Abstract
Recent advances in human genome studies have opened new avenues for the identification of susceptibility genes for many complex genetic disorders, especially in the field of rare cancers such as glioma. To date, eight glioma susceptibility loci have been identified by candidate gene-association studies: PRKDC G6721T, XRCC1 W399R, PARP1 A762V, MGMT F84L, ERCC1 A8092C, ERCC2 Q751K, EGF +61 A/G and IL13 R110G. Five loci have been identified by genome-wide association studies: TERT rs2736100, CCDC26 rs4295627, CDKN2A–CDKN2B rs4977756, PHLDB1 rs498872, and RTEL1 rs6010620. Using the Ingenuity Pathway Analysis tool, we investigated whether these 13 susceptibility genes are biologically related. Our data provide not only networks for understanding the biological properties of gliomagenesis but useful pathway maps for future understanding of disease.
Keywords: Glioma, Susceptibility, Pathway analysis
Introduction
While primary brain tumors (PBTs), are relatively rare (5%– 9% of all cancers), approximately 22,070 individuals are diagnosed each year in the US [1].Moreover, PBTs have very poor prognosis and are associated with considerable morbidity and mortality. They are a leading cause of cancer death in childhood, the second leading cause of cancer-related death in men aged 20–39, and the fifth leading cause of cancer-related death in women aged 20–39. Gliomas, the most common type of brain tumors, are derived from glial cells that surround and support neurons in the brain. Gliomas of astrocytic, oligodendroglial, and ependymal origin account for ~ 80% of malignant PBTs.
Diverse environmental factors have been implicated in epidemiological studies of glioma including radiation, occupational exposure and environmental carcinogens, Allergy and immunological conditions, and viral infection [2]. However, to date only therapeutic ionizing or high-dose radiation is an established risk factor [3]. A number of rare inherited cancer syndromes, melanoma-brain tumor (P16/CDKN2A mutations) [4], neurofibromatosis (NF1 mutations), tuberous sclerosis (APC mutations), and Li-Fraumeni (TP53 mutations) are recognized to confer substantive risk of glioma [5], however collectively these only explain a small proportion of cases [6–10]. Current evidence suggests that a model of glioma susceptibility based solely on high-risk mutations seems unlikely. Thus, as with other cancers, much of the inherited risk is likely to be a consequence of the co-inheritance of multiple low-risk variants, some of which will be common and hence identifiable through association-based analyses.
Completion of the Human Genome Project in 2000 has allowed complete annotation of genetic variation across the genome. This coupled with the development of analytical platforms capable of parallel processing of genotyping is allowing performing genome-wide association studies a cost-effective manner. In this brief review we will critically discuss the candidate genes and SNPs which have been linked to glioma risk. We also review emerging data on whole genome-association searches to identify risk loci. Finally, we have implemented an Ingenuity Pathways Analysis tool to study the inter-relationship between risk loci in a biologically meaningful fashion.
Candidate gene-association studies
Candidate gene-association studies for glioma have mainly focused on four hypothesized pathways: DNA repair, cell cycle, metabolism, and inflammation (including allergies and infections). These studies are continuously updated at The Genetic Association Database online (http://geneticassociationdb.nih.gov/). Although some promising findings have been reported, very few have been confirmed in subsequent independent replication studies. Because glioma is a rare cancer, there are serious limitations of small sample sizes (very few studies have had >500 cases and controls) and the characteristic heterogeneity of gliomas. Furthermore, studies have been seriously underpowered to establish interactions.
We recently published an overview of candidate genes associated with glioma susceptibility [11]. Based on those results, we identified eight susceptibility loci that each had been reliably associated with glioma in at least two populations from case-control studies. Table 1 presents a list of eight literature-defined putative functional polymorphisms. Additional criteria for inclusion included replication in at least one other population and replication with the same SNP. These eight genes and SNPs were: DNA repair genes PRKDC (also known as XRCC7) G6721T [12,13], XRCC1 W399R [13–15], PARP1 A762V [13,15], MGMT F84L [15,16], ERCC1 A8092C [17,18], and ERCC2 Q751K [13,18]; cell cycle gene EGF +61 A/G [19,20]; and inflammation gene IL-13 R110G [21,22]. Studies that examined the correlations between metabolism genes and glioma risk did not generate a clear pattern of association. The effect estimates (odds ratios [ORs]) associated with variants is modest, typical between 0.5 and 1.5.
