Abstract
Conserved serines of transmembrane segment (TM) five (TM5) are critical for the interactions of endogenous catecholamines with α1- and α2-adrenergic, β2-adrenergic, and D1, D2, and D3 dopamine receptors. The unique high-affinity interaction of the D4 dopamine receptor subtype with both norepinephrine and dopamine, and the fact that TM5 serine interactions have never been studied for this receptor subtype, led us to investigate the interactions of ligands with D4 receptor TM5 serines. Serine-to-alanine mutations at positions 5.42 and 5.46 drastically decreased affinities of dopamine and norepinephrine for the D4 receptor. The D4-S5.43A receptor mutant had substantially reduced affinity for norepinephrine, but a modest loss of affinity for dopamine. In functional assays of cAMP accumulation, norephinephrine was unable to activate any of the mutant receptors, even though the agonist quinpirole displayed wild-type functional properties for all of them. Dopamine was unable to activate the S5.46A mutant and had reduced potency for the S5.43A mutant and reduced potency and efficacy for the S5.42A mutant. In contrast, Ro10-4548 [RAC-2′-2-hydroxy-3-4-(4-hydroxy-2-methoxyphenyl)-1-piperazinyl-propoxy-acetanilide], a catechol-like antagonist of the wild-type receptor unexpectedly functions as an agonist of the S5.43A mutant. Other noncatechol ligands had similar properties for mutant and wild-type receptors. This is the first example of a dopamine receptor point mutation selectively changing the receptor's interaction with a specific antagonist to that of an agonist, and together with other data, provides evidence, supported by molecular modeling, that catecholamine-type agonism is induced by different ligand-specific configurations of intermolecular H-bonds with the TM5 conserved serines.
The D4 dopamine receptor has had a checkered history of popularity. It was for a time believed to be the ideal target for an atypical antipsychotic drug (for review see Schetz and Sibley, 2007; Schetz, 2009). The D4 receptor has also been pursued as a drug target for treating attention-deficit hyperactivity disorder (ADHD) and erectile dysfunction, but these possibilities remain controversial. A D4 polymorphic variant (D4.7) was reported to be hyporesponsive to dopamine and associated with a higher risk for ADHD (LaHoste et al., 1996). However, the relevance of a low-magnitude hyporesponsiveness is unclear, and the association of the D4.7 polymorphic variant has not always been replicated by different laboratories (for a review see Schetz and Sibley, 2007) with some studies even suggesting the D4.7 variant is associated with better clinical outcomes (Shaw et al., 2007) that no genetic linkage exists between the D4 receptor and ADHD regardless of the polymorphic variant (Johansson et al., 2008). A considerable amount of evidence suggests D4 receptors mediate penile erections (for a review see Schetz, 2009); however, recent reports dispute this (Collins et al., 2009, Depoortère et al., 2009). Less controversial has been the role of the D4 receptor in mediating adaptive ocular responses. Dopamine signals the eye to adjust to different levels of illumination through an adaptive response via D4 receptor-mediated changes in cAMP levels (Cohen et al., 1992; Nir et al., 2002). Densities of the D4 receptor are high in the retina, and drugs that mediate adaptive ocular responses by targeting D4 receptors might have clinical utility (Patel et al., 2003).
Although much has been learned pertaining to how ligands with high selectivity for the D4 receptor interact with specific D4 receptor microdomains (Simpson et al., 1999; Schetz et al., 2000; Kortagere et al., 2004; Ericksen et al., 2009), no studies have detailed the interaction of D4-selective ligands with the conserved serines of TM5. That TM5 serines in the D4 receptor have not been studied was surprising given the interest in D4 receptor agonists as potential therapeutics, evidence that conserved TM5 serines in biogenic amine G protein-coupled receptors (GPCRs) are sites of interaction for their amine neurotransmitters, and the conserved TM5 serines in the D1, D2, and D3 subtypes of dopamine receptor are key sites of catecholamine agonist interaction (for review see Floresca and Schetz, 2004). In addition, the high affinity of norepinephrine for only the D4 subtype of dopamine receptor, when coupled with the knowledge that the endogenous function of noradrenergic receptors relies on key interactions between conserved TM5 serines and norepinephrine (Peltonen et al., 2003), suggests that norepinephrine's affinity and efficacy at the D4 receptors may also involve the conserved serines of TM5. From the viewpoint of drug design and the tailoring of ligand selectivity and function, it would also be very informative to identify whether the interactions of TM5 serines, which are presumed to be important for the interaction of dopamine by analogy to the D1, D2, and D3 dopamine receptors, are necessary for D4 receptor activation by selective agonists.
We report here for the first time that, in contrast to D2 and D3 receptors, both S5.42 and S5.46 significantly affect the interactions of dopamine with the D4 receptor. However, for dopamine's β-hydroxylated counterpart, norepinephrine, each of the three conserved serines must be present for high affinity and function. Although no D4-selective noncatechol agonists had pronounced binding interactions with the conserved serines, one of the D4-selective antagonists, the catechol-like Ro10-4548 [RAC-2′-2-hydroxy-3–4-(4-hydroxy-2-methoxyphenyl)-1-piperazinyl-propoxy-acetanilide], had an unexpected agonist response at the D4-S5.43A mutant receptor. To our knowledge this is the first report of a single-point mutation in a dopamine receptor that converts the functional properties for a specific and highly selective ligand from an antagonist to an agonist. These findings suggest that catecholamine-type agonism at the D4 receptor is induced by a specific configuration of intermolecular H-bonds with the TM5 conserved serines that differs between specific catechol-like ligands and across the known catecholamine receptors where these interactions have been characterized.
Materials and Methods
Reagents.
Bovine calf serum and powdered F-12 cell culture media were purchased from HyClone Laboratories (Logan, UT) for use in cell culture. Sterile F-12 growth media consisted of F-12 cell culture media supplemented with 10% bovine calf serum and 100 μM sodium pyruvate. Hanks' balanced salt solution (HBSS; 10×) was purchased from Invitrogen (Carlsbad, CA) and diluted as needed for functional assays. [3H]Methylspiperone (NET-856; 80–85 Ci/mmol) was purchased from PerkinElmer Life Sciences and Analytical Sciences (Waltham, MA) for use in radioligand assessments. Tris buffer reagents were purchased from US Biological (Swampscott, MA). With the exception of Ro10-4548, PNU101,387G [4-[4-[2-[(1S)-3,4-dihydro-1H-isochromen-1-yl]ethyl]piperazin-1-yl]benzenesulfonamide (sonepiprazole)], and CP226,269 [5-fluoro-2-{[4-(2-pyridinyl)-1-piperazinyl]methyl}-1H-indole], all chemicals tested for binding or function at dopamine receptors were purchased from Sigma-Aldrich. (St. Louis, MO) or Tocris Bioscience, Inc. (Ellisville, MO).
Membrane Preparation.
Wild-type and mutant rat D4 dopamine receptors were stably expressed in Chinese hamster ovary (CHO) 10001 cells using the same methods as described previously for expression of these receptors in human embryonic kidney (HEK) 293 cells (Ericksen et al., 2009). In brief, mutant DNA containing the receptor and a resistance vector for G418 was transfected by CaPO4 precipitation into a low confluence of CHO10001 cells. Monoclonal colonies were isolated by challenging the transfection plates with F-12 media containing 2 mg/ml G418 for 1 to 2 weeks. Stable receptor expression was confirmed by saturation isotherm analysis. The CHO10001 cells that stably expressed mutant receptor were maintained (37°C, 5.0% CO2) in F-12 media containing 100 μg/ml G418 and used to prepare cell membranes for radioligand binding assays as described previously for HEK293 cells (Ericksen et al., 2009). In brief, CHO10001 cells expressing dopamine receptors were detached from 175-cm2 culture flasks by using 5 mM EDTA lifting buffer (Dulbecco's phosphate buffered saline without Ca2+ and Mg2+ and 5 mM EDTA). The cells were pelleted by centrifugation at 700g before resuspension in lysis buffer (5 mM Tris, 5 mM MgCl2, pH 7.4 at 4°C). After 5 to 10 min the cell lysate was homogenized (Dounce homogenizer, eight strokes) and centrifuged at 28,000g for 30 min. The pellet was resuspended in binding buffer (50 mM Tris, pH 7.4 at 4°C) and recentrifuged at 28,000g for 30 min. This purified membrane pellet was rehomogenized (Dounce homogenizer, four strokes) in binding buffer (50 mM Tris, pH 7.4 at 4°C) and stored on ice for use the same day. Binding buffers were pH-adjusted by using 1 N KOH and 1 N HCl.
Radioligand Binding Studies.
Similar to our studies in HEK293 cells, (Ericksen et al., 2009) CHO10001 membranes expressing the D4-WT, D4-S5.42A, D4-S5.43A, or D4-S5.46A receptors were challenged with [3H]methylspiperone alone and in competition with other D4 receptor ligands to characterize the affinity changes of serine-to-alanine mutations from the D4 background. In brief, dopaminergic ligands, nanomolar concentrations of [3H]methylspiperone, and purified CHO10001 cell membranes expressing dopamine receptors were equilibrated in binding buffer (50 mM Tris, pH 7.4 at 25°C) at room temperature for 90 min. After equilibration, the receptors were isolated by rapid filtration through GF/C filters pretreated for 10 min with 0.3% polyethyleneimine (Sigma-Aldrich) and three brief washes with ice-cold binding buffer (50 mM Tris, pH 7.4 at 0°C). Dried filters were cut into individual scintillation vials, filled with 3.5 ml of scintillation fluid, and counted in the scintillation counter. Nonspecific interactions of the radioligand were defined by the competition of [3H]methylspiperone with 5 μM (+)-butaclamol [3-(1,1-dimethylethyl)-2,3,4,4a,8,9,13b,14-octahydro-1H-benzo[6,7]cyclohepta[1,2,3-de]pyrido[2,1-a]isoquinolin-3-ol]. The amount of membrane protein determined by BCA assay (Thermo Fisher Scientific, Waltham, MA) ranged from 0.2 to 0.4 mg/ml for most assays.
