Abstract
Trophoblast stem cells (TS cells), derived from the trophectoderm (TE) of blastocysts, require transcription factors (TFs) and external signals (FGF4, INHBA/NODAL/TGFB1) for self-renewal. While many reports have focused on TF networks that regulate embryonic stem cell (ES cell) self-renewal and pluripotency, little is know about TF networks that regulate self-renewal in TS cells. To further understand transcriptional networks in TS cells, we used chromatin immunoprecipitation with DNA microarray hybridization (ChIP-chip) analysis to investigate targets of the TFs—TCFAP2C, EOMES, ETS2, and GATA3—and a chromatin remodeling factor, SMARCA4. We then evaluated the transcriptional states of target genes using transcriptome analysis and genome-wide analysis of histone H3 acetylation (AcH3). Our results describe previously unknown transcriptional networks in TS cells, including TF occupancy of genes involved in ES cell self-renewal and pluripotency, co-occupancy of TCFAP2C, SMARCA4, and EOMES at a significant number of genes, and transcriptional regulatory circuitry within the five factors. Moreover, RNAi depletion of Tcfap2c, Smarca4, and Eomes transcripts resulted in a loss of normal colony morphology and down-regulation of TS cell–specific genes, suggesting an important role for TCFAP2C, SMARCA4, and EOMES in TS cell self-renewal. Through genome-wide mapping and global expression analysis of five TF target genes, our data provide a comprehensive analysis of transcriptional networks that regulate TS cell self-renewal.
The trophoblast is the first cell lineage to emerge during mammalian development. Originating from a thin layer of trophectoderm surrounding the inner cell mass (ICM) of the blastocyst, trophoblast cells differentiate into epithelial cells types of the placenta. Trophoblast stem (TS) cells are derived from preimplantation stage embryos and are capable of self-renewing indefinitely in the presence of external signals, including FGF4 (Tanaka et al. 1998), INHBA, NODAL, and TGFB1 (Tanaka et al. 1998; Erlebacher et al. 2004), and of differentiating into fetal tissues of the placenta, including trophoblast giant cells, spongiotrophoblasts, glycogen trophoblast cells, and syncytiotrophoblasts (Cross et al. 2003). Removal of these external signals results in decreased proliferation and trophoblast cell differentiation, whereas targeted disruption of Fgf4 or Fgfr2 results in post-implantation lethality due to insufficient trophoblast proliferation (Feldman et al. 1995; Arman et al. 1998; Goldin and Papaioannou 2003).
TS cells, in contrast to pluripotent embryonic stem (ES) cells, are multipotent and are therefore only capable of differentiating into cells represented in the trophoblast lineage. TS cells and ES cells both share the capacity to self-renew indefinitely in vitro in the presence of appropriate external signals and transcriptional machinery. ES cell self-renewal and pluripotency requires core transcription factors (TFs) POU5F1 (formerly known as OCT4), SOX2, and NANOG (Nichols et al. 1998; Avilion et al. 2003; Chambers et al. 2003), while TS cell self-renewal and multipotency require TFs such as TCFAP2C, CDX2, EOMES, ESRRB, ETS2, SOX2, and TEAD4 (Chawengsaksophak et al. 1997; Luo et al. 1997; Russ et al. 2000; Auman et al. 2002; Avilion et al. 2003; Wen et al. 2007; Nishioka et al. 2008). Recently, it has been shown that forced expression of Pou5f1, Sox2, Klf4, and Myc (formerly known as c-Myc) is sufficient to reprogram mouse and human fibroblasts into pluripotent cells (Takahashi et al. 2007a,b; Wernig et al. 2007). It has also been demonstrated that Mycn and Esrrb aid in inducing pluripotency (Blelloch et al. 2007; Feng et al. 2009). While TS cells do not express Pou5f1 or Klf4 at significant levels, TS cells express high levels of the reprogramming factors Sox2, Myc, Mycn, and Esrrb, suggesting that common factors may promote self-renewal in both TS cells and ES cells. While these studies provide a foundation for understanding mechanisms of self-renewal, additional work is necessary to further understand transcriptional networks and epigenetic phenomena that contribute to TS cell self-renewal and induce pluripotency in somatic cells.
To further understand transcriptional networks that promote self-renewal in TS cells, we evaluated global promoter binding of five factors in TS cells TCFAP2C (formerly known as Ap-2γ), SMARCA4 (formerly known as BRG1), EOMES, ETS2, and GATA3 using genome-wide chromatin immunoprecipitation with DNA microarray hybridization (ChIP-chip) analysis. These factors have important roles in regulating TS cell self-renewal and placental development. TCFAP2C, SMARCA4, EOMES, ETS2, and GATA3 have been implicated in maintaining TS cell self-renewal and placental development (Ma et al. 1997; Bultman et al. 2000; Russ et al. 2000; Auman et al. 2002; Werling and Schorle 2002; Wen et al. 2007; Kidder et al. 2009). TCFAP2C is expressed in the TE of implantation stage embryos, and Tcfap2c-null embryos, which have reduced expression of CDX2 and EOMES, die around embryonic day (E) 8.5 (Werling and Schorle 2002). Moreover, Tcfap2c-null blastocysts are unable to form outgrowths in vitro (Werling and Schorle 2002), demonstrating that TCFAP2C is important in TS cell self-renewal and placental development. EOMES has also been shown to be essential for mouse trophoblast formation, where Eomes-null embryos arrest at the blastocyst stage, and TE from these embryos fails to differentiate into trophoblast cells, suggesting that EOMES is necessary for TS cell self-renew or differentiation (Russ et al. 2000). The ETS2 TF is also essential for placental development and TS cell self-renewal, where a conditional knock-out of Ets2 promotes TS cell differentiation and a loss of self-renewal (Wen et al. 2007). GATA3 and SMARCA4 are also important in TS cell maintenance and differentiation. Recently, it was shown that GATA3 is expressed in the TE but not the ICM of preimplantation embryos, and RNAi depletion of Gata3 transcripts inhibits morula to blastocyst development, demonstrating that GATA3 is also important in TS cell self-renewal and differentiation (Home et al. 2009). SMARCA4 may also be important in TS cell self-renewal, where Smarca4-null embryos die around the implantation stage and fail to form outgrowths (Bultman et al. 2000; Kidder et al. 2009). By mapping global targets of five factors TCFAP2C, SMARCA4, EOMES, ETS2, and GATA3, we aim to evaluate transcriptional networks that contribute to TS cell self-renewal. Our results describe previously unknown global maps of transcriptional networks in TS cells. TFs occupied genes uniquely expressed in TS cells, and genes involved in ES cell self-renewal and pluripotency. We also observed co-occupancy of multiple TFs at target genes and transcriptional regulatory circuitry within the five factors. Interestingly, TCFAP2C, SMARCA4, and EOMES co-occupied a significant number of genes. In addition, RNAi depletion of Tcfap2c, Smarca4, and Eomes transcripts resulted in a loss of self-renewal and down-regulation of TS cell– and ES cell–enriched genes, suggesting an important role for TCFAP2C, SMARCA4, and EOMES in maintenance of TS cell self-renewal. By comparing TF target genes with transcriptome data generated from undifferentiated and differentiated TS cells and with genome-wide AcH3 marks, we found that TF-bound genes are mainly active in TS cells, suggesting that the investigated TFs may support transcription of target genes. By mapping genome-wide targets of five TFs, our data provide insight into transcriptional networks that regulate TS cell self-renewal.
Results
Genome-wide mapping of TF binding profiles in TS cells
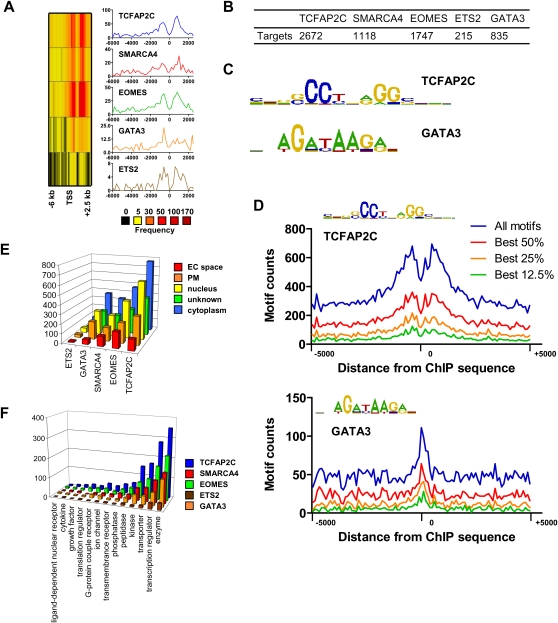
We used ChIP-chip analysis to interrogate whole-genome promoter binding of four TFs, TCFAP2C, EOMES, ETS2, and GATA3, and one chromatin remodeling factor, SMARCA4, in TS cells using antibodies specific to five proteins to enrich for factor bound DNA sequences. DNA-enriched sequences were amplified and hybridized to whole-genome promoter tiling arrays covering 28,000 mouse promoter regions (Affymetrix mouse promoter 1.0R tiling arrays; see methods). Binding peaks were analyzed using Tilemap (Ji and Wong 2005) and annotated to the nearest transcriptional start site (TSS) using CisGenome (see Methods) (Ji et al. 2008).
