Abstract
We have previously proposed that the pathogenesis of eosinophilic esophagitis (EE) is mediated by an IL-13–driven epithelial cell response associated with marked gene dysregulation including eotaxin-3 overproduction. Herein, we compared epithelial responses between normal (NL) and EE patients aiming to uncover molecular explanations for EE pathogenesis. Esophageal epithelial cells could be maintained up to 5 passages, with 67% and 62% of cell lines reaching confluence in NL and EE, respectively. Both sets of epithelial cells avidly responded to IL-13 at similar levels as assessed by eotaxin-3 production. Acidic pH increased cellular release of eotaxin-3 (4.6 ± 1.98 ng/mL vs. 12.46 ± 2.90 ng/mL at pH 7.4 and 4 respectively, p<0.05). Numerous epidermal differentiation complex (EDC) genes, such as filaggrin, and SPRR3 were downregulated both in IL-13-stimulated esophageal epithelial cells and in EE biopsies compared to NL. While the filaggrin loss of function mutation 2282del4 was overrepresented in EE compared to control individuals (6.1% vs. 1.3% respectively, p=0.0172), the decreased filaggrin expression was uniformly seen in all EE patients in vivo. Indeed, expression of the EDC genes filaggrin and involucrin was strongly decreased directly by IL-13. These results establish that the epithelial response in EE involves a cooperative interaction between IL-13 and expression of EDC genes.
Introduction
Eosinophilic esophagitis (EE) is a complex atopic disorder of the esophageal mucosa characterized by allergen-induced eosinophilic infiltration and epithelial cell hyperplasia (1). Yet the growth characteristics of normal (NL) and EE epithelial cells in vitro have not been studied and there is limited data regarding the intrinsic properties of esophageal epithelial cells in EE patients (1). EE patients typically present with symptoms that mimic gastroesophageal reflux disease (GERD), but GERD and EE are distinguished by the lack of histological response to acid suppression therapy in EE (2). The distinction between GERD and EE is not always clear, as GERD can have substantial esophageal eosinophilia and EE patients sometimes report clinical responses to acid neutralization therapy.
There is a striking overlap between the mucosa structure of the skin and the esophagus (3, 4). During the terminal differentiation of epidermal keratinocytes, cells undergo desquamation resulting in cornified cells at the surface. These dead cells comprise a major part of the epithelial barrier and are continuously renewed. Interestingly, the human esophageal epithelium is not normally cornified and expresses differentiation markers distinct from the skin (5–8). For example, involucrin is expressed in early differentiating cells in the esophagus, and in the late-differentiating cells in the skin (8). Proliferation/differentiation gene expression have been largely studied in the context of the skin (9) in normal and pathologic conditions (10); whereas, markers of differentiation of the esophageal epithelium have not been extensively studied. During skin keratinocyte differentiation, keratin filament organization is modified by the keratin-binding protein filaggrin in stratified epithelia (10, 11). Association of filaggrin or related proteins of the epidermal differentiation complex (EDC) genes clustered on chromosome 1q21 (12), (e.g. involucrin, NICE, loricrin, and SPRR) with keratin prevents the proteolytic destruction of keratin during epidermal cell terminal differentiation (13–15), which in turn forms an important part of the epithelial barrier of cornified cells (10, 13, 15, 16). The importance of this pathway in the development of atopy is underscored by the marked effect of loss of function mutations in the filaggrin gene, which predispose to atopic dermatitis (AD), a disease that co-exists in approximally 50% of EE patients. Indeed both EE and AD share molecular characteristics including food sensitization. Notably, defects in epithelial barrier function associated with filaggrin mutations have been associated with AD (17–23); such mutations are only present in 0–5 % of control individuals and 9–27% of AD individuals, depending upon the study (17, 24).
We have recently proposed that IL-13 is a key cytokine in EE disease pathogenesis. In particular, esophageal epithelial cells express all components of the IL-13 receptor including IL-4Rα, IL-13Rα1 and IL-13Rα2 (25). In addition, IL-13 induces prominent dysregulation of gene expression in the esophageal epithelium including marked overexpression of eotaxin-3, the most strongly induced IL-13 target gene, which is also highly overexpressed in vivo in the EE esophageal transcriptome (25). Furthermore, intratracheal IL-13 induces features of experimental EE in mice and IL-13 and STAT6 deficient mice are protected from the development of experimental EE (26–28). A recent study has shown that in AD patients, some EDC genes might be downregulated by IL-13 through a STAT6 dependent mechanism (29). In our initial analysis of the EE transcriptome, we have recently noted that filaggrin mRNA was dramatically decreased in the esophagus of EE compared to NL biopsies using microarray analysis (30), suggesting abnormal epithelial differentiation in EE. Herein, we aimed to determine whether there is an intrinsic defect in epithelial cell responses in EE, and to further determine which features of the disease may be mediated by IL-13. We report a profound dysregulation of EDC gene expression, identify cooperate effects between IL- 13 and EDC gene expression and the presence of gene variants in the EDC gene filaggrin in EE patients.
Material and Methods
Cell lines and primary cell culture
Primary esophageal epithelial cells were grown as previously described (25). Human esophageal epithelial cell line (TE) and squamous epithelial cells were provided by Dr. Hainault (IARC, Lyon France) and were maintained in RPMI medium (Invitrogen) supplemented with 10% FCS (Atlanta, J0138, Advantage) and 1% penicillin/streptomycin/amphotericin (Invitrogen). In preliminary studies, we optimized the culture conditions, particularly by modifying the source of the FBS which had a potential effect on cell growth and responses (see supplemental figure 1). For primary cell culture, one or two distal esophageal biopsies from EE patients were collected during routine endoscopy and with informed consent as approved by the Institutional Review Board. The eosinophil level in each patient was quantified and corresponds to the maximum or peak eosinophils found in one hpf. All sections of every biopsy available were assessed and counted. The samples were cultured in modified F-media (3:1 F-12/DMEM) supplemented with 5%, FBS (Atlanta, J0138, Advantage), adenine (24.2 µg/mL), cholera toxin (10−4 µM), insulin (5µg/mL), hydrocortisone (0.4 µg/mL), and human epidermal growth factor (10 ng/mL) in the presence of penicillin, streptomycin, and amphotericin (Invitrogen). Briefly, tissue was first digested twice with trypsin, followed by trypsin neutralization in F-media without epidermal growth factor. The cells were then plated onto 1–5×105 NIH 3T3 J2 feeder cells irradiated with 6000 rad. The next day, the media was exchanged with F-media containing epidermal growth factor (10 ng/mL) and fresh media was added every other day. NL and EE patient-derived cells were cultured for 2 weeks in this way, and feeder fibroblasts were then removed by differential trypsinization. Cells were stimulated with IL-13 (0, 1, 10, 100 ng/mL) obtained from Peprotech, as described (25). For acidification experiments, a pH 4 solution of HCl / NaCl (300 mosmol /kg H2O) or a pH 7.4 solution of NaCl (300 mosmol / kg H2O) was added for 4 hours. After IL-13 stimulation, 250 µl of saline or HCl/saline was added for 4 hours. Supernatants were harvested, neutralized, and volumes adjusted prior to eotaxin-3 quantification. Viable cells were counted using a hematocytometer and trypan blue staining.