TABLE 1.
Selected glioma susceptibility genes and SNPs observed in at least two studies
| Main pathways, genes | Associated SNPs | Effect | OR (95%CI) | Reference |
|---|---|---|---|---|
| DNA repair | ||||
| (1) Double strand break repair: PRKDC | G6721T (rs7003908) | Risk | GG vs TT, 1.82 (1.13–2.93); GG vs TT, 1.44 (1.13–1.84) |
[12] [13] |
| (2) Base excision repair | ||||
| XRCC1 | W399R (rs25487) | Risk | AA vs GG, 1.23 (0.96–1.57); AA vs GG, 1.32 (0.97–1.81); GA/AA VS GG, 1.44 (1.05–1.92) |
[13] [14] [15] |
| PARP1 | A762V (rs1136410) | Protective | CT/CC vs TT, 0.80 (0.67–0.95); CT/CC vs TT, 0.71 (0.52–0.97) |
[13] [15] |
| (3) Nucleotide excision repair | ||||
| ERCC1 | A8092C (rs3212981) | Risk | AA/AC vs CC, 4.41 (1.6–12.2); AA/AC vs CC, 1.67 (0.93–3.02) |
[17] [18] |
| ERCC2 | Q751K (rs13181) | Risk | CC vs AA, 1.19 (0.93–1.52); AA vs AC/CC, 1.66 (1.01–2.72) |
[13] [18] |
| (4) Direct reversal of damage: MGMT | F84L (rs12917) | Protective or risk? |
CT/TT vs CC, 0.66 (0.45–0.94); CT/TT vs CC, 1.26(0.90–1.75); |
[15] [16] |
| Cell cycle: EGF | +61 A/G (rs4444903) | Risk | P=0.032; AG/GG vs AA, 1.52(1.03–2.23); |
[19] [20] |
| Inflammation : IL13 | R110G (rs20541) | Protective | AG vs GG, 0.75 (0.48–1.17); TT vs CC/CT, 0.39 (0.16–0.93) |
[21] In press |
It has been reported that there is an inverse association between self-reported allergies and glioma risk [23]. The observation that IL13 R110G showed a protective effect with glioma lends further biological plausibility for such a relationship as variation in IL13 has been linked to atopy risk. Of these the eight susceptibility genes, six were from DNA repair pathway, which supports the importance of DNA repair genes in the pathogenesis of glioma. These observations suggest that evaluation of further polymorphisms mapping to these loci is likely to lead to the identification of additional novel risk variants.
Genome wide association studies
Two glioma genome wide association (GWA) studies have so far been reported. In our GWAS [24], we conducted a meta-analysis of two GWA studies which involved the genotyping of 454,576 tagging SNPs in a total of 1,878 glioma cases and 3670 controls (1,434 from the 1958 Birth Cohort controls, 2,236 from Cancer Genetic Markers of Susceptibility [CGEMS] breast controls), with validation in three additional independent series totaling 2545 cases and 2953 controls. We identified five risk loci for glioma at 5p15.33 (TERT rs2736100), 8q24.21 (CCDC26 rs4295627), 9p21.3 (CDKN2A–CDKN2B rs4977756), 11q23.3 (PHLDB1 rs498872), and 20q13.33 (RTEL1 rs6010620). The second independent GWA study of glioma [25] reported was based on an analysis of 275,895 SNPs in 692 adult patients with high-grade glioma and 3992 controls (602 from University of California – San Francisco and 3390 from Illumina iControlDB) with a replication series of 176 high-grade glioma cases and 174 controls. That analysis provided further evidence to implicate the 9p21 (CDKN2B rs1412829) and 20q13.3 (RTEL1 rs6010620) in glioma risk. Although the replication set of Wrensch et al. [25] did not reach formal statistical significance for 5p15.33 (TERT rs2736100), this SNP had the lowest p-value in their discovery set. Both GWA used publicly available controls in the discovery phase. While this reduces the cost of studies, it limits the ability to identify gene-environment interactions.
