Abstract
Isotope-edited difference Raman and FTIR studies complemented by ab initio calculations have been applied to the transition state analogue complex of HGPRT●ImmHP●MgPPi to determine the ionic states of the 5’-phosphate moiety of ImmHP and of PPi. These measurements characterize electrostatic interactions within the enzyme active site as deduced from frequency shifts of the phosphate groups. The bound 5’-phosphate moiety of ImmHP is di-anionic, and this phosphate group exists in two different conformations within the protein complex. In one conformation, a hydrogen bond between the 5’-phosphate of ImmHP and the OH group of Tyr104 in the catalytic loop appears to be stronger. With the stronger H-bond, the OH of Tyr104 approaches one of the P••O bonds from the bridging oxygen side to cause distortion of the PO3 moiety, as indicated by a lowered symmetric P••O stretch frequency. The asymmetric stretch frequencies are similar in both phosphate conformations. Bound PPi in this complex is fully ionized to P2O74−. Bond frequency changes for bound PPi indicate coordination to Mg2+ ions but show no indication of significant P••O bond polarization. Extrapolation of these results to reaction coordinate motion for HGPRT suggests that bond formation between C1’ of the nucleotide ribose and the oxygen of PPi is accomplished by migration of the ribocation toward immobilized pyrophosphate.
HGPRT belongs to the Type I phosphoribosyltransferase family of enzymes that catalyzes the displacement of pyrophosphate (PPi) from α-D-5-phosphoribosyl 1-pyrophophate (PRPP) by nitrogen nucleophiles including ammonia, adenine, hypoxanthine, guanine, xanthine, orotate or uracil, to produce a β-substituted ribose 5’-phosphate and PPi. Hypoxanthine-guanine phosphoribosyl transferases from humans and parasites catalyze the reversible Mg2+-dependent transfer of the phosphoribosyl group from PRPP to hypoxanthine or guanine to form IMP or GMP nucleotides, respectively (see Scheme 1). The substrates bind in a functionally ordered fashion, with PRPP binding first in the forward direction and IMP or GMP first in the reverse reaction (1). This enzyme has been studied as a chemotherapeutic target for malaria, giardiases, trypanosomiasis, and toxoplasmosis (2, 3). The feasibility to target Trypanosoma cruzi HGPRT, the agent of Chagas’ disease, has been demonstrated (4). In humans, genetically impaired purine salvage by HGPRT is the basis for heritable gouty arthritis and, in severe cases, Lesch-Nyhan syndrome (5).
Scheme 1.
Reaction catalyzed by HGPRT and its putative transition state.
A detailed understanding of the structure and function of HGPRT is provided from analysis of nearly thirty crystal structures of human or parasite isozymes, including those complexed with a variety of ligands that resemble Michaelis, transition state, and product complexes (1, 6–17). The crystal structures of HGPRT with a bound transition-state analogue show that a disordered catalytic loop moves up to 25 Å to cover the active site and becomes an ordered two-stranded, antiparallel β-sheet, as compared to its disordered state in the HGPRT●GMP●Mg2+ complex (8, 12, 15, 16). A schematic drawing of active site contacts in the transition state analogue complex of human HGPRT is shown in Scheme 2.
Scheme 2.
Active site contacts in the HGPRT/immucillinGP/PPi complex (taken from ref. 16).
Analysis of these crystal structures suggests a reaction coordinate with a relatively fixed purine ring and pyrophosphate but significantly different ribose ring conformations and positions on conversion of reactants to products. The pyrophosphate moiety is highly immobilized by its coordination with two Mg2+ ions and hydrogen bonds to active site Arg199, Lys68, and the backbone NH of Ser103. The position of the 5’-phosphate moiety is also fixed by numerous hydrogen bonds, including one from the conserved Tyr104 in the catalytic loop that closes the catalytic site upon substrate binding. The purine base is hydrogen bonded with conserved protein residues Asp137 and Lys165 and the carbonyl oxygen of Val187 (Scheme 2).
The transition state structure for phosphoribosyltransferases is based on results of kinetic isotope effect studies from the related orotate phosphoribosyltransferases from human, Plasmodium falciparum, and Salmonella typhimurium sources, using phosphonoacetate as a slowly reacting substrate analogue for pyrophosphate (Scheme 1, 18, 19). The transition state for S. typhimurim OPRTase is a dissociative structure with an elongated N1--C1’ bond (bond order of 0.3), minimal formation of the incoming C1’—O bond (bond order 0.02, from the pyrophosphate analogue), oxacarbenium ion character in the ribosyl group and partial positive charge near C-1’. Human and P. falciparium OPRTases are also dissociative but with complete loss of the N1—C1’ bond and low bond order to the attacking nucleophile (19). Ionic stabilization for the transition state is proposed to come from the nearby pyrophosphate anion. In the case of HGPRTase, activation of the base may come from protein or solvent mediated protonation of N-7. From the crystal structures and the putative transition state structures, it has been suggested that the reaction catalyzed by HGPRT proceeds by a mechanism involving nucleophilic displacement by electrophile migration; both the purine and pyrophosphate nucleophiles are relatively fixed in position and the ribosyl group is converted to an oxacarbenium ion that migrates toward the purine base in the forward reaction or toward the pyrophosphate in the reverse direction (20).
Despite these extensive studies on HGPRT, some important mechanistic questions related to proton movements during catalysis remain to be answered. For example, high resolution X-ray structural studies (at 1.05 Å) suggested a tight hydrogen bond between the OH group of Tyr104 in the catalytic loop and one of the 5’-phosphate oxygens of the substrate PRPP. The distance between the two oxygens is 2.67 Å in the HGPRT●9-deazaguanine●MgPRPP complex, and the hydrogen between the two oxygens was proposed to be part of a low-barrier H-bond (12). Thus, there is a possibility that the proton from the Tyr104 hydroxyl group may protonate the 5’-phosphate of the substrate at the transition state. The ionic states of PPi in the HGPRT complexes are also unknown. Moreover, the crystal structures of HGPRT with transition state analogues show the bound PPi with one of the three nonbridging P••O bonds on each phosphorus atom approximately 0.1 Å longer than the other two (PDB code 1BZY, 1CJB), suggesting a dianionic PPi in the complex; however, the 2.0 Å resolution of these structures can not be used with confidence to establish such small differences in PO bond lengths. These, and other important structural issues, can be addressed by vibrational spectroscopy. This approach is sensitive to the electronic nature of specific bonds. For example, bond vibrational frequencies are strongly affected by protonation state. In addition, the characteristic time constant for vibrational spectroscopy is on the femtosecond scale. Hence, structural conformations interconverting on slower time scales are resolved in the vibrational spectrum as isolated bands. These frequencies are diagnostic of specific conformations, and band intensity is proportional to the concentration of each specific conformation.
In this study, isotope edited FTIR and Raman difference techniques are used to determine the ionic states of the 5’-phosphate of Immucillin-HP (ImmHP) and of PPi in the HGPRT●ImmHP●MgPPi complex and to examine the extent of 'activation' of the pyrophosphate nucleophile. ImmHP binds to human HGPRT 14,000 times tighter than IMP, and this complex is considered to be a mimic of the HGPRT transition state (16). The evidence for this contention includes slow, tight binding behavior (21), the protonation states of N7 and N4' (22), and striking immobilization of the peptide backbone (23).