Real time PCR
Total cellular RNA was extracted with Trizol (Invitrogen) according to the manufacturer’s instructions. The samples were DNase treated and first strand cDNA was synthesized using Superscript II (Invitrogen). Total RNA from biopsy samples was extracted using the mini RNA extraction Kit (Qiagen) and reverse transcription was performed using Iscript (Biorad). Real-time PCR was performed by rapid-cycling using the IQ5 (Biorad) SYBR mix (Biorad) as a ready-to-use reaction mix according to the manufacturer’s instructions. The following primers set (5’3’) were: GAPDH (350bp): tggaaatcccatcaccatct and gtcttctgggtggcagtgat; Filaggrin (125 bp): gggaagttatcttttcctgtc and gatgtgctagccctgatgttg; Involucrin (378bp): gttcctcctccagtcaatacccatc and cttcattcccagttgctcatctctc; Eotaxin-3 (125bp): gtaactctgggaggaaacaccctctcc and aactccgaaacaattgtgactcagctg. PCR products were sequenced using the CCHMC sequencing Core facility.
Chromosome location
The 574 probe sets of the EE transcriptome were subjected to David ontology 2.0 DAVID (database for annotation, visualization and integrated discovery) and EASE analyses (expression analysis systematic explorer) a web-based (http://david.abcc.ncifcrf.gov/) application that allows access to a relational database of functional annotations (31, 32). Chromosome locations of the transcripts were obtained and p values were calculated based on the total number of genes present on each chromosome.
Filaggrin mutation genotyping
Patient DNA was isolated from saliva, peripheral blood leukocytes, or oral buccal swab samples. The history of allergic diseases (asthma, atopic dermatitis, atopic rhinitis) was recorded in our EE population. EE was defined as ≥ 24 eosinophils/hpf in at least one high power field of the esophageal biopsy as previously described (33). For R501X, the following primers were used 5’-CTG GAG GAA GAC AAG GAT CG-3’ and 5’-TTG TCT GCT TGC ACT TCT GG-3’ to amplify a 245 bp fragment as previously described (34). The PCR product was digested for one hour with NlaIII at 37° C and electrophoresis was performed on a 4% agarose gel with ethidium bromide. An allele with no mutation digested into 2 fragments of 213 bp and 32 bp. An allele with the R501X mutation digested into three fragments of 176 bp, 37 bp, and 32 bp. For 2282del4, the following primers 5’-AAT AGG TCT GGA CAC TCA GGT-3’ and 5’-GGG AGG ACT CAG ACT GTT T-3’ were used to amplify an 811 bp fragment as previously described (35). Two fragments (671 bp and 136 bp) were obtained for the mutated alleles and non-mutated allele did not contain the restriction site. The PCR products were digested for one hour with DraIII at 37° C and electrophoresis was performed on a 1% agarose gel with ethidium bromide as previously described (17–23).
Statistical analysis
Statistical significance comparing different treatments or groups was determined by the Student’s t test (normal distribution equal variance), Welch t test (normal distribution, unequal variances), Mann Whitney test (non parametric test, two groups) or ANOVA and Kruskal-Wallis test followed by a Dunn’s multiple comparison Test (non parametric test, three groups or more) using Prism 5 Software. For case control study, χ2 , odd ratio and 95% confidence interval were calculated using Prism 5 (GraphPad Software).
Results
Growth characteristics of primary epithelial cells from NL and EE patients
We aimed to determine if primary esophageal epithelial cells from NL and EE patients had similar growth characteristics. The percentage of cells able to reach 80–100% confluence by 35 days was similar with 67% and 62% of NL and EE cell lines, respectively (Figure 1A). No differences were found in the number of cells at confluence, or after 35 days in culture with 1.8 ± 1.3 × 106 and 1.3 ± 1.0 × 106 in NL and EE cells, respectively (Figure 1B). An average of 19 ± 8 days was sufficient to reach confluence (80–100%) in the NL group compared with 21 ± 8 days in the EE group (Figure 1C).
Figure 1. Growth characteristics and response to IL-13 of primary esophageal epithelial cells from normal (NL) and eosinophilic esophagitis (EE) patients.
Cells were grown in modified F-media for a maximum of 35 days. In A, the percentage of cells that reach 80-100% confluence is shown in the NL and EE group. In B, the number of cells at confluence or after 35 days of culture is presented. In C, the number of days to reach 80-100% confluence is shown. In D, the fold increase in eotaxin-3 mRNA expression following IL-13 stimulation at different doses is shown; note the Log10 scale. In D, data points are average ± SD, non significant (ns). In E, EE and NL derived cells were stimulated for 48h with IL-13 (100 ng/mL) then the media was acidified or heparinized for 4h and eotaxin-3 protein was quantified in the supernatant. Bars indicate mean values ± SD, * p<0.05.