Assessing Dopamine Receptor Function by cAMP Signaling.
CHO10001 cells stably expressing wild-type and mutant dopamine receptors were assessed for their ability to inhibit a forskolin [[(3R,4aR,5S,6S,6aS,10S,10aR,10bS)-3-ethenyl-6,10,10b-trihydroxy-3,4a,7,7,10a-pentamethyl-1-oxo-5,6,6a,8,9,10-hexahydro-2H-benzo[f] chromen-5-yl] acetate] stimulated cAMP signal in the presence of various agonists, while antagonists were tested for their ability to displace the full agonist (−)-quinpirole [(4aR,8aR)-5-propyl-1,4,4a,6,7,8,8a,9-octahydropyrazolo[3,4-g]quinoline] and therefore preserve the accumulation of intracellular cAMP. Intracellular cAMP concentration was determined with a cAMP Alphascreen detection kit (PerkinElmer Life and Analytical Sciences) and a PerkinElmer Fusion plate analyzer as described in our previous work (Ericksen et al., 2009). CHO10001 cells seeded at a density of 50,000 cells per well (200 μl of F-12 growth medium per well) were allowed to attach overnight to sterile 96-well poly(l-lysine)-coated microtiter plates (Sigma-Aldrich). After incubation for 16 to 18 h, the growth medium was removed and the cells were challenged for 25 min at 37°C (ambient CO2) with temperature-equilibrated drug dilutions containing 6 μM forskolin dissolved in stimulation buffer [1× HBSS, 50 mM HEPES, 100 μM sodium metabisulfite, 30 μM Ro20-1724 [4-(3-butoxy-4-methoxyphenyl)methyl-2-imidazolidone], pH 7.4 at 37°C]. No effort was made to pretreat the cell lines with antagonist before the addition of agonist. Microtiter plates were centrifuged at 1500g for 5 min after the drug incubation time had elapsed. The removal of the supernatant from each well was quickly followed by the addition of lysis buffer (100 μl of 0.3% Tween 20, 20 mM HEPES, 1 μg/μl bovine serum albumin, 30 μM Ro20-1724, pH 7.4 at 25°C) and lytic freezing at −80°C overnight. The lysates were thawed the next morning at 37°C (ambient CO2). The quantification of intracellular cAMP was determined by combining, in an opaque 96-well Costar plate (Corning Life Sciences, Lowell, MA), 10 μl of cell lysate with 10 μl of 0.5 unit (9.35 μg/ml) acceptor beads previously dark-adapted in bead buffer for 2 h (20 mM HEPES, 30 μM Ro20-1724, 1 μg/μl bovine serum albumin, 1× HBSS, pH 7.4 at 25°C). The Costar plates containing the lysate and acceptor bead mixture were centrifuged, while protected from light, at 5g for 2 min. Thirty minutes postcentrifugation, 10 μl of 0.5 unit (12.5 μg/ml) donor beads, equilibrated with 5 units of biotinylated cAMP (3.76 nM) in bead buffer for 2.5 h protected from light, was added to the dark-equilibrated lysate/acceptor bead mixture. The plates were carefully centrifuged for another 2 min at 5g while covered with aluminum foil. The donor bead/biotinylated cAMP complexes were allowed a minimum of 2 h to compete with cAMP for acceptor bead occupancy before quantification in the PerkinElmer Fusion plate analyzer.
Calculations and Data Analysis.
All data were analyzed and graphed by using GraphPad Prism version 4.0 (GraphPad Software Inc., San Diego, CA). Each data table reports the geometric mean and standard deviation for experiments repeated three times with two or more sample replicates per experiment except where noted. In contrast, the error margins depicted in the graphs are S.E.M. For radioligand competition assays, data from two or more sample replicates were averaged for each individual experiment and then normalized to the total amount of radioligand bound in the absence of drug minus the nonspecific binding as defined by 5 μM (+)-butaclamol. The data were then graphed to generate the individual IC50 values by extrapolating all dose-response curves to zero. The average IC50 value for a set of three radiolabeled competition experiments and the average equilibrium dissociation constant (KD) of [3H]methylspiperone at the tested receptor were used to find the inhibition constant (Ki) defined in the Cheng-Prusoff equation as Ki = IC50/(1 + [radioligand]/KD) (Cheng and Prusoff, 1973). Ki values were analyzed for significance by one-way analysis of variance (ANOVA) with a Dunnett's post hoc analysis. For cAMP assessments, the average cAMP accumulated per milligram of membrane protein (cAMP/mg protein) was determined by comparing the average raw counts per minute (cpm) of three replicates that contained cells, 6 μM forskolin, and experimental drugs, to the average cpm values generated by a cAMP standard curve. The magnitude of the cAMP change was obtained by normalizing the average cAMP/mg protein of each sample to the average cAMP/mg protein generated by unopposed 6 μM forskolin. The resultant value is plotted as a percentage of the maximal amount of cAMP that the cells should generate in this assay. For cells that were not stimulated by 6 μM forskolin, basal levels of cAMP accumulation are shown. Note that on cAMP graphs 0% represents the absence of detectable cAMP accumulation as observed for buffer controls that did not contain cells. Efficacy was determined by subtracting the lowest horizontal asymptote from the highest horizontal asymptote as defined by the graphical sigmoidal semilog dose-response plot. Half-maximal potency (EC50) and efficacy values generated from three cAMP functional experiments were analyzed by one-way ANOVA with a Dunnett's post hoc analysis, and significance was established at the 95% confidence level (p ≤ 0.05).
Ligand Docking and Analysis.
Wild-type rat D1–4 dopamine receptor homology models were constructed as described previously (Cummings et al., 2009) by using template structures bovine rhodopsin (Protein Data Bank code 1GZM), β2-adrenergic receptor (Protein Data Bank code 2RH1), and A2A adenosine receptor (Protein Data Bank code 3EML). To incorporate receptor backbone flexibility into the ligand docking procedure, a series of different conformers were generated as described (Cummings et al., 2009). In brief, an ensemble of 1000 receptor conformers was generated by applying random Cα displacements along a combination of the five lowest-frequency normal mode vectors computed for a Cα-only elastic network model of each of the D1–4 dopamine homology models. Normal modes were computed by using the Normal Mode Analysis Deformation and Refinement (NOMAD-Ref) web server (http://lorentz.dynstr.pasteur.fr/nomad-ref.php), and receptor conformers were obtained by using the decoy generation utility available on the server. The amplitude of displacement in the conformers (decoys) was set so the average Cα root mean square deviation (RMSD) for Cα displacements was 1.0 Å to explore only minor backbone displacements from the initial conformation. In cases where insufficient poses pass the filter (see below), this value was increased to 2.0 Å to induce larger receptor fluctuations that promoted more frequent TM5 serine contacts with the ligand. Backbone and side-chain atoms were rebuilt onto the various Cα scaffolds followed by energy minimization as described previously. The resulting 1000 receptor conformers were clustered by RMSD comparison to one another to obtain a smaller set of 26 representative conformers, one from each cluster.
Ligands were constructed with Maestro 8.5 (Schrödinger, Inc., Portland, OR) and geometry-optimized with Gaussian03 (Gaussian, Inc., Wallingford, CT) by using ab initio quantum mechanical calculations with the HF6-31G** basis set. Partial atomic charges were set according to the automated Gasteiger partial charge assignment of AutoDockTools 1.4.5 (The Scripps Research Institute, La Jolla, CA). The correct structure for Ro10-4548 that contains a 2-methoxy (Dr. Claus Riemer, personal communication) was used in the current study; the original (Simpson et al., 1999) and subsequent (Kortagere et al., 2004) reports mistakenly depicted a 3-methoxy. Each ligand was docked 20 times to each receptor conformation (26 distinct backbone conformers) by using the default Lamarckian genetic algorithm search routine in AutoDock 4.0 (The Scripps Research Institute) with nondefault parameters for increased genetic algorithm population size (300) and increased maximum number of energy evaluations (3.5 × 106, 5.5 × 106 for Ro10-4548). Side-chain torsions were allowed for residues D112 (D3.32), V113 (V3.33), C116 (C3.36), V188 (V5.39), S/A191 (5.42), S/A192 (5.43), S/A195 (5.46), F196 (F5.47), W327 (W6.48), F330 (F6.51), F331 (F6.52), and H334 (H6.55). These residues were selected because they are believed to line the binding cleft and be in close proximity to the conserved serines in TM5.
Resulting poses were filtered such that each exhibited both 1) an ionic interaction between the ligand's ammonium group and D3.32 and 2) any interaction between the ligands' catechol m- or p-hydroxyl (dopamine or norepinephrine) or Ro10-4548's o-methoxy or p-hydroxyl groups and any conserved TM5 serine/alanines (5.42, 5.43, and 5.46). The filtered pose subsets were then used to calculate the mean intermolecular contact maps (van der Waals or H-bonding interactions) based on contact counts from Ligplot version 4.4.2 (University College, London, United Kingdom). PyMOL version 0.99 (DeLano Scientific LLC, San Carlos, CA) was used for rendering figures, and Excel 2007 (Microsoft, Redmond, WA) was used to produce the colored contact matrices.
Results
Cloned wild-type or TM5 serine-to-alanine mutant D4 dopamine receptors were stably expressed in CHO10001 cells. The membrane densities of expressed receptors were determined by saturation isotherm analysis with the antagonist [3H]methylspiperone. Cell lines expressing similar receptor densities (Bmax = 1.3–4.6 pmol/mg membrane protein) of either the wild-type D4 or one of the D4-S5.42A, D4-S5.43A, or D4-S5.46A mutant receptors were selected for further comparative analysis (Table 1). All receptors were found to bind [3H]methylspiperone with high affinity (KD = 113–246 pM). These same cell lines served as a source of cloned receptor-containing cell membranes used to characterize the affinities of 13 additional ligands, including selective and nonselective agonists and antagonists, and an allosteric modulator (Schetz and Sibley, 2001) (Fig. 1).
TABLE 1.