MATERIALS AND METHODS
Mono-sodium and di-sodium glucose 6-phosphate, tetra-sodium and di-sodium PPi were purchased from Aldrich. ImmHP was prepared according published procedures (24). [18O3P]-labeled ImmHP was prepared using γ–18O3 labeled ATP as the phopsphoryl donor in ImmHP synthesis. 18O labeled PPi was prepared according to the published procedure except the amount of 18O water added was reduced to maximize PPi production (25). The product was purified through two cycles of AG1-X8, with elution by 150 mM HCl and neutralized with NaOH.
Enzyme samples
Recombinant human HGPRT was expressed and purified to homogeneity as described previously (26). The purification method yielded an enzyme with undetectable alkaline phosphatase activity, permitting incubations with nucleotide ligands without phosphate loss. Purified HGPRT was stored at 4 °C in 0.1 M Tris-HCl (pH 7.4) containing ammonium sulfate at 70% saturation until ready to use for spectroscopic studies. The enzyme samples for spectroscopic measurements were prepared by extensive dialysis in 10 mM tris at pH 7.5 with 10 mM MgCl2 and 5 mM DTT, followed by concentration with a Centricon30 centrifuge (Amicon, Lexington MA) to the desired concentration. In some cases, a second round of ion exchange chromatography was performed to remove a slight fluorescent impurity. The concentrations of the HGPRT were determined spectroscopically, using a molar extinction coefficient of 26,000 M−1 cm−1 at 280 nm. The HGPRT●ImmHP●MgPPi complex was prepared by adding a slight excess of ImmHP (or 18O labeled ImmHP) and PPi (or 18O labeled PPi) to the concentrated HGPRT sample. After two hours the samples were washed twice with buffer in a Centricon30 concentrator. The binding of the ImmHP●MgPPi complex is tight (Kd = 1 nM; 21), and stoichiometric amounts of ImmHP●MgPPi bind to the HGPRT to form the transition state analogue complex. The typical concentration of enzyme-complex samples was 2 mM.
Difference Raman spectroscopy
The Raman spectra were measured using an optical multichannel analyzer system consisting of a Triplemate spectrometer (Spex Industries, Metuchen, NJ) and a model DIDA-1000 reticon detector connected to an ST-100 detector (Princeton Instruments, Trenton, NJ, or a CCD detector (Princeton Instruments model LN/CCD-1152UV) connected to a ST-135 CCD controller. Details of the system can be found elsewhere (27). The 514.5 nm line from an argon ion laser or 568.2 nm line from a krypton ion laser was used to irradiate the sample (∼100 mW).
Separate spectra for the HGPRT●ImmHP●MgPPi complex with and without 18O labeling on ImmHP or PPi were measured using a special split cell (the volume of each side being about 30 µl) and a sample holder with a linear translator as previously described (27). The spectrum from one side of the split cuvette was taken, the split cell was translated, and the spectrum from the other side was taken. This sequence was repeated until sufficient signal to noise was achieved. A difference spectrum was generated by numerically subtracting the sum of the spectra obtained from each side. In general, the two summed spectra did not subtract to zero, as judged by the subtraction fidelity of know protein bands (for example, the amide-I, amide-III, and the sharp phenylalanine 1004 cm−1 bands, the latter band being especially useful since it is generally not affected by protein conformational changes). These protein bands were discerned from their band widths (generally much broader than those from spectra of bound substrates) and their characteristic positions. Hence, one summed spectrum was scaled by a small numerical factor, generally between 1.05-0.95, which was adjusted until the protein bands were nulled. The same control procedures were performed on all the difference spectra results herein. Resolution of the spectrometer is 8 cm−1 for the present results. Spectral calibration was done for each measurement using the known Raman lines of toluene, and absolute band positions are accurate to within ±2 cm−1. None of the spectra presented here have been smoothed.
Difference FTIR spectroscopy
FTIR spectroscopy was performed on a Magna 760 Fourier transform spectrometer (Nicolet Instrument Corp., WI) using a MCT (mercury-cadmium-telluride) detector. Enzyme ternary complexes with and without 18O labeled ImmHP or PPi were simultaneously loaded into a two-position dual cell shuttle accessory. The FTIR spectra were taken alternatively between these two samples with BaF2 windows and 15-µm Teflon spacers. Spectra were collected in the range of 900 to 4000 cm−1 with 2 cm−1 resolution. A Blackman-Harris three-term apodization and a Happ-Genzel apodization were applied respectively. Omnic 4.1a (Nicolet Instruments, Corp.) software was used for data collection and analysis. Since the sample and reference cells were assembled separately, their path lengths were not exactly equal. To correct for this, the subtracted spectrum was multiplied by a correction factor, typically in the range of 0.95–1.05.
Theoretical Calculations
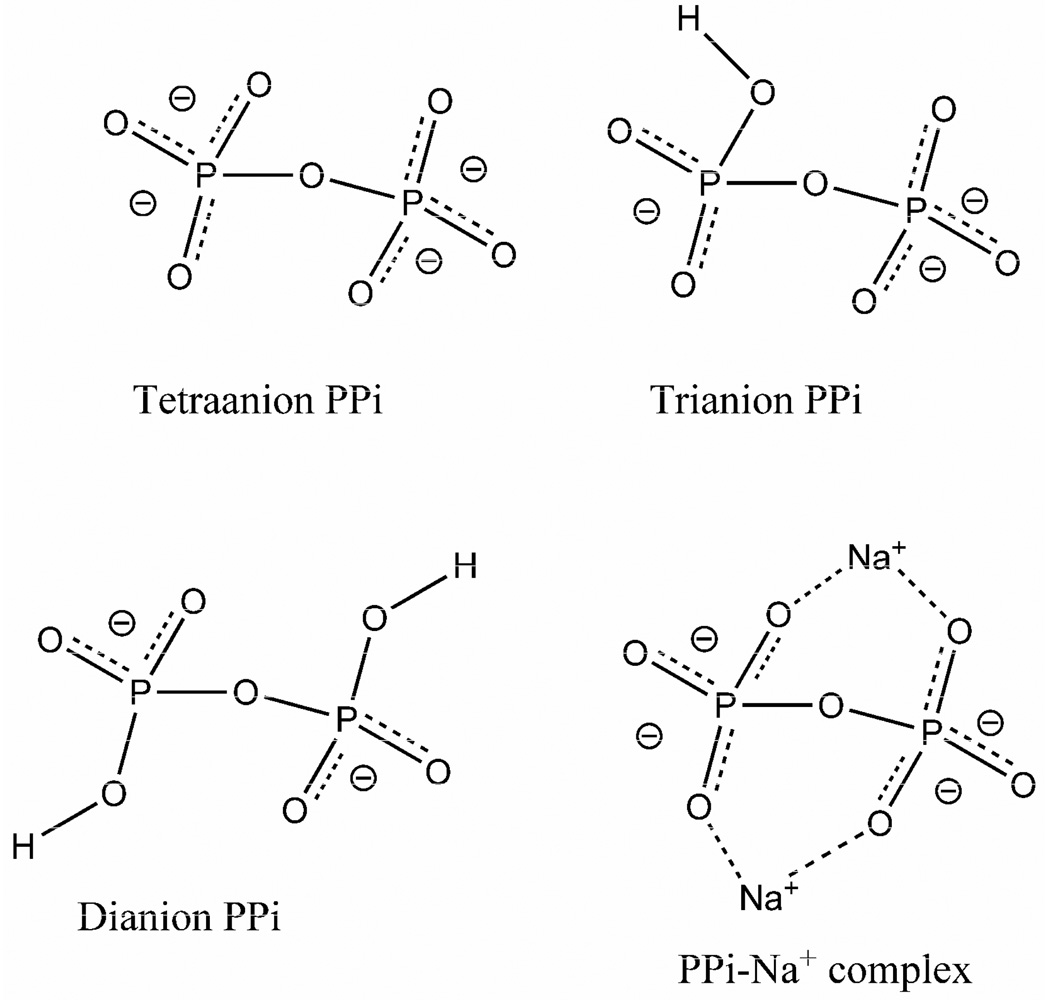
Ab initio calculations were carried out on PPi model compounds using the HF/6–31g** functional, as implemented in Guassian 98. The geometries of the model compounds of tetra-, tri- and di-anionic PPi were first optimized energetically and the vibrational normal modes with Raman and IR intensities were calculated using the same basis set. True local minimum on the potential surface of the complexes for the geometry-optimized complexes were verified from the vibrational frequency calculations in which no imaginary frequency was found. In all cases, a stable structure of the model compound was achieved without any geometry constraints. The same calculations were also performed for the tetra- and di-anionic PPi complexed with two Na+ ions to model the structure of PPi in the HGPRT●ImmHP●MgPPi complex and to determine the effect of Mg2+ ions on the nonbridging P••O stretch frequencies.