Epithelial cell responses to IL-13 and pH
In order to test if epithelial cells from NL and EE individuals have the same propensity to respond to IL-13, cells were treated with IL-13 (0 to 100 ng/mL). The fold-change of eotaxin-3 mRNA expression was 281 ± 513 and 64 ± 84-fold increase at 1 ng/mL, 7783 ± 14689 and 2893 ± 2381-fold increase with IL-13 at 10 ng/mL and 34174 ± 23974 and 22910 ± 24389-fold increase with IL-13 at 100 ng/mL in NL and EE cells, respectively (Figure 1D). Eotaxin-3 secretion was dependent upon the culture conditions, especially the type of serum present (Supplementary Figure 1). In addition, eotaxin-3 secretion into the supernatant was affected by proton concentrations. In particular, 4.6 ± 2.0 ng/mL and 12.4 ± 2.9 ng/mL were released into the supernatant of EE-derived cells stimulated with IL-13 (100 ng/mL) when cells were incubated 4 hours at pH 7.4 and pH 4, respectively, even though cell viability was not affected (data not shown). In addition, eotaxin-3 release was increased by heparin; as 12.9 ± 3.8 ng/mL eotaxin-3 was obtained following heparin treatment compared to the 4.6 ± 2.0 ng/mL in the absence of heparin.
Similar results were obtained in primary epithelial cell cultures derived from NL individuals (Figure 1E). These results suggest that the esophageal responses in EE may be a cell-extrinsic phenomenon in the presence of IL-13 and microenvironmental components, such as the levels of protons and heparin, affect eotaxin-3 levels.
Down regulation of EDC genes in eosinophilic esophagitis
The EE transcriptome, defined as the 574 probes significantly different (p<0.001) in esophageal biopsies of EE patients compared to NL patients (30), was analyzed for the chromosomal locations of the respective genes. Chromosome 1 was the most significantly (p=0.0002) affected chromosome with 48 genes (corresponding to 61 probes or 10.6% of the EE transcriptome) altered (Figure 2A). We noted the modulation of several EDC genes (such as filaggrin) clustered on chromosome 1q21 (12), a region linked with atopy (36). Using microarray results in NL and EE patient biopsies, we studied the expression pattern of the EDC genes at the location 1q21 (Figure 2B). Most of the genes in this cluster showed significant decreased expression or trends toward decreased expression (Figure 2B). Filaggrin mRNA was down regulated 16-fold in EE compared with normal individuals (Figure 3A). We verified our microarray findings using real-time PCR by examining normal and EE patients in order to determine the relative expression of filaggrin in different disease states. Using a cohort of 144 individuals, filaggrin mRNA level was down regulated by 16-fold in active EE patients compared to NL patients. The esophageal specific SPRR gene esophagin (SPRR3) was highly expressed in the esophagus and significantly decreased 6-fold in EE compared to NL patients (Figure 3B). In contrast, the EDC marker involucrin was not significantly down regulated in EE compared to NL patients (0.58 [0.017 – 0.086] and 0.13 [0.057 – 0.49], respectively) (Figure 3C). Taken together, these data show that the esophagus of EE patients has dyregulated expression of select EDC genes including filaggrin and SPRR3.
Figure 2. Chromosome involvement in the EE transcriptome and physical map of the epidermal differentiation complex gene cluster on the chromosome 1.
In A, the EE transcriptome was assigned to chromosomes using DAVID ontology. The number of genes and the p value are presented. In B, a schematics of the physical map of the 1q21 gene cluster and the corresponding microarray expression profile in NL and EE patients are shown.
Figure 3. Esophageal expression of filaggrin mRNA.
A. Filaggrin expression was assessed by real-time PCR and normalized to GAPDH in control patients (NL), active EE patients (EE active; eosinophil level ≥ 24), EE patients with an intermediate eosinophilia level (EE int; eosinophils level from 1 to 23) and inactive EE patients who responded successfully to their therapy (inactive; eosinophil level = 0). N = 48, 21, 18 2 and 9 individuals in EE active, EE intermediate, EE inactive and NL groups, respectively. Each data point corresponds to a patient and the lines show the median with interquartile range. C and D Involucrin and SPRR3 expression were assessed by realtime PCR and normalized to GAPDH as described in (A). Each data point corresponds to a patient and the lines show the median with interquartile range.
Effect of therapy on EDC gene expression
We examined filaggrin expression in treated EE patients in order to gain molecular insight into the reversibility of the EDC gene expression changes (Figure 3A). In EE patients who responded to therapy (glucocorticoids and/or diet modification), the filaggrin expression level was not significantly different compared to the control level and was significantly (p<0.01) different compared to active EE. The same trend as filaggrin expression was observed with SPRR3 gene expression; no significant decrease in involucrin gene expression in NL vs. active or inactive EE patients was observed (Figure 3B and C). These results show that abnormal EDC gene expression in EE is reversible following successful therapy.
Association of EE with filaggrin gene polymorphisms
We propose that the observed decrease in filaggrin mRNA levels could reduce filaggrin function and cause associated disease phenotypes; this is consistent with the finding that filaggrin levels normalized after successful therapy. However, two loss-of-function genetic variants (R501X and 2282del4) in the filaggrin gene have recently been associated with AD; these may not affect the level of filaggrin mRNA, but instead the resulting inactive truncated proteins may compromise epidermal integrity and barrier function. Interestingly, no significant difference in filaggrin mRNA expression was observed between allergic and non-allergic active EE patients (Figure 4 and Supplementary Table II).
Figure 4. Filaggrin expression in active EE patients with or without allergic symptoms or sensitization.
Filaggrin expression was assessed by real-time PCR and normalized to GAPDH in non-allergic (n=7) and allergic EE (n=25) patients. Each data point corresponds to a patient and the lines show the median with the interquartile range.