The majority of regions bound by the five factors are distal to the TSS of target genes (Fig. 1A). The number of promoters occupied by each gene can be found in Figure 1B, and the locations of annotated bound regions can be found in Supplemental Tables S1 through 5. The factors occupied varying numbers of promoters: TCFAP2C occupied more promoters (10% of 28,000 promoters) than the other four factors, while SMARCA4, EOMES, and GATA3 occupied a relatively similar number of promoters and ETS2 occupied a fewer number of promoters. Only a small number of bound regions are found within proximal promoter regions (<300 bp upstream or downstream of TSS; TCFAP2C [43 genes], SMARCA4 [24 genes], EOMES [34 genes], ETS2 [four genes], and GATA3 [18 genes]), while the majority of bound regions occupy distal regulatory regions (>300 bp upstream or downstream of TSS). To determine consensus sequences bound by the five factors, we performed de novo motif discovery using CisFinder (Sharov and Ko 2009) software. Using this approach, we obtained consensus-binding motifs for TCFAP2C (McPherson and Weigel 1999) and GATA3 (Fig. 1C; Merika and Orkin 1993) and overrepresented DNA motifs for SMARCA4, EOMES, and ETS2 (data not shown). TCFAP2C binds predominantly to the palindrome motif (GCCNNNGGC), and GATA3 binds mainly to the motif (GATA) (Fig. 1D).
Figure 1.
Analysis of TF binding in TS cells. (A) Distance of binding regions relative to the nearest transcriptional start site (TSS). (B) Number of target genes occupied by each TF or transcriptional regulator. (C) Enriched DNA binding motifs for TCFAP2C and GATA3 identified using CisFinder software (http://lgsun.grc.nia.nih.gov/CisFinder/). (D) TCFAP2C and GATA3 binding motifs are overrepresented in ChIP-enriched regions. Genomic sequences (10,000 bp) centered on ChIP-enriched TCFAP2C or GATA3 binding regions were extracted from the genome and evaluated for the number of TCFAP2C and GATA3 motifs. CisFinder software was used to generate position-frequency matrices (PFM) for TCFAP2C and GATA3 binding motifs. We assumed five false-positives per 10 kb of random sequence. (E) Gene Ontology (GO) functional annotation of bound genes was performed using ingenuity pathway analysis (IPA). GO terms for cellular location include extracellular space (EC), plasma membrane (PM), nucleus, unknown, and cytoplasm. (F) GO terms for biological function identified using IPA.
Next, we functionally annotated genes bound by the five factors using ingenuity pathway analysis (IPA). For each factor (TCFAP2C, SMARCA4, EOMES, ETS2, and GATA3), IPA identified several biological processes overrepresented in the list of bound genes such as gene expression, cell cycle, and cellular, organismal, or tissue development (data not shown). Additionally, for all factors, proteins encoded by bound genes are located with similar frequencies in cellular compartments, including extracellular space, plasma membrane, nucleus, and cytoplasm (Fig. 1E). Moreover, the five factors occupied genes encoding a similar percentage of protein types (e.g., ligand-dependent nuclear receptors, cytokines, and transcription regulator) (Fig. 1F). The DAVID tool (Dennis et al. 2003) was used to further functionally annotate genes bound by each factor. Similar to IPA analysis, we observed target genes encoding proteins with TF activity (data not shown). Additionally, TCFAP2C, SMARCA4, and EOMES bound genes shared enrichment of many Gene Ontology (GO) terms, suggesting that potential targets of these three factors may be more functionally related to one another than they are to potential targets of ETS2 and GATA3.
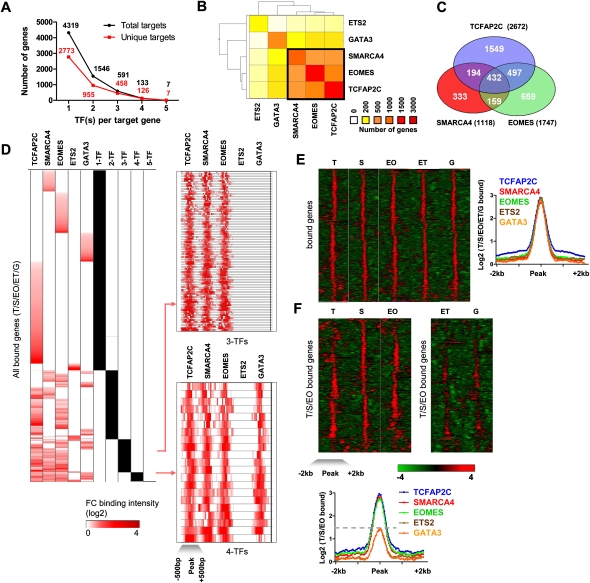
Multiple TF occupancy at target genes
Recent reports have shown that multiple pluripotency-related factors, including POU5F1, SOX2, and NANOG, co-occupy many target genes in ES cells (Boyer et al. 2005; Chen et al. 2008; Kim et al. 2008). To investigate whether multiple multipotency-related and self-renewal factors co-occupy target genes in TS cells, we evaluated the number of target promoters occupied by multiple TFs. The five factors were found to bind a total of 4319 genes (Fig. 2A). Interestingly, cobinding of multiple factors occurred at many target genes. We observed two TFs bound at 955 genes, three TFs bound at 458 genes, and four TFs bound at 126 genes (Fig. 2A). Moreover, the genomic binding patterns of genes with multiple TF occupancy are similar, suggesting that protein complexes may potentially act in concert to transcriptionally regulate target genes. To further investigate the relatedness of TF binding at common targets, we analyzed multiple factor co-occupancy at target gene loci (Fig. 2B). Among the five factors, TCFAP2C and EOMES co-occupied the greatest number of target genes. Additionally, TCFAP2C, SMARCA4, and EOMES cobound a significant number of promoters (432 genes) (Fig. 2B,C), with the number of genes bound by TCFAP2C/SMARCA4/EOMES accounting for 73% of the total genes bound by any three TFs examined in this study. Other pairwise associations of TF cobinding, including less frequent co-occupancy of 2-TF combinations, can be found in Figure 2B. Binding of multiple TFs to common targets suggests potential synergistic regulation of target gene expression.
Figure 2.
Multiple transcription factor occupancy at target genes. (A) Number of target genes occupied by multiple factors. X-axis represents the number of factors per target gene, and the y-axis represents the number of genes occupied by multiple factors. (Black dots) Total targets occupied by at least N-factors. (B) Factor co-occupancy. Heat map colors reflect co-occurrence of factor pairs, where red indicates a higher and yellow indicates a lower number of genes. One major cluster includes factors TCFAP2C, SMARCA4, and EOMES. (C) Venn diagram showing the relationship between TCFAP2C, SMARCA4, and EOMES bound genes. (D) Max fold-change binding profiles for accumulated target genes of all factors were clustered to reveal binding patterns. Representative binding profiles of 3-TF and 4-TF target genes were centered on enrichment peaks [−500 bp, +500 bp] and clustered using Spotfire. (E) TF binding profiles were centered on enrichment peaks [−2 kb, +2 kb] and clustered using Spotfire. Average binding profiles of 5-TFs are also shown. (F) Promoter profiles of TCFAP2C (T), SMARCA4 (S), and EOMES (EO) common target genes were clustered. Note the enriched binding of TCFAP2C, SMARCA4, and EOMES relative to ETS2 (ET) and GATA3 (G) at target loci. Average binding profiles of TCFAP2C, SMARCA4, and EOMES at common target loci are also shown.
To investigate a relationship between peak binding intensity at enriched loci and occupancy of multiple factors, we clustered peak TF binding intensities at target genes according to occupancy by single or multiple factors (Fig. 2D). While peak binding intensities were not identical at all target genes, we observed similar binding intensities between single and multiple factors at target genes. To further analyze binding intensities of genes bound by TCFAP2C, SMARCA4, EOMES, ETS2, and GATA3, we clustered binding profiles of the five factors using a 4-kbp window around the bound enrichment peak, and we found that the overall binding profiles for each factor were similar in overall intensity and length of binding (Fig. 2E). Next, we clustered binding profiles of genes cobound by TCFAP2C, SMARCA4, and EOMES (Fig. 2F). For genes bound by TCFAP2C, SMARCA4, and EOMES, binding profiles were stronger compared with ETS2 and GATA3 binding at the same genes, demonstrating enriched binding at factor-occupied target gene loci. Representative binding profiles of 4-TF, 3-TF, 2-TF, and 1-TF occupied target genes can be found in Figure 2.