RESULTS and DISCUSSION
Ionic state of the 5’-phosphate group of ImmHP in the complex with HGPRT
The ionic state of the ImmHP 5’-phosphate group and its interaction with HGPRT was determined in difference Raman and FTIR measurements. The phosphate vibrational spectra of bound ligand is compared with solution model compounds. Since ImmHP is poorly soluble in common solvents, G6P was used in the phosphomonester solution given the similarity of the phosphate groups in G6P and ImmHP. The Raman and FTIR spectra of G6P in aqueous solution at pH 8.5 characterize the dianionic phosphate moiety (spectra a in Figure 1 and Figure 2). The band in the Raman spectrum at 981 cm−1 (Figure 1) is assigned to the symmetric stretch mode of the PO32− moiety. This mode is also (weakly) IR active, appearing at 979 cm−1 (Fig. 2). Vibrational studies of more than twenty phosphate monoester compounds in solution indicate that the frequency of this mode is 981±5 cm−1 and is insensitive to the attached alcohol. An exception is glucose 1-phosphate, where it is 968 cm−1 (28). The bands at 1091 and 1110 cm−1 in the FTIR spectrum are assigned to the di-anionic phosphate asymmetric stretch modes (Fig. 2). In contrast to the symmetric stretch, the nearly degenerate asymmetric nonbridging P••O stretch modes are sensitive to the alcohol substituent, generally correlated with the pKa of their OH groups. They also show unusually large heterogeneously broadened band widths (28). These modes are strongly IR active but only weakly Raman active. The average frequencies for the asymmetric modes range from 1098 cm−1 for glycerol-2-P to 1112 cm−1 for phosphoenolpyruvate (28).
Figure 1.
(a) Raman spectrum of 100 mM dianionic G6P at pH 8.5. (b) Raman spectrum of 100mM monoanionic G6P at pH 4.0. (c) Raman difference spectrum between HGPRT●ImmHP●MgPPi and HGPRT● 18O-ImmHP●MgPPi complex, at a enzyme concentration of 2mM. The enzyme samples were prepared in 10 mM tris at pH 7.5 with 5 mM DTT. The 514.5 nm line from an argon ion laser or 568.2 nm line from Krypton ion laser was used to irradiate the sample (∼100 mW). The spectral resolution was 8 cm−1.
Figure 2.
(a) FTIR spectrum of 100 mM G6P at pH 8.5. (b) FTIR spectrum of 100mM G6P at pH 4.0. (c) FTIR difference spectrum between HGPRT●ImmHP●MgPPi complex and HGPRT● 18O-ImmHP●MgPPi complex. The enzyme samples were the same as described in Figure 1. The spectral resolution was 2 cm−1.
The Raman and FTIR spectra of G6P at pH 4.0 (where the phosphate moiety of G6P is monoanionic) show the stretch motions of the two P-O single bonds to be coupled, forming a symmetric stretch and an asymmetric stretch (spectra b, Fig. 1 and Fig. 2). The Raman active symmetric stretch mode is observed at 836 cm−1 (Fig. 1b), and the IR active asymmetric mode is observed at 1036 cm−1 (Fig. 2b). The two nonbridging P••O stretches couple to form a symmetric and an asymmetric stretch pair. The symmetric mode is both Raman and IR active, and is observed at 1088 cm−1 and 1086 cm−1 (Fig. 1b and Fig. 2b, respectively). The asymmetric mode is only IR active and is assigned to the band at 1182 cm−1 (Fig. 2b).
Since the vibrational spectra of mono and dianionic phosphate are different, the ionic state of phosphate monoester is readily identified by vibrational spectroscopy. The isotopically edited difference Raman and FTIR spectra for the HGPRT●ImmHP●MgPPi and HGPRT/18O-ImmHP●MgPPi complexes reveal only the vibrational modes that are sensitive to labeling, with the protein contributions eliminated (spectrum c in Fig. 1 and Fig. 2). Difference spectra are essential in these Raman studies since the band intensities from protein modes are comparable to or more intense than phosphate modes (29). The major Raman band at 979 cm−1 is assigned to the symmetric nonbridging P••O stretch mode of the PO32− group on ImmHP based on its shift upon 18O labeling (Figure 1c). The assignment is also supported by the difference IR spectrum in which this bands appears at 977 cm−1 (Fig. 2c). The 1082 cm−1 band in Fig. 2c is assigned to the asymmetric mode of the nonbridging P••O stretch mode of the PO32− moiety of ImmHP in the ternary complex. This mode shifts down to 1049 cm−1 upon 18O labeling.
Based on the vibrational spectroscopic measurements, the phosphate moiety of ImmHP in the HGPRT●ImmHP●MgPPi complex is dianionic. The symmetric nonbridging P••O stretch mode is near 980 cm−1, characteristic of the dianionic phosphate. X-ray crystallographic structures are consistent with this interpretation since the lengths of all three nonbridging P••O bonds of ImmHP in ternary complexes are within 0.03 Å of each other (PDB code 1BZY, 1CJB).