Affinities of [3H]N-methylspiperone for wild-type and mutant D4 receptors
Binding affinities (KD) and receptor densities (Bmax) are expressed as the mean ± S.D. of three separate experiments. Fold change relative to the wild-type D4 receptor is indicated in parentheses with down arrows (↓) indicating a decrease in KD value (higher relative affinity) and up arrows (↑) indicating an increase in Bmax value (increased relative receptor density). Significance relative to the wild-type D4 receptor was determined by one-way ANOVA with Dunnett's post hoc (*, P < 0.05).
| Receptor | [3H]N-methylspiperone |
|
|---|---|---|
| Bmax ± S.D. | KD ± S.D. | |
| fmol/mg protein | pM | |
| Wild-type D4 | 1336 ± 136 (1) | 246 ± 78 (1) |
| D4-S5.42A | 1468 ± 815 (1.1↑) | 113 ± 37* (2↓) |
| D4-S5.43A | 4636 ± 689* (3.5↑) | 168 ± 14 (1.5↓) |
| D4-S5.46A | 2073 ± 966 (1.6↑) | 115 ± 29* (2↓) |
Fig. 1.
Chemical structures of the ligands investigated.
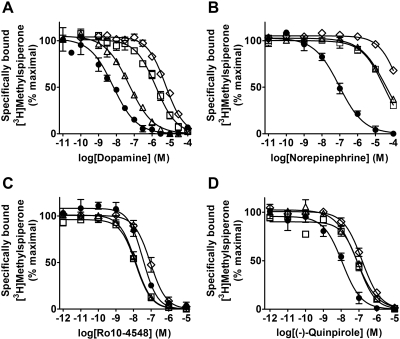
Dopamine had drastically reduced affinities for the D4-S5.42A (156-fold) and the D4-S5.46A (495-fold) mutant receptors, but moderately reduced affinity for the D4-S5.43A mutant (7-fold) (Fig. 2 and Table 2). Similar to dopamine, (−)-norepinephrine [4-[(1R)-2-amino-1-hydroxyethyl]benzene-1,2-diol] had drastically reduced affinity for D4-S5.42A (254-fold) and D4-S5.46A (1100-fold) mutant receptors, but in contrast to dopamine it also had drastically reduced affinity for the D4-S5.43A mutant (346-fold) (Fig. 2 and Table 2). Small to moderate (< 8-fold) increases or decreases in affinity were observed for the 11 other ligands: the D2-like receptor (i.e., D2, D3, and D4) agonist (−)-quinpirole; the D2-like receptor antagonists clozapine [8-chloro-11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine], and (−)-sulpiride [N-{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-2-methoxy-5-sulfamoylbenzamide]; the allosteric modulator methylisobutylamiloride (MIA); the D4-selective agonists PD168,077 [N-{[4-(2-cyanophenyl)piperazin-1-yl]methyl}-3-methylbenzamide] and CP226,269; the D4-selective antagonists PNU101,387G, Ro10-4548, and L-750,667 [3-{[4-(4-iodophenyl)piperazin-1-yl]methyl}-1H-pyrrolo[2,3-b]pyridine]; and finally, ABT-724 [2-[(4-pyridin-2-ylpiperazin-1-yl)methyl]-1H-benzimidazole] and Ro10-5824 [5-[(3,6-dihydro-4-phenyl-1(2H)-pyridinyl)methyl]-2-methyl-4-pyrimidinamine], two ligands previously reported to be partial agonists of the D4 receptor (Fig. 2 and Tables 2 and 3). Of this group of ligands, the largest improvements in affinity were for (−)-sulpiride and MIA (5.1- and 6.2-fold, respectively) for the D4-S5.42A mutant, and the largest reduction in affinity was for the agonist (−)-quinpirole (7.7-fold) for the D4-S5.46A mutant.
Fig. 2.
All three TM5 serine-to-alanine mutant receptors have significantly reduced affinities for both dopamine and (−)-norepinephrine. These graphs are the cumulative data of three separate experiments with dopamine (A), (−)-norepinephrine (B), the D4-selective ligand Ro10-4548 (C), or (−)-quinpirole (D) in competition with 0.5 nM [3H]methylspiperone at the wild-type D4 (●), D4-S5.42A (□), D4-S5.43A (▵), or D4-S5.46A (♢) receptors. Data points are graphed as the geometric mean ± S.E.M. Corresponding affinity values are listed in Table 2. A much greater reduction (49-fold more) in relative affinity was observed for (−)-norepinephrine (346- versus 7-fold for dopamine) at the D4-S5.43A mutant receptor.
TABLE 2.
Affinities for selective and nonselective D4 receptor agonists at wild-type and mutant D4 receptors
Affinities (Ki) of selective [(−)-norepinephrine, PD168,077, CP226,269, ABT-724, Ro10-5824], nonselective (dopamine), and D2-preferring [(−)-quinpirole] D4 receptor agonists are expressed as the mean ± S.D. (nM) of three separate experiments. Increased (↑) or decreased (↓) Ki values relative to the wild-type D4 receptor are listed in parentheses as fold changes. Significance relative to the wild-type D4 receptor was determined by one-way ANOVA with Dunnett's post hoc (*, P < 0.05). In cases where the fold changes in the data set exceed 100, two-tailed unpaired t tests were used to indicate significantly different means (†, P < 0.05) that would be otherwise missed because of excessive shift in the population mean.
| Receptor | Dopamine | Norepinephrine | (−)-Quinpirole | PD168,077 | CP226,269 | ABT-724 | Ro10-5824 |
|---|---|---|---|---|---|---|---|
| Wild-type D4 | 2.2 ± 0.16 (1) | 31 ± 9.2 (1) | 4.0 ± 1.6 (1) | 1.8 ± 1.8 (1) | 0.09 ± 0.054 (1) | 5.6 ± 3.1 (1) | 1.6 ± 0.51 (1) |
| D4-S5.42A | 337 ± 137* (156↑) | 7909 ± 2731† (254↑) | 21 ± 9.3 (5↑) | 0.68 ± 0.61 (2.6↓) | 0.12 ± 0.064 (1.4↑) | 3.7 ± 0.87 (1.5↓) | 1.6 ± 0.45 (1) |
| D4-S5.43A | 15 ± 4.2† (7↑) | 10,794 ± 1101† (346↑) | 19 ± 1.4 (4.8↑) | 2.1 ± 3.1 (1.2↑) | 0.17 ± 0.078 (2.0↑) | 3.4 ± 1.3 (1.7↓) | 1.4 ± 0.35 (1.2↓) |
| D4-S5.46A | 1070 ± 211* (495↑) | 34,313 ± 10,292* (1100↑) | 31 ± 11* (7.7↑) | 0.53 ± 0.53 (3.4↓) | 0.08 ± 0.070 (1.1↓) | 2.4 ± 0.15 (2.4↓) | 0.99 ± 0.33 (1.7↓) |
TABLE 3.
Affinities for nonselective and selective D4 receptor antagonists at wild-type and mutant D4 receptors
Nonselective [MIA, (−)-sulpiride, clozapine] and selective (Ro10-4548, PNU101,387G, L-750,667) D4 receptor antagonist affinities (Ki) are expressed as mean ± S.D. (nM) of three separate experiments. Increased (↑) or decreased (↓) Ki values relative to the wild-type D4 receptor are expressed as fold changes within parentheses. Significance relative to the wild-type D4 receptor was determined by one-way ANOVA with Dunnett's post hoc (*, P < 0.05). N.D., not determined.
| Receptor | MIA | (−)-Sulpiridea | Clozapine | Ro10-4548 | PNU101,387G | L-750,667 |
|---|---|---|---|---|---|---|
| Wild-type D4 | 190 ± 105 (1) | 1426 ± 439 (1) | 1.8 ± 0.20 (1) | 12 ± 2.2 (1) | 1.66 ± 0.091 (1) | 0.14 ± 0.056 (1) |
| D4-S5.42A | 31 ± 7.2* (6.2↓) | 282 ± 38 (5.1↓) | 0.41 ± 0.16* (4.3↓) | 2.6 ± 0.49 (4.5↓) | 3.58 ± 0.454* (2.2↑) | 0.15 ± 0.020 (1.1↑) |
| D4-S5.43A | 60 ± 26 (3.2↓) | 978 ± 444 (1.5↓) | 0.57 ± 0.38* (3.1↓) | 3.4 ± 0.70b (3.3↓) | 3.79 ± 0.414* (2.3↑) | N.D. |
| D4-S5.46A | 49 ± 31* (3.9↓) | 6701 ± 5220 (4.7↑) | 0.40 ± 0.21* (4.4↓) | 19 ± 10 (1.6↑) | 1.04 ± 0.204 (1.6↓) | 0.10 ± 0.014 (1.4↓) |
Note that the affinity values of all ligands, including sodium-sensitive (−)-sulpiride, were conducted in the absence of sodium.
Ro10-4548 becomes an agonist at the mutant D4-S5.43A receptor.
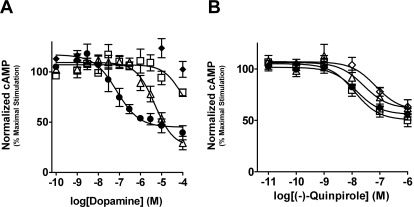
Functional responses for the wild-type and mutant D4 receptors were characterized by the ability of D4 receptor agonists to inhibit the forskolin-induced accumulation of intracellular cAMP in attached whole cells (Figs. 3–6). The potency of dopamine was decreased >118- and 41-fold, respectively for the D4-S5.42A and D4-S5.43A mutants (Fig. 3 and Table 4). No potency value was calculated for the D4-S5.46A mutant, because no significant efficacy was observed at the highest tested concentration of dopamine (100 μM). Whereas limited efficacy for dopamine was observed for the D4-S5.42A mutant at the highest concentration (100 μM), a reliable efficacy value could not be calculated (Fig. 3). Interestingly, no significant deviation from the wild-type efficacy was observed for dopamine for the D4-S5.43A mutant (Fig. 3 and Table 4). Potency and efficacy values for (−)-quinpirole were not significantly different between the wild-type and mutant receptors with the largest deviation being a 5-fold decrease in potency for the D4-S5.46A mutant (Fig. 3 and Table 4).
