This work uses functional genomics to delineate the role of the inositol synthesis genes in regulating growth, development, and cell death and reveals a connection between inositol, phosphatidylinositol, and sphingolipids.
Abstract
l-myo-inositol 1-phosphate synthase (MIPS; EC 5.5.1.4) catalyzes the rate-limiting step in the synthesis of myo-inositol, a critical compound in the cell. Plants contain multiple MIPS genes, which encode highly similar enzymes. We characterized the expression patterns of the three MIPS genes in Arabidopsis thaliana and found that MIPS1 is expressed in most cell types and developmental stages, while MIPS2 and MIPS3 are mainly restricted to vascular or related tissues. MIPS1, but not MIPS2 or MIPS3, is required for seed development, for physiological responses to salt and abscisic acid, and to suppress cell death. Specifically, a loss in MIPS1 resulted in smaller plants with curly leaves and spontaneous production of lesions. The mips1 mutants have lower myo-inositol, ascorbic acid, and phosphatidylinositol levels, while basal levels of inositol (1,4,5)P3 are not altered in mips1 mutants. Furthermore, mips1 mutants exhibited elevated levels of ceramides, sphingolipid precursors associated with cell death, and were complemented by a MIPS1-green fluorescent protein (GFP) fusion construct. MIPS1-, MIPS2-, and MIPS3-GFP each localized to the cytoplasm. Thus, MIPS1 has a significant impact on myo-inositol levels that is critical for maintaining levels of ascorbic acid, phosphatidylinositol, and ceramides that regulate growth, development, and cell death.
INTRODUCTION
In multicellular eukaryotes, myo-inositol becomes incorporated into phosphatidylinositol phosphate (PtdInsP), myo-inositol phosphate (InsP), and certain sphingolipid signaling molecules that function in many processes, such as regulation of gene expression (Alcazar-Roman and Wente, 2008), phosphorus storage (Raboy and Bowen, 2006), auxin receptor association (Tan et al., 2007), membrane tethering (Fujita and Jigami, 2008), stress tolerance (Taji et al., 2006), oligosaccharide synthesis (galactinol) (Karner et al., 2004; Michell, 2007), and regulation of cell death (sphingolipids) (Wang et al., 1996; Liang et al., 2003). The oxidation product of myo-inositol (d-glucuronic acid) is used for cell wall pectic noncellulosic compounds (Loewus et al., 1962; Loewus, 1965, 2006) and, in some organisms, for synthesis of ascorbic acid (Baig et al., 1970; Allison and Stewart, 1973; Banhegyi et al., 1997). Thus, myo-inositol synthesis and catabolism impact metabolites involved in many different and critical plant biochemical pathways.
The rate-limiting step of myo-inositol synthesis is catalyzed by l-myo-inositol 1-phosphate synthase (MIPS; EC 5.5.1.4) (Eisenberg et al., 1964; Loewus and Loewus, 1980; Loewus et al., 1980, 1984; Gumber et al., 1984). This reaction is followed by dephosphorylation of l-myo-inositol 1-phosphate to myo-inositol, which is catalyzed by the myo-inositol monophosphatase (IMP; EC 3.1.3.25) (Sherman et al., 1981; Loewus and Loewus, 1983; Torabinejad and Gillaspy, 2006; Torabinejad et al., 2009). These two reactions together are known as the Loewus pathway, which was first studied in plants and is the only known route for myo-inositol synthesis.
Although yeast and animal genomes contain a single gene encoding MIPS (GhoshDastidar et al., 2006), plants have multiple MIPS genes (Torabinejad and Gillaspy, 2006). Expression studies point to the possibility of specialized roles for individual enzyme isoforms (Smart and Fleming, 1993; Dean-Johnson and Wang, 1996; Ishitani et al., 1996; Yoshida et al., 1999, 2002; Hegeman et al., 2001; Mitsuhashi et al., 2008), although a complete characterization of the MIPS gene family in a single plant species has not been performed to date. The importance of examining how different MIPS isoforms function in plants is underscored by recent studies examining mutants in the Arabidopsis thaliana MIPS1 and MIPS2 genes (Murphy et al., 2008; Meng et al., 2009). It was shown that both mips1 and mips2 mutants contained lower levels of myo-inositol hexakisphosphate (InsP6) (Raboy, 2003), which is the major form of phosphorous stored in seeds. Interestingly, despite having similar depletion of InsP6 in leaves, these mips mutants have different phenotypes, with mips2 mutants being more susceptible to viral, fungal, and bacterial pathogens (Murphy et al., 2008), while mips1 mutants exhibit spontaneous cell death and enhanced basal resistance to pathogens (Meng et al., 2009). Thus, a specific pool of InsP6 may regulate defense pathways, illuminating the need for a broader understanding of MIPS function within plants.
To determine how the individual MIPS genes impact myo-inositol synthesis in plants, we took a functional genomics approach and examined the expression patterns and loss-of-function mutants for the three MIPS genes in Arabidopsis. Because the myo-inositol supplied by MIPS action is used by many pathways, it was critical to measure the impact of each gene on levels of myo-inositol and other metabolites. Our data indicate that the MIPS1 gene has a larger overall impact on myo-inositol levels in plants and is required for proper growth and development as well as suppression of spontaneous cell death. Our metabolite measurements indicate that decreased myo-inositol and phosphatidylinositol levels in mips1 mutants correlate with elevated ceramide levels, suggesting an important regulation by phosphatidylinositol on sphingolipid synthesis and cell death. Furthermore, this impact on metabolism and signaling is regulated at the level of transcription of specific MIPS genes, which are temporally and spatially restricted.
RESULTS
MIPS Gene Expression Is Temporally and Spatially Restricted
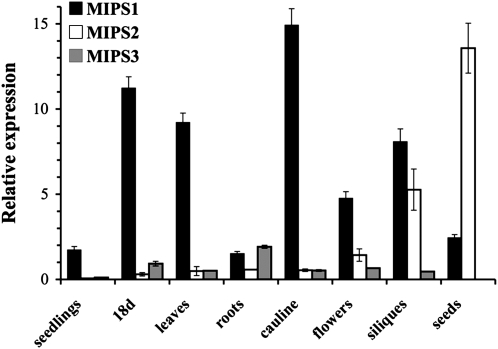
The Arabidopsis genome contains three genes encoding MIPS enzyme isoforms that are 89 to 93% identical to one another at the amino acid level (GhoshDastidar et al., 2006; Torabinejad and Gillaspy, 2006). To determine whether the MIPS genes are differentially regulated, we performed quantitative RT-PCR to compare relative expression of MIPS1, MIPS2, and MIPS3 in various tissues (Figure 1). We found that MIPS1 is expressed in all tissues examined, and levels are higher as compared with MIPS2 and MIPS3 in all tissues except for roots and seeds. In roots, MIPS3 expression is highest, and in seeds, MIPS2 expression is highest. Our data for siliques are similar to those reported previously that found expression of MIPS1 and MIPS2 (Mitsuhashi et al., 2008). In addition, our patterns are in keeping with those reported from microarray data in Genevestigator (see Supplemental Figure 1 online) and AtGenExpress visualization. Overall, these data indicate that MIPS1 expression is mostly constitutive, while low levels of MIPS2 and MIPS3 are found in all tissues, with highest expression in siliques and seeds (MIPS2) and roots (MIPS3).
Figure 1.
Relative Expression of MIPS Genes as Determined by Quantitative RT-PCR.
MIPS1, MIPS2, and MIPS3 gene expression was measured in 7-d-old wild-type seedlings grown on 0.5× MS-soaked filter paper under 16-h-light conditions (seedlings), soil-grown 18-d-old whole plants (18 d), young rosette leaves (leaves), roots, cauline leaves (cauline), and flowers from 25-d-old plants, immature siliques (siliques), and seeds imbibed in water for 3 d at 4°C. Real-time PCR amplification curves (see Methods) were compared with standard curves and PEX4 amplification to generate relative expression levels. Means of triplicate reactions ± se are presented.
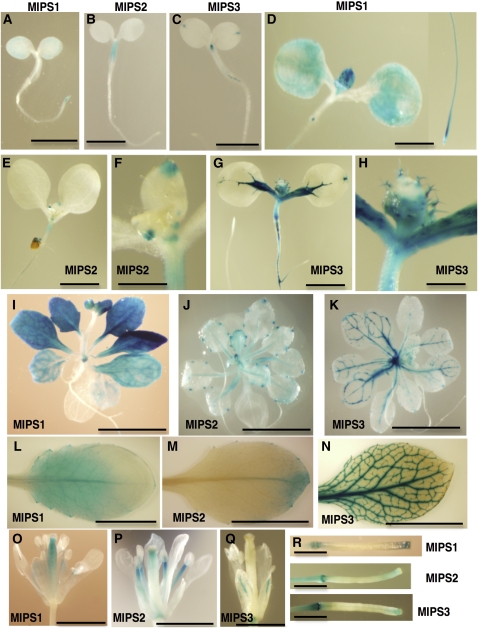
To investigate the spatial pattern of regulation of the MIPS genes, we generated transgenic plants expressing MIPS1, MIPS2, or MIPS3 promoters fused to the uidA gene. Two independent transgenic lines for each ProMIPS-uidA construct were analyzed, and consistent patterns were detected (Figure 2). In ProMIPS1-uidA 3-d-old seedlings, β-glucuronidase (GUS) activity was noted along the edges of cotyledons and in root tips (Figure 2A). By contrast, MIPS2 and MIPS3 are expressed in different areas of 3-d-old seedlings. Specifically, ProMIPS2-uidA seedlings have GUS activity in the hypocotyl only, while ProMIPS3-uidA seedlings have GUS activity in the hydathodes and vascular tissue in the shoot apex and at the hypocotyl/root junction (Figures 2B and 2C). These discrete domains of MIPS1-, MIPS2-, and MIPS3-specific patterns are maintained in 7-d-old seedlings (Figures 2D to 2H). In 7-d-old seedlings, MIPS1 expression has increased along the edges of cotyledons and is present in developing leaf primordia, root tips, and in a small area near the hypocotyl/root junction (Figure 2D). MIPS2 expression at 7 d is maintained in the hypocotyl, and expression in the tips of leaf primordia and stipules is evident (Figures 2D and 2E). At 7 d, MIPS3 expression has become evident throughout all shoot vascular tissue and in hydathodes and trichomes (Figures 2G and 2H). MIPS3 is also expressed in portions of the root vascular tissue, especially near lateral roots (Figure 2G). GUS activity in 19-d-old plants also reveals spatial restriction of the MIPS genes (Figures 2I to 2K). In 19-d-old plants, MIPS1 expression is maintained throughout all cells in young rosette leaves but is restricted to vascular tissue in older leaves (Figure 2I). MIPS2, by contrast, is only expressed in hydathodes and vascular tissue of 19-d-old plants (Figure 2J), while MIPS3 expression is confined to vascular tissue and hydathodes of leaves (Figure 2K). Examination of leaves from flowering plants grown on soil indicates that MIPS1 is confined to the proximal portion of the leaf (Figure 2L), MIPS2 is confined to the hydathodes and distal portion of leaves (Figure 2M), and MIPS3 is confined to the vascular tissue (Figure 2N). In flowers, both MIPS1 and MIPS2 are expressed in the pistil and in the stamen filaments (Figures 2O and 2P), while MIPS3 is confined to vascular tissue in the sepals (Figure 2Q). All three MIPS genes show expression in the tips and abscission zones of immature siliques (Figure 2R) and no expression in mature siliques. Together, these data indicate that the MIPS genes are both developmentally and spatially regulated, which may provide spatial regulation of myo-inositol synthesis. Specifically, while all three MIPS genes are expressed to various degrees in vascular tissues, MIPS1 is the only isoform expressed outside of vascular tissues beyond the seedling stage.