Interactions between HGPRT and the 5’-phosphate of ImmHP, a transition state analogue
The minor Raman band at 957 cm−1 (954 cm−1 in IR) can be assigned to the phosphate symmetric stretch mode since it is active in both Raman and IR and shifts down by about 50 cm−1 upon 18O labeling (Fig. 1c and Fig. 2c, respectively). As the stoichiometries of molecular species are proportional to signal strength in vibrational spectra, the simplest explanation for two 5’-phosphate symmetric stretch bands is two distinct conformations in the protein complex. Phosphate groups within the two conformations have similar asymmetric stretch frequencies but have different symmetric stretch frequencies. Previous studies of phosphate compounds based on molecular modeling with ab initio and empirical methods suggest that the symmetric stretch mode is sensitive to geometry (30). A small increase in the angle between adjacent P••O bonds would reduce the symmetric stretch frequency but leave the asymmetric stretch mode relatively unchanged (30). A structural explanation consistent with the data is for catalytic loops of the different protein subunits to have different hydrogen bonding patterns with the 5’-phosphate. Different hydrogen bonds to the 5’-phosphate from Tyr104 OH would result in a variation of the 5’-phosphate O••P••O angles. Small distance differences, below the resolution of X–ray crystallography (16, 24) can easily account for the observed spectral band pattern (30, 31).
The frequency of the 5’-phosphate asymmetric stretch mode of ImmHP in the HGPRT●ImmHP●MgPPi complex is 1082 cm−1, about 20 cm−1 lower than that of the solution model. Lowered stretch frequency correlates with reduced bond order of the nonbridging P••O bonds, indicating stronger hydrogen bonding to phosphate in the enzyme complex than in water. The enzyme environment also restricts the O••P••O angles compared to solution as shown by the narrow asymmetric P••O stretch band width in the complex (Fig. 2).
The ionic state of PPi in the presence of a transition state analogue
Difference spectroscopic studies were also performed to determine the stretch mode frequencies of PPi within the HGPRT●ImmHP●MgPPi complex, using 18O labeled and unlabeled PPi. The results were compared to the solution spectra to establish phosphate ionic states within the enzyme complex. Many PPi bands are broad in solution and are masked or appear shifted from their true position in difference spectra. Thus, Raman and FTIR spectra were analyzed for solution PPi data. In the HGPRT●ImmHP●MgPPi complex, the PPi bands are sharpened considerably. Here, difference Raman and FTIR spectra eliminate background protein bands and permit characterization of the enzyme complex.
The Raman spectrum of tetra-anionic PPi, obtained from tetra-sodium PPi at pH 11, shows a major band at 1023 cm−1, assigned to the in-phase combination of the two symmetric P••O stretch modes from both PO32− moieties (spectrum a in Figs. 3). This mode shifts down to 978 cm−1 upon 18O labeling (data not shown) and is only active in the Raman spectrum. The bands at 1108 and 1136 cm−1 in the FTIR spectrum of tetra-anionic PPi, are assigned to the asymmetric combinations of the P••O bond stretches (only IR active; spectrum a, Figure 4). The FTIR band at 915 cm−1 is assigned to the asymmetric P-O stretch mode of the bridging P-O-P bonds (IR active, Raman inactive; spectrum a, Figure 4). The assignments of the PPi modes are supported by ab initio calculations on related model compounds (discussed below).
Figure 3.
(a) Raman spectrum of 100 mM tetraanionic PPi (pH11). (b) Raman spectrum of trianionic PPi. This spectrum is obtained by subtracting appropriate amount of a and c from the spectrum of 100 mM PPi at pH7.5. (c) Raman spectrum of 100mM dianionic PPi (pH 4.5). (d) Raman difference spectrum between HGPRT●ImmHP●MgPPi complex and HGPRT●ImmHP●Mg 18O-PPi complex. The experimental conditions were the same as described in Figure 1.
Figure 4.
(a) FTIR spectrum of 100 mM tetraanionic PPi (pH11). (b) FTIR spectrum of 100 mM PPi. This spectrum is obtained by subtracting appropriate amount of a from the spectrum of 100 mM PPi at pH8.0. (c) FTIR spectrum of 100mM dianionic PPi (pH 4.5). (d) FTIR difference spectrum of HGPRT●ImmHP●MgPPi complex and HGPRT●ImmHP●Mg 18O-PPi complex. The experimental conditions were the same as described in Figure 2.
The Raman and FTIR spectra of dianionic PPi, prepared from di-sodium PPi at pH 4.5 are shown in spectrum c in Fig. 3 and Fig. 4, respectively. The major Raman band at 1104 cm−1 is assigned to the in-phase combination of the two symmetric nonbridging P••O stretch modes from the two PO2− moieties (Fig. 3c). This mode shifts down by 39 cm−1 to 1065 cm−1 upon 18O labeling (data not shown) and is also observed in IR spectrum (Fig 4c). One of the major IR bands at 1196 cm−1 is assigned to the out-of-phase combination of the two symmetric nonbridging P••O stretches from the two PO2− moieties (Fig. 4c). Another major IR band at 1082 cm−1 is assigned to an asymmetric stretch of the two terminal P-OH bond stretches. This mode is only IR-active and shifts down by about 37 cm−1 to 1046 cm−1 upon 18O labeling of PPi (data not shown). The shoulder band at ∼1240 cm−1 is assigned to an asymmetric combination of the nonbridging P••O stretch mode. The band at 920 cm−1 is assigned to the asymmetric P-O stretch mode of the bridging P-O-P bonds, and this mode is only IR-active (Fig. 4c).
The Raman and FTIR spectra of tri-anionic PPi are shown in spectrum b in Fig. 3 and Fig. 4, respectively. For PPi, pKa3 is 6.7, and pKa4 is 9.4, so that, at pH values between 7.5–8.0, PPi is >80% tri-anionic. However, contributions from tetra- and di-anionic PPi in the original spectra were apparent and were subtracted to obtain the spectrum of tri-anionic PPi presented here (spectrum b, Fig. 3 and Fig. 4). In tri-anionic PPi, there are five P••O bonds, and three of them are on one phosphorus and two on the other. The most prominent band in the Raman spectrum is at 1085 cm−1. This band is assigned to the symmetric stretch mode of the two nonbridging P••O bonds on one phosphorus since this mode is expected to have the strongest Raman intensity. The Raman band at 1006 cm−1 in Fig. 3b is tentatively assigned to the symmetric stretch of the three P••O bonds of the PO32− moiety on the other phosphorus. These two modes, only Raman active, do not appear in the FTIR spectrum, supporting the assignment. The Raman band at 958 cm−1 (Figure 3b) is tentatively assigned to a combination of stretch motions of the single P-O bonds. The IR bands in Figure 4b at 1008, 1138 and 1195 cm−1 are assigned to various asymmetric combinations of nonbridging P••O stretch vibrations.
The difference Raman and FTIR spectra between HGPRT●ImmHP●MgPPi and HGPRT●ImmHP●Mg 18O-PPi complexes are shown in spectra d of Fig. 3 and Fig. 4, respectively. Only one Raman band is observed at 1027 cm−1 which shifts down by 45 cm−1 to 982 cm−1 upon 18O labeling (spectrum d, Fig. 3). The derivative feature marked with * in this spectrum is an artifact from imperfect subtraction of the sharp protein phenylalanine peak. Two asymmetric nonbridging P••O stretch modes are observed in the FTIR difference spectrum: one at 1153 cm−1 which shifts down by 35 cm−1 to 1118 cm−1 upon 18O labeling, and the other at 1105 cm−1 shifting down to 1082 cm−1 (spectrum d, Fig. 4). The position of the second band can not be accurately determined due to its partial cancellation from the 1118 cm−1 band of the 18O labeled PPi. However, their band widths are much narrower than those of the model compounds in solution.