Fig. 3.
Dopamine, but not quinpirole, had drastically reduced function at TM5 serine-to-alanine mutants. Graphed are the cumulative data of three separate experiments that measure the ability of dopamine (A) or (−)-quinpirole (B) to activate wild-type D4 (●), D4-S5.42A (□), D4-S5.43A (▵), and D4-S5.46A (♢) receptors. Data points are graphed as the geometric mean ± S.E.M of accumulated cAMP that was normalized to the maximal cAMP accumulated in the presence of unopposed 6 μM forskolin. Corresponding potency values are listed in Table 2. Note that (−)-quinpirole-mediated D4 receptor inhibition of cAMP accumulation was not significantly affected by the TM5 serine-to-alanine mutations.
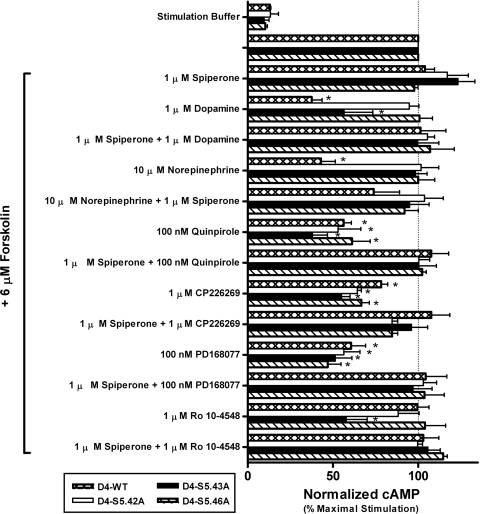
Fig. 4.
Screening ligand functional properties at the wild-type and mutant D4 receptors revealed an unexpected gain of function for Ro10-4548 at the D4-S5.43A receptor. Single high concentrations (10 μM) of dopamine, (−)-norepinephrine, (−)-quinpirole, CP226,269, PD168,077, and Ro10-4548 were tested for their ability to mediate cAMP functional responses at wild-type and mutant D4 receptors. At a concentration of 10 μM, (−)-norepinephrine acts as an agonist at the wild-type D4 receptor, but loses agonist activity at all TM5 mutant receptors. Dopamine has a pronounced loss of agonist activity at the S5.42A and S5.46A mutants and reduced activity at the S5.43A mutant. The agonist activity of CP226,269, PD168,077, and (−)-quinpirole for all of the mutant receptors is similar to the wild-type receptor. For all agonist–receptor pairings, the antagonist spiperone was able to inhibit the functional response. No functional response was observed in untransfected CHO10001 cells (data not shown). Data are plotted as the mean ± S.E.M. of three separate experiments normalized to the concentration of accumulated intracellular cAMP generated by unopposed 6 μM forskolin. Significance relative to the wild-type D4 receptor was determined by one-way ANOVA with Dunnett's post hoc (*, p < 0.05).
Fig. 5.
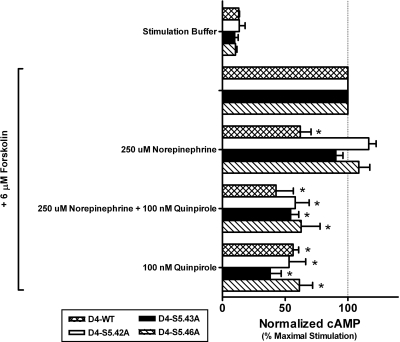
The conserved serine microdomain of TM5 is critical for (−)-norepinephrine-mediated activation of the D4 receptor. Functional assays with a concentration of (−)-norepinephrine (250 μM) that should saturate or nearly saturate even the mutant receptors, which have reduced affinity for (−)-norepinephrine, revealed that none of the mutant D4 receptors can be activated by (−)-norepinephrine. Surprisingly, (−)-norepinephrine was also unable to prevent 100 nM (−)-quinpirole from functionally activating the mutant receptors. Individual data sets were normalized to the concentration of accumulated intracellular cAMP generated by unopposed 6 μM forskolin and graphed as the geometric mean ± S.E.M. of three separate experiments. Significance relative to the wild-type D4 receptor was determined by one-way ANOVA with Dunnett's post hoc (*, p < 0.05).
Fig. 6.
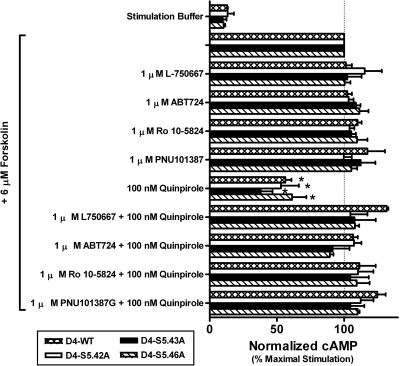
ABT-724, PNU-101,387G, L-750,667, and Ro-10-5824 exhibit antagonist properties at wild-type and mutant D4 receptors. Saturating concentrations of L-750,667, ABT-724, Ro10-5824, and PNU101,387G were tested, alone or in competition with the agonist (−)-quinpirole, for D4 receptor-mediated inhibition of forskolin-induced intracellular cAMP accumulation. In all cases, L-750,667, ABT-724, Ro10-5824, and PNU101,387G failed to elicit a change in forskolin-stimulated cAMP responses, but were able to block 100 nM (−)-quinpirole from activating the receptors. Individual data sets were normalized to the concentration of accumulated intracellular cAMP generated by unopposed 6 μM forskolin and graphed as the geometric mean ± S.E.M. of three separate experiments. Significance relative to the wild-type D4 receptor was determined by one-way ANOVA with Dunnett's post hoc (*, p < 0.05).
TABLE 4.
Potencies and efficacies for dopamine and (−)-quinpirole at wild-type and mutant D4 receptors
Data for the potencies (EC50, nM) and relative efficacies of dopamine and (−)-quinpirole are expressed as the mean ± S.D. of three separate experiments. Relative efficacy is defined as the percentage decrease in cAMP levels relative to the level of cAMP generated by 6 μM forskolin minus basal levels of cAMP. Increased (↑) or decreased (↓) values relative to the wild-type D4 receptor are expressed as fold changes within parentheses. Significance relative to the wild-type D4 receptor was determined by one-way ANOVA with Dunnett's post hoc (*, P < 0.05). N.D., not determined.
| Receptor | Dopamine |
(−)-Quinpirole |
||
|---|---|---|---|---|
| EC50 | Relative Efficacy | EC50 | Relative Efficacy | |
| nM | % | nM | % | |
| Wild-type D4 | 85 ± 72 (1) | 72 ± 18 (1) | 17 ± 7.7 (1) | 48 ± 13 (1) |
| D4-S5.42A | >10,000a (>118↑) | N.D.a (4.5↓) | 25 ± 19 (1.5↑) | 57 ± 6.6 (1.2↑) |
| D4-S5.43A | 3487 ± 561* (41↑) | 75 ± 28 (1) | 22 ± 11 (1.3↑) | 45 ± 6.9 (1.1↓) |
| D4-S5.46A | N.D. | N.D. | 86 ± 55 (5↑) | 46 ± 9.1 (1) |
Dopamine potency in the S5.42A mutant is estimated at more than 10 μM and the maximal effect cannot be determined.
Because very large reductions in (−)-norepinephrine binding affinity were observed in all three mutants (Fig. 2), no attempt was made to assess their (−)-norepinephrine-promoted activity with concentration-response curves. Single-point functional assays at a concentration of 10 μM confirmed that (−)-norepinephrine is an agonist of the D4 wild-type receptor as reported previously (Lanau et al., 1997). The same concentration of (−)-norepinephrine did not activate any of the mutant receptors (Fig. 4); however, it was possible that the functional responses, even if they were present, would not be observed because of the low affinity of (−)-norepinephrine for the mutant receptors (Fig. 2B). A subsequent screen of mutant receptor function used 250 μM (−)-norepinephrine, which is a concentration sufficient for saturating or nearly saturating all of the serine-to-alanine mutant receptors in spite of their reduced affinity. Yet, even at this very high concentration, not only was (−)-norepinephrine unable to activate any of the mutant receptors, the ligand was also unable to antagonize activation of these mutants by (−)-quinpirole (Fig. 5), suggesting that they do not have overlapping interaction sites.
Additional single-point cAMP functional assays indicated that, as for dopamine, (−)-norepinephrine, and (−)-quinpirole, PD168,077 and CP226,269 also acted as agonists at the wild-type D4 receptor by decreasing forskolin-stimulated cAMP responses (Fig. 4). However, ABT-724 and Ro10-5824 functional responses at the wild-type receptor were indistinguishable from the antagonists L750,667, PNU101,387G, and Ro10-4548 in that they did not activate the wild-type receptor but they did prevent (−)-quinpirole from activating it (Figs. 4 and 6). Like the wild-type receptors, all three mutant receptors retained agonist responses for (−)-quinpirole, PD168,077, and CP226,269, and in every case these agonist responses were reversed by the antagonist spiperone [8-[3-(p-fluorobenzoyl)propyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one] (Fig. 4). Furthermore, the functional properties of L750,667, PNU101,387G, ABT-724, and Ro10-5824 for the mutant receptors were the same as for the wild-type receptors: they were all antagonists because they did not activate the receptors and prevented activation by the agonist (−)-quinpirole. Two of the D4-selective ligands used in this study, ABT-724 and Ro10-5824, have been reported to possess partial agonist properties when using either [35S]GTPγS binding or FLIPR assays as functional end points (Powell et al., 2003; Cowart et al., 2004; Newman-Tancredi et al., 2008). However, using intracellular cAMP accumulation as a functional endpoint we found that ABT-724 and Ro10-5824 lacked agonist properties, but antagonized (−)-quinpirole activation in wild-type and mutant receptors. Although the reason for this discrepancy in functional activity is unclear, it could be an example of functional selectivity, meaning, in this case, that these drugs do not mediate changes in adenylyl cyclase activity.