We genotyped 164 controls and 365 EE patients for these two mutations. Reliable genotype data for 2282del4 were obtained in 329 out of the 365 EE patients genotyped and 157 out of all the 164 controls. We found 6.1% heterozygotes for 2282del4 in the EE group and 1.3% in the control group corresponding to 3% and 0.6% allele frequency in the EE and control group, respectively. Out of the heterozygote EE patients with the deletion, 60% had a history of eczema and 70% of food allergy (see Table III and Supplemental Table I). Significant associations (p=0.0172, odds ratio=5.016 and p=0.0185, odds ratio= 4.89) were found between this deletion (genotype and allele frequency, respectively) and EE (Table I). Reliable genotype data for R105X were obtained in 339 out of the 365 EE patients genotyped and all the 164 controls (see Table I). We identified 4.4% heterozygotes for R501X in the EE group and 3% in the control group corresponding to 2.2% and 1.5% allele frequency in the EE and control group, respectively (see Table I). No significant association was found between this SNP and EE. No overlap of mutations was found between heterozygote patients; a total of 11% of our EE population had one or the other mutation compared to 4.3% in the control population (p=0.0363, Table II). The prevalence of atopy (Table IIIB) was similar across the EE R105X positive, the EE 2282Del4 positive and EE WT groups (86.6%, 85.0% and 83.3%, respectively). The prevalence of atopic dermatitis was 33%, 60% and 44% (p=0.11) across the EE R105X positive, the EE 2282del4 positive and EE WT groups, respectively. Interestingly, 2282del4 mutation was still associated with EE when EE patients without any history of atopic dermatitis were analyzed (p=0.0315, Table IIIA). As such, while filaggrin mutations (2282del4 and R501X) were increased in EE compared to control individuals; they were only present in 11% of EE patients, suggesting that germline mutations in the filaggrin gene are not the primary etiology of filaggrin reduction.
Table III.
Frequency of atopic dermatitis and genotypes in the EE patients
| A | R501X positive |
2282del4 positive |
WT | P values Odds ratio |
|---|---|---|---|---|
| EE Without Atopic Dermatitis | 10 | 8 | 131 * | p=0.0315 (for 2282Del4) |
| Controls | 5 | 2 | 157 | 4.794 [1.00–23.98] |
| B |
R501X positive n=283 |
2282del4 positive n=283 |
WT n=283 |
P values |
| Atopic Dermatitis1 | 5 (33.3%) | 12 (60%) | 102 (44.3%)* | ns^ |
| Skin Prick Test Positive2 | 7 (46.7%) | 10 (50.0%) | 72 (69.9%)+ | ns |
| History of Atopic disease3 | 13 (86.6%) | 17 (85.0%) | 210 (83.3%)& | ns |
Past or present history of allergic dermatitis
History of at least one positive skin prick testing to food or aeroallergen
History of atopic dermatitis, atopic rhinitis, asthma or positive skin prick test
not significant
Information unavailable for 50 of the genotyped patients
Information unavailable for 180 of the genotyped patients
Information unavailable for 31 of the genotyped patients
Table I.
2282del4 and R501X mutations in the EE and control population
| Deletion 2282del4 | Patients with EE (n=329) |
Controls (n=157) | P values/ Odds ratio |
|---|---|---|---|
| Genotype frequency | |||
| WT | 309 (93.9%) | 155 (98.7%) | p= 0.0172 / |
| Het | 20 (6.1%) | 2 (1.3%) | 5.0 [1.157–21.74] |
| Allele frequency | |||
| No deletion | 638 (97.0%) | 312 (99.4%) | p=0.0185 / |
| Deletion | 20 (3.0%) | 2 (0.6%) | 4.89 [1.135- 21.06] |
| SNP R501X |
Patients with EE (n=339) |
Controls (n=164) |
p values |
| CC | 324 (96.2%) | 159 (97.0%) | p=0.4591 |
| CT | 15 (4.4%) | 5 (3.0%) | |
| C | 663 (97.8%) | 323 (98.5%) | p=0.4637 |
| T | 15 (2.2%) | 5 (1.5%) | |
Table II.
Combined analysis of R501X and 2282del4 mutation in EE and controls
| Mutation | EE Population (n=365) |
Controls (n=164) | p value / Odds ratio |
|---|---|---|---|
| R501X | 15 (4.7%) | 5 (3.0%) | ns1 |
| 2282del4 | 20 (6.3%) | 2 (1.2%) | ns |
| R501X and 2282del4 | 0 (0%) | 0 (0%) | ns |
| R501X or 2282del4 | 35 (11.0%) | 7 (4.3%) | p=0.0363 |
| 2.379 [1.033–5.475] | |||
| Unknown R501X | 26 (7.1%) | 0 (0%) | ns |
| Unknown 2282del4 | 36 (9.9%) | 7 (4.3%) | ns |
not significant
Involvement of IL-13 in epidermal differentiation complex gene expression
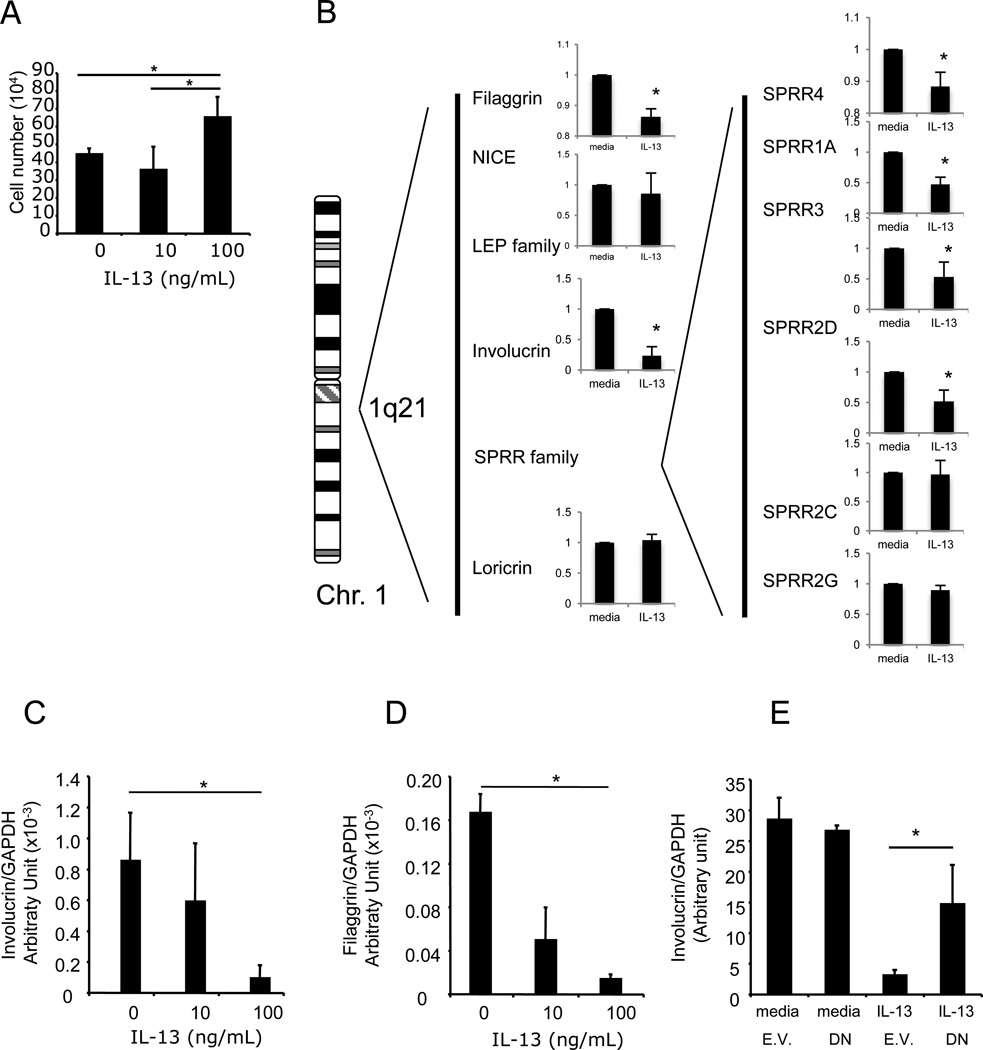
We next aimed to investigate the effect of IL-13 on epithelial cell growth and differentiation. Chronic exposure of esophageal epithelial cells to IL-13 (100ng/mL) for 9 days increased cell growth modestly (mean ± SD; 44.5 ± 3.1 to 65.2 ± 11.4 ×104 cells; p<0.05; Figure 5A). In contrast, a similar stimulation with IL-13 for 48h did not increase the cell number (data not shown). We aimed to determine whether the expression of the EDC genes such as involucrin, SPRRs and filaggrin were down regulated by IL-13 (Figure 5B–D). Microarray analysis revealed that filaggrin, involucrin and SPRR4, 1A and 3 were significantly down regulated in IL-13 treated cells. Using real time PCR, IL-13 (100 ng/mL) decreased filaggrin and involucrin mRNA expression by over 9-fold and 5fold in primary esophageal epithelial cells, respectively (Figure 5C–D). Interestingly, pretreatment with glucocorticoids was not able to inhibit IL-13 downregulation of filaggrin mRNA (data not shown). Using a dominant negative form of STAT6, we examined the role of STAT6 in the decreased expression observed following IL-13 stimulation. Expression of a dominant negative form of STAT6 partially rescued the IL-13 mediated repression of involucrin (Figure 5E), suggesting that STAT6 functionally contributes to IL-13-regulated involucrin expression in esophageal epithelial cells.