X-ray structures of HGPRT●ImmGP●MgPPi (where G represents 9-deazaguanine) and HGPRT●ImmHP●MgPPi are closely related and show two Mg2+ ions coordinated to bound PPi. The X-ray structures suggest that one of the PO bonds of PPi is about 0.1 Å longer than the other two PO bonds, but this difference is within the error limits for a structure at 2.0 Å resolution (PDF 1BZY, 16). This bond length difference is characteristic of dianionic PPi. However, vibrational spectroscopic studies of PPi in the HGPRT●ImmHP●MgPPi complex indicate that PPi is in the tetra-anionic state since the Raman active band at 1027 cm−1 (Fig. 3d) is close to the band at 1024 cm−1 from solution tetra-anionic PPi (Fig. 3a) but far from the major Raman band of other ionic forms. If PPi were in a bound tri/di-anionic form, PPi would be expected to show a nonbridging P••O stretch mode would have shifted down by 60∼80 cm−1 in the Raman data (compare spectrum b, c with d in Fig. 3). To determine if such a shift is possible for P••O stretching modes, studies on the PPi and Mg2+ ion interactions were performed by vibrational spectroscopic studies of PPi/Mg2+ model compounds in solution and by ab initio calculations.
When PPi is complexed with Mg2+, nonbridging P••O stretch mode frequencies increase. With GDP as an example, the symmetric stretch mode of the αPO2− moiety at 1089 cm−1 (analogous to the 1085 cm−1 band in tri-anionic PPi) is increased by 5 cm−1 in the MgGDP complex, and the two asymmetric stretch modes at 1115 cm−1 and 1205 cm−1 (analogous to the 1138 and 1195 cm−1 bands in tri-anionic PPi) are increased by 8 and 10 cm−1, respectively, in the MgGDP complex (32). Spectra of PPi complexed with Mg2+ in various ionic states show similar shifts of the nonbridging P••O stretch modes. For example, the in-phase symmetric P••O stretch mode of tetra-anionic PPi at 1023 cm−1 (spectrum a, Fig. 3) shifts up by 9 cm−1 in the MgPPi complex although the two asymmetric P••O stretch modes at 1108 and 1136 cm−1 (spectrum a, Fig. 4) shift up by less than 2 cm−1 (data not shown). Thus, the interaction between Mg2+ and PPi in solution causes a small upward shift (up to 15 cm−1) in the nonbridging P••O stretch modes. However, it is also clear that interactions similar to those in solution can not cause the 60∼80 cm−1 downward shift of the symmetric stretch mode of the tri-/di-anionic PPi if we assume the PPi bound in HGPRT●ImmHP●MgPPi is tri-/di-anionic.
Theoretical calculations
Vibrational analyses using ab initio methods on model compounds for PPi and their Mg2+ complexes were used to determine if the Mg2+-PPi interactions suggested by X-ray crystallographic studies can induce the observed nonbridging P••O stretch mode frequency changes from solution to the HGPRT complex. The tetra-, tri- and di-anionic forms of PPi were used to model their respective solution compounds, and the results of the calculations were then evaluated against the experimentally observed values. The tetra- and di-anionic PPi complexed with two sodium ions are used to model Mg2+-PPi interaction in HGPRT●ImmHP●MgPPi for reasons discussed below.
Since there is a high degree of symmetry in all ionic states of PPi, group theory can be applied to predict Raman and IR intensities of the PO stretch modes. Group theory predicts that the most prominent bands in the Raman spectrum should be the in-phase combination of the two symmetric nonbridging P••O stretch modes of the PO32− groups for tetra-anionic PPi and of the PO2− groups for dianionic PPi because the motions of these modes maintain the symmetry of the molecule. These modes are easily identified in the Raman spectra and from calculations. They are also sensitive to the ionic state of PPi, and thus we focus our attention on these modes (see Scheme 3).
Scheme 3.
Model PPi compounds used in vibrational normal mode calculations.
The PPi models were first optimized at the HF/6–31g** level and the vibrational frequencies with their Raman and IR intensities were calculated with the same basis set. These calculations overestimate the stretching force constants by about 20% and frequencies by about 10% (33, 34). The coupling constants among nonbridging P••O stretches are also overestimated, resulting in a larger frequency separation between symmetric and asymmetric stretch modes than is measured. Another source of computational of error arises from the neglect of solvent. The interaction of water with PPi decreases nonbridging P••O stretch frequencies. However, the errors are mostly uniform, and we are more interested in the frequency change when Mg2+ ions are complexed with PPi. The errors in the absolute frequencies yielded by the calculations do not affect our conclusions. Similar frequency calculations on the phosphate compounds at MP2/6–31g** or B3LYP/6–31g** levels yielded better agreement with the experimental values for asymmetric stretch but worse on the symmetric stretch frequencies, and, on average, no significant improvements over HF calculations were observed (22). Thus, they were not explored further.
The experimentally observed nonbridging P••O stretch frequencies in Raman and FTIR spectra of PPi in various ionic states and in the HGPRT●ImmHP●MgPPi complex are compared (Table 1, Fig. 3 and Fig. 4). The calculated frequencies of the nonbridging P••O stretch modes of various PPi model compounds are shown in Table 2. For tetra-anionic PPi, there are six nonbridging P••O stretch modes from the six P••O bonds. The calculations suggest that two of the modes do not have significant intensity in either Raman or IR spectra. The calculated Raman-active mode is the in-phase combination of the two symmetric P••O stretches of the two PO32− groups with a frequency of 1039 cm−1. The calculations also predict that three of the five asymmetric combinations of the P••O stretches at 1163, 1172 and 1178 cm−1 have significant IR intensity (Table 2). The calculated frequency of the in-phase P••O stretch mode is less that 20 cm−1 higher than the experimentally observed value but the calculated frequencies of the asymmetric modes are 40–50 cm−1 higher.
Table 1.
Experimentally observed nonbridging P••O stretch modes in tetra-, tri-, dianionic and HGPRT bound PPi.
| Normal Modes |
||||
|---|---|---|---|---|
| νs | νa1 | νa2 | νa3 | |
| P2O74− | 1023 | 1108 | 1136 | |
| P2O7H3− | 1006, 1085 | 1108 | 1138 | 1195 |
| P2O7H22− | 1104 | 1196 | 1240 | |
| In the complex | 1027 | 1105 | 1153 | |
νs is the Raman active symmetric nonbridginge P••O stretches observed in the Raman spectra.
νas are the IR active asymmetric combination of the nonbridgng P••O stretches. All frequencies are in cm−1.
Table 2.