However, the D4-selective ligand Ro10-4548 was different from the six other D4-selective ligands whose functional properties were unchanged by the mutations. Ro10-4548 is clearly an antagonist of the D4 wild-type receptor, but remarkably and unexpectedly it is able to activate the D4-S5.43A mutant receptor, and this activation could be blocked by the antagonist spiperone (Fig. 4). Competition binding experiments indicated a small and insignificant improvement (3.3-fold) in the affinity of Ro10-4548 for the D4-S5.43A mutant receptor. This minor improvement in affinity was comparable with that observed for the D4-S5.42A mutant (4.5-fold) (Table 3), and although Ro10-4548 appeared to have very weak agonist properties at the D4-S5.42A mutant, the effect was not significant (Fig. 4). No significant changes in Ro10-4548's affinity or function were observed for the D4-S5.46A mutant.
Early attempts to dock dopamine and norepinephrine to the original homology model of D4 failed to converge on poses with interactions at S5.43 or S5.46, although interactions with S5.42 were still achieved. Because side-chain torsions are allowed for cleft residues in the procedure, it is unlikely that side-chain rotamer state changes alone can account for the most favorable agonist-accommodating receptor state involving each of the serines, and thus some backbone position changes are probably required. Ligand interactions with these deeper serines (S5.43 and S5.46) are observed only after introducing receptor backbone flexibility to the docking procedure. Receptor conformational ensembles were produced from normal mode-based fluctuations that produced backbone movements averaging only 1 Å (or 2 Å if required). This requirement was not unexpected given that our homology models of receptor structure very closely resemble the crystal structure of the presumed low activity conformer of the β2-adrenergic receptor bound to its partial inverse agonist (S)-carazolol [1-(9H-carbazol-4-yloxy)-3-(propan-2-ylamino)propan-2-ol]. Therefore, it is likely that at least some backbone conformational change away from the original state is required to accommodate the expected catechol H-bonding interactions with S5.43 and/or S5.46. The particular interaction configuration between catechol hydroxyls and the serines is likely to depend on both the receptor's subtype structure and conformational state.
To examine the roles of the conserved TM5 serine positions 5.42, 5.43, and 5.46 in catecholamine agonist recognition by the D4 receptor, we docked dopamine and norepinephrine into models of wild type and each of the mutant D4 receptor constructs. After docking, poses were filtered from each ligand-receptor system based on experimentally suggested criteria (see Materials and Methods). The poses that pass the filter are consistently anchored by an H-bond-reinforced ionic interaction between the carboxylate group of D3.32 and ligand ammonium group, but vary significantly in their individual binding configurations between the ligand catechol's m- and p-hydroxyl groups and the conserved serines. Using the filtered pose subsets, the mean ligand–receptor residue contact distributions for a series of receptor conformations, which include both H-bonds and van der Waals contacts, were broadly similar across the D4 receptor constructs and between dopamine and norepinephrine. This “fingerprint” for catecholamine contacts at the D4 constructs frequently includes cleft positions M109 (M3.29), D112 (D3.32), V113 (V3.33), V166 (V4.61), Y187 (Y5.38), V188 (V5.39), S191 (S5.42), S192 (S5.43), S195 (S5.46), F196 (F5.47), W327 (W6.48), F330 (F6.51), F331 (F6.52), and H334 (H6.55), residues that correspond with those listed previously for phenethylamine ligands contacts at the α2A adrenoceptor (Nyrönen et al., 2001). However, some specific differences noted were that dopamine and norepinephrine additionally interact with C116 (C3.36) and Y358 (Y7.43) in D4 wild-type receptor and conversely lack the analogous contact with T118 (T3.37) made by the α2A adrenoceptor with phenylethylamine-type ligands.
Across the D4 constructs, the sets of filtered dopamine poses show substantial overlap in their respective positions of occupancy within the cleft. Variation within the filtered subset of poses for a given receptor construct was expected given 1) the relatively dynamic character of the extracellular region of TM5 as observed in fluctuations in our normal mode-based receptor conformations, 2) the most favored serine-ligand interaction configuration may depend on the variable conformational state of the receptor (e.g., G protein-coupled/uncoupled, etc.), and 3) the complexity of the TM5 serine H-bond network based on inspection of the β1- and β2-adrenergic receptor structures and D4 homology models that suggests each of the conserved serine side chains has at least three potential intramolecular H-bond donor/acceptor sites besides other potential intermolecular interactions sites (ligand/water). However, despite the inherent noise within the samples (variation in pose subsets) used for characterizing these interactions, each alanine substitution deprives the ligand of a potential H-bonding side chain and thus was hypothesized to alter the preferred H-bonding configuration and binding orientation of the catecholamine ligands. Indeed, some differences were apparent in the distributions of mode occupancy for the catecholamine ligands compared across the receptor constructs. Because H-bonding is likely to play a key role for the TM5 serine interactions with catecholamines (Strange, 1996), we refiltered poses more stringently to pass only those having at least a single H-bond between the catechol and a conserved serine side chain. We then computed the mean intermolecular H-bond contact maps from this subset of poses (Fig. 7A). At the S5.42A mutant, dopamine has a reduced tendency to H-bond to the side chains of S5.42A (obviously) and S5.43 but is afforded a significantly more robust H-bonding capacity with S5.46. This change in the H-bonding fingerprint is expected based on inspection of the dopamine poses in the S5.42A receptor (Fig. 8B) that tend to occupy a region slightly deeper in the primary cleft where S5.46 resides with the catechol ring more proximal to TM6 in comparison to those bound in the wild-type D4 receptor (Fig. 8A). This minor shift in dopamine position may account for the reduced affinity and efficacy in this mutant (Table 4). At the S5.43A mutant, the only mutant construct where dopamine retains full efficacy, H-bonding with the side chain of S5.42 is consistently maintained and even somewhat enhanced (1.57) with respect to the frequency observed in the wild type (0.79). H-Bonding is also observed (though infrequently) with the side chain of S5.46 in the same small fraction of poses as observed at D4WT (0.14). In addition to the similarities in conserved serine interactions, occasional H-bonding is observed between the ligand catechol hydroxyl group and H6.55 in this mutant (0.14) and the wild type (0.14), an interaction that was absent in the D4-S5.42A or D4-S5.46A mutants. At the D4-S5.46A mutant, the dopamine modes of occupancy are not clearly differentiated from those observed for D4WT (Fig. 8D). This is reflected to some extent in similar mean H-bond distributions (Fig. 7A) between D4WT and D4-S5.46A where the side chains of S5.42 and S5.43 are nearly equivalent in their H-bond accessibility to the catechol groups of dopamine (Fig. 8D). However, the H-bonding with the side chain of 5.46 is completely absent only in this construct, whereas it was occasionally observed at the dopamine-responsive constructs S5.43A and wild type and frequently observed at the weakly dopamine-responsive S5.42A mutant (Table 4).
Fig. 7.
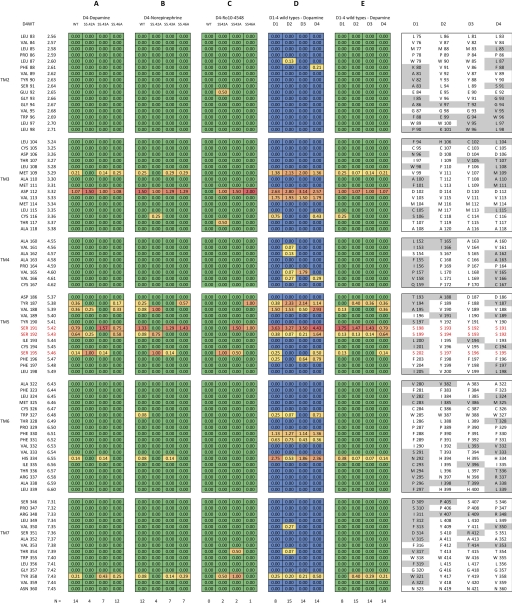
Mean intermolecular contact maps for catechol/catechol-like ligand poses at dopamine receptor constructs. Mean contact maps were computed from pose subsets that pass the filter for each ligand–receptor system examined by docking. The fingerprints of intermolecular contacts are shown as colored matrices for mean H-bonding frequency (A, B, C, and E) with color indicating the relative frequency of observing an intermolecular H bond at a particular cleft residue site from red (high) to yellow (moderate) to green (low). Alternatively, mean van der Waals contacts are displayed in matrix D using an alternative color spectrum from red (high) to yellow (moderate) to blue (low). Residue positions near the cleft are indexed on the left for D4 wild type. The homologous positions for rat D1–4 sequences are listed on the right with gray highlights to emphasize the regions of sequence heterogeneity. A, the mean H-bond map for dopamine at the D4 receptor constructs. B, the mean H-bond map for epinephrine at the D4 receptor constructs. C, the mean H-bond contact map for Ro10-4548 at the D4 receptor constructs. D, the mean van der Waals contact maps for dopamine at wild-type receptor subtypes D1–4. E, the mean H-bond map for dopamine at wild-type receptor subtypes D1–4. At the bottom of each column, N indicates the number of poses in the subset.
Fig. 8.
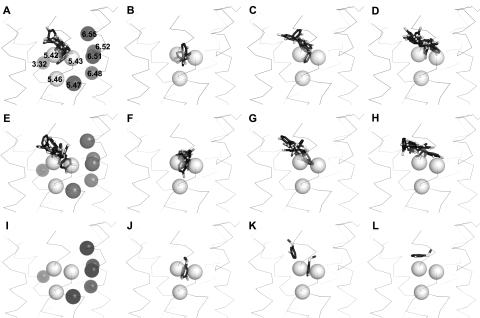
Filtered pose distributions for catechol and catechol-like ligands at the D4 receptor constructs. Shown are the positions of the catechol and catechol-like moieties from all poses that pass the filter in the following systems: dopamine at D4-WT (A), S5.42A (B), S5.43A (C), and S546A (D); norepinephrine at D4-WT (E), S5.42A (F), S5.43A (G), and S546A (H); and Ro10-4548 at D4-WT (I), S5.42A (J), S5.43A (K), and S5.46A (L). For wild type (A, E, and I), the aromatic microdomain residues 5.47, 6.48, 6.51, 6.52, and 6.55 are shown as dark spheres in addition to the critical D3.32 residue on TM3 that makes an H-bond-reinforced ionic interaction with the amine ligands. The light spheres represent the general positions of the conserved TM5 serines viewing from the bilayer. Only the initial backbone conformation is depicted as a ribbon for reference, although backbone flexibility was allowed in the docking procedure. Only ligands' catechol and catechol-like groups are displayed as dark sticks with white oxygen atoms; the remaining ligand atoms were removed for clarity. Note in J the two poses are almost superimposed.