Figure 5. Physical map and gene expression of the epidermal differentiation complex gene cluster on the chromosome 1 in IL-13-stimulated primary esophageal epithelial cells.
In A, the effect of chronic exposure of IL-13 on esophageal epithelial cell number is shown. Cells (10% confluence) were stimulated every 3 days with IL-13 (0, 10, 100 ng/mL) for 9 days (80-90% confluence). Cell numbers are presented as average ± SD. The results are representative of at least 3 independent experiments. In B, schematics of the physical map of the 1q21 gene cluster and the microarray expression profile primary esophageal epithelial cells stimulated with IL-13. In B and C, esophageal primary epithelial cells from EE patients were stimulated with IL-13 (100 ng/mL) for 48h. Total mRNA was extracted and subjected to real-time RT-PCR for involucrin (B) and filaggrin (C). In D, the esophageal epithelial cell line TE-7 (<10% confluence), was transfected with a dominant negative form of STAT6 (DN) or the empty vector (E.V.). Total mRNA was extracted and submitted to real-time RT-PCR for involucrin mRNA quantification. Results are normalized to a housekeeping gene (GAPDH). Bars are average ± SD. Experiments were performed in triplicate, * is p<0.05.
Discussion
In this study, we compared epithelial responses between NL and EE patients in order to uncover molecular explanations for EE pathogenesis. Esophageal epithelial cells from both sets grew with comparable characteristics and avidly responded to IL-13 at similar levels. Numerous EDC genes, such as filaggrin, and sprr3 were downregulated both in IL-13-stimulated esophageal epithelial cells and in EE compared to NL biopsies. While a filaggrin loss of function mutation (2282del4) was overrepresented in a subset of EE patients compared to control individuals, the acquired decrease in filaggrin expression was uniformly seen in all EE patients in vivo, suggesting a generalized importance. Indeed, EDC genes filaggrin and involucrin expression were differentially increased during esophageal epithelial cell differentiation (Supplemental Figure 2) and strongly decreased by IL-13.
In our cell culture condition, NL and EE derived biopsies had similar growth characteristics. These results suggest that the esophageal hyperplasia observed in EE may result from an extrinsic phenomenon. It has been shown that esophageal epithelial cells differentially express a multitude of genes in EE compared to NL (30). While the upregulated genes include proliferation markers such as Ki67, the down regulated genes include members of the EDC (33, 37). In EE patients, there is severe epithelial cell hyperplasia and the number of epithelial cells with early esophageal differentiation markers (e.g. involucrin) are increased. Additionally, there is a decrease in occurrence of well-differentiating cells. This may explain, in part, why the involucrin gene, expressed in early differentiating cells, is not significantly decreased in EE. In contrast, filaggrin, expressed in well differentiating cells, is highly downregulated in EE patients. While both are downregulated by IL-13, the increased number of involucrin expressing cells may be responsible for the involucrin mRNA expression level not significantly changing in EE biopsy samples. We have previously shown a strong overlap between the IL-13 stimulated esophageal epithelial cell transcriptome and the EE gene signature suggesting a major involvement of IL-13 in EE (30). Chronic exposure of esophageal epithelial cells to IL-13 induces a modest increase in cell number in vitro. IL-13 has indeed, in vivo, been shown, to induce esophageal epithelial hyperplasia in mice (38). Our results also demonstrate that NL and EE epithelial cells both avidly respond to IL-13 by overexpressing eotaxin-3. While these results are representative of only one gene, they suggest that the IL-13/IL-13 receptor/STAT6 pathway is not dramatically altered in NL vs. EE. Our data are thus in line with the increase in IL-13 levels in EE biopsies previously reported (25) and argues against the pathogenic involvement of keratinocytes-intrinsic defects. Additionally, filaggrin is restored to normal levels when patients respond to glucocorticoids. This glucocorticoid effect is most likely due to the decrease of IL-13 in vivo, as in vitro experiments have shown that glucocorticoids do not modify IL-13-induced filaggrin expression in esophageal epithelial cells (data not shown). Murine models have shown the potential important role of IL-13 in EE as IL-13 deficient mice have a decrease in eosinophil infiltration. Similar observations are seen in IL-4/IL-13 double deficient and Stat6 deficient mice (27). EE is thus a multifactorial disease that certainly involves IL-13, (at least in part), but also numerous other components (genetic, immunologic or environmental) that may partially overlap in their role and function in EE pathogenesis.