RTEL1 (regulator of telomere elongation helicase 1) is a DNA helicase critical for regulation of telomere length in mice, and its loss has been associated with shortened telomere length, chromosome breaks, and translocations [26]. Moreover, RTEL1 is important member of the DNA double-stand break-repair pathway, and it maintains genomic stability directly by suppressing homologous recombination [27]. Coincidentally, TERT (telomerase reverse transcriptase) is also a telomerase-related gene that is essential for telomerase activity in maintaining telomeres and cell immortalization. TERT has an established role in glioma grade and prognosis [28,29]. CCDC26 is a retinoic acid modulator of differentiation and death that induces caspase 8 transcription and increases apoptosis in response to death stimuli in neuroblastoma cells and in glioblastoma cells by down-regulating telomerase activity [30,31]. CDKN2A is a well-known tumor suppressor gene. CDKN2A encoding p16(INK4A) a negative regulator of cyclin-dependent kinases and p14(ARF1), an activator of p53. Regulation of p16/p14ARF is important for sensitivity to ionizing radiation, the only environmental factor strongly linked to gliomagenesis [32–34]. CDKN2B also is a cyclin-dependent kinase inhibitor that regulates cell growth and cell cycle G1 progression. In glioblastoma cells, overexpression of CDKN2B in a CDKN2A-deficient background has been shown to inhibit cell growth, induce replicative senescence, and inhibit telomerase activity [35]. Although there is no direct evidence for a role of PHLDB1 in glioma, intriguingly, 11q23.3 is commonly deleted in neuroblastoma [36].
It is very interesting to note that the function of the five glioma susceptibility genes identified by GWA studies (RTEL1, TERT, CCDC26, CDKN2A–CDKN2B, and PHLDB1), four (TERT, RTEL1, CCDC26, and CDKN2B) have very close links to telomerase. TERT is the telomerase catalytic subunit, and RTEL1 is directly involved in telomerase length, whereas the other two are related to telomerase activity. Taken together, these data provide strong evidence that common variation in telomerase-related genes contributes to glioma predisposition.
While the effect estimates of glioma associated with each of these five risk genes and variants are modest (ORs 1.18–1.36) in GWA the risk variants are common in the population and hence they play a significant role in the development of glioma in general. Because there is substantial genetic heterogeneity exists within glioma tumors of the same histological subtype, it is generally believed that there are multiple pathways and genes of genetic alterations leading to the pathogenesis of gliomas. Thus, it remains to be discovered how these genes (and SNPs) work in networks, and the susceptibility genes must be examined together to determine the true meaning of how these genes form functional biological networks and pathways that confer risk for glioma.