For norepinephrine, the H-bonding patterns are generally quite similar to the patterns exhibited for dopamine across the various D4 constructs (Fig. 7, A and B). Interestingly, the pose clusters for either epinephrine or dopamine appear to occupy approximately the same region for each given D4 construct (Fig. 8, compare A–D with E–H, respectively). However, because norepinephrine is not an agonist at any of these mutants, the H-bonding pattern in the wild type was expected to deviate from those patterns observed in each of the serine mutants. Inspection of the mean H-bond maps (Fig. 7B) shows that only in the D4WT construct does norepinephrine H-bond robustly with S5.42 and S5.43 (less often). Thus, it is possible that intermolecular H bonds with these positions are required for norepinephrine agonism because none of the mutants show interactions at both S5.42 and S5.43 sites, and binding studies indicating interaction strength demonstrate that the interactions at S5.43 are more critical for the docking of norepinephrine than dopamine (Table 2 and Fig. 2, A and B). This mode of interaction contrasts with that obtained for dopamine at D4WT (and mutant S5.43A).
A similar docking analysis was applied to the D4 antagonist Ro10-4548 because its pharmacophore shared a likeness to the catecholamines (Fig. 1), and surprisingly it demonstrated agonism in the D4 S5.43A mutant, thus suggesting some interactions with the TM5-conserved serines. With Ro10-4548, however, no poses passed the filtering criteria applied to the catecholamines. Therefore, to enhance sampling, we docked Ro10-4548 into a second set of receptor conformations for each D4 construct obtained from a second round of receptor conformational sampling by using larger fluctuations in backbone amplitude (2-Å average RMSD).
None of the poses obtained for Ro10-4548 at the wild-type D4 receptor H-bond to any the conserved serine side chains and thus no poses pass the filter (Figs. 7C and 8I). At the S5.42A mutant receptor, however, two poses pass the filter, H-bonding only with S5.46 and with the catechol-like ring occupying a deep cleft position consistent with some of the poses observed for the catecholamine ligands, dopamine and norepinephrine. This deep cleft position is also occupied by the lowest energy-filtered pose obtained at the S5.43A mutant where H bonds are observed with the side chains of both S5.42 and S5.46. Of note, at the S5.46A mutant, only one pose was obtained. However, the catechol-like ring rides noticeably higher in the pocket, allowing H bonds only with S5.42. This elevated mode was also observed in some cases for dopamine and norepinephrine, resembling the pose described for models of salbutamol [(RS)-4-[2-(tert-butylamino)-1-hydroxyethyl]-2-(hydroxymethyl)phenol] binding at the β2 adrenoceptor (Swaminath et al., 2005). On the other hand, the predominant region of occupancy for the catechol-like moiety of Ro10-4548 at the S5.42A and S5.43A mutants is substantially lower in the cleft and closer to TM6 (Fig. 8, J and K), similar to the deeper poses observed for the catecholamines (Fig. 8, A–C, E, and F). This deeper ring position resembles that expected for the more efficacious agonist, isoproterenol, at the β2 adrenoceptor (Swaminath et al., 2005).
With one exception, none of the seven D4-selective ligands were strongly influenced by mutation of any of the conserved serines in TM5. The exception was for Ro10-4548, which antagonizes the D4 wild-type and D4-S5.42A and D4-S5.46A mutants, but was discovered to have agonist properties at the D4-S5.43A mutant receptor. To our knowledge this is one of a very limited number of examples of a ligand-specific gain-of-function mutation resulting from single-point mutation of a biogenic amine GPCR, i.e., one that enabled a specific antagonist to act as an agonist. In the other example, the wild-type β2-adrenergic receptor antagonists alprenolol [(RS)-1-(2-allylphenoxy)-3-(isopropylamino)propan-2-ol] and pindolol [(RS)-1-(1H-indol-4-yloxy)-3-(isopropylamino)propan-2-ol] act as partial agonists (30–35% efficacy) of a β2-adrenergic receptor D3.32E mutant (Strader et al., 1989b). The switch in functional properties for Ro10-4548 at the D4-S5.43A mutant receptor was not accompanied by any major changes in its affinity. With the exception of reduced affinity and potency for dopamine and norepinephrine, the D4-S5.43A mutant is, with respect to 10 other ligands, not remarkably different from the wild-type receptor.
To discriminate the subtype-specific dopamine receptor interactions with dopamine, the docking procedure was repeated on the wild-type D1–3 dopamine receptor subtypes. The mean van der Waals contact and H-bond maps were computed for dopamine and compared across receptor subtypes (Fig. 7D). Here, of the conserved serines, S5.42 H-bonding was robust across the subtypes. S5.43 was accessible but infrequently contacted in D1–3 and noticeably more accessible in D4. Dopamine H-bonding to the side chain of S5.46 was observed only in D1 and D4, indicating, perhaps, a slightly deeper region of dopamine occupancy in these subtypes, and is consistent with large reductions in dopamine binding affinity for S5.46A mutants of D1 and D4, but not the D2 and D3 subtypes.
Discussion
Previous studies of D1–D3 dopamine receptors indicate the interactions between the conserved serines in TM5 and dopamine appear to vary by subtype (for a review see Floresca and Schetz, 2004); however, analyses of such subtype variations are noticeably deficient with regard to the pharmacologically relevant D4 subtype. In fact, our basic findings (Table 5) reveal some key differences. At the D4 receptor, the affinity of dopamine was drastically reduced by S5.42A and S5.46A mutations (>150-fold) but moderately reduced by S5.43A (< 8-fold). This loss in dopamine's affinity for the D4-S5.43A mutant is consistent with changes for D2-S5.43A (average 7.5-fold reduction, range 1.2- to 20-fold), D3-S5.43A (2.1-fold reduction), and D1-S5.43A mutants (9.9-fold reduction) (Cox et al., 1992; Pollock et al., 1992; Woodward et al., 1996; Wiens et al., 1998; Sartania and Strange, 1999; Wilcox et al., 2000). Although the reduced affinity of dopamine for the D4-S5.42A mutant was consistent with that observed for other D2-like receptors (average 101-fold reduction for D2-S5.42A across different studies, range 43- to 250-fold, and 62-fold reduction for D3-S5.42A), the large reduction in dopamine affinity for the D4-S5.46A mutant was not (average 1.5-fold reduction for D2-S5.42A, range 0.8- to 8.4-fold, and 1.2-fold increase for D3-S5.46A) (Cox et al., 1992; Woodward et al., 1996; Wiens et al., 1998; Sartania and Strange, 1999; Wilcox et al., 2000) and surprisingly more similar to the D1 subtype (47-fold reduction for D1-S5.46A) (Pollock et al., 1992).
TABLE 5.
Summary of the most salient results of the present study concerning the TM5 serine-to-alanine mutations in the D4 receptor
Serine-to-alanine mutations of the TM5 conserved serines lead to:
|
Previous D1-S5.42A functional data indicate a reduction in dopamine potency (52-fold) (Pollock et al., 1992). Similar potency shifts were observed here for the D4-S5.42A mutant (>118-fold) and even larger shifts for D2-S5.42A (444-fold reduction, range 220- to 667-fold) (Cox et al., 1992; Wiens et al., 1998). S5.42 thus appears to be generally important for dopamine affinity and potency across the subtypes studied to date (D1–D4). The roles of S5.43 and S5.46 are, however, more variable amongst subtypes. Both D4-S5.43A and D1-S5.43A mutant receptors (Pollock et al., 1992) had similar decreases in dopamine potency (41- and 23-fold, respectively). Remarkably, in the D2 receptor, dopamine was unable to activate the S5.43A mutant (Cox et al., 1992; Wiens et al., 1998), although other agonists 7-OH-DPAT (7-hydroxy-N,N-dipropyl-2-aminotetralin), (−)-quinpirole, and propylnorapomorphine could, albeit at reduced potencies for (−)-quinpirole and propylnorapomorphine (Wiens et al., 1998). Functional responses were not attainable in corresponding studies of the D3 subtype (Sartania and Strange, 1999), so no comparison with the analogous mutants of D4 is possible. The D4-S5.46A mutant has no demonstrable efficacy at a concentration of dopamine sufficient for full receptor occupancy yet it was still activated by a variety of other agonists including (−)-quinpirole, PD168,077, and CP226,269. In contrast, the D2-S5.46A mutant was reported to have only modest changes in potency for dopamine (1.7- to 18-fold) and no change in efficacy (Cox et al., 1992; Wiens et al., 1998). The D1-S5.46A mutant has a large decrease in potency (48-fold reduction) and a significant reduction in efficacy (>50%) with dopamine (Pollock et al., 1992). In general, the D4 receptor appears more D1-like than D2-like with respect to the interactions between conserved TM5 serines and dopamine.
These experimentally observed distinctions between subtypes suggest different specific modes of interaction between the dopamine catechol and the TM5 serines. Subtype-dependent modes are also supported by our docking data that show subtype differences in the H-bonding tendencies with dopamine (Fig. 7E). Dopamine H bonds to S5.46 were sometimes achieved in the D1/4 subtypes but they were completely absent in the D2/3 subtypes. This reflects a deeper cleft accessibility for dopamine at the D1 and D4 subtypes than for the D2 and D3 subtypes, where H bonds are achieved only with TM5 serines S5.42 and S5.43.