Figure 2.
Spatial Expression Patterns of MIPS Genes.
The promoters from MIPS1, MIPS2, or MIPS3 were used to drive GUS expression in transgenic plants.
(A) to (C) Three-day-old seedlings grown on 0.5× MS. Bars = 2 mm.
(D) to (H) Seven-day-old seedlings grown on 0.5× MS plus 1% sucrose. Bars = 2 mm in (D), (E), and (G) and 0.5 mm in (F) and (H).
(I) to (K) Nineteen-day-old plants grown on 0.5× MS plus 1% sucrose. Bars = 5 mm.
(L) to (R) Organs from soil-grown plants.
(L) to (N) Leaves. Bars = 1 cm.
(O) to (Q) Flowers. Bars = 2 mm.
(R) Immature siliques. Bars = 2 mm.
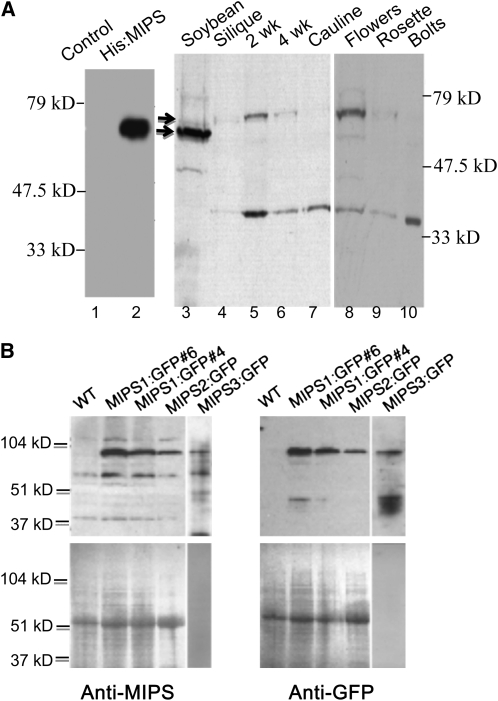
To examine accumulation of MIPS proteins in plant extracts, we produced polyclonal antisera within rabbits, using a soybean (Glycine max) His-tagged MIPS fusion protein (Hegeman et al., 2001) as the immunogen. The resulting anti-MIPS antisera recognizes the recombinant soybean His-tagged MIPS protein produced in Escherichia coli (Figure 3A, lane 2) as well as MIPS1-, MIPS2-, and MIPS3-green fluorescent protein (GFP) fusions expressed in transgenic plants (Figure 3B). Thus, our anti-MIPS antisera are useful for detecting all three Arabidopsis MIPS proteins. In wild-type Arabidopsis extracts, we detected two immunoreactive bands (Figure 3A). The upper band corresponds well to the predicted size of cytosolic MIPS1, 2, and 3 proteins from Arabidopsis (56.5, 56.3, and 56.4 kD, respectively), while the lower band (∼35 kD) has been predicted by others to represent a chloroplastic MIPS produced by differential splicing (Dean-Johnson and Wang, 1996). The larger MIPS isoforms are most abundant in 2-week-old seedlings and flowers, with less abundant expression present in older (4-week-old) seedlings, siliques, leaves, and bolts. The smaller immunoreactive band was detectable in all tissues from light-grown plants examined, with expression highest in seedlings and bolts. We tried to resolve the three MIPS protein isoforms with native gels (see Supplemental Figures 2A and 2B online) but have not been successful in separating these three, very similar proteins with gel electrophoresis.
Figure 3.
Expression of MIPS Proteins.
(A) Denaturing SDS-PAGE and protein gel blot analysis of bacterial (lanes 1 and 2) and plant extracts (lanes 3 to 10) with anti-MIPS antibody. The arrows mark migration of soybean (lane 3) and Arabidopsis MIPS proteins (lanes 4 to 10).
(B) The MIPS antibody cross-reacts with all three MIPS-GFP fusion proteins. Protein extracts of wild-type and MIPS1-GFP seedlings, MIPS2-GFP rosette leaves, and MIPS3-GFP roots. The anti-MIPS antibody was used in the left panel, and the anti-GFP antibody in the right panel. Ponceau S staining of the blots is shown in the bottom two panels.
Characterization of mips Mutants
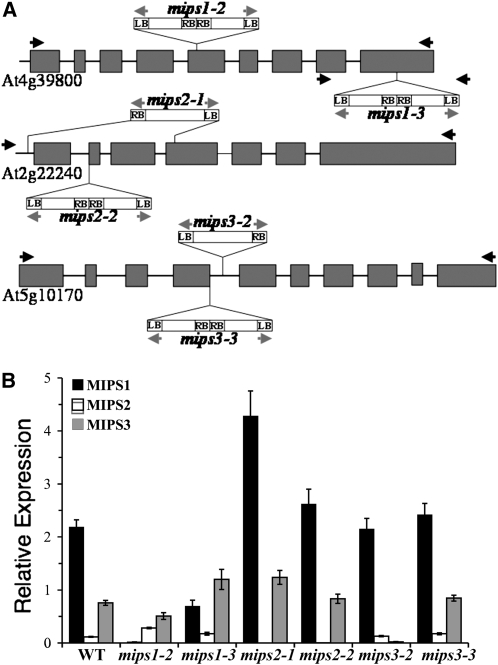
To determine how the different MIPS genes impact inositol synthesis and plant growth and development, T-DNA insertion mutants were obtained from the SALK T-DNA insertion collection (Alonso et al., 2003). Seed for mips1-2 (SALK_023626), mips1-3 (SAIL_676_D08), mips2-1 (SALK_031685), mips2-2 (SALK_108779), mips3-2 (SALK_120131), and mips3-3 (SAIL_425_F09) were obtained, and homozygous mutants were verified by diagnostic PCR screening and DNA sequencing (Figure 4A), as described in the Supplemental Methods online. MIPS gene expression was verified in the mutants by quantitative RT-PCR (Figure 4B). Our data indicate a large reduction of MIPS1 expression in mips1-2, while mips1-3 still retains 25% of wild-type expression, indicating that the mips1-3 mutant is a partial loss-of-function mutant. Interestingly, MIPS2 expression is increased in both of these mips1 mutants, and the mips1-3 mutant also contains elevated MIPS3 expression. Our analyses of mips2 mutants showed that both mips2-1 and mips2-2 mutants retain little, if any MIPS2 expression, and both can be considered loss-of-function mutants. The mips3-2 mutant expresses very little MIPS3, while the mips3-3 mutant we characterized has wild-type levels of MIPS3 expression. From this analysis, we conclude that these mutants are suitable for examining the consequences of eliminating expression of specific MIPS isoforms.
Figure 4.
T-DNA Insertions and Mutant Gene Expression.
(A) Schematic of T-DNA insertion sites in the mips1-2, mips2-2, mips2-1, mips2-2, mips3-2, and mips3-3 mutants. Exons are shown as dark-gray boxes; the gray arrows indicate primers used to amplify the right border (RB) and left border (LB) of the T-DNA; black arrows indicate the positions of gene-specific primers.
(B) Expression levels of MIPS1, MIPS2, and MIPS3 genes in 21-d-old wild-type and mutant plants. Real-time PCR amplification curves (see Methods) were compared with standard curves and PEX4 amplification to generate relative expression levels. Before normalization to PEX4, wild-type levels were as follows: MIPS1, 1.6 ± 0.03 fmol per μg RNA; MIPS2, 0.087 ± 0.025; MIPS3, 0.57 ± 0.009; and PEX4, 0.75 ± 0.013 fmol per μg RNA. Means of quadruplicate reactions ± se are represented. Asterisks indicate significant difference from the wild type (P < 0.01) in a Student's t test.
We also tried to examine whether MIPS1, 2, and 3 proteins were reduced in the mips mutants. Using the previously described anti-MIPS antibody, we could not effectively separate and identify the endogenous MIPS protein isoforms using SDS- or native-PAGE followed by protein gel blotting of 18-d-old plants, in leaves (see Supplemental Figure 2A online), seedlings, or flowers. Thus, we cannot draw any conclusions about MIPS protein levels in the mips mutants.