TFs bind pluripotency and self-renewal related genes in TS cells
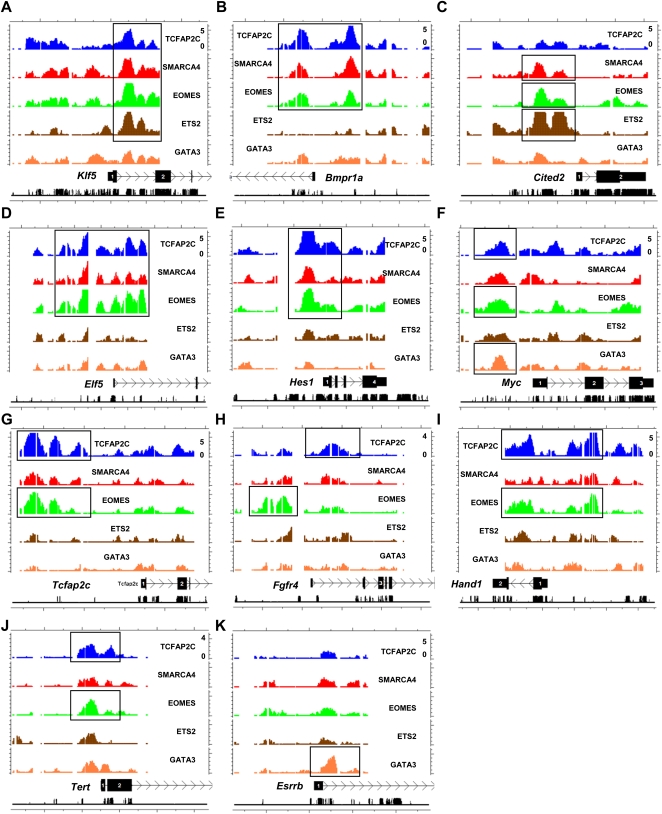
Strikingly, our genome-wide promoter analysis revealed that TFs occupy genes important in ES cell self-renewal and pluripotency, including Bmp4, Esrrb, Klf5, Klf6/7/9, Lif, Lifr, Rest, Sall4, Stat3, and Tert (Fig. 3; Supplemental Tables S1–S5). TFs also occupied genes highly expressed in ES cells whose role in promoting self-renewal is less clear, such as Fbxo15 and Zfp42 (formerly known as Rex1). Moreover, TFs occupied epigenetic regulators, including the de novo DNA methyltransferase Dnmt3b, Dnmt3l, and histone deacetylases (Hdac6/9/10) (Supplemental Tables S1–S5). Binding of TFs to epigenetic regulators may contribute to regulation of chromatin structure in TS cells. Furthermore, by occupying genes involved in ES cell self-renewal, these TFs may control TS cell self-renewal through shared mechanisms with ES cells to promote transcription of target genes. For example, ESRRB is highly expressed and is essential for maintaining self-renewal in both ES cells and TS cells (Luo et al. 1997; Zhang et al. 2008). While it is surprising that TFs occupy pluripotency-related genes in TS cells, it is noteworthy to mention that three TFs investigated in this study, including Tcfap2c, Smarca4, and Ets2, are highly expressed at the mRNA level in both TS cells and ES cells (Sharova et al. 2007), which lends support to the idea that these TFs may have common self-renewal functions in TS cells to promote multipotency and in ES cells to promote pluripotency. The remaining TFs, Eomes and Gata3, are expressed at elevated levels in TS cells compared with ES cells, suggesting that these TFs have unique functions in TS cell maintenance and differentiation. Expression of Gata3 in TS cells and trophoblast cells is consistent with TH2 dominance that prevails at the placental–decidual interface (Yang et al. 1994).
Figure 3.
Genomic view of TF binding in TS cells. TF binding site analysis at target gene loci. Representative profiles of TS cell–enriched genes occupied by four (A), three (B–F), two (G–J), and one (K) factor(s). MA enrichment values adjusted to log2 and conservation are shown on the plot.
TF binding was also found at additional genes enriched in TS cells, important in TS cell self-renewal and differentiation, such as Acvr2a, Elf5, Fgfr2, Fgfr4, Bmpr1a, Bmp4, Bmp8b, Cdh1/3, Cited2, Hand1, Hes1, Notch1-4, Smad2-7, Tead4, Transforming growth factor-beta receptors (Tgfbr1/3), and Wnt signaling genes (Wnt2b/3a/5a/7a/7b/9a, and Smo) (Fig. 3; Supplemental Tables S1–S5). Canonical WNT-CTNNB1 signaling has been suggested to play a role in TS cell self-renewal versus differentiation (Xie et al. 2008), where inhibition of WNT signaling perturbs normal blastocyst implantation and TS cell differentiation. NOTCH signaling components such as Notch1-4, Jag2, Dll1, Dtx2/3, and Rbpj, bound by at least one factor in this study, have been shown to be expressed during mouse preimplantation and in ES cells and TS cells (Cormier et al. 2004). In human ES cells, NOTCH signaling is required for differentiation into the three germ layers but is dispensable for trophoblast commitment (Yu et al. 2008), suggesting that NOTCH signaling, while not necessary for differentiation, may be involved in TS cell self-renewal. Disruption of NOTCH signaling genes is lethal around midgestation or results in no observed phenotype possibly due to functional redundancy or maternal expression (Swiatek et al. 1994; Conlon et al. 1995; de la Pompa et al. 1997). FGF4/INHBA/NODAL/TGFB1 signals are sufficient to maintain TS cell self-renewal. TF occupancy of FGF and TGFB signaling genes suggests a positive-feedback loop where TFs occupy FGF receptors (Fgfr2/4), Bmpr1a, TGFB receptors (Tgfbr1/3), Activin receptor (Acvr2a), and SMAD genes (Smad2-7), which transduce external signals (FGF4/INHBA/NODAL) that are required to maintain TS cell self-renewal. These results suggest that multiple signaling pathways are involved in maintaining the TS cell phenotype.
We also observed transcriptional regulatory and autoregulatory circuitry within the five factors. Four factors were found to bind at least one promoter of the other three genes (Supplemental Tables S1–S5). For example, TCFAP2C bound its own promoter in addition to the Gata3 promoter, while EOMES exhibited self-promoter binding and also bound Tcfap2c, Ets2, and Gata3 promoters. These results that showed self-promoter binding are consistent with previous reports in ES cells where POU5F1, SOX2, and NANOG have been shown to bind to their own promoter (Boyer et al. 2005). Self-promoter binding may serve to reinforce expression of genes that are important in maintaining TS cells self-renewal.
Transcriptome analysis of undifferentiated and time-course differentiated TS cells
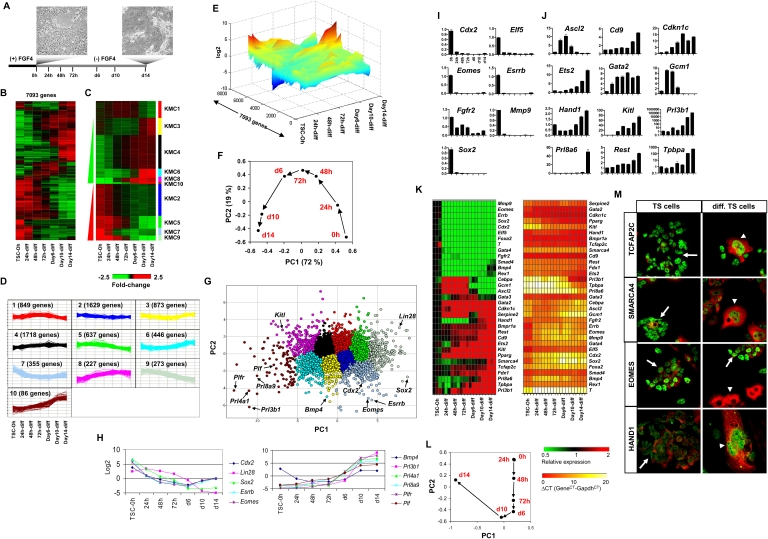
To evaluate the expression state of target genes of the investigated five TFs, we performed whole-genome expression analysis of undifferentiated and differentiated TS cells through a time-course of 14 d (Fig. 4A). Total RNA was harvested at several time points from TS cells cultured without FGF4 over 2 wk for transcriptome analysis using Affymetrix Mouse 430 v2.0 microarrays. Genes whose expression differed by at least twofold between any two groups with a false discovery rate (FDR) adjusted P-value < 5% were considered differentially expressed. We used hierarchical clustering analysis (HCA) to identify major patterns of gene expression variability during TS cell differentiation (Fig. 4B). We further refined patterns of gene expression variability using k-means clustering analysis (KMC) (Fig. 4C). KMC2/5/7/9 include genes whose expression is down-regulated during TS cell differentiation, while KMC4/6/8/10 include genes whose expression is up-regulated during TS cell differentiation; KMC1/3 includes additional patterns of gene expression observed during differentiation (Fig. 4C,D). These patterns of gene expression variability are distinctly visible on a landscape plot (Fig. 4E). Genes whose expression was down-regulated during differentiation include TS cell–enriched genes such as Cdx2, Elf5, and Eomes and pluripotency-related genes such as Dnmt3l, Dppa2/4, Esrrb, Lin28, Sall4, Sox2, Utf1, and Tbx3. Genes whose expression was up-regulated include genes enriched in differentiated trophoblast and placenta tissue such as Arnt, Cdkn1c, Cited1/2, Hand1, Kitl, Plf, Plfr, Prl3b1, Prl8a6, and Tpbpa. Interestingly, we observed up-regulation of pluripotency-related genes such as Klf4 and Rest and genes enriched in TS cells such as Ets2, suggesting that these genes may function in self-renewal and differentiation. Expression of Egfr and Gcm1 was up-regulated early during differentiation and then down-regulated later during TS cell differentiation. Gcm1 expression demonstrates initiation of differentiation into the labyrinthine/syncytiotrophoblast lineage (Cross 2005). Our transcriptome data recapitulates the in vivo expression profile of Gcm1, where Gcm1 expression is enriched beginning at E7.5 in the chorion (Cross 2005) and peaks around E13.5 in mid-gestation placentas (Knox and Baker 2008).