Network analysis
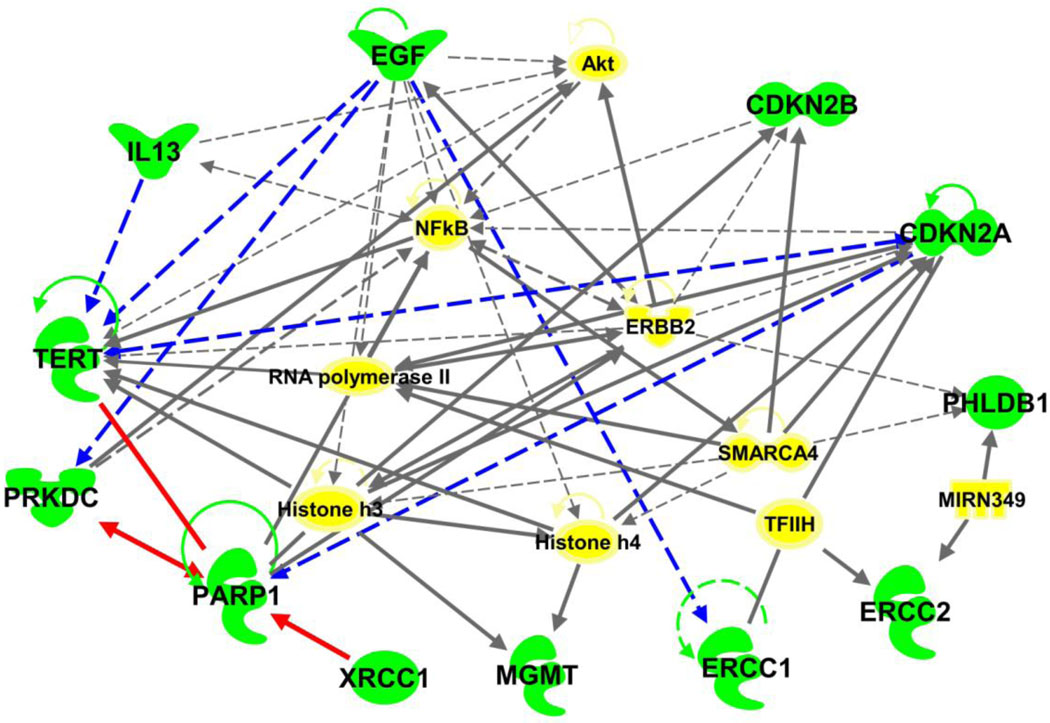
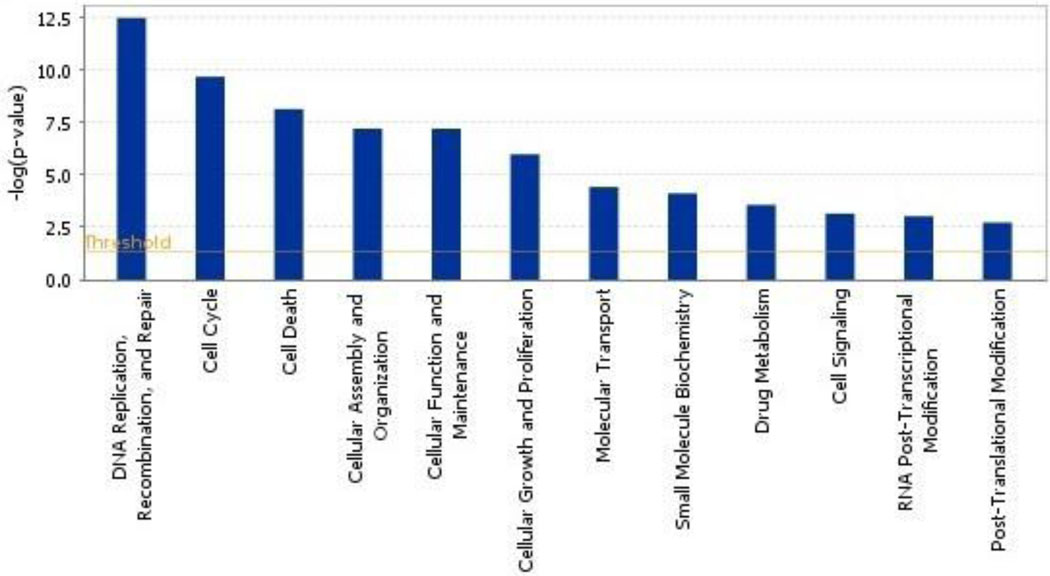
Ingenuity Pathway Analysis software (Ingenuity® Systems, www.ingenuity.com) were utilized to investigate the functional epistatic relationships among the genes identified by the candidate gene and GWAS approaches. Figure 1 shows how these genes relate to one or another and form pathways. Of these 14 genes, 12 (all except RTEL1 and CCDC26) have established direct or indirect interactions, with 21 network-eligible genes involved. These networks were identified around several major genes, including cell cycle genes CDKN2A, ERBB2, and EGF; telomerase maintenance gene TERT; and inflammation gene NFKB. Filtering the interaction data to focus on these genes revealed 3 pairs of direct two-way interactions: (1) PARP1 and XRCC1, (2) PARP1 and PRKDC, (3) PARP1 and TERT; and 6 pairs of indirect two-way interactions: (1) TERT and CDKN2A, (2) TERT and IL13, (3) TERT and EGF, (4) EGF and ERCC1, (5) EGF and PRKDC, and (6) PARP1 and CDKN2A. The key bio-functions of these genes are related to (1) DNA replication, recombination, and repair; (2) the cell cycle; and (3) cell death (Fig.2). Glioma signaling was the most significant disease specific canonical pathway.