Norepinephrine has been reported to be a high-affinity agonist at the D4 dopamine receptor (Lanau et al., 1997), but until now no reports have shown the specific sites of norepinephrine interaction with the D4 receptor. Like dopamine, the affinity of norepinephrine for the D4-S5.42A and D4-S5.46A mutant receptors was greatly weakened. However, in contrast to dopamine, where affinity at the D4-S5.43A mutant was moderately decreased, norepinephrine's affinity for the D4-S5.43A mutant was severely reduced. Furthermore, unlike dopamine, which retained the ability to activate two of the three D4 mutants, norepinephrine had no efficacy with any of the mutants. However, none of the conserved serines are required for activation by the structurally unrelated agonists (−)-quinpirole, PD168,077, and CP226,269. These noncatechol agonists thus appear to activate the receptor by promoting different initial events in an activation mechanism that do not necessarily involve the TM5 serines.
The side-chain rotamer-state equilibria of the conserved TM5 serines are likely to be sensitive to a catechol-like ligand's preferred mode of binding. Perturbations to the H-bonding networks involved in these equilibria may, in turn, propagate more global changes through influence on TM5 helical geometry and stability. Catecholamine agonist-induced signal propagation may arise from the dependence of helical conformations on these serine rotamer states (along with cysteines and threonines), especially within transmembrane helices (Deupi et al., 2010). In addition, the most conserved position in the TM5 of GPCRs, P5.50, is one turn below S5.46 and may further destabilize the TM5 helix, allowing the helix to swivel or kink to dynamically accommodate ligand associations (Del Carmine et al., 2004). To identify the receptor states stabilized by catecholamine binding, we are currently examining the specific TM backbone changes that are required to achieve proper ligand contacts. However, it is likely that catecholamine agonists of different efficacies promote different active receptor conformers (Seifert et al., 2001; Swaminath et al., 2004).
To help develop a molecular mechanism that accounts specifically for catechol-type agonist recognition and function at the D4 constructs, we used a docking approach to obtain a range of possible binding modes and then compared how the interaction fingerprints vary among the constructs. In the case of both dopamine and norepinephrine at each of the D4 mutants, only in the S5.43A mutant were these ligands capable of H-bonding to both S5.42 and S5.46. Because dopamine retains full efficacy in only the wild-type and S5.43A mutant receptors, it appears that for maximal efficacy dopamine likely interacts with both S5.42 and S5.46. Interestingly, dopamine does remain weakly efficacious at S5.42A (with >118-fold weaker potency) and also demonstrates robust H-bonding with S5.46 (and S5.43) in the model. For norepinephrine at the D4 wild type, the only construct for which norepinephrine retains efficacy, the H-bonding is strongest with S5.42, much less frequent with the side chain of S5.43, and absent with S5.46. Here, the role for S5.46 is unclear because the S5.46A mutant abolishes binding and efficacy. This could suggest that none of the receptor conformations generated for the docking procedure resemble a norepinephrine-activated state, or perhaps the role for S5.46 in norepinephrine binding and function is not through direct H-bonding with the catechol group but through intramolecular interactions that enhance other interactions with the ligand. It has been demonstrated in the β2 receptor that S5.43 and S5.46 are energetically coupled to the basal conformational equilibrium (Ambrosio et al., 2000) and thus play a role in function without interaction by agonist. In addition, dopamine and norepinephrine differ somewhat in their H-bonding patterns with the serines and other cleft residues within the same D4 constructs.
Interestingly, although a serine at position 5.46 in the wild-type D4 receptor is required for both strong binding and efficacy with either norepinephrine or dopamine, these ligands do not show frequent contacts with this position in our molecular models. The β-hydroxyl group in norepinephrine appears to shift the distribution of poses, reducing the accessibility of the catechol hydroxyls to the deeper TM5 serines (S5.43 and S5.46). From docking, we observed a distribution of poses with the β-hydroxyl H-bonding to H6.55, D3.32, and potentially even S5.42. Findings from multiple groups indicate a variable role for the β-hydroxyl interaction in the adrenergic receptors. In the α-adrenergic receptors, D3.32 appears to anchor the β-hydroxyl when present in the ethylamine ligands (Nyrönen et al., 2001). However, in the β-adrenergic receptors, the β-hydroxyl group appears to interact with N6.55 (Zuurmond et al., 1999; Liapakis et al., 2004). This interaction with TM6 appears to promote a secondary, spectroscopically distinct conformational state that is internalized (Swaminath et al., 2004). This residue in the dopamine receptors, H/N6.55, could potentially play a similar role with β-hydroxylated catecholamines. Clearly, the specific interactions of catecholamine receptors with catecholamine agonists vary substantially across receptor types and catecholamine structures.
Although the precise molecular interactions between Ro10-4548 and D4-S5.43A that confer agonism are not clear, it is possible that, in this cell line, agonist properties are only evident for this ligand at higher levels of receptor expression. Alternatively, it is possible that the catechol-like moiety of Ro10-4548 is shifted in its configuration with the TM5 serines that in turn promotes a functional binding mode. Such a shift was observed in the S5.43A mutant where the Ro10-4548 distribution of occupancy appears to favor a deeper binding site noted by H-bonding with S5.42 and S5.46 (Figs. 7C and 8K). This pose was also observed for the S5.42A mutant (Fig. 8J), but obviously lacks the stabilizing S5.42 H bond. Again, the experimental and modeling data suggest that the preferred mode of interaction between the catechol or catechol-like moieties with the conserved serines of TM5 determines the catechol-like agonists' specific tendencies to elicit a functional response at the D4 dopamine receptor and thus tunes the efficacy.
Acknowledgments
We thank Angela Chao and Brian Weiss for technical assistance and Dr. Claus Riemer and Hoffman-La Roche (Basel, Switzerland) for Ro10-4548.
This work was supported in part by the National Institutes of Health [Grants R01-MH063162, DBI-0649889 (to J.A.S.), P01-DA012923 (to S.S.E.)]; the National Institutes of Health National Institute on Drug Abuse [Grant T32-DA007274] (to S.S.E.); and the Cofrin Center for Biomedical Information in the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine at Weill Medical College of Cornell University (S.S.E.).
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.109.164962.
- TM
- transmembrane segment
- ABT-724
- 2-[(4-pyridin-2-ylpiperazin-1-yl)methyl]-1H-benzimidazole
- CP226,269
- 5-fluoro-2-{[4-(2-pyridinyl)-1-piperazinyl]methyl}-1H-indole
- L-750,667
- 3-{[4-(4-iodophenyl)piperazin-1-yl]methyl}-1H-pyrrolo[2,3-b]pyridine
- MIA
- methylisobutylamiloride
- PD168,077
- N-{[4-(2-cyanophenyl)piperazin-1-yl]methyl}-3-methylbenzamide
- PNU101,387G
- 4-[4-[2-[(1S)-3,4-dihydro-1H-isochromen-1-yl]ethyl]piperazin-1-yl]benzenesulfonamide (sonepiprazole)
- RMSD
- root mean square deviation
- Ro10-4548
- RAC-2′-2-hydroxy-3–4-(4-hydroxy-2-methoxyphenyl)-1-piperazinyl-propoxy-acetanilide
- Ro10-5824
- 5-[(3,6-dihydro-4-phenyl-1(2H)-pyridinyl)methyl]-2-methyl-4-pyrimidinamine
- Ro20-1724
- 4-(3-butoxy-4-methoxyphenyl)methyl-2-imidazolidone
- ANOVA
- analysis of variance
- HBSS
- Hanks' balanced salt solution
- GPCR
- G protein-coupled receptor
- CHO
- Chinese hamster ovary
- HEK
- human embryonic kidney.