Acid reflux and reflux diseases are confounders in EE diagnosis and acid neutralization resolves esophageal eosinophilia in some patients (2). The release of eotaxin-3 by protons may explain why proton pump inhibitors often have partial clinical effects in EE (1, 2). Protons are known to modify barrier function of the squamous epithelium by disrupting intercellular desmosomes. Interestingly, epithelial permeability to protons differs between healthy subjects and patients with active GERD (39). It is interesting to note that eotaxin-3 is associated with the outer surface of the cell membrane, possibly via CCR3 which has been reported to be expressed by skin keratinocytes (40, 41), Notably, heparin releases bound-eotaxin-3 from the cell surface. Chemokines are known to bind to glycosaminoglycan side chains of proteoglycans such as heparin and heparin potentiates eotaxin-induced eosinophil recruitment in vivo (42). The role of heparin in eotaxin-3 release has been previously demonstrated in bronchoalveolar epithelial cells (43) and in blood where the level of eotaxin-3 in heparin-plasma is higher compared to EDTA-plasma (33, 44). These findings emphasize the potential role of mast cell-derived heparin in eosinophil recruitment in eosinophilic esophagitis. Consistent with the ability of external factors to regulate eotaxin-3 release, eotaxin-3 recovery in the cell media was dependent on the type of sera employed. Taken together, these results demonstrate that the external components in the microenvironment likely regulate eotaxin-3 levels and the propensity of eosinophils to infiltrate the tissue.
EDC genes and filaggrin are strongly down regulated in EE patients (30). It has also been shown that a strong association exists between filaggrin mutations and atopic dermatitis (45). Since a large part of the EE population (~43%) has eczema or a history of eczema, we hypothesized that these mutations would be associated with EE. While the filaggrin mutations would be expected to be enriched in an atopic population, the prevalence of atopic diseases was the same in EE R105X positive, the EE 2282del4 positive and EE WT groups; in addition, EE patients without atopic dermatitis had a significantly increased frequency of the 2282del4 mutation compared with controls (Table III) suggesting that the association of the 2282del4 mutation with EE was not dependent upon the high rate of atopic dermatitis in EE. The significant association found between EE and 2282del4 suggests enhanced percutaneous sensitization in EE (21). It is interesting to note that the frequency of the R501X was below that expected in such a highly atopic population and this needs further investigation. Taken together, our results demonstrate that mutations in filaggrin may separately contribute to EE but are only present in a small subset of the EE population despite the profound down-regulation of filaggrin gene expression seen in nearly all patients.
In conclusion, we report that esophageal epithelial cells from EE patients are functionally normal compared with cells isolated from control individuals as assessed by growth properties ex vivo. IL-13 responsiveness based on eotaxin-3 production. While a filaggrin loss of function mutation (2282del4) was overrepresented in EE compared to control individuals, the acquired decrease in filaggrin expression was uniformly seen in all EE patients in vivo. Indeed, EDC gene (filaggrin and involucrin) expressions were strongly decreased directly by IL-13. As such, these results establish that the primary defect in epithelial responses in EE is not intrinsic to the epithelium, but rather likely secondary to the effects of IL-13, which regulates expression of EDC genes. Taken together, our results support a model of coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis.
Supplementary Material
Acknowledgements
The authors would like to thank Lawanda Bryant, Katherine Henderson, Kiran Mudambi, and Tova Holowinko for their assistance.
Abbreviations
- NL
normal
- EE
eosinophilic esophagitis
- EDC
epidermal differentiation complex
- SPRR3
small proline-rich protein 3
- AD
atopic dermatitis
- GERD
gastro esophageal reflux disease
Footnotes
This work was supported by in part by the Thrasher Research Fund NR-0014 (C.B.), the 2009 APFED HOPE research grant (C.B.), the PHS Grant P30 DK0789392 (C.B.) the NIH AI079874-01 (C.B.), AI070235, AI45898, and DK076893 (M.E.R.), the Food Allergy and Anaphylaxis Network (M.E.R.), Campaign Urging Research for Eosinophil Disorders (CURED), the Buckeye Foundation (M.E.R.) and the Food Allergy Project (M.E.R).