Figure 1. Biological networks for selected glioma susceptibility genes.
Interactions among the glioma susceptibility genes (green emblems) were analyzed using Ingenuity Pathway Analysis (http://www.ingenuity.com). This diagram shows the direct (solid lines) and indirect (dashed lines) interactions among the 21 genes. The red solid lines are the direct two-way interactions, and blue dashed lines are the indirect two-way interactions. For clarity, we do not show all of the interactions identified but focus primarily on direct interactions that link glioma.
Figure 2. Functional annotations of the glioma susceptibility genes.
The annotations show that the key bio-function of these susceptibility genes is related to DNA replication, recombination and repair pathway. The 21 network genes associated with biological functions in the Ingenuity Pathways Knowledge Base were considered for the analysis. Fisher’s exact test was used to calculate a p-value determining the probability that each biological function assigned to that network is due to chance alone. Threshold bar shows cut-off point of significance P < 0.05, −log(P-value) of 1.3.
It should be noted that there are certain limitations to our in silico analysis: it is exploratory and hypothesis generating. Since there are many different gene-gene interactions resulting from various cellular/experimental conditions, the gene relationships denoted in our network may not represent the actual causal relationship between genes. It does, however, provide exploration of the connectivity between genes and disease development. Hence despite its limitations, this preliminary analysis suggests interconnectedness of several currently well known gene-gene interactions, base excision repair pathway genes XRCC1 interacts with PARP1 [37,38]; and TERT interacts with PARP1[39]. It also uncovered new information for direct interactions between PARP1 and PRKDC.
Conclusions and future directions
While the candidate gene-association studies have thus far identified variation in DNA repair pathway genes as most promising basis of inherited susceptibility this may in part be a consequence of a self fulfilling prophecy as such genes have been the major subject of evaluation. Adopting an agnostic approach GWA studies have implicated cell cycle– and telomerase-related genes as important risk determinants. The development and progression of gliomas are likely due to a multistep process that involves functional inactivation of tumor suppressor genes (i.e., CDKN2A and CDKN2B) and DNA repair genes (i.e., PARP1, PRKDC, and ERCC2) as well as activation of oncogenes and proto-oncogenes (i.e., EGF and ERBB2). It is possible that a profile of genetic alterations including the cell cycle control, DNA repair pathway, and telomerase change–related genes will provide the most informative approach for molecular epidemiological studies aimed at understanding variations in glioma risk.
Compared with many other cancers, little is known about the lifestyle or environment factors that influence the risk of developing brain tumors. Identifying these environment factors and investigations of not only gene-gene but also gene-environment interactions are important. Besides the GLIOGENE familial glioma study assembled in 2007, we are now launching another international consortium, the GLIOGENE case-control prospective study in the begging of 2010, to examine the complex interplay of gene -gene and gene-environment interactions in 6000 glioma patients and 6000 healthy controls. With this, we will be able to identify the potential risk and protective factors in a much larger study, and ultimately find improved ways to diagnose and treat the disease.
Acknowledgements
We thank V. Mohlere for scientific editing. This work was supported by National Institutes of Health grants 5R01CA119215, 5R01CA070917 and 1R01CA139020-01A1. Additional support was obtained from the American Brain Tumor Association and the National Brain Tumor Society. The Wellcome Trust provided principal funding for the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests statement
The authors declare no competing financial interests
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2. Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. • This review article provides a broad and detailed overview of the epidemiology and etiology of gliomas.