Figure 4.
Microarray expression analysis of undifferentiated and time-course differentiated TS cells. (A) Experimental design. TS cells were cultured in MEF-conditioned medium in the presence of FGF4. TS cell differentiation proceeded in the absence of FGF4 over 14 d. RNA was collected from TS cells at 0 h, 24 h, 48 h, and 72 h and on day 6, day 10, and day 14. (B) Hierarchical clustering analysis (HCA) of differentially expressed genes (greater than twofold). (C) HCA of k-means-clustered differentially expressed genes. (D) Patterns of gene expression identified using k-means clustering. (E) Landscape view of k-means-clustered genes. (F) PCA plot of the first two principal components describing most of the data variability. (G) PCA plot of differentially expressed genes clustered according to k-means. (H) Log2 adjusted expression values of genes expressed in TS cells and differentiated cells. (I) Genes expressed in TS cells were down-regulated during differentiation. qRT-PCR expression analysis of genes enriched in undifferentiated TS cells or bound by at least one factor (Cdx2, Elf5, Eomes, Esrrb, Fgfr2, Mmp9, and Sox2). Data was normalized to Gapdh and then to the expression of TS cells cultured in the presence of FGF4 at 0 h. (J) Genes expressed in differentiated trophoblast and placental cells were up-regulated during TS cell differentiation. qRT-PCR expression analysis of trophoblast lineage-specific genes (Ascl2, Cd9, Cdkn1c, Ets2, Gata2, Gcm1, Hand1, Kitl, Prl3b1, Prl8a6, Rest, and Tpbpa), including genes also bound by at least one factor (Ets2, Gata2, Gcm1, Hand1, and Rest). (K) HCA of qRT-PCR gene expression data. Fold-change expression values were clustered with a green/black/red color scale. Expression values relative to Gapdh (Gene CT – Gapdh CT) were clustered with a red/orange/yellow color scale. (L) PCA of fold-change gene expression data obtained from qRT-PCR. Data plotted in the first two components reveal a stepwise progression of undifferentiated TS cells at 0 h to differentiated TS cells at day 14. (M) Confocal immunofluorescence analysis of TCFAP2C, SMARCA4, EOMES, and HAND1 expression in undifferentiated and differentiated TS cells. TCFAP2C and SMARCA4 expression is enriched in both undifferentiated and differentiated TS cells, EOMES expression is enriched in undifferentiated TS cells, and HAND1 expression is enriched in differentiated TS cells. TCFAP2C, SMARCA4, EOMES, and HAND1 are labeled in green, and Cytokeratin is labeled in red. Arrows indicate undifferentiated TS cells, and arrowheads indicate differentiated trophoblast cells.
Next, we used principal component analysis (PCA) to reduce the dimensionality of the data set to a two-dimensional space. PCA revealed a stepwise progression within the first two components along the PC1 axis beginning with TS cells undergoing differentiation at 0 h through 14 d (Fig. 4F). Genes plotted in the first two components and clustered according to k-means include genes that are enriched in either TS cells or differentiated cells (Fig. 4G). Examples of genes that are enriched in undifferentiated TS cells include Eomes, Esrrb, Lin28, and Sox2, while genes that are enriched in differentiated TS cells include Kitl, Pfn2, Prl3b1, Prl4a1, and Prl8a6 (Fig. 4G,H).
qRT-PCR was used to confirm the expression of a subset of differentially expressed genes. TS cells cultured in the absence of FGF4 down-regulated expression of Cdx2 and other TS cell–enriched genes, also occupied by TFs, including Elf5, Eomes, Esrrb, Fgfr2, Mmp9, and Sox2 (Fig. 4I,K). We also observed up-regulation of genes expressed in differentiated giant cells and placental tissue, including Ascl2, Cd9, Cdkn1c, Kitl, Prl3b1, Prl8a6, and Tpbpa, and additional placental enriched genes, also bound by TFs, including Ets2, Gata2, Gcm1, Hand1, and Rest (Fig. 4J,K). Interestingly, several TS cell–enriched genes were up-regulated upon differentiation, including Ets2 and Rest, while other TS cell–enriched genes were expressed at relatively similar levels in undifferentiated and differentiated TS cells such as Tcfap2c, Smarca4, and Gata3 (Fig. 4K), suggesting that these genes may support both TS cell self-renewal and differentiation. To further evaluate differences in gene expression during differentiation, we performed PCA using data obtained from qRT-PCR (Fig. 4L). Prl3b1 was removed from this analysis because its change in expression was much greater than any other gene analyzed. PCA demonstrated that gene expression profiles move in a step-wise fashion during differentiation, which is consistent with our microarray expression data. Confocal immunofluorescence analysis showed that TCFAP2C, SMARCA4, and EOMES protein is expressed in undifferentiated TS cells while TCFAP2C, SMARCA4, and HAND1 protein is expressed in differentiated TS cells (Fig. 4M), which suggests that TCFAP2C and SMARCA4 may be involved in TS cell self-renewal and differentiation.
Expression of TF occupied genes in TS cells and ES cells
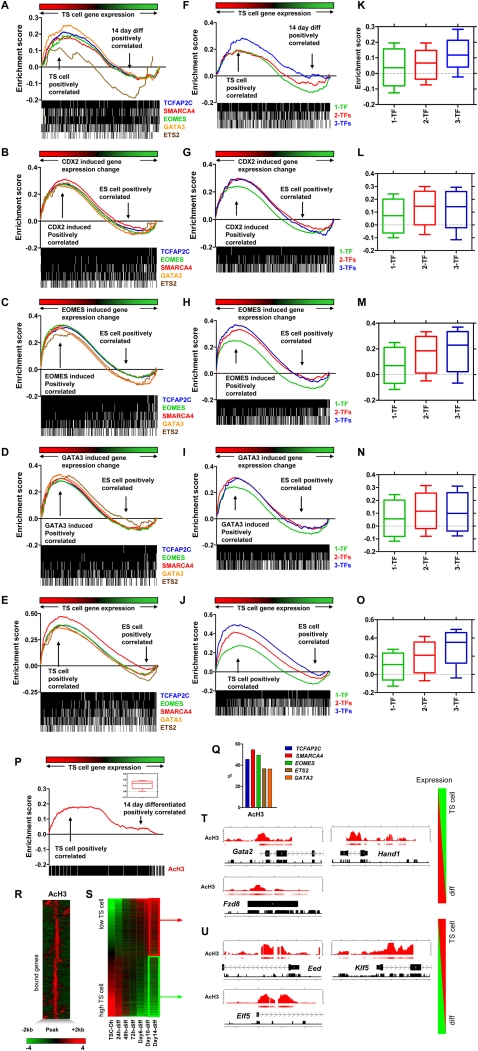
Our results demonstrate that the investigated five TFs occupy a subset of genes that are implicated in self-renewal in both TS cells and ES cells. However, to further understand the expression states of other factor-occupied genes, we compared these genes with transcriptome data from this study and public data sets, including expression data from induction of Cdx2, Eomes, and Gata3 in ES cells (Nishiyama et al. 2009) and TS cells and ES cells (Sharova et al. 2007), using gene set enrichment analysis (GSEA) (Subramanian et al. 2005). We first compared factor-occupied genes with time-course transcriptome data generated in this study. With the exception of ETS2 target genes, which are active and inactive in TS cells, these GSEA results show that the expression of the majority of factor-occupied genes is enriched in undifferentiated TS cells. We did, however, observe enrichment of a subset of TCFAP2C, SMARCA4, EOMES, and GATA3 target genes in differentiated TS cells, suggesting that TFs occupy some inactive genes in TS cells (Fig. 5A). Next, by comparing factor-occupied genes identified in this study with global expression data generated from induction of Cdx2, Eomes, and Gata3 in ES cells (Nishiyama et al. 2009), we found that the majority of TF occupied genes are active following overexpression of the TE-specific genes Cdx2, Eomes, and Gata3 (Fig. 5B–D). Moreover, target gene expression is enriched in TS cells compared with ES cells (Fig. 5E). While TFs occupy several genes that promote self-renewal in ES cells and TS cells as described earlier, these results demonstrate that expression of the majority of occupied genes is enriched in TS cells compared with ES cells.