References
- 1.Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, Bonis P, Hassall E, Straumann A, Rothenberg ME. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Molina-Infante J, Ferrando-Lamana L, Mateos-Rodriguez JM, Perez-Gallardo B, Prieto-Bermejo AB. Overlap of reflux and eosinophilic esophagitis in two patients requiring different therapies: a review of the literature. World J Gastroenterol. 2008;14:1463–1466. doi: 10.3748/wjg.14.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lierse W. The physiology and pathology of the esophagus. Eur J Pediatr Surg. 1992;2:323–326. doi: 10.1055/s-2008-1063470. [DOI] [PubMed] [Google Scholar]
- 4.De Benedetto A, Qualia CM, Baroody FM, Beck LA. Filaggrin expression in oral, nasal, and esophageal mucosa. J Invest Dermatol. 2008;128:1594–1597. doi: 10.1038/sj.jid.5701208. [DOI] [PubMed] [Google Scholar]
- 5.Crish JF, Bone F, Banks EB, Eckert RL. The human involucrin gene contains spatially distinct regulatory elements that regulate expression during early versus late epidermal differentiation. Oncogene. 2002;21:738–747. doi: 10.1038/sj.onc.1205038. [DOI] [PubMed] [Google Scholar]
- 6.Crish JF, Gopalakrishnan R, Bone F, Gilliam AC, Eckert RL. The distal and proximal regulatory regions of the involucrin gene promoter have distinct functions and are required for in vivo involucrin expression. J Invest Dermatol. 2006;126:305–314. doi: 10.1038/sj.jid.5700019. [DOI] [PubMed] [Google Scholar]
- 7.Crish JF, Howard JM, Zaim TM, Murthy S, Eckert RL. Tissue-specific and differentiation-appropriate expression of the human involucrin gene in transgenic mice: an abnormal epidermal phenotype. Differentiation. 1993;53:191–200. doi: 10.1111/j.1432-0436.1993.tb00708.x. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi M, Naomoto Y, Nobuhisa T, Okawa T, Takaoka M, Shirakawa Y, Yamatsuji T, Matsuoka J, Mizushima T, Matsuura H, Nakajima M, Nakagawa H, Rustgi A, Tanaka N. Heparanase regulates esophageal keratinocyte differentiation through nuclear translocation and heparan sulfate cleavage. Differentiation. 2006;74:235–243. doi: 10.1111/j.1432-0436.2006.00072.x. [DOI] [PubMed] [Google Scholar]
- 9.Marenholz I, Zirra M, Fischer DF, Backendorf C, Ziegler A, Mischke D. Identification of human epidermal differentiation complex (EDC)-encoded genes by subtractive hybridization of entire YACs to a gridded keratinocyte cDNA library. Genome Res. 2001;11:341–355. doi: 10.1101/gr.114801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proksch E, Folster-Holst R, Jensen JM. Skin barrier function, epidermal proliferation and differentiation in eczema. J Dermatol Sci. 2006;43:159–169. doi: 10.1016/j.jdermsci.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Mischke D. The complexity of gene families involved in epithelial differentiation. Keratin genes and the epidermal differentiation complex. Subcell Biochem. 1998;31:71–104. [PubMed] [Google Scholar]
- 12.Marenholz I, Volz A, Ziegler A, Davies A, Ragoussis I, Korge BP, Mischke D. Genetic analysis of the epidermal differentiation complex (EDC) on human chromosome 1q21: chromosomal orientation, new markers, and a 6-Mb YAC contig. Genomics. 1996;37:295–302. doi: 10.1006/geno.1996.0563. [DOI] [PubMed] [Google Scholar]
- 13.Regnier M, Caron D, Reichert U, Schaefer H. Reconstructed human epidermis: a model to study in vitro the barrier function of the skin. Skin Pharmacol. 1992;5:49–56. doi: 10.1159/000211017. [DOI] [PubMed] [Google Scholar]
- 14.Poumay Y, Dupont F, Marcoux S, Leclercq-Smekens M, Herin M, Coquette A. A simple reconstructed human epidermis: preparation of the culture model and utilization in in vitro studies. Arch Dermatol Res. 2004;296:203–211. doi: 10.1007/s00403-004-0507-y. [DOI] [PubMed] [Google Scholar]
- 15.Presland RB, Coulombe PA, Eckert RL, Mao-Qiang M, Feingold KR, Elias PM. Barrier function in transgenic mice overexpressing K16, involucrin, and filaggrin in the suprabasal epidermis. J Invest Dermatol. 2004;123:603–606. doi: 10.1111/j.0022-202X.2004.23226.x. [DOI] [PubMed] [Google Scholar]
- 16.Vickery BP. Skin barrier function in atopic dermatitis. Curr Opin Pediatr. 2007;19:89–93. doi: 10.1097/MOP.0b013e328012315a. [DOI] [PubMed] [Google Scholar]
- 17.Morar N, Cookson WO, Harper JI, Moffatt MF. Filaggrin mutations in children with severe atopic dermatitis. J Invest Dermatol. 2007;127:1667–1672. doi: 10.1038/sj.jid.5700739. [DOI] [PubMed] [Google Scholar]
- 18.Hanifin JM. Filaggrin mutations and allergic contact sensitization. J Invest Dermatol. 2008;128:1362–1364. doi: 10.1038/sj.jid.5701253. [DOI] [PubMed] [Google Scholar]
- 19.Novak N, Baurecht H, Schafer T, Rodriguez E, Wagenpfeil S, Klopp N, Heinrich J, Behrendt H, Ring J, Wichmann E, Illig T, Weidinger S. Loss-of-function mutations in the filaggrin gene and allergic contact sensitization to nickel. J Invest Dermatol. 2008;128:1430–1435. doi: 10.1038/sj.jid.5701190. [DOI] [PubMed] [Google Scholar]
- 20.Weidinger S, O'Sullivan M, Illig T, Baurecht H, Depner M, Rodriguez E, Ruether A, Klopp N, Vogelberg C, Weiland SK, McLean WH, von Mutius E, Irvine AD, Kabesch M. Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J Allergy Clin Immunol. 2008;121:1203–1209. doi: 10.1016/j.jaci.2008.02.014. e1201. [DOI] [PubMed] [Google Scholar]
- 21.Fallon PG, Sasaki T, Sandilands A, Campbell LE, Saunders SP, Mangan NE, Callanan JJ, Kawasaki H, Shiohama A, Kubo A, Sundberg JP, Presland RB, Fleckman P, Shimizu N, Kudoh J, Irvine AD, Amagai M, McLean WH. A homozygous frameshift mutation in the mouse Flg gene facilitates enhanced percutaneous allergen priming. Nat Genet. 2009;41:602–608. doi: 10.1038/ng.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marenholz I, Kerscher T, Bauerfeind A, Esparza-Gordillo J, Nickel R, Keil T, Lau S, Rohde K, Wahn U, Lee YA. An interaction between filaggrin mutations and early food sensitization improves the prediction of childhood asthma. J Allergy Clin Immunol. 2009;123:911–916. doi: 10.1016/j.jaci.2009.01.051. [DOI] [PubMed] [Google Scholar]