- 3. Bondy ML, Scheurer ME, Malmer B, Barnholtz-Sloan JS, Davis FG, Il'yasova D, Kruchko C, McCarthy BJ, Rajaraman P, Schwartzbaum JA, et al. Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113:1953–1968. doi: 10.1002/cncr.23741. •• This is a key review article to critically discuss the current state of epidemiologic literature on brain tumors and presents the Brain Tumor Epidemiology Consortium (BTEC) group’s consensus on research priorities needs to drive the science forward over the next decade.
- 4.Randerson-Moor JA, Harland M, Williams S, Cuthbert-Heavens D, Sheridan E, Aveyard J, Sibley K, Whitaker L, Knowles M, Bishop JN, et al. A germline deletion of p14(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. 2001;10:55–62. doi: 10.1093/hmg/10.1.55. [DOI] [PubMed] [Google Scholar]
- 5.Melean G, Sestini R, Ammannati F, Papi L. Genetic insights into familial tumors of the nervous system. Am J Med Genet C Semin Med Genet. 2004;129C:74–84. doi: 10.1002/ajmg.c.30022. [DOI] [PubMed] [Google Scholar]
- 6.Malmer B, Gronberg H, Andersson U, Jonsson BA, Henriksson R. Microsatellite instability, PTEN and p53 germline mutations in glioma families. Acta Oncol. 2001;40:633–637. doi: 10.1080/028418601750444196. [DOI] [PubMed] [Google Scholar]
- 7.Tachibana I, Smith JS, Sato K, Hosek SM, Kimmel DW, Jenkins RB. Investigation of germline PTEN, p53, p16(INK4A)/p14(ARF), and CDK4 alterations in familial glioma. Am J Med Genet. 2000;92:136–141. doi: 10.1002/(sici)1096-8628(20000515)92:2<136::aid-ajmg11>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 8.Zhou XP, Sanson M, Hoang-Xuan K, Robin E, Taillandier L, He J, Mokhtari K, Cornu P, Delattre JY, Thomas G, et al. The ANOCEF Group. Association des NeuroOncologues d'Expression Francaise. Germline mutations of p53 but not p16/CDKN2 or PTEN/MMAC1 tumor suppressor genes predispose to gliomas. Ann Neurol. 1999;46:913–916. doi: 10.1002/1531-8249(199912)46:6<913::aid-ana15>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 9.van Meyel DJ, Ramsay DA, Chambers AF, Macdonald DR, Cairncross JG. Absence of hereditary mutations in exons 5 through 9 of the p53 gene and exon 24 of the neurofibromin gene in families with glioma. Ann Neurol. 1994;35:120–122. doi: 10.1002/ana.410350120. [DOI] [PubMed] [Google Scholar]
- 10.Paunu N, Syrjakoski K, Sankila R, Simola KO, Helen P, Niemela M, Matikainen M, Isola J, Haapasalo H. Analysis of p53 tumor suppressor gene in families with multiple glioma patients. J Neurooncol. 2001;55:159–165. doi: 10.1023/a:1013890022041. [DOI] [PubMed] [Google Scholar]
- 11. Gu J, Liu Y, Kyritsis AP, Bondy ML. Molecular epidemiology of primary brain tumors. Neurotherapeutics. 2009;6:427–435. doi: 10.1016/j.nurt.2009.05.001. • This review article summarizes most of the glioma susceptibility candidate genes and variations from DNA repair, cell cycle, metabolism, and inflammation pathways.