References
- Ambrosio et al., 2000.Ambrosio C, Molinari P, Cotecchia S, Costa T. (2000) Catechol-binding serines of β2-adrenergic receptors control the equilibrium between active and inactive receptor states. Mol Pharmacol 57:198–210 [PubMed] [Google Scholar]
- Cheng and Prusoff, 1973.Cheng Y, Prusoff WH. (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22:3099–3108 [DOI] [PubMed] [Google Scholar]
- Cohen et al., 1992.Cohen AI, Todd RD, Harmon S, O'Malley KL. (1992) Photoreceptors of mouse retinas possess D4 receptors coupled to adenylate cyclase. Proc Natl Acad Sci USA 89:12093–12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins et al., 2009.Collins GT, Truccone A, Haji-Abdi F, Newman AH, Grundt P, Rice KC, Husbands SM, Greedy BM, Enguehard-Gueiffier C, Gueiffier A, et al. (2009) Proerectile effects of dopamine D2-like agonists are mediated by the D3 receptor in rats and mice. J Pharmacol Exp Ther 329:210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart et al., 2004.Cowart M, Latshaw SP, Bhatia P, Daanen JF, Rohde J, Nelson SL, Patel M, Kolasa T, Nakane M, Uchic ME, et al. (2004) Discovery of 2-(4-pyridin-2-ylpiperazin-1-ylmethyl)-1H-benzimidazole (ABT-724), a dopaminergic agent with a novel mode of action for the potential treatment of erectile dysfunction. J Med Chem 47:3853–3864 [DOI] [PubMed] [Google Scholar]
- Cox et al., 1992.Cox BA, Henningsen RA, Spanoyannis A, Neve RL, Neve KA. (1992) Contributions of conserved serine residues to the interactions of ligands with dopamine D2 receptors. J Neurochem 59:627–635 [DOI] [PubMed] [Google Scholar]
- Cummings et al., 2009.Cummings DF, Ericksen SS, Schetz JA. (2009) Three amino acids in the D2 dopamine receptor regulate selective ligand function and affinity. J Neurochem 110:45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Carmine et al., 2004.Del Carmine R, Molinari P, Sbraccia M, Ambrosio C, Costa T. (2004) “Induced-fit” mechanism for catecholamine binding to the β2-adrenergic receptor. Mol Pharmacol 66:356–363 [DOI] [PubMed] [Google Scholar]
- Depoortère et al., 2009.Depoortère R, Bardin L, Rodrigues M, Abrial E, Aliaga M, Newman-Tancredi A. (2009) Penile erection and yawning induced by dopamine D2-like receptor agonists in rats: influence of strain and contribution of dopamine D2, but not D3 and D4 receptors. Behav Pharmacol 20:303–311 [DOI] [PubMed] [Google Scholar]
- Deupi et al., 2010.Deupi X, Olivella M, Sanz A, Dölker N, Campillo M, Pardo L. (2010) Influence of the g-conformation of Ser and Thr on the structure of transmembrane helices. J Struct Biol 169:116–123 [DOI] [PubMed] [Google Scholar]
- Ericksen et al., 2009.Ericksen SS, Cummings DF, Weinstein H, Schetz JA. (2009) Ligand selectivity of D2 dopamine receptors is modulated by changes in local dynamics produced by sodium binding. J Pharmacol Exp Ther 328:40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floresca and Schetz, 2004.Floresca CZ, Schetz JA. (2004) Dopamine receptor microdomains involved in molecular recognition and the regulation of drug affinity and function. J Recept Signal Transduct Res 24:207–239 [DOI] [PubMed] [Google Scholar]
- Johansson et al., 2008.Johansson S, Halleland H, Halmøy A, Jacobsen KK, Landaas ET, Dramsdahl M, Fasmer OB, Bergsholm P, Lundervold AJ, Gillberg C, et al. (2008) Genetic analyses of dopamine-related genes in adult ADHD patients suggest an association with the DRD5-microsatellite repeat, but not with DRD4 or SLC6A3 VNTRs. Am J Med Genet B Neuropsychiatr Genet 147B:1470–1475 [DOI] [PubMed] [Google Scholar]
- Kortagere et al., 2004.Kortagere S, Gmeiner P, Weinstein H, Schetz JA. (2004) Certain 1,4-disubstituted aromatic piperidines and piperazines with extreme selectivity for the dopamine D4 receptor interact with a common receptor microdomain. Mol Pharmacol 66:1491–1499 [DOI] [PubMed] [Google Scholar]
- LaHoste et al., 1996.LaHoste GJ, Swanson JM, Wigal SB, Glabe C, Wigal T, King N, Kennedy JL. (1996) Dopamine D4 receptor gene polymorphism is associated with attention-deficit hyperactivity disorder. Mol Psychiatry 1:121–144 [PubMed] [Google Scholar]
- Lanau et al., 1997.Lanau F, Zenner MT, Civelli O, Hartman DS. (1997) Epinephrine and norepinephrine act as potent agonists at the recombinant human dopamine D4 receptor. J Neurochem 68:804–812 [DOI] [PubMed] [Google Scholar]
- Liapakis et al., 2004.Liapakis G, Chan WC, Papadokostaki M, Javitch JA. (2004) Synergistic contributions of the functional groups of epinephrine to its affinity and efficacy at the β2 adrenergic receptor. Mol Pharmacol 65:1181–1190 [DOI] [PubMed] [Google Scholar]
- Newman-Tancredi et al., 2008.Newman-Tancredi A, Heusler P, Martel JC, Ormière AM, Leduc N, Cussac D. (2008) Agonist and antagonist properties of antipsychotics at human dopamine D4.4 receptors: G-protein activation and K+ channel modulation in transfected cells. Int J Neuropsychopharmacol 11:293–307 [DOI] [PubMed] [Google Scholar]
- Nir et al., 2002.Nir I, Harrison JM, Haque R, Low MJ, Grandy DK, Rubinstein M, Iuvone PM. (2002) Dysfunctional light-evoked regulation of cAMP in photoreceptors and abnormal retinal adaptation in mice lacking dopamine D4 receptors. J Neurosci 22:2063–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyrönen et al., 2001.Nyrönen T, Pihlavisto M, Peltonen JM, Hoffrén AM, Varis M, Salminen T, Wurster S, Marjamäki A, Kanerva L, Katainen E, et al. (2001) Molecular mechanism for agonist-promoted α(2A)-adrenoceptor activation by norepinephrine and epinephrine. Mol Pharmacol 59:1343–1354 [DOI] [PubMed] [Google Scholar]
- Patel et al., 2003.Patel S, Chapman KL, Marston D, Hutson PH, Ragan CI. (2003) Pharmacological and functional characterization of dopamine D4 receptors in the rat retina. Neuropharmacology 44:1038–1046 [DOI] [PubMed] [Google Scholar]
- Peltonen et al., 2003.Peltonen JM, Nyrönen T, Wurster S, Pihlavisto M, Hoffrén AM, Marjamäki A, Xhaard H, Kanerva L, Savola JM, Johnson MS, et al. (2003) Molecular mechanisms of ligand–receptor interactions in transmembrane domain V of the α2A-adrenoceptor. Br J Pharmacol 140:347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock et al., 1992.Pollock NJ, Manelli AM, Hutchins CW, Steffey ME, MacKenzie RG, Frail DE. (1992) Serine mutations in transmembrane V of the dopamine D1 receptor affect ligand interactions and receptor activation. J Biol Chem 267:17780–17786 [PubMed] [Google Scholar]
- Powell et al., 2003.Powell SB, Paulus MP, Hartman DS, Godel T, Geyer MA. (2003) RO-10-5824 is a selective dopamine D4 receptor agonist that increases novel object exploration in C57 mice. Neuropharmacology 44:473–481 [DOI] [PubMed] [Google Scholar]
- Sartania and Strange, 1999.Sartania N, Strange PG. (1999) Role of conserved serine residues in the interaction of agonists with D3 dopamine receptors. J Neurochem 72:2621–2624 [DOI] [PubMed] [Google Scholar]
- Schetz, 2009.Schetz JA. (2009) Dopamine receptors: introduction, IUPHAR database. Available from: http://www.iuphar-db.org/DATABASE/FamilyIntroductionForward?familyID=20
- Schetz et al., 2000.Schetz JA, Benjamin PS, Sibley DR. (2000) Nonconserved residues in the second transmembrane-spanning domain of the D(4) dopamine receptor are molecular determinants of D(4)-selective pharmacology. Mol Pharmacol 57:144–152 [PubMed] [Google Scholar]
- Schetz and Sibley, 2001.Schetz JA, Sibley DR. (2001) The binding-site crevice of the D4 dopamine receptor is coupled to three distinct sites of allosteric modulation. J Pharmacol Exp Ther 296:359–363 [PubMed] [Google Scholar]
- Schetz and Sibley, 2007.Schetz JA, Sibley D. (2007) Dopaminergic neurotransmission, in Handbook of Contemporary Neuropharmacology (Sibley D, Hanin I, Kuhar MJ, Skolnick P. eds) pp 221–256, John Wiley & Sons, Inc, Hoboken, NJ [Google Scholar]
- Seifert et al., 2001.Seifert R, Wenzel-Seifert K, Gether U, Kobilka BK. (2001) Functional differences between full and partial agonists: evidence for ligand-specific receptor conformations. J Pharmacol Exp Ther 297:1218–1226 [PubMed] [Google Scholar]
- Shaw et al., 2007.Shaw P, Gornick M, Lerch J, Addington A, Seal J, Greenstein D, Sharp W, Evans A, Giedd JN, Castellanos FX, et al. (2007) Polymorphisms of the dopamine D4 receptor, clinical outcome, and cortical structure in attention-deficit/hyperactivity disorder. Arch Gen Psychiatry 64:921–931 [DOI] [PubMed] [Google Scholar]
- Simpson et al., 1999.Simpson MM, Ballesteros JA, Chiappa V, Chen J, Suehiro M, Hartman DS, Godel T, Snyder LA, Sakmar TP, Javitch JA. (1999) Dopamine D4/D2 receptor selectivity is determined by a divergent aromatic microdomain contained within the second, third, and seventh membrane-spanning segments. Mol Pharmacol 56:1116–1126 [DOI] [PubMed] [Google Scholar]
- Strader et al., 1989b.Strader CD, Candelore MR, Hill WS, Dixon RA, Sigal IS. (1989b) A single amino acid substitution in the β-adrenergic receptor promotes partial agonist activity from antagonists. J Biol Chem 264:16470–16477 [PubMed] [Google Scholar]
- Strange, 1996.Strange PG. (1996) The energetics of ligand binding at catecholamine receptors. Trends Pharmacol Sci 17:238–244 [DOI] [PubMed] [Google Scholar]
- Swaminath et al., 2005.Swaminath G, Deupi X, Lee TW, Zhu W, Thian FS, Kobilka TS, Kobilka B. (2005) Probing the β2 adrenoceptor binding site with catechol reveals differences in binding and activation by agonists and partial agonists. J Biol Chem 280:22165–22171 [DOI] [PubMed] [Google Scholar]
- Swaminath et al., 2004.Swaminath G, Xiang Y, Lee TW, Steenhuis J, Parnot C, Kobilka BK. (2004) Sequential binding of agonists to the β2 adrenoceptor. Kinetic evidence for intermediate conformational states. J Biol Chem 279:686–691 [DOI] [PubMed] [Google Scholar]
- Wiens et al., 1998.Wiens BL, Nelson CS, Neve KA. (1998) Contribution of serine residues to constitutive and agonist-induced signaling via the D2S dopamine receptor: evidence for multiple, agonist-specific active conformations. Mol Pharmacol 54:435–444 [DOI] [PubMed] [Google Scholar]
- Wilcox et al., 2000.Wilcox RE, Huang WH, Brusniak MY, Wilcox DM, Pearlman RS, Teeter MM, DuRand CJ, Wiens BL, Neve KA. (2000) CoMFA-based prediction of agonist affinities at recombinant wild type versus serine to alanine point mutated D2 dopamine receptors. J Med Chem 43:3005–3019 [DOI] [PubMed] [Google Scholar]
- Woodward et al., 1996.Woodward R, Coley C, Daniell S, Naylor LH, Strange PG. (1996) Investigation of the role of conserved serine residues in the long form of the rat D2 dopamine receptor using site-directed mutagenesis. J Neurochem 66:394–402 [DOI] [PubMed] [Google Scholar]
- Zuurmond et al., 1999.Zuurmond HM, Hessling J, Blüml K, Lohse M, Ijzerman AP. (1999) Study of interaction between agonists and asn293 in helix VI of human β(2)-adrenergic receptor. Mol Pharmacol 56:909–916 [DOI] [PubMed] [Google Scholar]