Figure 5.
GSEA analysis of TF occupied genes and histone H3 acetylation (AcH3) in TS cells. GSEA analysis of TF target genes in undifferentiated and differentiated TS cells (A); Cdx2- (B), Eomes- (C), and Gata3-overexpressing (D) ES cells; and TS cells and ES cells (E). The majority of target genes are expressed higher in undifferentiated compared with differentiated TS cells, and Cdx2-, Eomes-, and Gata3-overexpressing ES cells compared with control ES cells, and TS cells compared with ES cells. The x-axis represents enrichment of gene expression in TS cells at 0 h through 14 d of differentiation (A); Cdx2- (B), Eomes- (C), or Gata3-overexpressing (D) ES cells compared with control ES cells; or TS cells compared with ES cells (E). Target genes are represented as a black and white heat map below the plot. (F–J) GSEA analysis of multiple versus single factor-occupied genes in TS cells. Expression of genes that are common targets of three and two factors is more enriched in TS cells (F); Cdx2- (G), Eomes- (H), and Gata3-overexpressing (I) ES cells; and TS cells compared with genes occupied by a single factor (J). (K–O) Distribution of enrichment scores of multiple factor-occupied genes in TS cells (K); Cdx2- (L), Eomes- (M), and Gata3-overexpressing (N) ES cells; and TS cells (O). (P) GSEA of AcH3 associated genes in undifferentiated and differentiated TS cells. Expression of AcH3 associated genes is enriched in undifferentiated TS cells. The x-axis represents enrichment of gene expression in TS cells at 0 h through 14 d of differentiation. Inset shows the distribution of enrichment scores of AcH3 associated genes in TS cells. (Q) Association between AcH3 marks and binding of TFs. (R) Profiles of AcH3 marks were centered on enrichment peaks [−2 kb, +2 kb] and clustered. (S) Heat map of TS cell transcriptome data sorted by the level of gene expression in TS cells at 0-h genomic profiles of AcH3 marks at genes expressed at low (T) and high (U) levels in undifferentiated versus differentiated TS cells. MA enrichment values adjusted to log2 and conservation are shown on the plot.
Previous reports have suggested that multiple-factor occupancy at target genes is correlated with gene activity, while genes bound by fewer or single factors are less active (Kim et al. 2008). Binding of single factors may support repression of target genes, possibly in association with corepressor complexes, while promoter binding of multiple TFs may drive expression of target genes. To investigate a relationship between multiple factor occupancy and the expression state of target genes, we categorized genes based on the number of factors occupied at their promoters. Due to the low number of genes occupied by four and five factors, these data were excluded from this analysis. GSEA was then used to evaluate the expression states of genes in undifferentiated and time-course–differentiated TS cells (Fig. 5F), ES cells overexpressing TE-specific genes Cdx2, Eomes, and Gata3 (Fig. 5G–I), and TS cells and ES cells (Fig. 5J). We found that genes occupied by multiple factors were more highly expressed in TS cells relative to differentiated TS cells or ES cells, compared with genes occupied by single factors (Fig. 5F–O). Binding of multiple factors may support active transcription through the cumulative function of each factor or through cooperation with other cofactors, while binding of single factors may be less supportive of active transcription through association with repressive cofactors.
Genome-wide histone H3 acetylation in TS cells
Chromatin modifications play important roles in regulating mammalian gene expression (Kouzarides 2007). For example, histone H3 acetylation (AcH3) is associated with actively transcribed regions. To evaluate genes with acetylated histone H3 marks in TS cells also bound by TCFAP2C, SMARCA4, EOMES, ETS2, or GATA3, we performed ChIP-chip analysis in TS cells using an antibody specific to AcH3 (see Methods). GSEA was then used to evaluate the expression state of target genes in undifferentiated and differentiated TS cells (Fig. 5P). Genes associated with AcH3 marks were enriched in TS cells (Fig. 5P). By evaluating AcH3 enrichment near promoter regions occupied by TCFAP2C, SMARCA4, EOMES, ETS2, or GATA3, we found that TCFAP2C, SMARCA4, and EOMES bound regions were more associated with AcH3 marks than ETS2 and GATA3 bound regions (Fig. 5Q). Moreover, SMARCA4 bound regions were more associated with AcH3 marks than the other four TFs. This result is supported by the known role of SMARCA4 in associating through its bromodomain with acetylated lysine residues on histone H3 and H4 tails (Chandrasekaran and Thompson 2007; Shen et al. 2007). We then evaluated AcH3 profiles near TSSs (Fig. 5R), sorted gene expression data from undifferentiated TS cells to identify low and high expression genes (Fig. 5S), and analyzed AcH3 states of genes with high and low enrichment in TS cells. We did not observe significant AcH3 enrichment near promoters of genes with low expression in TS cells such as Gata2, Hand1, and Fzd8 (Fig. 5T). However, we observed AcH3 enrichment near promoters of genes highly expressed in TS cells, including Eed, Elf5, and Klf5 (Fig. 5U), thus supporting our findings that the investigated TFs occupy active and inactive genes in TS cells.