Calculated nonbridging P••O stretch modes in tetra-, tri-, and dianionic PPi. As well as the tetra- and dianionic PPi complexed with 2 Na+.
| Model | Normal Modes |
||||
|---|---|---|---|---|---|
| νs | νa1 | νa2 | νa3 | ||
| P2O74− | 1039 | 1163 | 1172 | 1178 | |
| + 2Na+ | 1042 | 1070 | 1279 | 1294 | |
| P2O7H3− | 1052, 1161 | 1209 | 1258 | 1292 | |
| P2O7H22− | 1179 | 1180 | 1343 | 1360 | |
| + 2Na+(1) | 1213 | 1177 | 1360 | 1438 | |
| + 2Na+(2) | 1230 | 1175 | 1320 | 1354 | |
| + 2Na+(3) | 1207 | 1130 | 1442 | 1461 | |
νs is the in-phase combination of the two symmetric P••O stretches from the two PO32− and two PO2− groups in tetra- and di- anionic PPi, respectively. For tri-anionic PPi, one symmetric P••O stretch from PO32− and one from PO2− are listed as νs. νas are the asymmetric P••O stretch modes with the highest IR intensities as calculated by ab initio methods. All frequencies are in cm−1. The two Na+s in the model were positioned approximately at equal distances from the two phosphorus atoms. In model 1 of the dianionic NaPPi complex, one P-OH bond pointed away from the two Na+ ions and the other P-OH bond pointed approximately towards the Na+ ions. In model 2, both P-OH bonds pointed away from Na+ ions. In model 3, both P-OH bonds pointed approximately towards Na+ ions.
In trianionic PPi, there are five nonbridging P••O stretch modes. The calculations suggest two Raman-active modes from the two symmetric P••O stretch modes at 1052 and 1161 cm−1 of PO32− and PO2− moiety, respectively. The calculations also predict that all three asymmetric combinations of the P••O stretches at 1209, 1259 and 1293 cm−1 have significant IR intensity (listed in Table 2). The calculated frequency of the symmetric P••O stretch mode from PO32− is ∼40 cm−1 higher than the experimentally observed frequency (1006 cm−1) but all other calculated frequencies are ∼100 cm−1 higher. Since the errors are quite uniform among different vibrational modes, we can make the assignments of the observed Raman and IR bands based on these calculations.
In dianionic PPi, there are four nonbridging P••O stretch modes. The calculated Raman active mode is the in-phase combination of the two symmetric P••O stretches of two PO2− moiety and its frequency is at 1179 cm−1. The calculated frequencies for asymmetric P••O stretch modes are at 1180, 1343 and 1360 cm−1, respectively (Table 2). The calculated frequency of the in-phase P••O stretch mode is about 70 cm−1 higher than the experimentally observed value and the calculated frequencies of the asymmetric modes are 20 to 100 cm−1 higher.
Although the calculations overestimate the non-bridging P••O stretch frequencies of PPi, the order of the frequencies are maintained not only within the same ionic state, but also amongst different ionic states. Thus, additional calculations on tetra- and dianionic PPi complexed with two Na+ ions are expected to provide reliable information about the effect of Mg2+-PPi interactions on the non-bridging P••O stretch modes. Na+ ions rather than Mg2+ ions were used because both of the two Mg2+ ions in the HGPRT complex are also coordinated with four other ligands (see Scheme 2), and its electronic effects on PPi are reduced as a consequence.
Two Na+ ions were placed near the bridging oxygens between the two phosphorus atoms of PPi, similar to the positions of Mg2+ found in the X-ray structures. The geometry of the complex was then optimized and the frequencies were calculated from the optimized geometry. In the optimized geometry for dianionic PPi complexed with Na+, the P-O(H) bond lengths are about 0.1 Å longer than the nonbriding P••O bonds, the same as the difference in the bond lengths of the PO bonds in the X-ray structure of the HGPRT●ImmGP●MgPPi complex (2.0 Å resolution, 1BZY, 16). In the tetra-anionic PPi-Na+ complex, the length differences among the nonbriding P••O bonds were less than 0.04 Å, similar to that of the 5’-phosphate group of ImmGP in the X-ray structure of the HGPRT●ImmGP●MgPPi complex. Calculations on the tetra-anionic PPi/Na+ complex indicate that the Raman active in-phase symmetric P••O stretch mode would shift up by only 3 cm−1 while the IR active asymmetric stretch modes shift by about 100 cm−1 in both up and down directions (Table 2). Three different models were used for dianionic PPi/Na+ complexes: the geometry of the first was constructed with one P-OH bond pointing away from both Na+ ions and another P-OH bond pointing approximately toward one Na+. A second model had both P-OH bonds pointed away from Na+ ions. The third had each P-OH pointing approximately towards a Na+ ion. In each of the three models, the in-phase symmetric P••O stretch mode was shifted up by 30 cm−1 or more compared to isolated dianionic PPi. The asymmetric modes were mostly shifted up as well (Table 2).
Changes of the PPi P••O stretch modes upon binding to HGPRT●ImmHP●MgPPi are best simulated by the tetra-anionic NaPPi model. The frequency of the in-phase symmetric stretch mode shifts up by a few cm−1 and the highest frequency asymmetric stretch mode shifts up by more. Dianonic PPi in the HGPRT●ImmHP●MgPPi complex can be eliminated as the in-phase P••O stretch mode would be expected to shift down by about 70 cm−1 compared to that in solution (Table 1). This shift is not possible in the complex with two Mg2+ ions since experiments and calculations indicated that the effect of the Mg2+ on dianionic PPi would be to shift the in-phase P••O stretch mode to higher frequency. Thus, we conclude that tetra-ionic PPi is bound in the HGPRT●ImmHP●MgPPi complex.
Conclusion
Isotope edited difference Raman and FTIR spectroscopy permit the assignment of the ionic states of the phosphate groups in the HGPRT●ImmHP●MgPPi complex. The 5’-phosphate group of ImmHP is dianionic and exists in two different conformations. One explanation is that the hydrogen bond between the 5’-phosphate of ImmHP and the OH of Tyr104 in the catalytic loop is slightly stronger in half of the subunits. As the Tyr-OH approaches one of the P••O bonds from the bridging oxygen side, it likely causes some flattening of the PO3 moiety, as indicated by a lowered symmetric P••O stretch frequency and a lowered average P••O frequency. Bound PPi is tetra-anionic. The frequency changes for enzyme bound PPi are consistent with coordination to Mg2+ ions but show no indication of significantly polarized P••O bonds. In this structural mimic of the transition state for HGPRT, PPi bond polarization by interaction with the cationic iminoribitol group of ImmHP is negligible.