The mips1 Mutants Are Altered in Growth and Development
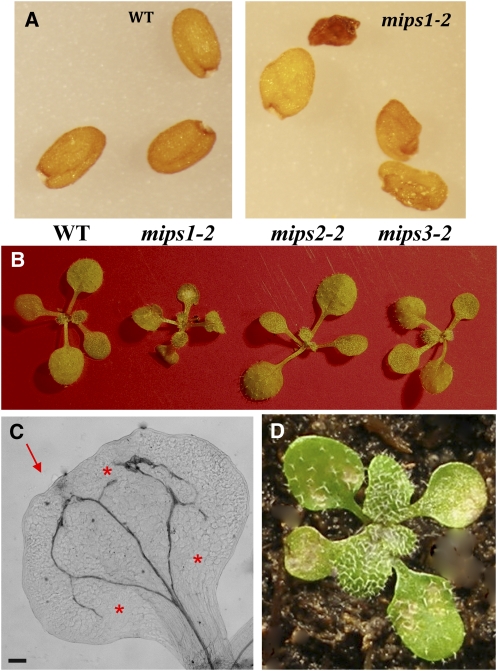
The widespread expression of MIPS1 in many different cell types suggested that MIPS1 catalyzes the majority of myo-inositol synthesis required for different aspects of plant growth and development. We compared the growth and development of mips mutants and noted seed phenotypes in mips1 mutants (Figure 5A; see Supplemental Figure 3 online). Seeds of mips1-2 and mips1-3 have a significant percentage of wrinkled (48.5 ± 9.9 and 54.9 ± 10.8, respectively) and empty seeds (6.8 ± 6.1 and 15.8 ± 18, respectively) compared with their corresponding wild-type accessions (wrinkled, 12.5 ± 7.3 and 5.3 ± 4.1; empty, 0.82 ± 1.3 and 2.3 ± 3.5, respectively). By contrast, there was no difference in seed production, with mips1-2 containing 50.7 ± 5.2 seed/siliques compared with 49.2 ± 10.4 seed/siliques in its wild-type accession (n = 10). Similarly, mips1-3 had 41 ± 5.9 seed/siliques compared with its wild type with 36.2 ± 11 (n = 10). Both the wrinkled and empty seed phenotypes can be complemented in the mips1-2 background with a wild-type copy of the MIPS1 gene fused to GFP (7.6 ± 5.2 wrinkled and 0.5 ± 1 empty), indicating the functionality of the transgene and confirming that the phenotype was due to the loss of MIPS1 (see Supplemental Figure 3 online; expression of MIPS1-GFP is shown in Supplemental Figure 2B online). Expression of MIPS1-GFP in the wild-type background (MIPS1-GFP#6 and MIPS1-GFP#4) did not affect the development or morphology of the seeds.
Figure 5.
Alterations in Seed and Cotyledon Morphology in mips1 Mutants.
(A) Seed phenotypes of the wild type and mips1 mutants.
(B) Seedling phenotypes of the wild type and mips mutants.
(C) Cleared cotyledon from the mips1-2 mutant. The arrow indicates the irregular cotyledon margin. The asterisks indicate areas where cotyledon vascular loops have not closed. Bar = 125 μm.
(D) mips1-3 seedling containing irregular cotyledons and lesions.
After germination of mips mutant seeds, we noted significant differences in mips1 seedling development that were not present in mips2 or mips3 mutants (Figure 5B). Homozygous mips1 mutants were overall smaller than the wild type (Figure 5B; see Supplemental Figure 3 online). Cotyledons from mips1 mutants had irregular margins, altered vascular patterning, and contained necrotic lesions (Figures 5B to 5D). These phenotypes are similar to those recently noted for mips1-2 mutants (Meng et al., 2009). Cleared cotyledons were observed by microscopy and found to contain areas of smaller, unexpanded cells that could result in the irregular margins (see Supplemental Figure 4A online). Veins in mips1 cotyledons did not close properly, resulting in open veins (Figure 5C). In addition, mips1 cotyledons sometimes contained areas of extra vascular tissue (see Supplemental Figure 4B online).
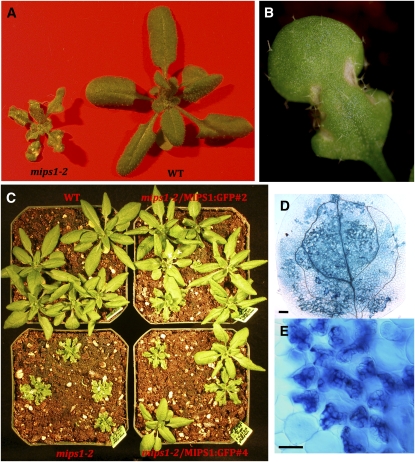
The decrease in size and presence of lesions in mips1 mutants remained apparent throughout development (Figures 6A and 6B), while mips2 and mips3 mutants showed no variation from the wild type (Figure 5B; see Supplemental Figure 3 online). Expression of MIPS1-GFP complemented the growth and cotyledon and lesion phenotypes, indicating that these phenotypes are due to loss of the MIPS1 gene (see Supplemental Figure 3 online; Figure 6C).
Figure 6.
A MIPS1-GFP Gene Complements the Cell Death–Associated Phenotypes of mips1 Mutants.
(A) Soil-grown mips1-2 (left) and wild-type (CS60000) plants.
(B) Leaf containing lesions from mips1-2 plants.
(C) Segregation of progeny from heterozygous mips1-2 plants containing a 35Spromoter-MIPS1-GFP transgene.
(D) and (E) Trypan blue staining of mips1-2 cotyledons. Bar = 100 μm in (D) and 40 μm in (E).
We next investigated the lesions on mips1 mutant cotyledons and leaves and potential factors that could contribute to their presence. We used trypan blue staining to determine that the lesions on mips1 mutant leaves and cotyledons were comprised of dead cells (Figures 6D and 6E). Since reactive oxygen species (ROS) and light are factors stimulating lesion development in other lesion mimic mutants, we investigated whether mips1 mutants were more sensitive to the herbicide paraquat, a redox-active compound that generates superoxide anion (Tsang et al., 1991) and stimulates lesion development in wild-type plants. When 33-d-old plants were treated with paraquat, mips1-2 mutants developed more lesions per leaf than the wild type or mips2-2 mutants, indicating increased sensitivity to paraquat when MIPS1 is lacking (see Supplemental Figure 5A online). Lesion development in mips1 mutants was also affected by the intensity of light, as noted previously (Meng et al., 2009), with high light promoting faster development of lesions in mips1-2 mutants (see Supplemental Figures 5B and 5C online). Lastly, we attempted to chemically rescue the development of lesions in mips1 mutants by watering mutants and wild-type plants with either myo-inositol or other polyols, such as mannitol or sorbitol. The results show that continuous watering with 10 mM myo-inositol, but not 10 mM mannitol or sorbitol, alleviated the growth and lesion-promoting effects of the mips1-2 mutation (see Supplemental Figure 6 online). We conclude that ROS, light, and myo-inositol are each involved in the aberrant development of lesions in the mips1 mutants.
MIPS1 Is Required for Physiological Responses
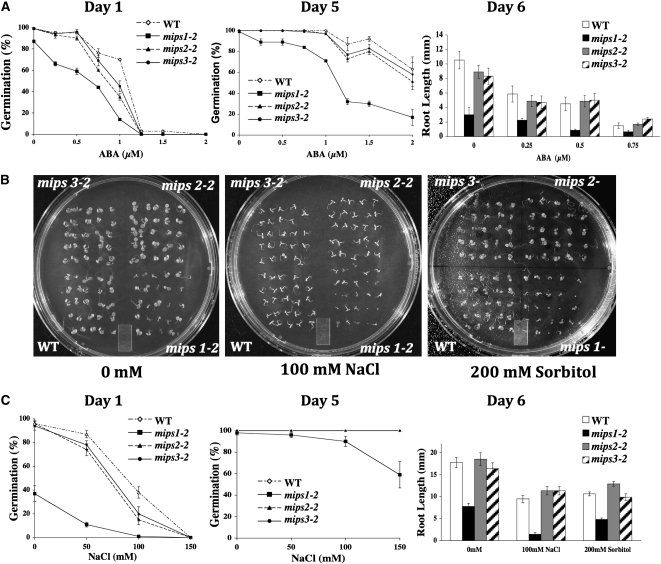
We have previously shown that a loss of function in another gene required for myo-inositol synthesis, Vitamin C-4 (VTC4), results in sensitivity to abscisic acid (ABA), salt, and cold (Torabinejad et al., 2009). To test whether known stress physiological pathways that use myo-inositol signaling (Xiong et al., 2001; Taji et al., 2006) were altered in mips mutants, we produced age-matched seed populations that had been harvested from plants grown at the same time. Control and mutant age-matched seeds were plated on Murashige and Skoog (MS) medium in the presence of various concentrations of ABA, NaCl, or mannitol and stratified for 3 d at 4°C. Germination was scored, and seedling root lengths were measured. Our results indicate that mips1 mutants germinate slower under control conditions and also have increased sensitivity to ABA during seed germination and in root growth (Figure 7A). By contrast, mips2 and mips3 mutants are not significantly altered in their response to ABA in either assay (Figure 7A). mips1 mutants are also sensitive to NaCl and sorbitol as measured in both germination and root growth assays (Figures 7B and 7C). This is in contrast with mips2 and mips3 mutants, which do not differ in their responses to NaCl and sorbitol compared with the wild type (Figures 7B and 7C). Together, these experiments indicate that mips1, but not mips2 or mips3 mutants, have impaired germination and root growth in response to ABA, NaCl, and mannitol, which is indicative of stress sensitivity.
Figure 7.
Physiological Responses in mips Mutants.
(A) Effects of ABA on germination and root length of the wild type and mips mutants grown on agar plates.
(B) Photos of 7-d-old wild-type and mips mutant seedlings grown for germination studies on agar plates with the indicated additions.
(C) Effects of NaCl and sorbitol on germination and root length of the wild type and mips mutants grown on agar plates. Presented are means ± se of four experiments of n = 25 (germination) and five experiments of n = 6 (root length).