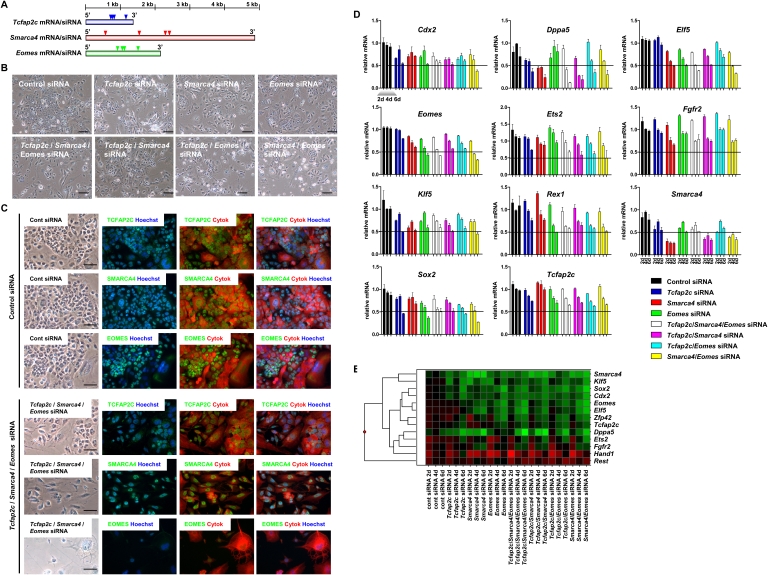
TCFAP2C, SMARCA4, and EOMES are required for TS cell self-renewal
While TCFAP2C, SMARCA4, and EOMES have previously been shown to have important roles in TE specification in vivo (Bultman et al. 2000; Russ et al. 2000; Auman et al. 2002; Kidder et al. 2009), their loss-of-function phenotypes have not been thoroughly evaluated in TS cells. Because our ChIP-chip results show that TCFAP2C, SMARCA4, and EOMES co-occupy many genes in TS cells, we were interested in evaluating the roles of TCFAP2C, SMARCA4, and EOMES in TS cell self-renewal. Therefore, we used RNAi to deplete transcripts of Tcfap2c, Smarca4, and Eomes in TS cells. TS cells were nucleofected with pooled siRNAs, targeting multiple regions, or scrambled control siRNAs, every 2 d for 6 d (Fig. 6A). Following 6 d of RNAi knockdown, we observed a loss of normal colony morphology in Tcfap2c siRNA, Smarca4 siRNA, and Eomes siRNA nucleofected TS cells compared with control siRNA nucleofected TS cells (Fig. 6B). Specifically, Tcfap2c siRNA, Smarca4 siRNA, and Eomes siRNA nucleofected TS cells lost tight cell contacts, indicative of differentiation. Because TCFAP2C, SMARCA4, and EOMES co-occupy many genes, we reasoned that combinatorial RNAi depletion of these genes may have a greater impact on TS cell self-renewal and gene expression compared with RNAi depletion of individual genes. To this end, we conucleofected Tcfap2c/Smarca4/Eomes siRNA, Tcfap2c/Smarca4 siRNA, Tcfap2c/Eomes siRNA, or Smarca4/Eomes siRNA in TS cells. Following conucleofection of these siRNAs, we observed a greater loss of normal colony morphology relative to TS cells nucleofected with siRNAs targeting individual genes or control siRNAs (Fig. 6B,C). Immunofluorescence analysis revealed that protein levels of TCFAP2C, SMARCA4, and EOMES were down-regulated in Tcfap2c/Smarca4/Eomes siRNA nucleofected TS cells relative to control-siRNA nucleofected TS cells (Fig. 6C). In addition, we observed a down-regulation of TS cell expressed genes (Cdx2, Elf5, Eomes, and Fgfr2) and genes expressed in TS cells and ES cells (Smarca4, Dppa5, Ets2, Klf5, Rex1, Sox2, and Tcfap2c) (Fig. 6D,E) in Tcfap2c/Smarca4//Eomes RNAi depleted TS cells. Elf5, occupied by TCFAP2C, SMARCA4, and EOMES (Fig. 3), was down-regulated more in TS cells nucleofected with Smarca4 siRNA, Eomes siRNA, or Smarca4/Eomes siRNA compared with TS cells nucleofected with control siRNA, suggesting that Elf5 may be a functional target of SMARCA4 and EOMES. While Elf5 was down-regulated in TS cells conucleofected with Tcfap2c/Smarca4/Eomes siRNA, the decrease in Elf5 expression was similar to that of TS cells nucleofected with Smarca4 siRNA or Eomes siRNA alone, suggesting that TCFAP2C, although bound to the Elf5 promoter, may not regulate Elf5 expression. Other genes whose expression was down-regulated following conucleofection of siRNAs compared with nucleofection of individual siRNAs or control siRNA include Dppa5, Eomes, and Sox2 (Fig. 6D,E). In summary, these results suggest an important role for TCFAP2C, SMARCA4, and EOMES in maintaining TS cell self-renewal.
Figure 6.
TCFAP2C, SMARCA4, and EOMES are required for TS cell self-renewal. (A) Tcfap2c, Smarca4, and Eomes siRNAs used to deplete Tcfap2c, Smarca4, and Eomes mRNA in TS cells. Control siRNA, Tcfap2c siRNA, Smarca4 siRNA, Eomes siRNA, Tcfap2c/Smarca4/Eomes siRNA, Tcfap2c/Smarca4 siRNA, Tcfap2c/Eomes siRNA, or Smarca4/Eomes siRNA were nucleofected into TS cells every 2 d for 6 d. (B) Normal colony morphology was lost in TS cells nucleofected with Tcfap2c siRNA, Smarca4 siRNA, Eomes siRNA, Tcfap2c/Smarca4/Eomes siRNA, Tcfap2c/Smarca4 siRNA, Tcfap2c/Eomes siRNA, or Smarca4/Eomes siRNA compared with TS cells nucleofected with control siRNA. (C) Immunofluorescence analysis of TS cells conucleofected with Tcfap2c/Smarca4/Eomes siRNA. TCFAP2C, SMARCA4, and EOMES expression was down-regulated in TS cells nucleofected with Tcfap2c/Smarca4/Eomes siRNA compared with TS cells nucleofected with control siRNA. Cytoplasm was stained with an antibody specific to Cytokeratin (Cytok) and nuclei were stained with Hoechst 33258. Scale bars, 100 μM. (D) qRT-PCR expression analysis of Tcfap2c, Smarca4, and Eomes siRNA TS cells with control TS cells. Expression of TS cell enriched genes were down-regulated following nucleofection of Tcfap2c, Smarca4, and Eomes siRNA. (E) Heat map summary of qRT-PCR results.
Transcriptional regulation of the chromatin remodeling factor SMARCA4 in TS cells and ES cells
Our studies and those of others have previously shown that Smarca4 is required for normal blastocyst development in mice (Bultman et al. 2000; Kidder et al. 2009), where Smarca4 siRNA injected into one-cell embryos or a Smarca4-null mutation impedes normal blastocyst development, hatching, and outgrowth. SMARCA4 is expressed in both the TE and ICM of blastocysts, as well as ES cells and TS cells, suggesting that SMARCA4 may regulate transcription in these cell types. Previously, we showed that RNAi-mediated depletion of Smarca4 resulted in a loss of ES cell self-renewal (Kidder et al. 2009). Moreover, we showed that SMARCA4 occupied promoters of key pluripotency-related genes such as Pou5f1, Sox2, Nanog, Sall4, and Rest, and epigenetic regulators including polycomb group (PcG) proteins. Because SMARCA4 is important in blastocyst development and ES cell self-renewal, identifying downstream targets of SMARCA4 in TS cells and investigating the role of SMARCA4 in TS cell self-renewal, may provide insight into mechanisms of TS cell self-renewal. In this study we found that RNAi-mediated depletion of Smarca4 transcripts results in a loss of normal TS cell morphology and down-regulation of TS cell–enriched genes, suggesting that SMARCA4 is required for TS cell self-renewal. We also found that SMARCA4 occupied the promoters of genes expressed highly in TS cells such as Bmp8b, Bmpr1a, Elf5, Klf5, Smad7, Rbpsuh, and Smo and genes expressed highly during placental development such as Cited2, Gata2, and Smad2 (Supplemental Table S2). To identify common targets of SMARCA4 in TS cells and ES cells, we compared genes bound by SMARCA4 in TS cells (this study) with genes bound by SMARCA4 in ES cells (Kidder et al. 2009). We found 282 common genes bound by SMARCA4 in TS cells and ES cells including Hes1, Klf5, Phc1, Serpine2, and Tcf4. By clustering the binding profiles of SMARCA4-occupied genes in TS cells (this study) with SMARCA4 binding profiles of the same genes in ES cells (Kidder et al. 2009), we found that binding profiles of SMARCA4 are enriched at target gene loci in TS cells compared with ES cells (data not shown), demonstrating that while SMARCA4 binds a common subset of target genes in TS cells and ES cells, SMARCA4 likely serves unique functions in the two cell types. In other words, SMARCA4 binds more unique versus common targets between TS cells and ES cells. Overall, SMARCA4 promoter binding to genes important for TS cell and ES cell self-renewal suggests that SMARCA4 occupancy of common targets may support self-renewal in both TS cells and ES cells, albeit through mechanisms involving potentially different cofactors.
Discussion
The trophoblast, which differentiates into placental tissues, is the first lineage to be specified during mammalian development. TS cells, like ES cells, are derived from preimplantation stage embryos in the presence of appropriate external signals. While recent efforts have focused on TF networks in ES cells that regulate pluripotency, little is know about transcriptional networks that maintain TS cell self-renewal. To further understand transcriptional networks in TS cells, we used genome-wide tiling array analysis (ChIP-chip) to interrogate promoter binding of five factors (TCFAP2C, SMARCA4, EOMES, ETS2, and GATA3). Our data show that TFs occupy genes involved in self-renewal, pluripotency, and epigenetic regulation of chromatin. To understand the expression state of these targets, we performed transcriptome analysis of undifferentiated and differentiated TS cells, and we showed that TFs occupy active and inactive genes in TS cells, which is consistent with reported activity of other TFs such as POU5F1, SOX2, and NANOG in ES cells (Boyer et al. 2005). To gain insight into the role of TCFAP2C, SMARCA4, and EOMES in TS cell self-renewal, we used RNAi to deplete Tcfap2c, Smarca4, and Eomes transcripts in TS cells. Our results show that Tcfap2c, Smarca4, and Eomes siRNA TS cells exhibit a loss of normal colony morphology and down-regulate expression of TS cell–enriched genes, suggesting that TCFAP2C, SMARCA4, and EOMES are required for TS cell self-renewal. Overall, these data offer a novel view into transcriptional regulatory mechanisms of TS cells.
