil H, Gonen L. Citrate condensing enzyme of pigeon breast muscle and moth flight muscle. Acta Chem Scand. 1963;17:129–134. doi: 10.3891/acta.chem.scand.17s-0129. [DOI] [Google Scholar]
- Torres GE, Sweeney AL, Beaulieu JM, Shashidharan P, Caron MG. Effect of torsinA on membrane proteins reveals a loss of function and a dominant-negative phenotype of the dystonia-associated DeltaE-torsinA mutant. Proc Natl Acad Sci USA. 2004;101(44):15650–15655. doi: 10.1073/pnas.0308088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky A. Physiology and pathophysiology of the calcium store in the endoplasmic reticulum of neurons. Physiol Rev. 2005;85(1):201–279. doi: 10.1152/physrev.00004.2004. [DOI] [PubMed] [Google Scholar]
- Walker RH, Brin MF, Sandu D, Good PF, Shashidharan P. TorsinA immunoreactivity in brains of patients with DYT1 and non-DYT1 dystonia. Neurology. 2002;58(1):120–124. doi: 10.1212/wnl.58.1.120. [DOI] [PubMed] [Google Scholar]
- Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995;14(9):1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Bukau B, Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J Biol Chem. 2003;278(35):32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8(12):1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- Wickner S, Gottesman S, Skowyra D, Hoskins J, Mckenney K, Maurizi MR. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA. 1994;91(25):12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, DeCuypere M, LeDoux MS. Abnormal motor function and dopamine neurotransmission in DYT1 DeltaGAG transgenic mice. Exp Neurol. 2008;210(2):719–730. doi: 10.1016/j.expneurol.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Li SH, Li XJ. Chaperone suppression of cellular toxicity of Huntingtin is independent of polyglutamine aggregation. J Biol Chem. 2001;276(51):48417–48424. doi: 10.1074/jbc.M104140200. [DOI] [PubMed] [Google Scholar]
- Zhu L, Wrabl JO, Hayashi AP, Rose LS, Thomas PJ. The torsin-family AAA+ protein OOC-5 contains a critical disulfide adjacent to sensor-II that couples redox state to nucleotide binding. Mol Biol Cell. 2008;19(8):3599–3612. doi: 10.1091/mbc.E08-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. J Biol Chem Comm. 1999;274(40):28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]