MIPS1 Impacts Inositol Synthesis
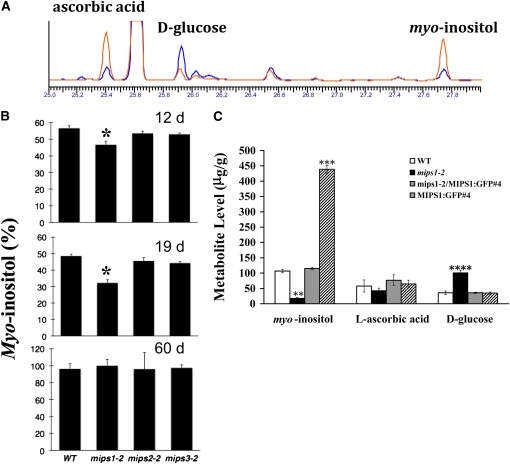
To determine if a loss in MIPS1, MIPS2, or MIPS3 function impacts myo-inositol synthesis, we used our previously established gas chromatography (GC) assay (Torabinejad et al., 2009) to quantify myo-inositol levels in the wild type and mips mutants (Figure 8A). To investigate the timing of myo-inositol changes in mips mutants, we analyzed leaves of 12-, 19-, and 60-d-old wild-type, mips1-2, mips2-2, and mips3-2 mutant plants (Figure 8B). Our results indicate that loss of MIPS1 has a larger impact on myo-inositol levels in leaves than loss of MIPS2 or MIPS3 (Figure 8B). In addition, the mips1-2 mutant has the largest decline in myo-inositol at the 19-d time point, which corresponds well with the development of lesions and other growth defects in this mutant (Figure 8B).
Figure 8.
Myo-Inositol Metabolic Alterations in mips Mutants.
(A) Overlay of representative GC traces from mips1-2 (blue) and wild-type plants (red).
(B) Leaves of 12-, 19-, and 60-d-old wild-type, mips1-2, mips2-2, and mips3-2 plants were harvested, and myo-inositol levels were quantified with GC as described in Methods. Values are percent of maximal levels. Standard error is indicated (n = 3). Asterisk indicates a P value < 0.05.
(C) Whole 18-d-old plants were harvested, and the indicated metabolites were quantified with GC as described in Methods. **, P value < 0.001; ***, P value < 0.0005.
Evidence exists that a minor pathway to ascorbic acid synthesis could proceed via myo-inositol synthesis (Baig et al., 1970; Radzio et al., 2003; Lorence et al., 2004; Torabinejad et al., 2009). We have previously shown that the vtc4 mutant (Conklin et al., 2006) has reduced myo-inositol and ascorbic acid levels, suggesting such a connection between these pathways (Torabinejad et al., 2009). Thus, we quantified ascorbic acid in leaves from 19-d-old mips1-2 mutants and found 190 ± 20 μg/g tissue in the wild type compared with 49 ± 9 μg/g tissue in mips1-2 mutants. We further profiled myo-inositol, ascorbic acid, and d-glucose in the wild type, mips1-2 mutants, complemented mips1-2, and MIPS1-GFP overexpressors (Figure 8C). These data show that the changes in myo-inositol, ascorbic acid, and d-glucose in mips1-2 plants is complemented by a MIPS1-GFP construct resulting in wild-type levels for each of these compounds (Figure 8C). This indicates that these metabolite alterations are due to a loss of MIPS1 function. We also examined metabolite levels in MIPS1-GFP plants and found wild-type levels for each metabolite, except for myo-inositol, which was increased by almost 4.5-fold (Figure 8C). The ascorbic acid precursor, l-galactonic acid γ-lactone, was also measured, and we found an almost fivefold increase in mips1-2 mutants (4.8 ± 1.5 μg/g tissue) compared with the wild type (0.8 ± 0.3 μg/g tissue), complemented mutants (1.3 ± 0.6 μg/g tissue), and MIPS1-GFP plants (1.6 ± 0.2 μg/g tissue).
We next investigated whether the reduction in myo-inositol in mips1-2 mutants resulted in alterations in Ins(1,4,5)P3 second messenger levels. For this work, we used a purified bovine Ins(1,4,5)P3 receptor in radiolabeled competition binding assays that are well established and used by several plant laboratories (Carland and Nelson, 2004; Zhong et al., 2004; Gunesekera et al., 2007; Perera et al., 2008). We found no differences, with 19-d-old wild-type plants having 404 ± 50 pg Ins(1,4,5)P3/g tissue and 19-d-old mips1-2 plants having 463 ± 91 pg Ins(1,4,5)P3/g tissue. In addition, plants expressing the MIPS1-GFP construct, which resulted in a 4.5-fold increase in myo-inositol levels (Figure 8C), did not contain significantly altered Ins(1,4,5)P3 levels [397 ± 56 pg Ins(1,4,5)P3/g tissue]. Our data indicate that there is no change in basal Ins(1,4,5)P3 in the mips1-2 mutant.
MIPS1 Impacts Phosphatidylserine (PtdIns or PI) Synthesis
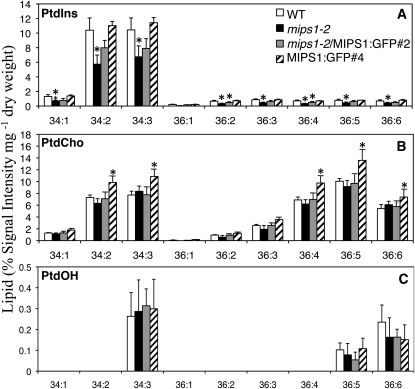
To determine if phospholipids are affected in mips1 mutants, we analyzed the phospholipid profile of 19-d-old mips1-2 and wild-type plants (Figure 9). Analysis of lipid extracts by mass spectrometry indicated a significant (almost 50% reduction) in several different PtdIns molecules in mips1-2 compared with the wild type (Figure 9A). Complemented mips1-2 plants contained intermediate levels of PtdIns, but these differed significantly from wild-type levels for only the 36:2 and 36:4 species (Figure 9A). This indicates that expression of the MIPS1-GFP transgene did not fully restore these two PtdIns species in complemented mips1-2 mutants. However, since these two species are present in minor amounts, the total PtdIns levels in complemented plants does not differ significantly from those in wild-type plants (19.5% ± 3.3% for complemented, 26% ± 4.3% for the wild type, and 15% ± 3.3% signal intensity/mg dry weight for mips1-2). Interestingly, overexpression of MIPS1-GFP, which increased myo-inositol levels by 4.5-fold, did not result in a corresponding increase in PtdIns; however, phosphatidylcholine levels were significantly increased for some species (Figure 9B). This suggests that levels of PtdIns are tightly regulated in plants. In contrast with the impact on PtdIns, levels of other measured phospholipids were not significantly altered in mips1-2 plants (Figures 9B and 9C). We conclude that the reduction in myo-inositol in mips1-2 mutants is accompanied by a decrease in PtdIns, which could impact functions that require PtdIns.
Figure 9.
Lipid Levels in mips Mutants, Complemented Mutants, and MIPS-GFP Plants.
Mass spectrometry was used to measure different species of PtdIns (A), phosphatidylcholine (PtdCho) (B), and phosphatidic acid (PtdOH) (C). Phospholipids are listed according to the carbon number followed by the number of double bonds. Data from three independent biological replicates were averaged. The standard error is indicated. Asterisk indicates a P value < 0.05 compared with the wild type.
MIPS1 Impacts Ceramide Accumulation
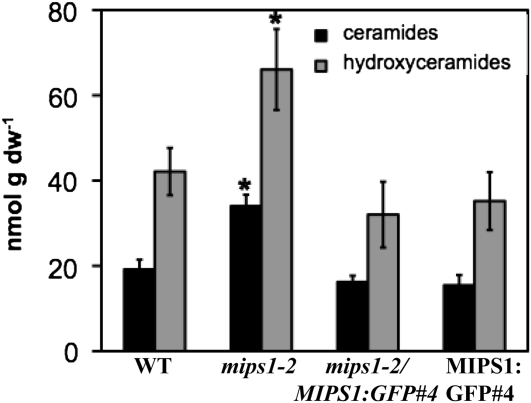
PtdIns is used during sphingolipid synthesis in plants (Wang et al., 2008). It has been shown that the plant inositol phosphorylceramide synthase catalyzes the transfer of the inositol phosphate headgroup from PtdIns to ceramide, resulting in inositol phosphorylceramide (Wang et al., 2008). Inositol phosphorylceramide synthase mutants expressing the resistance gene Resistance to Powdery Mildew8 (RPW8) have enhanced cell death and elevated ceramide and hydroxyceramide levels (Wang et al., 2008). To test whether the cell death in mips1 mutants is associated with altered ceramide and/or more complex sphingolipid levels, we measured total amounts of sphingolipids from 18-d-old plants (Figure 10; see Supplemental Figure 7 online). Total ceramides and hydroxyceramides were significantly elevated in mips1-2 mutants but not in complemented or MIPS1-GFP plants (Figure 10). In addition, there were no significant differences in other sphingolipids in mips1-2 mutants (see Supplemental Figure 7 online). We conclude that the elevated levels of ceramides and hydroxyceramides correlate with the decline of myo-inositol and PtdIns and the occurrence of cell death in mips1-2 mutants.
Figure 10.
Ceramide and Hydroxyceramide Levels Are Increased in mips Mutants.
Sphingolipids were extracted from leaves of 18-d-old plants of the indicated genotypes and measured essentially as described previously (Markham and Jaworski, 2007). Means and se are presented. Data from three independent biological replicates were averaged. The sd is indicated. Asterisk indicates a P value < 0.05 compared with the wild type. dw, dry weight.
The MIPS Proteins Are Biochemically Similar and Are Located in the Cytoplasm
To determine if the MIPS1, MIPS2, and MIPS3 genes encode enzymes with different kinetic properties, we expressed recombinant versions of all three MIPS proteins containing an N-terminal His-tag in E. coli and purified these proteins using immobilized metal ion affinity chromatography (see Supplemental Figure 8A online). The catalytic activity of the recombinant MIPS proteins was measured under steady state conditions (see Supplemental Figure 8B online; Table 1). It was determined that MIPS activity was enhanced by 10% glycerol and 20 mM NH4Cl, as previously observed for characterized MIPS enzymes (Mauck et al., 1980; RayChaudhuri et al., 1997; Ju et al., 2004). Therefore, NH4Cl and glycerol were included in our standard assay buffer. The dependence of MIPS activity on the concentration of the substrate Glc-6-P and cofactor NAD+ was measured, and the Michaelis-Menten equation was fit to the initial rates to obtain the steady state parameters (Km, kcat, kcat/Km). The results from these experiments show that there is no significant difference (≤2-fold) in the steady state parameters for the recombinant MIPS enzymes. Thus, we conclude that the MIPS1, MIPS2, and MIPS3 enzymes are likely to act in a similar fashion in vivo.
Table 1.