Multipotent TS cells share similar characteristics as pluripotent ES cells, including their ability to self-renew for an extended period of time in vitro and high expression of pluripotency-related genes such as Esrrb, Klf5, Lin28, Rest, Sall4, Sox2, Stat3, Tbx3, and Tert. It is of interest to understand common and divergent mechanisms of self-renewal in ES cells and TS cells to further our understanding of pluripotency and multipotency respectively. Using ChIP-chip analysis to map protein–DNA interactions of five TFs to promoters of all genes in mouse TS cells, we identified novel binding patterns of TFs at genes involved in ES cell renewal such as Esrrb, Klf5, Rest, Sall4, Stat3, and Tert (Fig. 3; Supplemental Tables S1–S5). These genes are highly expressed in TS cells and ES cells, suggesting that a subset of factor-occupied genes may support self-renewal in ES cells as well as TS cells.
While TS cells and ES cells coexpress a number of self-renewal related genes, TS cells do not express key pluripotency genes Pou5f1 and Nanog, and ES cells do not express Cdx2, a key determinant of TE fate. Cdx2 and Pou5f1 play critical roles in specifying TE and ICM and therefore determine TS cell and ES cell fates, respectively. CDX2 and POU5F1 form transcriptional regulatory networks that include reciprocal transcriptional repression of Pou5f1 in the TE and Cdx2 in the ICM (Niwa et al. 2005). Moreover, forced expression of CDX2 converts ES cells into trophoblast cells, and reduced levels of POU5F1 promote trophectoderm differentiation in ES cells (Niwa et al. 2000). TE fate also involves expression of Eomes and Elf5, where Elf5 is activated through FGF–FGFR signaling (Metzger et al. 2007), subsequently leading to promoter binding and activation of Cdx2 and Eomes expression (Ng et al. 2008). We showed that TCFAP2C, SMARCA4, and EOMES bind to the Elf5 promoter (Fig. 3), and TCFAP2C and EOMES bind promoters of genes involved in FGF signaling, including Fgfr2/4, implying the possibility of a positive-feedback regulatory loop between ELF5 and EOMES in TS cells (Supplemental Tables S1–S3). We also observed binding of EOMES to the promoter of Tcfap2c, and TCFAP2C and EOMES self-promoter binding, suggesting further potential feedback regulatory loops in TS cells. This functional relationship between ELF5 and EOMES is further strengthened by our data, where RNAi depletion of Eomes in TS cells resulted in down-regulation of Elf5 and Eomes expression (Fig. 6). In addition, Tead4, a gene necessary for trophectoderm specification, that has been shown to activate Eomes (Nishioka et al. 2008), was bound by TCFAP2C and EOMES in this study (Supplemental Tables S1, S3). In summary, these transcriptional regulatory loops in TS cells provide a view into mechanisms of the first lineage specification in mammals.
To investigate the expression state of factor-occupied genes, we performed transcriptome analysis of TS cells differentiated through 14 d in the absence of FGF4. KMC was used to identify major patterns of gene expression variability during TS cell differentiation (Fig. 4). Using this transcriptome data set and GSEA, we evaluated expression of the transcriptional targets of TCFAP2C, SMARCA4, EOMES, ETS2, and GATA3, and we found that these five TFs bind mainly active genes in TS cells. Moreover, we observed enriched expression of genes bound by multiple targets, suggesting that multiple factors positively regulate expression compared with single factors. We also observed differential expression of genes during TS cell differentiation that are involved in multiple signaling pathways, including FGF, TGFB, NOTCH, and WNT-signaling, also bound by at least one factor, suggesting that TS cell self-renewal versus differentiation decisions may involve transduction of external signals and downstream TF activity.
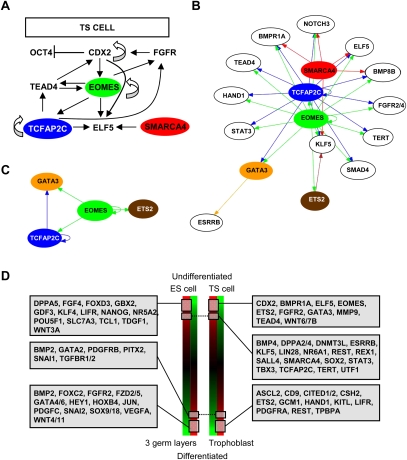
Our genome-wide ChIP-chip and transcriptome data shown here describe a novel view of transcriptional networks that may regulate self-renewal in TS cells. We show that five factors (TCFAP2C, SMARCA4, EOMES, ETS2, and GATA3) occupy genes involved in maintaining the TS cell phenotype (Fig. 7A,B). We also demonstrate transcriptional regulation within the factors, including autoregulation of TCFAP2C and EOMES through self promoter binding (Fig. 7C). Moreover, we identified genes involved in self-renewal that are coexpressed or uniquely expressed in TS cells and ES cells relative to their differentiated progeny (Fig. 7D). These findings provide a network for further evaluation of transcriptional regulatory mechanisms of TS cells, ES cells, and preimplantation embryos.
Figure 7.
Transcriptional regulation of target genes in TS cells. (A) Model for integration of TCFAP2C, SMARCA4, and EOMES with core transcriptional machinery in TS cells. Colored circles represent the investigated factors, and arrows represent direction of transcriptional regulation. (B) Extended transcriptional network in TS cells integrating the investigated five factors with downstream target genes. Open circles represent target genes. (C) Transcriptional regulation within the five factors, including autoregulation of TCFAP2C and EOMES. (D) Summary of genes coexpressed or uniquely expressed in TS cells, ES cells, and their differentiated progeny.
Methods
TS cell derivation and culture
TS cells were derived and cultured as previously described (Tanaka et al. 1998). Briefly, Bl6D2/F1 male and female mice were mated, and resulting blastocysts were flushed in M2 medium and transferred to a 24-well dish containing gamma-irradiated MEFs and RPMI 1640 medium supplemented with 20% FBS (GIBCO), 1 mM sodium pyruvate (GIBCO), 100 μM β-mercaptoethanol (Chemicon), 2 mM L-glutamine (GIBCO), 25 ng/mL recombinant human FGF4 (1×; R&D Systems), and 1 μg/mL heparin (1×; Sigma). The TSC-BK12 cell line was used in subsequent experiments. For ChIP-chip experiments, TS cells were transitioned to dishes containing TS cell medium, 70% iMEF-conditioned medium, and 1.5× FGF4 and heparin and were cultured at 37°C with 5% CO2. TS cells were passed twice to remove iMEFs, fed every 2 d, and harvested for ChIP-chip experiments. For differentiation experiments, TS cells were cultured in feeder-free dishes containing TS cell medium without FGF4 and heparin and were fed every 2–3 d for 14 d. Animals were treated according to an approved Institution Animal Care and Use Committee guidelines protocol for these studies at EMD Serono Research Institute.
siRNA nucleofection
TS cells were harvested by trypsinizing with 0.05% trypsin (Invitrogen), inactivated with DMEM media containing 10% FCS, and pelleted. TS cells were resuspended in 100 μL of ES cell nucleofector solution (Amaxa), and program A-23 on the Amaxa Nucleofector II was used to nucleofect 100 nM control siRNA or Tcfap2c, Smarca, or Eomes SMARTPool siRNA (Dharmacon). Immediately following nucleofection, TS cells were added to prewarmed TS cell media and incubated as described above. TS cells were nucleofected every 48 h for 6 d.
qRT-PCR expression profiling of TS cells
RNA isolation and qRT-PCR were performed as previously described with minor modifications (Kidder et al. 2009). TS cells were harvested using 0.25% trypsin (Invitrogen) to dissociate cells. Total RNA was extracted from TS cells using an RNeasy Mini Kit (Qiagen) and DNase treated using Turbo DNA-free (Ambion) for 30 min at 37°C. Reverse transcription was performed using 1 μg of total RNA and a Superscript III kit with random hexamer primers (Invitrogen). qPCR was performed using TaqMan probes and TaqMan Universal PCR Master Mix reagents (Applied Biosystems) or nonlabeled primers and SYBR Green PCR Master Mix reagents (Applied Biosystems). Primers used for Q-PCR with SYBR green reagents were designed using Primer3 (http://frodo.wi.mit.edu/).
Immunofluorescence analysis of TS cells
TS cells were fixed with 4% paraformaldehyde for 15 min at room temperature, washed three times with 0.1% Triton X-100 (Sigma), and blocked in 1% BSA/0.01% Tween-20/PBS for 30 min. The fixed cells were then incubated with a primary antibody (1:100) overnight at 4°C in blocking buffer. The next day, the cells were washed three times with blocking buffer for 15 min. Samples were then incubated with Alexa Fluor 488 secondary antibodies (1:1000; Molecular Probes) and Hoechst 33258 (1:2000) in 0.1% Triton X-100/PBS for 1 h at room temperature, washed three times with blocking buffer, and mounted in ProLong Gold anti-fade reagent (Invitrogen) overnight at room temperature. For immunofluorescence captured using confocal miscroscopy, Hoechst 33258 was omitted.