- 12.Wang LE, Bondy ML, Shen H, El-Zein R, Aldape K, Cao Y, Pudavalli V, Levin VA, Yung WK, Wei Q. Polymorphisms of DNA repair genes and risk of glioma. Cancer Res. 2004;64:5560–5563. doi: 10.1158/0008-5472.CAN-03-2181. [DOI] [PubMed] [Google Scholar]
- 13.McKean-Cowdin R, Barnholtz-Sloan J, Inskip PD, Ruder AM, Butler M, Rajaraman P, Razavi P, Patoka J, Wiencke JK, Bondy ML, et al. Associations between polymorphisms in DNA repair genes and glioblastoma. Cancer Epidemiol Biomarkers Prev. 2009;18:1118–1126. doi: 10.1158/1055-9965.EPI-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiuru A, Lindholm C, Heinavaara S, Ilus T, Jokinen P, Haapasalo H, Salminen T, Christensen HC, Feychting M, Johansen C, et al. XRCC1 and XRCC3 variants and risk of glioma and meningioma. J Neurooncol. 2008;88:135–142. doi: 10.1007/s11060-008-9556-y. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Scheurer ME, El-Zein R, Cao Y, Do KA, Gilbert M, Aldape KD, Wei Q, Etzel C, Bondy ML. Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol Biomarkers Prev. 2009;18:204–214. doi: 10.1158/1055-9965.EPI-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felini MJ, Olshan AF, Schroeder JC, North KE, Carozza SE, Kelsey KT, Liu M, Rice T, Wiencke JK, Wrensch MR. DNA repair polymorphisms XRCC1 and MGMT and risk of adult gliomas. Neuroepidemiology. 2007;29:55–58. doi: 10.1159/000108919. [DOI] [PubMed] [Google Scholar]
- 17.Chen P, Wiencke J, Aldape K, Kesler-Diaz A, Miike R, Kelsey K, Lee M, Liu J, Wrensch M. Association of an ERCC1 polymorphism with adult-onset glioma. Cancer Epidemiol Biomarkers Prev. 2000;9:843–847. [PubMed] [Google Scholar]
- 18.Wrensch M, Kelsey KT, Liu M, Miike R, Moghadassi M, Sison JD, Aldape K, McMillan A, Wiemels J, Wiencke JK. ERCC1 and ERCC2 polymorphisms and adult glioma. Neuro Oncol. 2005;7:495–507. doi: 10.1215/S1152851705000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhowmick DA, Zhuang Z, Wait SD, Weil RJ. A functional polymorphism in the EGF gene is found with increased frequency in glioblastoma multiforme patients and is associated with more aggressive disease. Cancer Res. 2004;64:1220–1223. doi: 10.1158/0008-5472.can-03-3137. [DOI] [PubMed] [Google Scholar]
- 20.Costa BM, Ferreira P, Costa S, Canedo P, Oliveira P, Silva A, Pardal F, Suriano G, Machado JC, Lopes JM, et al. Association between functional EGF+61 polymorphism and glioma risk. Clin Cancer Res. 2007;13:2621–2626. doi: 10.1158/1078-0432.CCR-06-2606. [DOI] [PubMed] [Google Scholar]
- 21.Schwartzbaum J, Ahlbom A, Malmer B, Lonn S, Brookes AJ, Doss H, Debinski W, Henriksson R, Feychting M. Polymorphisms associated with asthma are inversely related to glioblastoma multiforme. Cancer Res. 2005;65:6459–6465. doi: 10.1158/0008-5472.CAN-04-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.E Amirian YL, Scheurer MichaelE, El-Zein Randa, Gilbert MarkR, Bondy MelissaL. Interaction among Genetic Variants in Inflammation Pathway Genes and Asthma in Glioma Susceptibility. Neuro-Oncology. 2009 doi: 10.1093/neuonc/nop057. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linos E, Raine T, Alonso A, Michaud D. Atopy and risk of brain tumors: a meta-analysis. J Natl Cancer Inst. 2007;99:1544–1550. doi: 10.1093/jnci/djm170. [DOI] [PubMed] [Google Scholar]
- 24. Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. •• This GWAS study includes 1,878 glioma cases and 3670 controls, and validated in additional 2545 cases and 2953 controls, the authors obtain support for five risk loci for glioma at 5p15.33 TERT, 8q24.21 CCDC26, 9p21.3 CDKN2A–CDKN2B, 11q23.3 PHLDB1, and 20q13.33 RTEL1. This is a ground-breaking study and it was the first have a sufficiently large sample to understand the genetic risk for glioma.