Steady State Kinetic Parameters for Arabidopsis MIPS
| Enzymea | kcat (min−1)b | KmGlc-6-P (mM)b | kcat/Km (mM−1min−1)b | KmNAD+ (μM)c | kcat (min−1)c |
| MIPS1 | 6.4 ± 0.2 | 0.68 ± 0.08 | 9.4 ± 0.9 | 0.46 ± 0.1 | 5.0 ± 0.4 |
| MIPS2 | 4.0 ± 0.1 | 0.45 ± 0.06 | 8.8 ± 0.9 | 0.30 ± 0.1 | 3.6 ± 0.3 |
| MIPS3 | 5.2 ± 0.2 | 0.31 ± 0.04 | 17 ± 2 | 0.13 ± 0.07 | 4.8 ± 0.4 |
The initial rate for MIPS-catalyzed activity was determined at 30°C (50 mM Tris, pH 7.5, 1 mM MgCl2, 20 mM NH4Cl, and 10% glycerol) with the substrate Glc-6-P. The kinetic parameters were obtained from the initial velocities as described in Methods.
The concentration of Glc-6-P varied (0 to 10 mM), and the concentration of NAD+ was held constant (20 μM).
The concentration of NAD+ varied (0 to 50 μM), and the concentration of Glc-6-P was held constant (5 mM).
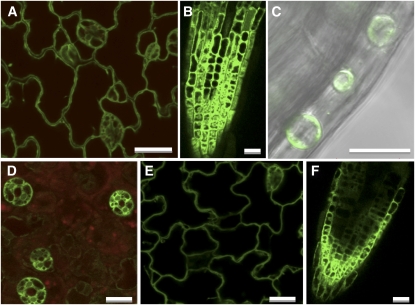
To investigate the subcellular location of MIPS proteins, we performed imaging experiments on mips1-2 mutants complemented with MIPS1-GFP and wild-type plants expressing MIPS2-GFP or MIPS3-GFP. As the MIPS1-GFP construct we used allowed for complementation of all phenotypes in mips1-2 mutants, it is likely that this fusion protein undergoes the same posttranslational modifications and subcellular localization as the native MIPS1 protein. We analyzed T2 progeny from two different complemented lines with confocal microscopy and found a similar pattern in these lines and in MIPS1-GFP overexpressors. GFP fluorescence was predominantly associated with the cytoplasm in 2-d-old light-grown seedling epidermis, roots, and hypocotyls (Figures 11A to 11C). To determine if the MIPS1-GFP protein was associated with the cell wall, 2-d-old light-grown seedlings were treated with 800 mM NaCl to stimulate plasmolysis (Figure 11C). In plasmolyzed cells, GFP fluorescence retracted from the cell wall, consistent with a cytoplasmic location. We also used a plasma membrane dye (FM4-64) with dual imaging to determine if any GFP was associated with the plasma membrane. Since no GFP was seen to colocalize with FM4-64, we conclude that MIPS1 is most likely located exclusively in the cytoplasm. In MIPS2-GFP seedlings, fluorescence was highest in guard cells (Figure 11D) and appeared to be confined to the cytoplasm as judged from plasmolysis experiments. MIPS3-GFP accumulation was apparent in the cytoplasm of epidermal and root cells (Figures 11E and 11F), and this localization was confirmed in plasmolysis experiments. We conclude that MIPS1-, MIPS2-, and MIPS3-GFP fusion proteins accumulate in a manner consistent with a similar cytoplasmic location for each.
Figure 11.
Subcellular Location of MIPS1-, MIPS2-, and MIPS3-GFP Proteins.
Single optical sections of transgenic plants expressing MIPS1-GFP ([A] to [C]), MIPS2-GFP (D), and MIPS3-GFP ([E] and [F]). Epidermal cells in cotyledon ([A], [D], and [E]), root ([B] and [F]), and differential interference contrast overlay of plasmolyzed cells within the hypocotyl with GFP fluorescence (C). (D) also includes autofluorescence from chlorophyll (red). Bars = 20 μm.
DISCUSSION
MIPS Genes Are Developmentally and Spatially Regulated
Our work delineates the function of the MIPS gene family in the model plant Arabidopsis and points to the greater impact of a single MIPS isoform (MIPS1) in this plant. We show that all three MIPS genes encode similar enzymes with similar kinetic constants (Table 1). In addition, we show that all three MIPS proteins are cytosolic (Figure 11). MIPS1 had been previously localized to both the cytoplasm and nucleus using transient expression in tobacco (Nicotiana tabacum) BY2 cells (Meng et al., 2009). Thus, a potential explanation for the exclusive cytoplasmic location seen in our studies is the difference in using stably transformed Arabidopsis plants versus tobacco BY2 cells. We also showed that all three MIPS genes are expressed in various tissues throughout development; however, MIPS1 expression is highest in most tissues, and its boundary of expression is larger than MIPS2 or MIPS3 throughout early development (Figures 1 and 2). Specifically, while MIPS2 and MIPS3 expression is confined to vascular or vascular-associated cells, the MIPS1 gene is expressed in cells throughout tissues such as leaves. These data point to a specialized or limited role for MIPS2 and MIPS3 compared with MIPS1. As all three MIPS genes are expressed in vascular tissues, this location may facilitate myo-inositol transport after synthesis. This corresponds well with previously published data showing that MIPS proteins accumulate in vascular-associated cells (Ishitani et al., 1996), with similar results being reported for the VTC4 or myo-inositol monophosphatase proteins, which function downstream of MIPS in the myo-inositol synthesis pathway (Gillaspy et al., 1995).
MIPS1 Has a Greater Impact on Myo-Inositol Levels Than MIPS2 or MIPS3
Our results show that higher expression levels of MIPS1 in wild-type plants correlate with a greater reduction in myo-inositol levels in mips1 mutants than in the mips2 and mips3 mutants (Figures 1 and 8). We have shown that mips1 mutants are dramatically impacted during growth and development, while mips2 and mips3 mutants appear normal in phenotype (Figures 5 to 7). Specifically, seed morphology, seedling growth, vascular development, cell death, and physiological responses were altered in mips1 mutants. This pleiotropism is not surprising given the multiple pathways and processes in plants that require myo-inositol. Furthermore, recent data correlate MIPS1 expression with biomass in Arabidopsis and suggest that MIPS1or its products may regulate carbon partitioning and growth (Sulpice et al., 2009).
Myo-Inositol, PtdIns, Ceramide, and Cell Death
The connection we found between myo-inositol synthesis and cell death (Figure 6) has been recently reported (Meng et al., 2009); however, our data offer additional and critical insights into the appearance of spontaneous cell death in mips1 mutants. Like Meng et al. (2009), we found that spontaneous cell death and the formation of lesions on leaves of mips1 mutants paralleled the progressive decline of myo-inositol in mips1 mutants throughout development (Figure 8) and can be rescued by myo-inositol supplementation (see Supplemental Figure 6 online). Overall, the mips1 mutants share characteristics with other lesion-mimic mutants (Lorrain et al., 2003) in that light potentiates the development of lesions (Bowling et al., 1997; Jambunathan et al., 2001; Lorrain et al., 2004), and the overall gene expression pattern is similar to lesion mimics as well as plants exposed to abiotic stress (Meng et al., 2009). Meng et al. (2009) also noted a dependence on salicyclic acid (SA) synthesis for lesion formation in mips1 mutants and a reduction in the galactinol levels, which requires myo-inositol for synthesis and has been speculated to function as a general ROS scavenger. We documented another connection to ROS in that mips1 mutants contain reduced ascorbic acid levels, making it logical to speculate that one factor involved in the spontaneous cell death in mips1 mutants is the reduced ability to withstand ROS and abiotic stress. In support of this, we found that mips1, but not mips2 and mips3 mutants, contained increased sensitivity to oxidative and certain abiotic stresses (Figure 7; see Supplemental Figure 5 online), which is also a characteristic phenotype of lesion mimics. This differs from the previously reported lack of stress sensitivity of mips1 mutants (Meng et al., 2009); however, we feel that our ability to document alterations in stress sensitivity is likely a result of using different growth regimes (i.e., high light) for mips1 mutants.
An even more critical factor in the spontaneous cell death of mips1 mutants, however, may be the alterations in lipids we found in these mutants. We documented lower levels of several species of PtdIns (Figure 9). In yeast, the single MIPS gene (ino1) is required for synthesis of PtdIns and normal growth (Greenberg et al., 1982). Changes in PtdIns in mips1 mutants could lead to alterations in membrane structure/fluidity, PtdInsP and Ins(1,4,5)P3 signaling, and/or vesicular trafficking. Since we found no differences in basal Ins(1,4,5)P3 levels in mips1 mutants, this suggests that signaling via phospholipase C hydrolysis of PtdIns(4,5)P2 is not impaired. In addition, the increased sensitivity of mips1 mutants to ABA, salt, and sorbitol is not consistent with a lack of phospholipase C signaling, since mutants in phospholipase C signaling with increased sensitivity to these stimuli most often contain elevated Ins(1,4,5)P3 (Xiong et al., 2001; Williams et al., 2005; Gunesekera et al., 2007). So while we cannot rule out a complex alteration in phospholipase C signaling in mips1 mutants, we feel it is more likely that the reduced capacity to handle ROS (described above) in mips1 mutants results in increased sensitivity to oxidative stress, ABA, salt, and sorbitol.
While the reduction in PtdIns in mips1 mutants may impact signaling and or trafficking functions of PtdInsPs in plants, one critical and known role of PtdIns is in the synthesis of sphingolipids. Specifically, PtdIns is a precursor to the sphingolipid, inositolphosphorylceramide (IPC). During synthesis of IPC, an IPC synthase transfers the inositol phosphate from PtdIns to ceramide (Wang et al., 2008). Thus, any decline in PtdIns could limit substrates for the IPC synthase and result in elevated ceramide, a signaling molecule known to induce cell death in plants (Wang et al., 1996; Liang et al., 2003). Our sphingolipid analyses confirmed that the cell death in mips1 mutants was accompanied by elevated ceramides and hydroxyceramides. This increase in ceramides and hydroxyceramides is similar to results noted for an IPC synthase mutant called Enhanced Resistance to HR Death1 carrying a specific RPW8 allele (S5) that increases SA signaling (Wang et al., 2008). In this pathway, elevated SA is required for elevated ceramide levels, as a knockout in the IPC synthase alone does not result in elevated ceramide or cell death (Wang et al., 2008). Given this, we speculate that mips1 mutants undergo spontaneous cell death as a result of altered oxidative stress sensitivity induced by changes in myo-inositol, galactinol, and ascorbic acid, along with elevated ceramides and hydroxyceramides that result from decreased PtdIns availability for sphingolipid production. Since complemented mips1-2 plants do not contain elevated ceramides (Figure 10) and also do not undergo cell death, we speculate that the intermediate levels of PtdIns in these plants provide a sufficient amount of PtdIns for sphingolipid synthesis. Together, these findings suggest a dynamic and critical interplay between PtdIns, sphingolipids, and regulation of cell death in plants.