ChIP-chip analysis
ChIP-chip experiments were performed as previously described with minor modifications (Kidder et al. 2009). The polyclonal EOMES (ab23345) antibody was obtained from Abcam, and the TCFAP2C (H-77, SC-8977), SMARCA4 (H-88, SC-10768), ETS2 (H-140, SC-22803), and GATA3 (D-16, SC-22206) antibodies were obtained from Santa Cruz Biotechnology. Briefly, 1 × 108 mouse TS cells (TSC-BK12) (feeder-free) were harvested and chemically cross-linked with 1% formaldehyde (Sigma) for 20 h at 4°C and subsequently sonicated. Sonicated cell extracts equivalent to 2 × 106 cells were used for immunoprecipitation assays. Five micrograms of purified ChIP-enriched DNA (TCFAP2C-chip, SMARCA4-chip, EOMES-chip, ETS2-chip, GATA3-chip, and Input) was amplified, DNase treated, labeled, and hybridized to Affymetrix mouse promoter 1.0R tiling arrays. Mouse promoter 1.0R DNA tiling arrays contain 4.6 million 25-mer oligonucleotide probes spanning a distance of −6 kb to +2.5 kb relative to the TSS for 28,000 mouse promoter regions, providing a resolution of 35 bp with 10-bp gaps between probes. Some genes have tiling array coverage exceeding 8.5 kb (for more information, see the Affymetrix bed file Mm_PromP_NCBIv36.accession.bed). At least three biological replicates were used for each ChIP-chip analysis. Quantile normalization, including probe intensity computation and log2 adjustment, was applied to the tiling array data using CisGenome (Ji et al. 2008). Peak detection and gene annotation were performed as previously described (Kidder et al. 2008, 2009). Briefly, peak detection was performed using the TileMap (Ji and Wong 2005) application in CisGenome (Ji et al. 2008). MA statistics was applied to analyze the tiling array data. Post-filtering included discarding peaks if the total length was <100 bp, or if there were less than three continuous probes passing the cutoff, and merging two adjacent peaks if the gap between the two peaks was <300 bp and if there were less than five probes that did not pass the cutoff between the two peaks. Peaks with an FDR of <0.2 were also discarded. Enrichment peaks were annotated with the closest gene using build 36 of the mouse genome.
Clustering analysis of promoter binding data
Promoter binding patterns for the five TFs (TCFAP2C, SMARCA4, EOMES, ETS2, and GATA3) were centered on enrichment peaks [−2 kb, +2 kb] and visualized by HCA. Promoter regions where greater than 25% of probes were absent were not included in this analysis. Hierarchical clustering was performed using Spotfire software.
Global expression profiling of undifferentiated and differentiated TS cells
Total RNA was isolated at several time points from TS cells cultured without FGF4 for 2 wk using an RNeasy Mini Kit (Qiagen). For each duplicate biological replicate, 100 ng of RNA was amplified using a MessageAmp Premier RNA Amplification kit (Ambion), and 14 μg of fragmented aRNA was hybridized to Affymetrix mouse 430 v2.0 microarrays. Analysis was performed as previously described with some modifications (Kidder et al. 2009). Briefly, CEL files were loaded into ArrayAssist (Stratagene), and probes were normalized using the GC-RMA algorithm. ANOVA was performed on all groups using a Benjamini and Hochberg FDR correction. Genes whose expression differed by at least twofold between any two groups with a P-value < 5% were considered differentially expressed. Validation of microarrays was performed using qRT-PCR with TaqMan probes as described above.
ChIP-chip microarray processing
TF target genes were compared with transcriptome data generated from this study and a published microarray data set to evaluate the expression of bound genes in TS cells, ES cells, and their differentiated progeny. Microarray expression data (Agilent) from TS cells and ES cells (Sharova et al. 2007) was analyzed using the NIA Array Analysis Tool software (Sharov et al. 2005). ANOVA and log-transformation were performed. Differentially expressed genes (FDR < 5%) were clustered using Spotfire software (Spotfire).
Acknowledgments
This work was funded by an internal grant at EMD Serono Research Institute, Inc.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE18507.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101469.109.
References
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T. Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development. 2002;129:2733–2747. doi: 10.1242/dev.129.11.2733. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes & Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1:245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran R, Thompson M. Polybromo-1-bromodomains bind histone H3 at specific acetyl-lysine positions. Biochem Biophys Res Commun. 2007;355:661–666. doi: 10.1016/j.bbrc.2007.01.193. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Cormier S, Vandormael-Pournin S, Babinet C, Cohen-Tannoudji M. Developmental expression of the Notch signaling pathway genes during mouse preimplantation development. Gene Expr Patterns. 2004;4:713–717. doi: 10.1016/j.modgep.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Cross JC. How to make a placenta: Mechanisms of trophoblast cell differentiation in mice—a review. Placenta. 2005;26:S3–S9. doi: 10.1016/j.placenta.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JC. Genes, development and evolution of the placenta. Placenta. 2003;24:123–130. doi: 10.1053/plac.2002.0887. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Erlebacher A, Price KA, Glimcher LH. Maintenance of mouse trophoblast stem cell proliferation by TGF-beta/activin. Dev Biol. 2004;275:158–169. doi: 10.1016/j.ydbio.2004.07.032. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Goldin SN, Papaioannou VE. Paracrine action of FGF4 during periimplantation development maintains trophectoderm and primitive endoderm. Genesis. 2003;36:40–47. doi: 10.1002/gene.10192. [DOI] [PubMed] [Google Scholar]
- Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009;284:28729–28737. doi: 10.1074/jbc.M109.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wong WH. TileMap: Create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Baker JC. Genomic evolution of the placenta using co-option and duplication and divergence. Genome Res. 2008;18:695–705. doi: 10.1101/gr.071407.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Luo J, Sladek R, Bader JA, Matthyssen A, Rossant J, Giguere V. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature. 1997;388:778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- Ma GT, Roth ME, Groskopf JC, Tsai FY, Orkin SH, Grosveld F, Engel JD, Linzer DI. GATA-2 and GATA-3 regulate trophoblast-specific gene expression in vivo. Development. 1997;124:907–914. doi: 10.1242/dev.124.4.907. [DOI] [PubMed] [Google Scholar]
- McPherson LA, Weigel RJ. AP2α and AP2γ: A comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 1999;27:4040–4049. doi: 10.1093/nar/27.20.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger DE, Xu Y, Shannon JM. Elf5 is an epithelium-specific, fibroblast growth factor-sensitive transcription factor in the embryonic lung. Dev Dyn. 2007;236:1175–1192. doi: 10.1002/dvdy.21133. [DOI] [PubMed] [Google Scholar]
- Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell. 2009;5:420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Ko MS. Exhaustive search for over-represented DNA sequence motifs with CisFinder. DNA Res. 2009;16:261–273. doi: 10.1093/dnares/dsp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Piao Y, Shaik N, Sullivan T, Stewart CL, Hogan BL, Ko MS. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev Biol. 2007;307:446–459. doi: 10.1016/j.ydbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Xu C, Huang W, Zhang J, Carlson JE, Tu X, Wu J, Shi Y. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46:2100–2110. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes & Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007a;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007b;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Wen F, Tynan JA, Cecena G, Williams R, Munera J, Mavrothalassitis G, Oshima RG. Ets2 is required for trophoblast stem cell self-renewal. Dev Biol. 2007;312:284–299. doi: 10.1016/j.ydbio.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling U, Schorle H. Transcription factor gene AP-2γ essential for early murine development. Mol Cell Biol. 2002;22:3149–3156. doi: 10.1128/MCB.22.9.3149-3156.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Xie H, Tranguch S, Jia X, Zhang H, Das SK, Dey SK, Kuo CJ, Wang H. Inactivation of nuclear Wnt-β-catenin signaling limits blastocyst competency for implantation. Development. 2008;135:717–727. doi: 10.1242/dev.015339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Gu L, Romeo PH, Bories D, Motohashi H, Yamamoto M, Engel JD. Human GATA-3 trans-activation, DNA-binding, and nuclear localization activities are organized into distinct structural domains. Mol Cell Biol. 1994;14:2201–2212. doi: 10.1128/mcb.14.3.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zou J, Ye Z, Hammond H, Chen G, Tokunaga A, Mali P, Li YM, Civin C, Gaiano N, et al. Notch signaling activation in human embryonic stem cells is required for embryonic, but not trophoblastic, lineage commitment. Cell Stem Cell. 2008;2:461–471. doi: 10.1016/j.stem.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang J, Wang T, Esteban MA, Pei D. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. J Biol Chem. 2008;283:35825–35833. doi: 10.1074/jbc.M803481200. [DOI] [PubMed] [Google Scholar]