Vibrational studies of purine nucleoside phosphorylase (PNP), a related enzyme, with the nonphosphorylated transition state analogue ImmH and phosphate have shown that the phosphate resembles a highly activated nucleophile with distorted P-O bond geometry similar to the transition state. One of the nonbridging P••O bonds is sufficiently polarized by enzyme active site interactions to cause its stretch motions to become decoupled from the motions of the other two P••O bonds (22). X-ray structures of the PNP complex suggest that the strong P••O bond polarization of the phosphate nucleophile is induced by combined electrostatic and hydrogen bonding interactions from the 3’-OH of the analogue and the positively charged iminoribitol cation of ImmH (35, 36). In the X-ray structures of the HGPRT●ImmHP●MgPPi complex, one of the pyrophosphate P••O bonds also forms hydrogen bonding and electrostatic interactions with the 3’-OH and with the iminoribitol cation of ImmHP. The oxygen of this P••O bond also coordinates with one of the active site Mg2+ ions (see Scheme 2). Thus, a strong polarization of this P••O bond might be expected. However, the vibrational spectra do not support a polarized P••O bond in the HGPRT●ImmHP●MgPPi complex. All P••O stretch frequencies from PPi shift to somewhat higher values compared with the solution frequencies, consistent with coordination to Mg2+ ions without additional polarization (see above). The X-ray structures of HGPRT●ImmHP●MgPPi revealed two factors to explain the lack of polarization. The Mg2+ ion coordinated with the P••O bond is almost perpendicular to the P••O bond orientation, and the iminoribitol cation is 3.3 Å from the O2A oxygen of PPi (see Scheme 2). This distance is 0.3 Å longer than the similar ionic interaction between iminoribitol cation of ImmH and the activated oxygen of phosphate bound in the PNP●ImmH●phosphate complex (37). These features indicate that the HGPRT●ImmHP●MgPPi complex in HGPRT differs from the PNP●ImmH●phosphate complex. The nonpolarized P••O bond of the HGPRT complex suggests more leaving group activation or weaker similarity to the transition state in the HGPRT complex than that in the PNP complex phosphate and ImmH. This hypothesis is supported by a Km/Ki ratio of 690,000 for PNP but only 14,000 for human HGPRT in complexes with ImmH (26, 38).
Acknowledgment
We are grateful to Dr. Beth L. Pietrak for making the enzyme used in this study.
Abbreviations
- HGPRT
hypoxanthine-guanine phosphoribosyltransferase
- OPRTase
orotate phosphoribosyltransferases
- PNP
purine nucleoside phosphorylase
- G6P
glucose-6-phosphate
- PPi
pyrophosphate
- PRPP
α-D 5-phosphoribosyl 1-pyrophosphate
- IMP
inosine 5’-monophosphate
- GMP
guanosine 5’-monophosphate
- ImmHP
Immucillin-H 5’-phosphate
- ImmGP
Immucillin-G 5’-phosphate
Footnotes
This work was supported in part by Grants EB001958 (HD), GM068036 (RC), GM41916 (VLS), and GM52125 (CG) from the National Institutes of Health.
References
- 1.Xu Y, Eads J, Sacchettini JC, Grubmeyer C. Kinetic mechanism of human hypoxanthine-guanine phosphoribosyltransferase: rapid phosphoribosyl transfer chemistry. Biochemistry. 1997;36:3700–3712. doi: 10.1021/bi9616007. [DOI] [PubMed] [Google Scholar]
- 2.Ullman B, Carter D. Hypoxanthine-guanine phosphoribosyltransferase as a therapeutic target in protozoal infections. Infectious Agents & Disease. 1995;4:29–40. [PubMed] [Google Scholar]
- 3.Wang CC. Parasite enzymes as potential targets for antiparasitic chemotherapy. Journal of Medicinal Chemistry. 1984;27:1–9. doi: 10.1021/jm00367a001. [DOI] [PubMed] [Google Scholar]
- 4.Freymann DM, Wenck MA, Engel JC, Feng J, Focia PJ, Eakin AE, Craig SP. Efficient identification of inhibitors targeting the closed active site conformation of the HPRT from Trypanosoma cruzi. Chem Biol. 2000;7:957–968. doi: 10.1016/s1074-5521(00)00045-4. [DOI] [PubMed] [Google Scholar]
- 5.Sculley DG, Dawson PA, Emmerson BT, Gordon RB. A review of the molecular basis of hypoxanthine-guanine phosphoribosyltransferase (HPRT) deficiency. Human Genetics. 1992;90:195–207. doi: 10.1007/BF00220062. [DOI] [PubMed] [Google Scholar]
- 6.Balendiran GK, Molina JA, Xu Y, Torres-Martinez J, Stevens R, Focia PJ, Eakin AE, Sacchettini JC, Craig SP., 3rd Ternary complex structure of human HGPRTase, PRPP, Mg2+, and the inhibitor HPP reveals the involvement of the flexible loop in substrate binding. Protein Science. 1999;8:1023–1031. doi: 10.1110/ps.8.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guddat LW, Vos S, Martin JL, Keough DT, de Jersey J. Crystal structures of free, IMP-, and GMP-bound Escherichia coli hypoxanthine phosphoribosyltransferase. Protein Sci. 2002;11:1626–1638. doi: 10.1110/ps.0201002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canyuk B, Focia PJ, Eakin AE. The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography. Biochemistry. 2001;40:2754–2765. doi: 10.1021/bi001195q. [DOI] [PubMed] [Google Scholar]
- 9.Kanaani J, Somoza JR, Maltby D, Wang CC. Probing the active site of Tritrichomonas foetus hypoxanthine-guanine-xanthine phosphoribosyltransferase using covalent modification of cysteine residues. European Journal of Biochemistry. 1996;239:764–772. doi: 10.1111/j.1432-1033.1996.0764u.x. [DOI] [PubMed] [Google Scholar]
- 10.Somoza JR, Chin MS, Focia PJ, Wang CC, Fletterick RJ. Crystal structure of the hypoxanthine-guanine-xanthine phosphoribosyltransferase from the protozoan parasite Tritrichomonas foetus. Biochemistry. 1996;35:7032–7040. doi: 10.1021/bi953072p. [DOI] [PubMed] [Google Scholar]
- 11.Munagala N, Basus VJ, Wang CC. Role of the flexible loop of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Tritrichomonas foetus in enzyme catalysis. Biochemistry. 2001;40:4303–4311. doi: 10.1021/bi0026932. [DOI] [PubMed] [Google Scholar]
- 12.Heroux A, White EL, Ross LJ, Kuzin AP, Borhani DW. Substrate deformation in a hypoxanthine-guanine phosphoribosyltransferase ternary complex: the structural basis for catalysis. Structure. 2000;8:1309–1318. doi: 10.1016/s0969-2126(00)00546-3. [DOI] [PubMed] [Google Scholar]
- 13.Heroux A, White EL, Ross LJ, Davis RL, Borhani DW. Crystal structure of Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase with XMP, pyrophosphate, and two Mg(2+) ions bound: insights into the catalytic mechanism. Biochemistry. 1999;38:14495–14506. doi: 10.1021/bi990508i. [DOI] [PubMed] [Google Scholar]