Myo-Inositol Synthesis and InsP6
One major use of myo-inositol during seed development is incorporation into InsP6, a storage form of phosphorous (Raboy, 2003). InsP6 has also been implicated in other processes, including mRNA export in yeast and animals (Alcazar-Roman et al., 2006), while in plants, it is associated with the Transport Inhibitor Response 1 auxin receptor (Tan et al., 2007). Murphy et al. (2008) reported lower leaf InsP6 levels in mips1 and mips2 mutants, which corresponds to the mips1-2 and mips2-1 mutants we describe here. This group showed that mips2 is more susceptible to bacterial, viral, and fungal pathogens but found no change in susceptibility in bacterial or viral resistance in mips1 mutants. These authors speculated that certain subcellular pools of InsP6 are required for pathogen resistance and that the decrease in InsP6 in mips2 mutants could preferentially impact susceptibility (Murphy et al., 2008). By contrast, Meng et al. (2009) recently reported that mips1 mutants contain enhanced resistance to Hyaloperonospora parasitica, a fungal pathogen. Given that mips1 mutants have altered ceramide levels, one possible explanation for the opposing resistance phenotypes of mips1 and mips2 mutants may be a difference in MIPS1 and MIPS2 impact on PtdIns and ceramide levels. Evidence exists that the two Arabidopsis PtdIns synthases channel different species of PtdIns into different physiological pools (Lofke et al., 2008); thus, metabolic channeling of myo-inositol derived from MIPS1 catalysis could be preferentially routed into PtdIns and then sphingolipid synthesis.
Reduction of InsP6 in cereal grains fed to nonruminant animals to reduce phosphorous pollution of certain watersheds has been a pressing goal of plant biotechnology (Raboy, 2007). Some of the first low phytic acid (lpa) mutants identified were defective in a single MIPS gene, and these mutants have nondesirable alterations in seed dry weight and/or germination, reminiscent of the mips1 mutant described here (Li et al., 2000; Raboy et al., 2000; Dorsch et al., 2003; Liu et al., 2007; Yuan et al., 2007). Our work on the MIPS genes in the model plant Arabidopsis points to specialized roles for individual MIPS genes, with the MIPS1 gene providing the greatest impact, overall, on myo-inositol, growth, development, and physiology. Our work also reveals the complexity of downstream pathways and processes impacted by myo-inositol synthesis, including crosstalk with sphingolipid synthesis.
METHODS
Plant Materials
Arabidopsis thaliana ecotype Columbia plants were maintained in Sunshine Mix #1 in a growth room set at 22°C. Visible radiation (100 to 320 μmol m−2 s−1 for 16 h) was provided with fluorescent/incandescent lamps. For seed germination and root growth assays, seeds were sterilized with 10% Clorox, rinsed, and plated on 0.8% agar plates containing 0.5× MS medium. As indicated, plates contained ABA, NaCl, or d-Sorbitol (all from Sigma-Aldrich). A seed was considered as germinated when the radical protruded from the seed coat. Four plates each of 25 seeds per line were scored in germination assays. Five plates with six seeds per line were scored for root growth. All plant material was harvested from late morning to late afternoon. Mutants were identified from the Salk T-DNA lines (Alonso et al., 2003) through the analysis of the SiGnAL database (http://www.signal.salk.edu/cgi-bin/tdnaexpress). Seeds for mips1-2 (SALK_023626), mips1-3 (SAIL_676_D08, CS829610), mips2-1 (SALK_031685), mips2-2 (SALK_108779), mips3-2 (SALK_120131), mips3-3 (SAIL_425_F09, CS819634), and the corresponding wild-type plants CS60000 and CS908 were obtained from the ABRC at Ohio State University. Genomic DNA from segregating plants was screened by PCR using the primers noted in Supplemental Table 1 online and then sequenced to verify T-DNA insertion sites. Details on the insertion sites is in the Supplemental Methods online.
Expression Analyses
RNA was purified using the Qiagen RNeasy kit with DNase treatment from soil-grown plants and 7-d-old seedlings grown on 0.5× MS-soaked filter paper with a 16-h day. Mature seeds, imbibed with water for 3 d at 4°C, were freeze-dried, followed by initial RNA extraction and LiCl precipitation (Vicente-Carbajosa and Carbonero, 2005). cDNA was synthesized from equal amounts of RNA using a Bio-Rad iScript cDNA synthesis kit, loaded into 96-well plates containing Sybr Green PCR MasterMix (Applied Biosystems) and primers for MIPS1, MIPS2, MIPS3, or PEX4 (At5g25760) (see Supplemental Table 1 online). Reactions in quadruplicate were monitored with Applied Biosystems 7300 Real-Time PCR instrumentation outfitted with SDS software version 1.3.1. Primers designed to span an intron were optimized for specificity: MIPS1 at 62°C, MIPS2 at 64°C, and MIPS3 and PEX4 at 58°C. Standard curves were generated from primer-pair amplifications of calibrated amounts of plasmids containing single amplicons of MIPS1, MIPS2, MIPS3, or PEX4 and their threshold cycle numbers. Comparison of the real-time data with the standard curves allowed the calculation of femtomoles of target cDNA per milligram of total RNA.
Constructs and Imaging
Intergenic regions containing promoters for MIPS1 (1907 bp), MIPS2 (1106 bp), and MIPS3 (1907 bp) were amplified from CS60000 genomic DNA by PCR and cloned via the Gateway system into pBGWFS7 (Karimi et al., 2002) containing a Egfp:uidA gene fusion. GUS staining of 3- to 19-d-old plants grown on 0.5× MS agar plates with 1% sucrose or of plant tissues from soil-grown plants was as described (Styer et al., 2004), and staining was observed with an Olympus SZX16 microscope. MIPS1, MIPS2, and MIPS3 open reading frames without stop codons were amplified by PCR from CS60000 cDNA and cloned into vector pK7FWG2 (Karimi et al., 2002) for expression of GFP fusions and into vector pDEST17 (Invitrogen) for expression of His-tagged proteins. Transformation of Arabidopsis was as described (Bechtold et al., 1993). GFP fluorescence was detected with a Zeiss LSM 510 laser scanning microscope (Carl Zeiss) using excitation with a 488-nm argon laser and a 505- to 550-nm band-pass emission filter. Chlorophyll autofluorescence was imaged using excitation with a 543-nm HeNe laser and 560-nm band-pass emission filter. Slides were examined with a ×40 C-Apochromat water immersion objective lens. Conditions for trypan blue staining of soil-grown seedlings has been previously described (Koch and Slusarenko, 1990).
Protein Blot Analyses
A recombinant soybean (Glycine max) MIPS protein (Hegeman et al., 2001) was purified in the presence of urea using Ni-NTA affinity beads (Qiagen) (see Supplemental Methods online) and injected into a rabbit to produce the anti-MIPS polyclonal antibody (Cocalico Biologicals). Plant extracts were made by freezing in N2(l), followed by grinding in Laemmle gel-loading buffer containing SDS and β-mercaptoethanol for denaturing gel fractionation, or alone for native gels. Lysates were analyzed by protein gel blotting as described previously (Burnette et al., 2003; Ercetin and Gillaspy, 2004) except that nonfat dry milk was used for blocking and a 1:20,000 dilution of anti-MIPS antibody or 1:1000 dilution of anti-GFP antibody (Invitrogen) was used.
Metabolite Measurements
General conditions for GC and GC–mass spectrometry were as described (Torabinejad et al., 2009) with minor modifications. One analyte peak was found for d-chiro-inositol, ascorbic acid, and l-galactonic acid γ-lactone, while under these conditions d-glucose is represented by two peaks (two isomers). Myo-inositol was found in three peaks; however, two of these peaks had a negligible area, so the single dominant peak was used. Additionally, peaks were identified by comparison of mass spectral data using GC–mass spectrometry. Plant samples and standards were separated by a 6890-N GC on an HP-5MS capillary column 30 m × 0.25 mm i.d. (Agilent Technologies) with helium as the carrier gas with pressure-controlled flow set at 9.1 p.s.i. The injection port was set at 250°C, the oven was set on a gradient from 75 to 274°C at 6.5C°/min, and compounds were submitted to electrospray ionization and detected by a 5975 mass spectrometer (Agilent Technologies). For compound quantification, levels were calculated based on standard curves for each of the compounds and recovery of the internal standard. Two to five different, independent extracts were analyzed and averaged for each tissue. For Ins(1,4,5)P3 measurements, 18-d-old plants were analyzed as previously described (Gunesekera et al., 2007). The assays from three experiments were performed in duplicate. d-myo-Ins(1,4,5)P3 standards (American Radiolabeled Chemicals) and 3H-labeled d-myo-Ins(1,4,5)P3 (Perkin-Elmer) were used. For phospholipid analysis, we followed a modified protocol supplied by the Kansas State Lipidomics Facility (Devaiah et al., 2006). Extraction of phospholipids from 18-d-old soil-grown plants (50 to 60 mg fresh weight) was performed as described by Devaiah et al. (2006). Postextraction plant material was dried and massed to determine dry weight. Biological triplicates for each plant line were analyzed. Extracted phospholipids were quantified essentially as described (Devaiah et al., 2006). The dried phospholipid extract was dissolved in 1 mL chloroform and 25 μL was mixed with 1 mL chloroform/methanol/300 mM ammonium acetate in water (300/665/35) containing 0.66 nmol 14:0-16:0 phosphatidylcholine (PC) (Avanti Polar Lipids), 0.66 nmol di24:0 PC (Avanti), 0.24 nmol di14:0 phosphatidylserine (PS) (Avanti), 0.24 nmol di16:0 PS (Avanti), 0.16 nmol di8:0 PI (Cayman Chemical), and 0.16 nmol di16:0 PI (Cayman). Samples were introduced into an Applied Biosystems 4000 Q-Trap mass spectrometer at 30 μL per minute via a Harvard syringe pump and a Turbo V electrospray ion source. Mass spectrometer settings were as described (Devaiah et al., 2006) except the mass-to-charge scan ranges for PC and PI were adjusted to acquire data corresponding to the internal standards listed above. Multiple continuum scans were averaged in multiple channel acquisition mode; 8, 160, 18, and 78 scans were averaged per sample for scans specific to PC, PI, phosphatidic acid/phosphatidylglycerol, and PS, respectively. Analyst software (version 1.4.2) was used for data processing. Averaged spectra were smoothed when necessary, and the centroid of each peak was determined. Peaks were binned based on isotope distribution and compared using Excel software from Microsoft. Peaks were normalized both to (1) the sum of the intensities of both internal standards to provide percentage of intensity and (2) dry weight of sample, resulting in percentage of intensity mg−1 dry weight. Statistical analysis of P values for peaks was generated by two-tailed t tests.