- 25. Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. • A concurrent GWAS paper which focuses on high grade glioma. This work provided further evidence to implicate the 9p21 CDKN2B and 20q13.3 RTEL1 in glioma risk.
- 26.Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 27. Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O'Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. • This study demonstrates clearly that human RTEL1 suppresses homologous recombination and is required for DNA repair.
- 28.Wager M, Menei P, Guilhot J, Levillain P, Michalak S, Bataille B, Blanc JL, Lapierre F, Rigoard P, Milin S, et al. Prognostic molecular markers with no impact on decision-making: the paradox of gliomas based on a prospective study. Br J Cancer. 2008;98:1830–1838. doi: 10.1038/sj.bjc.6604378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Wei Q, Wang LE, Aldape KD, Cao Y, Okcu MF, Hess KR, El-Zein R, Gilbert MR, Woo SY, et al. Survival prediction in patients with glioblastoma multiforme by human telomerase genetic variation. J Clin Oncol. 2006;24:1627–1632. doi: 10.1200/JCO.2005.04.0402. [DOI] [PubMed] [Google Scholar]
- 30.Jiang M, Zhu K, Grenet J, Lahti JM. Retinoic acid induces caspase-8 transcription via phospho-CREB and increases apoptotic responses to death stimuli in neuroblastoma cells. Biochim Biophys Acta. 2008;1783:1055–1067. doi: 10.1016/j.bbamcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das A, Banik NL, Ray SK. Differentiation decreased telomerase activity in rat glioblastoma C6 cells and increased sensitivity to IFN-gamma and taxol for apoptosis. Neurochem Res. 2007;32:2167–2183. doi: 10.1007/s11064-007-9413-y. [DOI] [PubMed] [Google Scholar]
- 32.Little MP, De Vathaire F, Shamsaldin A, Oberlin O, Campbell S, Grimaud E, Chavaudra J, Haylock RGE, Muirhead CR. Risks of brain tumour following treatment for cancer in childhood: Modification by genetic factors, radiotherapy and chemotherapy. Int J Cancer. 1998;78:269–275. doi: 10.1002/(SICI)1097-0215(19981029)78:3<269::AID-IJC1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Bondy ML, Wang LE, El-Zein R, de Andrade M, Selvan MS, Bruner JM, Levin VA, Alfred Yung WK, Adatto P, Wei Q. Gamma-radiation sensitivity and risk of glioma. J Natl Cancer Inst. 2001;93:1553–1557. doi: 10.1093/jnci/93.20.1553. [DOI] [PubMed] [Google Scholar]
- 34.Neglia JP, Robison LL, Stovall M, Liu Y, Packer RJ, Hammond S, Yasui Y, Kasper CE, Mertens AC, Donaldson SS, et al. New primary neoplasms of the central nervous system in survivors of childhood cancer: a report from the childhood cancer survivor study. J Natl Cancer Inst. 2006;98:1528–1537. doi: 10.1093/jnci/djj411. [DOI] [PubMed] [Google Scholar]
- 35.Fuxe J, Akusjarvi G, Goike HM, Roos G, Collins VP, Pettersson RF. Adenovirus-mediated overexpression of p15INK4B inhibits human glioma cell growth, induces replicative senescence, and inhibits telomerase activity similarly to p16INK4A. Cell Growth Differ. 2000;11:373–384. [PubMed] [Google Scholar]
- 36.Guo C, White PS, Weiss MJ, Hogarty MD, Thompson PM, Stram DO, Gerbing R, Matthay KK, Seeger RC, Brodeur GM, et al. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene. 1999;18:4948–4957. doi: 10.1038/sj.onc.1202887. [DOI] [PubMed] [Google Scholar]
- 37.Heale JT, Ball AR, Jr, Schmiesing JA, Kim JS, Kong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell. 2006;21:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cao Y, Li H, Deb S, Liu JP. TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene. 2002;21:3130–3138. doi: 10.1038/sj.onc.1205419. *. This works shows that hTERT interacts with p53 and PARP1.