- 14.Heroux A, White EL, Ross LJ, Borhani DW. Crystal structures of the Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase-GMP and -IMP complexes: comparison of purine binding interactions with the XMP complex. Biochemistry. 1999;38:14485–14494. doi: 10.1021/bi990507q. [DOI] [PubMed] [Google Scholar]
- 15.Focia PJ, Craig SP, 3rd, Nieves-Alicea R, Fletterick RJ, Eakin AE. A 1.4 A crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi. Biochemistry. 1998;37:15066–15075. doi: 10.1021/bi981052s. [DOI] [PubMed] [Google Scholar]
- 16.Shi W, Li CM, Tyler PC, Furneaux RH, Grubmeyer C, Schramm VL, Almo SC. The 2.0 A structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor. Nature Structural Biology. 1999;6:588–593. doi: 10.1038/9376. [DOI] [PubMed] [Google Scholar]
- 17.Eads JC, Scapin G, Xu Y, Grubmeyer C, Sacchettini JC. The crystal structure of human hypoxanthine-guanine phosphoribosyltransferase with bound GMP. Cell. 1994;78:325–334. doi: 10.1016/0092-8674(94)90301-8. [DOI] [PubMed] [Google Scholar]
- 18.Tao W, Grubmeyer C, Blanchard JS. Transition state structure of Salmonella typhimurium orotate phosphoribosyltransferase. Biochemistry. 1996;35:14–21. doi: 10.1021/bi951898l. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Luo M, Schramm VL. Transition state of Plasmodium falciparem and human orotate phosphoribosyltransferases. J. Am. Chem. Soc. 2009;131:4685–4694. doi: 10.1021/ja808346y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schramm VL, Grubmeyer C. Phosphoribosyltransferase mechanisms and roles in nucleic Acid metabolism. Prog Nucleic Acid Res Mol Biol. 2004;78:261–304. doi: 10.1016/S0079-6603(04)78007-1. [DOI] [PubMed] [Google Scholar]
- 21.Li CM, Tyler PC, Furneaux RH, Kicska G, Xu Y, Grubmeyer C, Girvin ME, Schramm VL. Transition-state analogs as inhibitors of human and malarial hypoxanthine-guanine phosphoribosyltransferases. Nature Structural Biology. 1999;6:582–587. doi: 10.1038/9367. [DOI] [PubMed] [Google Scholar]
- 22.Deng H, Lewandowicz A, Schramm VL, Callender R. Activating the phosphate nucleophile at the catalytic site of purine nucleoside phosphorylase: a vibrational spectroscopic study. J Am Chem Soc. 2004;126:9516–9517. doi: 10.1021/ja049296p. [DOI] [PubMed] [Google Scholar]
- 23.Wang F, Shi W, Nieves E, Angeletti RH, Schramm VL, Grubmeyer C. A transition state analogue reduces protein dynamics in hypoxanthine-guanine phosphoribosyltransferase. Biochemistry. 2001;40:8043–8054. doi: 10.1021/bi010203f. [DOI] [PubMed] [Google Scholar]
- 24.Shi W, Li CM, Tyler PC, Furneaux RH, Cahill SM, Girvin ME, Grubmeyer C, Schramm VL, Almo SC. The 2.0 A structure of malarial purine phosphoribosyltransferase in complex with a transition-state analogue inhibitor. Biochemistry. 1999;38:9872–9880. doi: 10.1021/bi990664p. [DOI] [PubMed] [Google Scholar]
- 25.Hackney DD, Stempel KE, Boyer PD, Daniel LP. Methods in Enzymology. Academic Press; 1980. [3] Oxygen-18 probes of enzymic reactions of phosphate compounds; pp. 60–83. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Grubmeyer C. Catalysis in human hypoxanthine-guanine phosphoribosyltransferase: Asp 137 acts as a general acid/base. Biochemistry. 1998;37:4114–4124. doi: 10.1021/bi972519m. [DOI] [PubMed] [Google Scholar]
- 27.Callender R, Deng H. Nonresonance Raman difference spectroscopy: a general probe of protein structure, ligand binding, enzymatic catalysis, and the structures of other biomacromolecules. Annual Review of Biophysics & Biomolecular Structure. 1994;23:215–245. doi: 10.1146/annurev.bb.23.060194.001243. [DOI] [PubMed] [Google Scholar]
- 28.Cheng H, Nikolic-Hughes I, Wang JH, Deng H, O'Brien PJ, Wu L, Zhang ZY, Herschlag D, Callender R. Environmental effects on phosphoryl group bonding probed by vibrational spectroscopy: implications for understanding phosphoryl transfer and enzymatic catalysis. Journal of the American Chemical Society. 2002;124:11295–11306. doi: 10.1021/ja026481z. [DOI] [PubMed] [Google Scholar]
- 29.Deng H, Ray WJ, Jr, Burgner JW, 2nd, Callender R. Comparison of vibrational frequencies of critical bonds in ground-state complexes and in a vanadate-based transition-state analog complex of muscle phosphoglucomutase. Mechanistic implications. Biochemistry. 1993;32:12984–12992. doi: 10.1021/bi00211a006. [DOI] [PubMed] [Google Scholar]
- 30.Deng H, Wang J, Ray WJ, Callender R. Relationship Between Bond Stretching Frequencies and Internal Bonding for [16O4] and [18O4] Phosphates in Aqueous Solution. J. Phys. Chem. B. 1998;102:3617–3623. [Google Scholar]
- 31.Ray WJ, Jr, Burgner JW, 2nd, Deng H, Callender R. Internal chemical bonding in solutions of simple phosphates and vanadates. Biochemistry. 1993;32:12977–12983. doi: 10.1021/bi00211a005. [DOI] [PubMed] [Google Scholar]
- 32.Wang JH, Xiao DG, Deng H, Callender R, Webb MR. Vibrational study of phosphate modes in GDP and GTP and their interaction with magnesium in aqueous solution. Biospectroscopy. 1998;4:219–227. doi: 10.1002/(SICI)1520-6343(1998)4:4%3C219::AID-BSPY1%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 33.Hehre WJ, Radom L, Schleyer PvR, Pople JA. ab-initio Molecular Orbital Theory. New York: Wiley; 1986. [Google Scholar]
- 34.Pulay P, Fogarasi G, Pongor G, Boggs JE, Vargha A. Combination of Theoretical ab initio and Experimental Information To Obtain Reliable Harmonic Force Constants. Scaled Quantum Mechanical (SQM) Force Fields for Glyoxal, Acrolain, Butadiene, Formaldehyde, and Ethylene. J. Am. Chem. Soc. 1983;105:7037–7047. [Google Scholar]
- 35.Shi W, Basso LA, Santos DS, Tyler PC, Furneaux RH, Blanchard JS, Almo SC, Schramm VL. Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces. Biochemistry. 2001;40:8204–8215. doi: 10.1021/bi010585p. [DOI] [PubMed] [Google Scholar]
- 36.Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, Almo SC. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 37.Lewandowicz A, Shi W, Evans GB, Tyler PC, Furneaux RH, Basso LA, Santos DS, Almo SC, Schramm VL. Over-the-barrier transition state analogues and crystal structure with Mycobacterium tuberculosis purine nucleoside phosphorylase. Biochemistry. 2003;42:6057–6066. doi: 10.1021/bi0343830. [DOI] [PubMed] [Google Scholar]
- 38.Murkin AS, Birck MR, Rinaldo-Matthis A, Shi W, Taylor EA, Almo SC, Schramm VL. Neighboring Group Participation in the transition state of human purine nucleoside phosphorylase. Biochemistry. 2007;46:5039–5049. doi: 10.1021/bi700147b. [DOI] [PMC free article] [PubMed] [Google Scholar]