Extraction of sphingolipids from 18-d-old plants was performed essentially as described by Markham and Jaworski (2007), except that 3 mL of extraction solvent (organic phase of isopropanol:hexane:water, 55:20:25) containing 10 μL of internal standards (Avanti LM-6005 Ceramide/Sphingoid Mixture II; 25 μM each standard) was used to homogenize ground material. Methods for separation and quantification were based on protocols detailed by Merrill et al. (2005) and Markham and Jaworski (2007) (details are in the Supplemental Methods online).
MIPS Activity Assays
Recombinant vectors containing genes encoding the MIPS proteins and an N-terminal His-tag (pHis-MIPS1, pHis-MIPS2, and pHis-MIPS3) were transformed into BL21(DE3) cells. pHis-MIPS-BL21(DE3) cells were grown in Luria-Bertani medium to OD600 ≈ 0.6, and expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 8 h. Cell pellets were resuspended in 50 mL Buffer A (1 mM tris(2-carboxyethyl) phosphine, 30 mM HEPES, and 150 mM NaCl, pH 8) containing 0.5 mM imidazole and 10% glycerol and stored at −80°C. Frozen cells were thawed, lysed by high-pressure homogenization, and centrifuged (18,000 rpm × 45 min). Cleared lysate was loaded onto a pre-equilibrated (Buffer A + 0.5 mM imidazole) immobilized metal ion affinity chromatography column. The column was washed with Buffer A containing increasing concentrations of imidazole; MIPS proteins were eluted with Buffer A + 250 mM imidazole. Fractions were pooled and concentrated and dialyzed in Buffer B (30 mM HEPES, 100 mM KCl, 3 mM MgCl2, and 1 mM tris(2-carboxyethyl) phosphine, pH 7.5). Protein concentrations were determined using the Bradford assay. The typical yield was 4 to 20 mg protein/liter culture. The conversion of glucose-6-phosphate (Glc-6-P) to inositol-1-phosphate was measured using a coupled assay (continuous) to detect phosphate formation following reaction with IMP (Sigma-Aldrich) using the EnzCheck phosphate assay kit (Invitrogen). The inorganic phosphate (Pi) released by IMP was measured following reaction with 2-amino-6-mercapto-7-methylpurine riboside and purine nucleoside phosphorylase, which results in an increase in absorbance at 360 nm. The assay mixtures (50 mM Tris, 1 mM MgCl2, pH 7.5, 20 mM NH4Cl, 10% glycerol, 1 μM MIPS, 20 μM NAD+, 0.6 mg IMP, 0.2 mM 2-amino-6-mercapto-7-methylpurine riboside, and 4 units/mL purine nucleoside phosphorylase) were preincubated at 30°C (96-well UV plates; Corning), and the reactions were initiated with the addition of Glc-6-P (0 to 10 mM). For determination of the KmNAD+, the concentration of Glc-6-P was held constant and the concentration of NAD+ was varied (0 to 50 μM). The time course for Pi production was monitored by measuring the A360 at various time points using a SpectraMax 5Me plate reader. The amount of Pi production was calculated from a Pi standard curve. The steady state parameters (kcat, Km, and kcat/Km) were obtained by fitting the Michaelis-Menten equation to the initial linear velocities measured at the various substrate concentrations using the curve-fitting program, Kaleidagraph (Synergy Software), which also calculates the asymptotic standard errors.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_120143 (MIPS1; At4g39800), NM_127790 (MIPS2; At2g22240), NM_121055 (MIPS3; At5g10170), and NM_122477 (PEX4; At5g25760).
Author Contributions
J.L.D. constructed all transgenic plants, characterized the T-DNA insertions, assisted in all metabolite measurements, and performed GUS staining, complementation experiments, and real-time PCR analyses. S.R.A. optimized and performed GC analyses and assisted in metabolite analyses. J.T. identified the mips mutants and performed physiological experiments. R.E.K. performed trypan blue staining experiments and assisted in mutant characterization, W.K.R. performed the lipid measurements, M.H. and X.H. performed enzyme assays, A.N. performed the confocal imaging, B.M.L. constructed and analyzed the MIPS promoter-GUS plants, P.P.H. performed physiology experiments, and G.E.G. directed the work, assisted in imaging experiments, and prepared the figures and manuscript.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genevestigator Expression Analysis of MIPS1, MIPS2, and MIPS3.
Supplemental Figure 2. Expression of MIPS Proteins in mips Mutants, Complemented Mutants, and Pro35S-MIPS1-GFP Plants.
Supplemental Figure 3. Seed and Seedling Phenotypes in mips1-2 Mutants and Complemented and Overexpression Plants.
Supplemental Figure 4. Irregular Cotyledons and Altered Vascular Development in mips 1-2 Mutants.
Supplemental Figure 5. MIPS Mutants, Reactive Oxygen, and Light.
Supplemental Figure 6. mips1-2 Mutants Can Be Rescued by myo-Inositol Supplementation.
Supplemental Figure 7. Levels of Sphingolipids in mips1-2 Mutants.
Supplemental Figure 8. Purified Recombinant MIPS Enzymes and Sample Kinetic Data.
Supplemental Table 1. Primers Used in This Work.
Supplemental Methods.
Supplemental References.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Rachael Elizabeth Hill, who wanted to pursue a PhD in biochemistry. We acknowledge David Schmale for help with real-time PCR, Kim Harrick and Richard Helm for mass spectrometry analyses, John McDowell and Ryan Anderson for advice and help with trypan blue staining, Eva Collakova for help with seeds, and Elizabeth Grabau and Carla Hegeman for the soybean MIPS plasmid. This work was supported by awards from the National Science Foundation (MCB316705) to G.E.G. and by the Hatch Pproject (VA-135583).
References
- Alcazar-Roman A.R., Tran E.J., Guo S., Wente S.R. (2006). Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8: 711–716 [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman A.R., Wente S.R. (2008). Inositol polyphosphates: A new frontier for regulating gene expression. Chromosoma 117: 1–13 [DOI] [PubMed] [Google Scholar]
- Allison J.H., Stewart M.A. (1973). Myo-inositol and ascorbic acid in developing rat brain. J. Neurochem. 20: 1785–1788 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baig M.M., Kelly S., Loewus F. (1970). L-ascorbic acid biosynthesis in higher plants from L-gulono-1, 4- lactone and L-galactono-1, 4-lactone. Plant Physiol. 46: 277–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banhegyi G., Braun L., Csala M., Puskas F., Mandl J. (1997). Ascorbate metabolism and its regulation in animals. Free Radic. Biol. Med. 23: 793–803 [DOI] [PubMed] [Google Scholar]
- Bechtold N., Ellis J., Pelletier G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. III, Sci. Vie 316: 1194–1199 [Google Scholar]
- Bowling S.A., Clarke J.D., Liu Y., Klessig D.F., Dong X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette R.N., Gunesekera B.M., Gillaspy G.E. (2003). An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiol. 132: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland F.M., Nelson T. (2004). Cotyledon vascular pattern2-mediated inositol (1,4,5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. Plant Cell 16: 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin P.L., Gatzek S., Wheeler G.L., Dowdle J., Raymond M.J., Rolinski S., Isupov M., Littlechild J.A., Smirnoff N. (2006). Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J. Biol. Chem. 281: 15662–15670 [DOI] [PubMed] [Google Scholar]
- Dean-Johnson M., Wang X. (1996). Differentially expressed forms of 1L-myo-inositol-1-phosphate synthase in Phaseolus vulgaris. J. Biol. Chem. 271: 17215–17218 [DOI] [PubMed] [Google Scholar]
- Devaiah S.P., Roth M.R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Dorsch J.A., Cook A., Young K.A., Anderson J.M., Bauman A.T., Volkmann C.J., Murthy P.P., Raboy V. (2003). Seed phosphorus and inositol phosphate phenotype of barley low phytic acid genotypes. Phytochemistry 62: 691–706 [DOI] [PubMed] [Google Scholar]
- Eisenberg F., Bolden A.H., Loewus F.A. (1964). Inositol formation by cyclization of glucose chain in rat testis. Biochem. Biophys. Res. Commun. 14: 419–424 [DOI] [PubMed] [Google Scholar]
- Ercetin M.E., Gillaspy G.E. (2004). Molecular characterization of an Arabidopsis gene encoding a phospholipid-specific inositol polyphosphate 5-phosphatase. Plant Physiol. 135: 938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Jigami Y. (2008). Lipid remodeling of GPI-anchored proteins and its function. Biochim. Biophys. Acta 1780: 410–420 [DOI] [PubMed] [Google Scholar]
- GhoshDastidar K., Chatterjee A., Chatterjee A., Majumder A.L. (2006). Evolutionary divergence of L-myo-inositol 1-phosphate synthase: Significance of a “core catalytic structure”. Subcell. Biochem. 39: 315–340 [PubMed] [Google Scholar]
- Gillaspy G.E., Keddie J.S., Oda K., Gruissem W. (1995). Plant inositol monophosphatase is a lithium-sensitive enzyme encoded by a multigene family. Plant Cell 7: 2175–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M.L., Reiner B., Henry S.A. (1982). Regulatory mutations of inositol biosynthesis in yeast: Isolation of inositol-excreting mutants. Genetics 100: 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumber S.C., Loewus M.W., Loewus F.A. (1984). myo-Inositol-1-phosphate synthase from pine pollen: Sulfhydryl involvement at the active site. Arch. Biochem. Biophys. 231: 372–377 [DOI] [PubMed] [Google Scholar]
- Gunesekera B., Torabinejad J., Robinson J., Gillaspy G.E. (2007). Inositol polyphosphate 5-phosphatases 1 and 2 are required for regulating seedling growth. Plant Physiol. 143: 1408–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman C.E., Good L.L., Grabau E.A. (2001). Expression of D-myo-inositol-3-phosphate synthase in soybean. Implications for phytic acid biosynthesis. Plant Physiol. 125: 1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Majumder A.L., Bornhouser A., Michalowski C.B., Jensen R.G., Bohnert H.J. (1996). Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. Plant J. 9: 537–548 [DOI] [PubMed] [Google Scholar]