- 23.Stemmler S, Nothnagel M, Parwez Q, Petrasch-Parwez E, Epplen JT, Hoffjan S. Variation in genes of the epidermal differentiation complex in German atopic dermatitis patients. Int J Immunogenet. 2009;36:217–222. doi: 10.1111/j.1744-313X.2009.00858.x. [DOI] [PubMed] [Google Scholar]
- 24.Barker JN, Palmer CN, Zhao Y, Liao H, Hull PR, Lee SP, Allen MH, Meggitt SJ, Reynolds NJ, Trembath RC, McLean WH. Null Mutations in the Filaggrin Gene (FLG) Determine Major Susceptibility to Early-Onset Atopic Dermatitis that Persists into Adulthood. J Invest Dermatol. 2006 doi: 10.1038/sj.jid.5700587. [DOI] [PubMed] [Google Scholar]
- 25.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, Collins MH, Putnam PE, Wells SI, Rothenberg ME. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 26.Akei HS, Brandt EB, Mishra A, Strait RT, Finkelman FD, Warrier MR, Hershey GK, Blanchard C, Rothenberg ME. Epicutaneous aeroallergen exposure induces systemic TH2 immunity that predisposes to allergic nasal responses. J Allergy Clin Immunol. 2006;118:62–69. doi: 10.1016/j.jaci.2006.04.046. [DOI] [PubMed] [Google Scholar]
- 27.Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology. 2005;129:985–994. doi: 10.1053/j.gastro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology. 2003;125:1419–1427. doi: 10.1016/j.gastro.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Kim BE, Leung DY, Boguniewicz M, Howell MD. Loricrin and involucrin expression is down-regulated by Th2 cytokines through STAT-6. Clin Immunol. 2008;126:332–337. doi: 10.1016/j.clim.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, Jameson SC, Kirby C, Konikoff MR, Collins MH, Cohen MB, Akers R, Hogan SP, Assa'ad AH, Putnam PE, Aronow BJ, Rothenberg ME. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 32.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchard C, Durual S, Estienne M, Emami S, Vasseur S, Cuber JC. Eotaxin-3/CCL26 gene expression in intestinal epithelial cells is up-regulated by interleukin-4 and interleukin-13 via the signal transducer and activator of transcription 6. Int J Biochem Cell Biol. 2005;37:2559–2573. doi: 10.1016/j.biocel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Enomoto H, Hirata K, Otsuka K, Kawai T, Takahashi T, Hirota T, Suzuki Y, Tamari M, Otsuka F, Fujieda S, Arinami T, Noguchi E. Filaggrin null mutations are associated with atopic dermatitis and elevated levels of IgE in the Japanese population: a family and case-control study. J Hum Genet. 2008;53:615–621. doi: 10.1007/s10038-008-0293-z. [DOI] [PubMed] [Google Scholar]
- 35.Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, Liao H, Evans AT, Goudie DR, Lewis-Jones S, Arseculeratne G, Munro CS, Sergeant A, O'Regan G, Bale SJ, Compton JG, DiGiovanna JJ, Presland RB, Fleckman P, McLean WH. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 36.Sharma M, Mehla K, Batra J, Ghosh B. Association of a chromosome 1q21 locus in close proximity to a late cornified envelope-like proline-rich 1 (LELP1) gene with total serum IgE levels. J Hum Genet. 2007;52:378–383. doi: 10.1007/s10038-007-0118-5. [DOI] [PubMed] [Google Scholar]
- 37.Noel RJ, Putnam PE, Collins MH, Assa'ad AH, Guajardo JR, Jameson SC, Rothenberg ME. Clinical and immunopathologic effects of swallowed fluticasone for eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2004;2:568–575. doi: 10.1016/s1542-3565(04)00240-x. [DOI] [PubMed] [Google Scholar]
- 38.Zuo L, Fulkerson PC, Blanchard C, Rothenberg ME. Esophageal IL-13 accumulation, eosinophilia, and tissue remodeling induced by the lung IL-13 transgene. J Allergy Clin Immunol. 2007 Abstract, AAAAI 2007 meeting. [Google Scholar]
- 39.Carlsson R, Fandriks L, Jonsson C, Lundell L, Orlando RC. Is the esophageal squamous epithelial barrier function impaired in patients with gastroesophageal reflux disease? Scand J Gastroenterol. 1999;34:454–458. doi: 10.1080/003655299750026155. [DOI] [PubMed] [Google Scholar]
- 40.Petering H, Kluthe C, Dulkys Y, Kiehl P, Ponath PD, Kapp A, Elsner J. Characterization of the CC chemokine receptor 3 on human keratinocytes. J Invest Dermatol. 2001;116:549–555. doi: 10.1046/j.1523-1747.2001.01302.x. [DOI] [PubMed] [Google Scholar]
- 41.Amerio P, Frezzolini A, Feliciani C, Verdolini R, Teofoli P, De Pita O, Puddu P. Eotaxins and CCR3 receptor in inflammatory and allergic skin diseases: therapeutical implications. Curr Drug Targets Inflamm Allergy. 2003;2:81–94. doi: 10.2174/1568010033344480. [DOI] [PubMed] [Google Scholar]
- 42.Ellyard JI, Simson L, Bezos A, Johnston K, Freeman C, Parish CR. Eotaxin selectively binds heparin. An interaction that protects eotaxin from proteolysis and potentiates chemotactic activity in vivo. J Biol Chem. 2007;282:15238–15247. doi: 10.1074/jbc.M608046200. [DOI] [PubMed] [Google Scholar]
- 43.Yuan Q, Campanella GS, Colvin RA, Hamilos DL, Jones KJ, Mathew A, Means TK, Luster AD. Membrane-bound eotaxin-3 mediates eosinophil transepithelial migration in IL-4-stimulated epithelial cells. Eur J Immunol. 2006;36:2700–2714. doi: 10.1002/eji.200636112. [DOI] [PubMed] [Google Scholar]
- 44.Konikoff MR, Blanchard C, Kirby C, Buckmeier BK, Cohen MB, Heubi JE, Putnam PE, Rothenberg ME. Potential of Blood Eosinophils, Eosinophil-Derived Neurotoxin, and Eotaxin-3 as Biomarkers of Eosinophilic Esophagitis. Clin Gastroenterol Hepatol. 2006;4:1328–1336. doi: 10.1016/j.cgh.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 45.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, O'Regan GM, Watson RM, Cecil JE, Bale SJ, Compton JG, DiGiovanna JJ, Fleckman P, Lewis-Jones S, Arseculeratne G, Sergeant A, Munro CS, El Houate B, McElreavey K, Halkjaer LB, Bisgaard H, Mukhopadhyay S, McLean WH. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.