- Jambunathan N., Siani J.M., McNellis T.W. (2001). A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13: 2225–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju G., Shamir A., Agam G., Greenberg M.L. (2004). Human 1-D-myo-inositol-3-phosphate synthase is functional in yeast. J. Biol. Chem. 279: 21759–21765 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karner U., Peterbauer T., Raboy V., Jones D.A., Hedley C.L., Richter A. (2004). myo-Inositol and sucrose concentrations affect the accumulation of raffinose family oligosaccharides in seeds. J. Exp. Bot. 55: 1981–1987 [DOI] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Ledoux D.R., Veum T.L., Raboy V., Ertl D.S. (2000). Effects of low phytic acid corn on phosphorus utilization, performance, and bone mineralization in broiler chicks. Poult. Sci. 79: 1444–1450 [DOI] [PubMed] [Google Scholar]
- Liang H., Yao N., Song J.T., Luo S., Lu H., Greenberg J.T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17: 2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.L., Xu X.H., Ren X.L., Fu H.W., Wu D.X., Shu Q.Y. (2007). Generation and characterization of low phytic acid germplasm in rice (Oryza sativa L.). Theor. Appl. Genet. 114: 803–814 [DOI] [PubMed] [Google Scholar]
- Loewus F. (1965). Inositol metabolism and cell wall formation in plants. Fed. Proc. 24: 855–862 [PubMed] [Google Scholar]
- Loewus F., Kelly S., Neufeld E. (1962). Metabolism of myo-inositol in plants: Conversion to pectin, hemicellulose, D-xylose, and sugar acids. Proc. Natl. Acad. Sci. USA 48: 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus F.A. (2006). Inositol and plant cell wall polysaccharide biogenesis. Subcell. Biochem. 39: 21–45 [DOI] [PubMed] [Google Scholar]
- Loewus M.W., Bedgar D.L., Loewus F.A. (1984). 1L-myo-inositol 1-phosphate synthase from pollen of Lilium longiflorum. An ordered sequential mechanism. J. Biol. Chem. 259: 7644–7647 [PubMed] [Google Scholar]
- Loewus M.W., Loewus F.A. (1980). The C-5 hydrogen isotope-effect in myo-inositol 1-phosphate synthase as evidence for the myo-inositol oxidation-pathway. Carbohydr. Res. 82: 333–342 [DOI] [PubMed] [Google Scholar]
- Loewus M.W., Loewus F.A. (1983). Myo-inositol-1-phosphatase from the pollen of Lilium longiflorum thunb. Plant Physiol. 70: 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus M.W., Loewus F.A., Brillinger G.U., Otsuka H., Floss H.G. (1980). Stereochemistry of the myo-inositol-1-phosphate synthase reaction. J. Biol. Chem. 255: 11710–11712 [PubMed] [Google Scholar]
- Lofke C., Ischebeck T., Konig S., Freitag S., Heilmann I. (2008). Alternative metabolic fates of phosphatidylinositol produced by phosphatidylinositol synthase isoforms in Arabidopsis thaliana. Biochem. J. 413: 115–124 [DOI] [PubMed] [Google Scholar]
- Lorence A., Chevone B.I., Mendes P., Nessler C.L. (2004). myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 134: 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Lin B., Auriac M.C., Kroj T., Saindrenan P., Nicole M., Balague C., Roby D. (2004). Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16: 2217–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Vailleau F., Balague C., Roby D. (2003). Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Markham J.E., Jaworski J.G. (2007). Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21: 1304–1314 [DOI] [PubMed] [Google Scholar]
- Mauck L., Wong Y.H., Sherman W.R. (1980). L-myo-inositol-1-phosphate synthase from bovine testis: Purification to homogeneity and partial characterization. Biochemistry 19: 3623–3629 [DOI] [PubMed] [Google Scholar]
- Meng P.H., Raynaud C., Tcherkez G., Blanchet S., Massoud K., Domenichini S., Henry Y., Soubigou-Taconnat L., Lelarge-Trouverie C., Saindrenan P., Renou J.P., Bergounioux C. (2009). Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS One 4: e7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill A.H., Jr., Sullards M.C., Allegood J.C., Kelly S., Wang E. (2005). Sphingolipidomics: High-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 36: 207–224 [DOI] [PubMed] [Google Scholar]
- Michell R.H. (2007). Evolution of the diverse biological roles of inositols. Biochem. Soc. Symp. 74: 223–246 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N., Kondo M., Nakaune S., Ohnishi M., Hayashi M., Hara-Nishimura I., Richardson A., Fukaki H., Nishimura M., Mimura T. (2008). Localization of myo-inositol-1-phosphate synthase to the endosperm in developing seeds of Arabidopsis. J. Exp. Bot. 59: 3069–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A.M., Otto B., Brearley C.A., Carr J.P., Hanke D.E. (2008). A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 56: 638–652 [DOI] [PubMed] [Google Scholar]
- Perera I.Y., Hung C.Y., Moore C.D., Stevenson-Paulik J., Boss W.F. (2008). Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20: 2876–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V. (2003). myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64: 1033–1043 [DOI] [PubMed] [Google Scholar]
- Raboy V. (2007). The ABCs of low-phytate crops. Nat. Biotechnol. 25: 874–875 [DOI] [PubMed] [Google Scholar]
- Raboy V., Bowen D. (2006). Genetics of inositol polyphosphates. Subcell. Biochem. 39: 71–101 [DOI] [PubMed] [Google Scholar]
- Raboy V., Gerbasi P.F., Young K.A., Stoneberg S.D., Pickett S.G., Bauman A.T., Murthy P.P., Sheridan W.F., Ertl D.S. (2000). Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 124: 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzio J.A., Lorence A., Chevone B.I., Nessler C.L. (2003). L-Gulono-1,4-lactone oxidase expression rescues vitamin C-deficient Arabidopsis (vtc) mutants. Plant Mol. Biol. 53: 837–844 [DOI] [PubMed] [Google Scholar]
- RayChaudhuri A., Hait N.C., Dasgupta S., Bhaduri T.J., Deb R., Majumder A.L. (1997). L-myo-inositol 1-phosphate synthase from plant sources (characteristics of the chloroplastic and cytosolic enzymes). Plant Physiol. 115: 727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman W.R., Loewus M.W., Pina M.Z., Wong Y.H. (1981). Studies on myo-inositol-1-phosphate from Lilium longiflorum pollen, Neurospora crassa and bovine testis. Further evidence that a classical aldolase step is not utilized. Biochim. Biophys. Acta 660: 299–305 [DOI] [PubMed] [Google Scholar]
- Smart C., Fleming A. (1993). A plant gene with homology to D-myo-inositol-3-phosphate synthase is rapidly and spatially up-regulated during ABA-induced morphogenic response in Spirodela polrrhiza. Plant J. 4: 279–293 [DOI] [PubMed] [Google Scholar]
- Styer J.C., Keddie J., Spence J., Gillaspy G.E. (2004). Genomic organization and regulation of the LeIMP-1 and LeIMP-2 genes encoding myo-inositol monophosphatase in tomato. Gene 326: 35–41 [DOI] [PubMed] [Google Scholar]
- Sulpice R., et al. (2009). Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T., Takahashi S., Shinozaki K. (2006). Inositols and their metabolites in abiotic and biotic stress responses. Subcell. Biochem. 39: 239–264 [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Torabinejad J., Donahue J.L., Gunesekera B.N., Allen-Daniels M.J., Gillaspy G.E. (2009). VTC4 is a bifunctional enzyme that affects myo-inositol and ascorbate biosynthesis in plants. Plant Physiol. 150: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabinejad J., Gillaspy G.E. (2006). Functional genomics of inositol metabolism. Subcell. Biochem. 39: 47–70 [DOI] [PubMed] [Google Scholar]
- Tsang E.W., Bowler C., Herouart D., Van Camp W., Villarroel R., Genetello C., Van Montagu M., Inze D. (1991). Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell 3: 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J., Carbonero P. (2005). Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 49: 645–651 [DOI] [PubMed] [Google Scholar]
- Wang H., Li J., Bostock R.M., Gilchrist D.G. (1996). Apoptosis: A functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8: 375–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2008). An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20: 3163–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.E., Torabinejad J., Cohick E., Parker K., Drake E.J., Thompson J.E., Hortter M., Dewald D.B. (2005). Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol. 138: 686–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Lee B., Ishitani M., Lee H., Zhang C., Zhu J.K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K.T., Fujiwara T., Naito S. (2002). The synergistic effects of sugar and abscisic acid on myo-inositol-1- phosphate synthase expression. Physiol. Plant. 114: 581–587 [DOI] [PubMed] [Google Scholar]
- Yoshida K.T., Wada T., Koyama H., Mizobuchi-Fukuoka R., Naito S. (1999). Temporal and spatial patterns of accumulation of the transcript of Myo-inositol-1-phosphate synthase and phytin-containing particles during seed development in rice. Plant Physiol. 119: 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F.J., Zhao H.J., Ren X.L., Zhu S.L., Fu X.J., Shu Q.Y. (2007). Generation and characterization of two novel low phytate mutations in soybean (Glycine max L. Merr.). Theor. Appl. Genet. 115: 945–957 [DOI] [PubMed] [Google Scholar]
- Zhong R., Burk D.H., Morrison W.H., III, Ye Z.H. (2004). FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. Plant Cell 16: 3242–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.